Abstract
Cell misidentification and cross-contamination have plagued biomedical research for as long as cells have been employed as research tools. Examples of misidentified cell lines continue to surface to this day. Efforts to eradicate the problem by raising awareness of the issue and by asking scientists voluntarily to take appropriate actions have not been successful. Unambiguous cell authentication is an essential step in the scientific process and should be an inherent consideration during peer review of papers submitted for publication or during review of grants submitted for funding. In order to facilitate proper identity testing, accurate, reliable, inexpensive, and standardized methods for authentication of cells and cell lines must be made available. To this end, an international team of scientists is, at this time, preparing a consensus standard on the authentication of human cells using short tandem repeat (STR) profiling. This standard, which will be submitted for review and approval as an American National Standard by the American National Standards Institute, will provide investigators guidance on the use of STR profiling for authenticating human cell lines. Such guidance will include methodological detail on the preparation of the DNA sample, the appropriate numbