Abstract
Obstetric complications play a role in the pathophysiology of schizophrenia. However, the biological consequences during neurodevelopment until adulthood are unknown. Microarrays have been used for expression profiling in four brain regions of a rat model of neonatal hypoxia as a common factor of obstetric complications. Animals were repeatedly exposed to chronic hypoxia from postnatal (PD) day 4 through day 8 and killed at the age of 150 days. Additional groups of rats were treated with clozapine from PD 120–150. Self-spotted chips containing 340 cDNAs related to the glutamate system (“glutamate chips”) were used. The data show differential (up and down) regulations of numerous genes in frontal (FR), temporal (TE) and parietal cortex (PAR), and in caudate putamen (CPU), but evidently many more genes are upregulated in frontal and temporal cortex, whereas in parietal cortex the majority of genes are downregulated. Because of their primary presynaptic occurrence, five differentially expressed genes (CPX1, NPY, NRXN1, SNAP-25, and STX1A) have been selected for comparisons with clozapine-treated animals by qRT-PCR. Complexin 1 is upregulated in FR and TE cortex but unchanged in PAR by hypoxic treatment. Clozapine downregulates it in FR but upregulates it in PAR cortex. Similarly, syntaxin 1A was upregulated in FR, but downregulated in TE and unchanged in PAR cortex, whereas clozapine downregulated it in FR but upregulated it in PAR cortex. Hence, hypoxia alters gene expression regionally specific, which is in agreement with reports on differentially expressed presynaptic genes in schizophrenia. Chronic clozapine treatment may contribute to normalize synaptic connectivity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00406-010-0159-1) contains supplementary material, which is available to authorized users.
Keywords: Hypoxia, Microarray, Presynaptic genes, Clozapine, PPI, Animal model
Introduction
In addition to genetic predisposition, the neurodevelopmental hypothesis of schizophrenia includes a number of environmental factors like obstetric and birth complications with ensuing consequences on the pathophysiology of the disease. Hence, the risk of schizophrenia increases with the number and severity of hypoxia-associated obstetric complications [5, 6, 22]. However, the pathogenetic implications of obstetric complications are unclear. The neurodevelopmental hypothesis of schizophrenia suggests that the onset of abnormal brain development and neuropathology occurs perinatally, whereas symptoms of the disease appear in early adulthood [32].
Schizophrenia patients show deficits in attention and processing of sensory information. A reliable test of sensorimotor gating in vertebrates is prepulse inhibition (PPI) of the acoustic startle reflex, which is disrupted in schizophrenia patients and restored by antipsychotics, mainly clozapine [2]. Several cortical brain regions and striatum are involved in PPI-modulating circuitry [30]. We previously reported on a PPI deficit in rats exposed to neonatal hypoxia appearing only during adulthood. This deficit was prevented and restored after chronic clozapine treatment [12].
In our animal model of chronic neonatal hypoxia, rats exhibited increased gene expression of the N-methyl-D-aspartate (NMDA) receptor subunit NR1 in hippocampal and cortical subregions [28], which suggests an involvement of the NMDA receptor in the pathophysiology of hypoxia-induced alterations. In schizophrenia, there is indeed strong support for disturbances of the glutamatergic and gamma-amino-butyric acid (GABA)ergic microcircuitry, mainly affecting the expression of presynaptic vesicle proteins including synaptosome-associated protein 25 (SNAP-25) and syntaxin, which form the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex [9, 10, 15, 20, 29]. Complexin (CPX) 1 and complexin 2 operate in synaptic vesicle exocytosis by stabilizing the SNARE complex [4]. Neuropeptide Y (NPY) and neurexins (NRXN) play a role in cell adhesion and are hypothesized to play a role in the pathophysiology of schizophrenia [14, 20, 23, 26].
There is only little information about the influence of antipsychotic treatment on gene expression, as has been shown for NPY and CPX in normal rats [8, 16]. Thus, the present study investigates on the impact of neonatal hypoxia and long-term treatment with the antipsychotic clozapine on the expression of presynaptic genes in different cortical and subcortical brain regions in an animal model. Furthermore, correlations between differentially expressed genes and PPI deficits have been evaluated.
Materials and methods
Animals
A total of 12 Sprague–Dawley rats (Animal Facility, Department of Animal Physiology, University of Tübingen) were used for the experiments. The rats were housed in groups of 3 animals under a 12-h light/dark cycle with food and drinking water available ad libitum. The experiments were carried out in accordance with the international ethical guidelines for the care and use of laboratory animals for experiments (declaration of Helsinki, NIH guidelines) and were approved by the local animal care committee (Regierungspräsidium Tübingen, ZP 3/02).
Neonatal hypoxia
Chronic hypoxia (11% O2, 89% N2) was imposed from postnatal day (PD) 4–8 on six animal pups and their mothers by placing them in an air-tight plastic chamber for a period of 6 h per day. The six other pups were subjected to identical handling conditions and were placed in identical chambers but with regular oxygen concentrations (21% O2 = normoxia) [12, 28].
Clozapine treatment
Between PD 120 and PD 150, half of the rats were being orally treated over a period of 4 weeks with 45 mg/kg/day clozapine (Novartis Pharma AG, Basel, Switzerland) [12].
Test on prepulse inhibition and tissue preparation
The startle magnitude was measured as described in [12]. Since a deficit in prepulse inhibition had been shown to surface on postnatal days 120 and 150, animals were killed at postnatal day 150. Brains were removed and immersed in −78°C cold isopentane and stored at −80°C until further use. Slices of 120 um thickness were cut on a cryostat and used to excise respective brain regions according to the atlas of Paxinos and Watson [25] using a micropunching tool. Resultant, cylindrical tissue blocks were collected on dry ice and stored at −80°C until further processing.
Molecular investigations
RNA isolation and reverse transcription
One milliliter of Trizol was added to excised tissue, and tissue was homogenized by 30 strokes of trituration. After addition of 200 ul chloroform, the solution was transferred to Phase-Lock tubes (Eppendorf) for phase separation. Aqueous supernatants were purified and concentrated by RNeasy mini columns (Qiagen). Quality control was performed by OD measurements in a NanoDrop ND-1000 spectrophotometer (260/280 nm >1.8) and by electrophoretic separation using an Agilent 2100 Bioanalyzer (R.I.N. values >7.0).
Reverse transcription of purified total RNA was carried out using oligo-dT primers with T7 promoter sequence and Superscript II (Invitrogen).
In vitro transcription and labeling
Reverse-transcribed cDNAs were treated with RNAse and with DNA polymerase to obtain double-stranded DNA. Purified DNA was used for amplification reactions using RNAMaxx High Yield Transcription Kit (Stratagene) with T7 polymerase. Typically, this in vitro transcription resulted in 40- to 50-fold amplification of mRNA/cDNA.
Subsequently, cRNA was reverse-transcribed. Labeled DNA was purified and dried.
Preparation of DNA (“glutamate”) Chips
The chips represent a hypothesis driven selection of 340 cDNA sequences of genes related to glutamatergic and GABAergic neurotransmission and synaptic vesicle proteins (Online Resource 1a–f), obtained from clones purchased from Deutschen Ressourcenzentrum für Genomfoschung (RZPD, Berlin). The cDNA-inserts of bacterial clones were PCR-amplified directly from aliquots of bacterial cultures grown overnight (LB medium, containing antibiotic selection). After electrophoretic quality control, PCR mixtures were dried, resuspended in betaine-containing spotting buffer [7], and spotted on amino silane slides (Nexterion, PeqLab), using a MicroGrid II Arrayer. Each DNA was double-spotted, and each array was spotted three times on the same slide. The following control DNAs were used: salmon sperm DNA, human COT-1 DNA, 4 Arabidopsis thaliana DNAs (for spike-ins), spotting buffer, water, and a series of DNAs derived from artificial genes (library of random external controls = LOREC) [19].
Altogether, a chip contained 4,608 spots, including markers at the array corners to facilitate finding the correct orientation after scanning.
After spotting, DNAs were UV-cross-linked with a radiation of 250 mJ/cm2. Then, they were baked at 80°C for 2 h and stored at RT.
Hybridization and scanning of slides
Spotted slides were sequentially washed in 1% SDS solution in distilled water, immersed in 95°C distilled water, and incubated in 55°C prehybridization solution for 45 min. Then, they were washed in distilled water, in isopropanol (20 s, each), and dried by a stream of nitrogen or air. Hybridizations were done in SlideHyb Glass Array Hybridization buffer #1 (Ambion) overnight at 56°C. All subsequent washing steps at increasing stringency were performed in the dark.
Scanning was carried out in a Packard ScanArray 5,000 scanner. Each slide was scanned twice at laser strengths of 90 and 100% to obtain a larger spectrum of fluorescence intensities.
Data processing
Data quality was assessed by a number of procedures, including visual inspection of intensities, MA-plots, and correlation analysis of replicates. Within-array normalization was performed according to locally weighted scatterplot smoothing (LOESS) method [34]. Only rank-invariant probes were used for model generation during this procedure [27]. For between-array normalization, scaling [34] was used. After assessing normal distribution of replicate spots using the Kolmogorov–Smirnov test, replicates were pooled by their arithmetic means. To identify differentially expressed genes, two-tailed t-tests and the RankProduct (RP) [3] method were used. The false discovery rate (FDR) method was employed to avoid accumulation of alpha-error. Corrected P-values <0.05 were considered significant. To limit the number of differentially expressed genes, only those were used which were found by both t-test and RP methods. Finally, to profile overall regulation, only genes that were differentially regulated in at least two regions were selected for further analyses.
Quantitative real-time PCR (qRT-PCR)
Quantitative RT-PCR was carried out in 384-well plates in an ABI Prism 7900 HT Fast Real-Time PCR cycler (Applied Biosystems, Foster City, Ca., USA). The primer pairs used for the respective genes are listed in Table 1. Beta actin was used as a reference (“house-keeping”) gene.
Table 1.
Primers used in qRT-PCR
| Sequences reading from 5′ -end | ||
|---|---|---|
| Gene | Forward primer | Reverse primer |
| CPLX1 | GCAGGGCATACGAGATAAGT | TGGTGGGGTCAGTGATGGCAGTA |
| NPY | CGCTCTGCGACACTACAT | CTCAGGGCTGGATCTCTTG |
| NRXN1 | AGGGCGTCAGCTCACAATCTTCAA | TCTGCCGAGCCTGGGTATGGT |
| SNAP25 | AGTGGCGTTTGCTGAATGAC | TGCTGGTATGACTTAATCTTGACA |
| STX1A | TGTCCCGAAAGTTTGTG | GCGTTCTCGGTAGTCT |
Results
Microarray experiments
After statistical evaluations of the 768 genes spotted on the glass slides by Students t tests, many of them turned out to be significantly regulated.
Table 2 provides an overview of differentially expressed genes in frontal (FR), parietal (PAR), and temporal (TE) cortex as well as in caudate putamen (CPU). At first sight, there are apparently large regional differences in terms of up- or downregulation, i.e. many more genes are upregulated in FR [44 vs. 4 (ratio11)] and in TE [51 vs. 22 (ratio 2.32)] compared with PAR [10 vs. 33 (ratio 0.3)]. A more balanced situation seems to occur in CPU.
Table 2.
Differentially expressed genes in some regions of normoxic and hypoxic rat brains
| CPU | FR | PAR | TE | ||||
|---|---|---|---|---|---|---|---|
| Genelist t-test fdr correction | Genelist t-test fdr correction | Genelist t-test fdr correction | Genelist t-test fdr correction | ||||
| The factor “hypoxia” regulated the following genes in comparison with “normoxia” | The factor “hypoxia” regulated the following genes in comparison with “normoxia” | The factor “hypoxia” regulated the following genes in comparison with “normoxia” | The factor “hypoxia” regulated the following genes in comparison with “normoxia” | ||||
| UP | DOWN | UP | DOWN | UP | DOWN | UP | DOWN |
| ATF2 | ATF2 | CAPN2 | ADCY3 | ||||
| ATF4 | AURKB | CAPON | ADCY6 | ||||
| ATP2A2 | CALM2 | CASK | ATP2A2 | ||||
| CPX1 | CAP2 | EAAT3 | BDNF | ||||
| CPX2 | CAPN8 | GABBR1 | BNPI | ||||
| GAP43 | CPX1 | GAPDH | CAPN2 | ||||
| GAPDH | DEAD BOX | GNA12 | CAPN6 | ||||
| GATA4 | DLGAP2 | LOC294679 | CASQ2 | ||||
| GRM2 | DUSP1 | LOC362704 | CPX1 | ||||
| ITPR1 | FYN | MAPK3 | DAT1 | ||||
| MARK1 | GAPDH | PIPS | EAAT3 | ||||
| MPPS7 | GNAO | PLD2 | ERK7 | ||||
| MYD116 | MAPK14 | PLEKHC1 | GABRG3 | ||||
| NM_017261 | MAPK3 | POU3F4 | GAD67 | ||||
| NOS2 | MRPS7 | PPP1R14A | GADD45A | ||||
| NRXN1 | NELL2 | PPP2R2C | GAP43 | ||||
| PKN1 | NPY | PPP5C | GATA4 | ||||
| PLD1 | NOS2-1149 | PREFOLDIN5 | GNAO | ||||
| PLEKHC1 | NRXN1 | PRKCZ | GPSM1 | ||||
| PPM1B | PACSIN2 | PTPRA | GRM8 | ||||
| PPP1CC | PIM3 | PTPRR | ID3 | ||||
| PPKCC | PLAT | RAB14 | ITPR3 | ||||
| PTP4A1 | PPP1R14B | RAB3A | LOC294679 | ||||
| PTPRA | PPP1R14C | RAB3B | LOC362704 | ||||
| PTPRN | PTP4A2 | RN.25045 | MGC105762 | ||||
| PTPRN2 | PTPRD | RN.28284 | NPY | ||||
| RAB10 | PTPRN | SHANK1 | NR2F2 | ||||
| RN.28284 | PTPRN2 | SLC2A1 | NRXN3 | ||||
| SACM1L | RAB3A | SLC6A12 | NUP54 | ||||
| SLC1A3 | SLC1A3 | SLK | PIPS | ||||
| SLC1A3 | SLC6A12 | SRC | PLD2 | ||||
| SLC1A6 | SNAP25 | STAT3 | PNOC | ||||
| SLC32A1 | SOD1 | SYT1-1511 | PPM1G | ||||
| SNAP25 | SQSTM1 | TRANSC.LOCUS | PPP1R10 | ||||
| STAT3 | SRC | PPP2R2C | |||||
| SYN1 | STK39 | PPP2R5B | |||||
| VAMP2 | STX1A | PPP4R1 | |||||
| TRANSC.LOCUS | PPP5C | ||||||
| PRKCH | |||||||
| PRKCZ | |||||||
| RAB10 | |||||||
| RGS3 | |||||||
| RN.137813 | |||||||
| RN.15926 | |||||||
| RN.7389 | |||||||
| SLC1A6 | |||||||
| SLC32A1 | |||||||
| SLC6A12 | |||||||
| SMAGP | |||||||
| SNAP25 | |||||||
| SP1 | |||||||
| SQSTM1 | |||||||
| STAT3 | |||||||
| STK6 | |||||||
| STX1A | |||||||
| SYN1 | |||||||
| SYN2 | |||||||
| TCF12 | |||||||
| VAMP1 | |||||||
Many more genes are upregulated in frontal (FR) and temporal (TE) cortex compared with parietal (PAR) cortex
Clozapine treatment and qRT-PCR results
To obtain a clearer picture, the focus was laid upon genes primarily being expressed presynaptically. Those were further investigated by qRT-PCR in the four brain regions in normoxic and hypoxic animals treated with clozapine (Table 3).
Table 3.
“Clozapine”-induced effects in normoxic and hypoxic animals (empty = no effect)
| Normoxic | Hypoxic | ||||||
|---|---|---|---|---|---|---|---|
| Region | Gen | Reg.-Dir. | P-value | Region | Gen | Reg.-Dir. | P-value |
| CPU | CPX1 | CPU | CPX1 | up | 0.026 | ||
| FR | CPX1 | down | 0.043 | FR | CPX1 | ||
| PAR | CPX1 | PAR | CPX1 | up | 0.012 | ||
| TE | CPX1 | TE | CPX1 | ||||
| CPU | NPY | CPU | NPY | ||||
| FR | NPY | FR | NPY | ||||
| TE | NPY | TE | NPY | ||||
| CPU | NRXN1 | CPU | NPY | ||||
| FR | NRXN1 | FR | NRXN1 | ||||
| PAR | NRXN1 | PAR | NRXN1 | ||||
| TE | NRXN1 | TE | NRXN1 | ||||
| CPU | SNAP25 | CPU | NRXN1 | ||||
| FR | SNAP25 | FR | SNAP25 | ||||
| PAR | SNAP25 | PAR | SNAP25 | ||||
| TE | SNAP25 | TE | SNAP25 | ||||
| FR | STX1A | down | 0.015 | FR | SNAP25 | ||
| PAR | STX1A | up | 0.048 | PAR | STX1A | up | 0.002 |
| TE | STX1A | TE | STX1A | ||||
Evidently, CPX1 is upregulated in frontal and temporal cortex as well as in caudate putamen, but not in parietal cortex in hypoxic animals (Table 2). Treatment with clozapine resulted in an upregulation also in parietal cortex. NPY was upregulated by hypoxia in frontal but downregulated in temporal cortex. Neurexin1 was upregulated in both frontal cortex and CPU in hypoxic animals. SNAP25 was also upregulated in frontal cortex but downregulated in CPU. Additionally, it was upregulated in temporal cortex. Another counterregulation was observed with syntaxin1A in frontal and temporal cortex (Table 2). Interestingly, in parietal cortex, where hypoxia had no effect, STX1A was upregulated by clozapine (Table 3).
Discussion
Complexin1
Upregulation of CPX1 upon postnatal hypoxia in frontal, temporal cortex, and CPU has not been reported yet (Fig. 1). In the light that clozapine additionally upregulated the gene in parietal cortex, it may be assumed that hypoxia-induced upregulation may be a compensatory mechanism to ensure proper vesicular transmitter release. Associations of complexins with schizophrenia have been repeatedly reported [9, 10, 13]. A recent report comes to the conclusion that complexins facilitate neurotransmitter release [33]. Specifically, it has been suggested that CPX1 downregulation during perinatal hypoxia in GABAergic neurons facilitates glutamate release and loss of overstimulated neurons—which subsequently results in reduced glutamatergic projections [24]. The upregulation of CPX1 in parietal cortex by clozapine is supported by the findings of Eastwood [8]. Our results showing the very strong correlation of CPX1 with PPI in hypoxia-treated animals (Table 4) support the view that this gene/protein is a key player in the development of Schizophrenia-like symptoms.
Fig. 1.
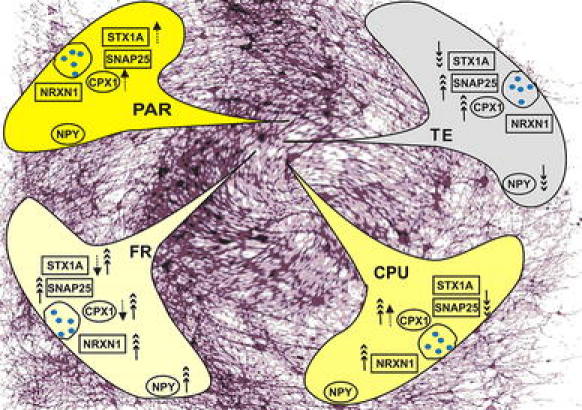
Hypoxia- and clozapine-mediated differential regulation of presynaptic transcripts in some rat brain regions. Triple arrows : hypoxia : chip results; dotted arrows : clozapine : qRT-PCR results
Table 4.
Correlations of qRT-PCR expression data with prepulse inhibition (PPI)
| Gene | Region | Entity | Normoxia | Normoxia | Normoxia | Hypoxia | Hypoxia | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Entity | Without Clozapine | With Clozapine | Without Clozapine | With Clozapine | |||||||||||||||
| N | rho | P | N | rho | P | N | rho | P | N | rho | P | N | rho | P | N | rho | P | ||
| CPX1 | CPU | 23 | 0.35 | 0.11 | 12 | −0.38 | 0.23 | 6 | −0.89 | 0.02 | 6 | 0.03 | 0.96 | 5 | 1 | 0 | 12 | −0.38 | 0.23 |
| NPY | TE | 23 | 0.1 | 0.65 | 12 | 0.000 | 1.000 | 6 | 0.89 | 0.02 | 6 | −0.31 | 0.54 | 5 | 0.3 | 0.62 | 12 | 0.000 | 1.000 |
| NRXN1 | CPU | 23 | 0.07 | 0.77 | 12 | 0.7 | 0.01 | 6 | 0.600 | 0.21 | 6 | 0.66 | 0.16 | 5 | 0.2 | 0.75 | 12 | 0.7 | 0.01 |
| SNAP25 | FR | 20 | 0.02 | 0.93 | 9 | 0.02 | 0.97 | 4 | −1.000 | 0.000 | 5 | 0.1 | 0.87 | 5 | −0.3 | 0.62 | 9 | 0.02 | 0.97 |
Levels of gene expression in individual animals were correlated with the performance shown in the behavioral testing of prepulse inhibition (PPI). Four out of the five genes investigated turned out to show significant correlations (P-value smaller than 0.05) in specific brain regions under specific conditions. As shown in the table, in normoxic, non-medicated animals, PPI negatively correlates with complexin 1 level in CPU (P = 0.02) and SNAP25 (P < 0.01) level in the frontal cortex. A positive correlation between PPI and neuropeptide Y can be seen in the temporal cortex (P = 0.02). In contrast, in caudate putamen of hypoxic animals, complexin 1 shows a positive correlation with PPI. A comparable correlation is found also in CPU of clozapine-treated hypoxic animals with neurexin1 (P = 0.01)
Neurexin1
Neurexins and neuroligins are emerging as central organizing molecules for excitatory glutamatergic and inhibitory GABAergic synapses. They function as cell adhesion molecules, bridging the synaptic cleft. Neurexin 1beta and neuroligin normally form trans-synaptic complexes [1]. Finally, there is a recent report showing that disruption of the neurexin 1 gene is associated with schizophrenia [26].
Preconditioning ischemia can downregulate the increased neurexin–neuroligin1–PSD-95 interaction induced by ischemia injury and exerts a neuroprotective effect. These results indicate that the neurexin-neuroligin1 is an important signaling module in hypoxic injury and a novel possible target in therapeutics of brain ischemia [21].
SNAP25
Recent genetic studies implicate alterations in SNAP-25 gene structure, expression, and/or function in contributing directly to neuropsychiatric and neurological disorders. Protein-by-protein analyses indicated a significant reduction in SNAP-25 immunoreactivity in the schizophrenia non-suicide group [11, 15].
In another study, however, there were no differences in the immunoreactivity of proteins measured in schizophrenic patients. Apparently prefrontal cortical dysfunction in schizophrenia may exist without gross changes in the abundance of many synaptic proteins [13].
There were significant decreases in tissue levels of these proteins in prefrontal cortex of schizophrenic patients relative to controls, whereas the mRNAs encoding these proteins were not decreased [18]. Normal levels of SNAP-25 are noted in schizophrenia patients in Brodman area 17, decreased levels in areas 10 and 20, and an elevated level in area 9 [31].
In comparison with our results (Fig. 1), those findings strongly support the notion that regionally distinct expressions prevail and have always to be taken into consideration.
NPY
Regionally different gene expression of NPY is observed also in our report (Fig. 1). Patients with schizophrenia showed deficits in NPY mRNAs [14, 23]. Olanzapine and clozapine administration acutely decreased NPY mRNA levels in the nucleus accumbens, striatum, and anterior cingulate cortex of rats [16, 20]. The proportion of NPY neurons in the upper cortical layers was low, but it was high in the deep white matter in the disorganized and paranoid form [17].
Syntaxin 1
This presynaptic protein is another good example for regionally different gene expression. It is upregulated in FR but downregulated in TE, whereas there is no change in CPU and PAR by hypoxia treatment. Clozapine upregulates its expression in PAR but downregulates it in FR (Fig. 1). From literature reports, we know that there were significant negative correlations between age and mRNA levels of syntaxin 1A in schizophrenic patients. They were increased in the younger (58–79 years) subgroup of schizophrenic patients—which suggests that they reflect overall neurodevelopmental abnormalities in synaptic connectivity in the temporal cortex of schizophrenic patients [29].
Conclusions
Presynaptic proteins have been reported to be differentially regulated in schizophrenia. In our animal model of perinatal hypoxia, alterations in presynaptic genes in adulthood during the time period of behavioral deficits have been shown and may be involved in the pathophysiology leading to schizophrenia symptoms. However, we focused only on genes of the glutamatergic system, and all conclusions are limited to this domain. Our results reveal that chronic clozapine treatment may differentially influence gene expression and in this way contribute to the treatment of symptoms. Animal models of risk factors of schizophrenia are warranted to elucidate disease mechanisms and specific effects of antipsychotic treatment in disease states.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Supported by a research grant from the Central Institute of Mental Health, Mannheim, Germany.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The authors J. U. Sommer and A. Schmitt contributed equally to this article.
References
- 1.Berninghausen O, Rahman MA, Silva JP, Davletov B, Hopkins C, Ushkaryov YA. Neurexin ibeta and neuroligin are localized on opposite membranes in mature central synapses. J Neurochem. 2007;103(5):1855–1863. doi: 10.1111/j.1471-4159.2007.04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braff DL. Reply to cognitive therapy and schizophrenia. Schizophr Bull. 1992;18(1):37–38. doi: 10.1093/schbul/18.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 4.Brose N. Altered complexin expression in psychiatric and neurological disorders: cause or consequence? Mol Cells. 2008;25(1):7–19. [PubMed] [Google Scholar]
- 5.Cannon TD, Rosso IM, Bearden CE, Sanchez LE, Hadley T. A prospective cohort study of neurodevelopmental processes in the genesis and epigenesis of schizophrenia. Dev Psychopathol. 1999;11(3):467–485. doi: 10.1017/S0954579499002163. [DOI] [PubMed] [Google Scholar]
- 6.Dalman C, Thomas HV, David AS, Gentz J, Lewis G, Allebeck P. Signs of asphyxia at birth and risk of schizophrenia. Population-based case-control study. Br J Psychiatry. 2001;179:403–408. doi: 10.1192/bjp.179.5.403. [DOI] [PubMed] [Google Scholar]
- 7.Diehl F, Grahlmann S,Beier M, Hoheisel JD (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res 29 Apr 2001 Nr. 7, S. E38 [DOI] [PMC free article] [PubMed]
- 8.Eastwood SL, Burnet PW, Harrison PJ (2000) Expression of complexin I and II mRNAs and their regulation by antipsychotic drugs in the rat forebrain. Synapse. 1 Jun 2000;36(3):167–177 [DOI] [PubMed]
- 9.Eastwood SL, Harrison PJ. Synaptic pathology in the anterior cingulate cortex in schizophrenia and mood disorders. A review and a Western blot study of synaptophysin, GAP-43 and the complexins. Brain Res Bull. 2001;55(5):569–578. doi: 10.1016/S0361-9230(01)00530-5. [DOI] [PubMed] [Google Scholar]
- 10.Eastwood SL, Harrison PJ. Decreased expression of vesicular glutamate transporter 1 and complexin II mRNAs in schizophrenia: further evidence for a synaptic pathology affecting glutamate neurons. Schizophr Res. 2005;73(2–3):159–172. doi: 10.1016/j.schres.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Fatemi SH, Earle JA, Stary JM, Lee S, Sedgewick J. Altered levels of the synaptosomal associated protein SNAP-25 in hippocampus of subjects with mood disorders and schizophrenia. Neuroreport. 2001;12(15):3257–3262. doi: 10.1097/00001756-200110290-00023. [DOI] [PubMed] [Google Scholar]
- 12.Fendt M, Lex A, Falkai P, Henn FA, Schmitt A. Behavioural alterations in rats following neonatal hypoxia and effects of clozapine: implications for schizophrenia. Pharmacopsychiatry. 2008;41(4):138–145. doi: 10.1055/s-2008-1058107. [DOI] [PubMed] [Google Scholar]
- 13.Halim ND, Weickert CS, McClintock BW, Hyde TM, Weinberger, Kleinman JE, Lipska BK. Presynaptic proteins in the prefrontal cortex of patients with schizophrenia and rats with abnormal prefrontal development. Mol Psychiatry. 2003;8(9):797–810. doi: 10.1038/sj.mp.4001319. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto T, Arion D, Unger T, Maldonado-Avilés JG, Morris HM, Volk DW, Mirnics K, Lewis DA. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13(2):147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honer WG, Falkai P, Bayer TA, Xie J, Hu L, Li HY, Arango V, Mann JJ, Dwork AJ, Trimble WS. Abnormalities of SNARE mechanism proteins in anterior frontal cortex in severe mental illness. Cereb Cortex. 2002;12(4):349–356. doi: 10.1093/cercor/12.4.349. [DOI] [PubMed] [Google Scholar]
- 16.Huang XF, Deng C, Zavitsanou K. Neuropeptide Y mRNA expression levels following chronic olanzapine, clozapine and haloperidol administration in rats. Neuropeptides. 2006;40(3):213–219. doi: 10.1016/j.npep.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda K, Ikeda K, Iritani S, Ueno H, Niizato K. Distribution of neuropeptide Y interneurons in the dorsal prefrontal cortex of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28(2):379–383. doi: 10.1016/j.pnpbp.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Karson CN, Mrak RE, Schluterman KO, Sturner WQ, Sheng JG, Griffin WS. Alterations in synaptic proteins and their encoding mRNAs in prefrontal cortex in schizophrenia: a possible neurochemical basis for ‘hypofrontality’. Mol Psychiatry. 1999;4(1):39–45. doi: 10.1038/sj.mp.4000459. [DOI] [PubMed] [Google Scholar]
- 19.Koschmieder A, Ma D, Hoheisel J, Frohme M (2005) Construction of libraries of random external controls (LOREC) for application in microarray experiments. DKFZ, Degree Dissertation, Heidelberg
- 20.Kuromitsu J, Yokoi A, Kawai T, Nagasu T, Aizawa T, Haga S, Ikeda K. Reduced neuropeptide Y mRNA levels in the frontal cortex of people with schizophrenia and bipolar disorder. Brain Res Gene Expr Patterns. 2001;1(1):17–21. doi: 10.1016/S1567-133X(01)00003-5. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Han D, Zhang F, Zhou C, Yu HM, Zhang GY. Preconditioning ischemia attenuates increased neurexin-neuroligin1-PSD-95 interaction after transient cerebral ischemia in rat hippocampus. Neurosci Lett. 2007;426(3):192–197. doi: 10.1016/j.neulet.2007.08.065. [DOI] [PubMed] [Google Scholar]
- 22.McNeil TF, Cantor-Graae E, Ismail B. Obstetric complications and congenital malformation in schizophrenia. Brain Res Brain Res Rev. 2000;31(2–3):166–178. doi: 10.1016/S0165-0173(99)00034-X. [DOI] [PubMed] [Google Scholar]
- 23.Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S (2008) Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry.31 Dec 2008 [DOI] [PubMed]
- 24.Muller DJ, Klempan TA, De Luca V, Sicard T, Volavka J, Czobor P, Sheitman BB, Lindenmayer JP, Citrome L, McEvoy JP, Lieberman JA, Honer WG, Kennedy JL. The SNAP-25 gene may be associated with clinical response and weight gain in antipsychotic treatment of schizophrenia. Neurosci Lett. 2005;379:81–89. doi: 10.1016/j.neulet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 25.Paxinos G, Watson C (1986) The rat brain in stereotaxic coordinates, 2nd edn. Academic Press
- 26.Rujescu D, Ingason A, Cichon S, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18(5):988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt A, Fendt M, Zink M, Ebert U, Starke M, Berthold M, Herb A, Petroianu G, Falkai P, Henn F. Altered NMDA receptor expression and behaviour following postnatal hypoxia: potential relevance to schizophrenia. J Neural Transm. 2007;114:239–248. doi: 10.1007/s00702-006-0440-7. [DOI] [PubMed] [Google Scholar]
- 29.Sokolov BP, Tcherepanov AA, Haroutunian V, Davis KL. Levels of mRNAs encoding synaptic vesicle and synaptic plasma membrane proteins in the temporal cortex of elderly schizophrenic patients. Biol Psychiatry. 2000;48(3):184–196. doi: 10.1016/S0006-3223(00)00875-1. [DOI] [PubMed] [Google Scholar]
- 30.Swerdlow NR, Geyer MA, Braff DL. Neural circuit regulation of prepulse inhibition of startle in the rat: current knowledge and future challenges. Psychopharmacology (Berl) 2001;156(2–3):194–215. doi: 10.1007/s002130100799. [DOI] [PubMed] [Google Scholar]
- 31.Thompson PM, Sower AC, Perrone-Bizzozero NI. Altered levels of the synaptosomal associated protein SNAP-25 in schizophrenia. Biol Psychiatry. 1998;43(4):239–243. doi: 10.1016/S0006-3223(97)00204-7. [DOI] [PubMed] [Google Scholar]
- 32.Weinberger DR. On the plausibility of “the neurodevelopmental hypothesis” of schizophrenia. Neuropsychopharmacology. 1996;14(3):1S–11S. doi: 10.1016/0893-133X(95)00199-N. [DOI] [PubMed] [Google Scholar]
- 33.Xue M, Stradomska A, Chen H, Brose N, Zhang W, Rosenmund C, Reim K. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc Natl Acad Sci U S A. 2008;105(22):7875–7880. doi: 10.1073/pnas.0803012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang YH, Dudoit S, Luu P, Speed TP (2001) Normalization for cDNA microarray data. In: Bittner ML, Chen Y, Dorsel AN, Dougherty ER (eds) Microarrays: optical technologies and informatics. Proc SPIE, vol 4266, pp 141–152
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


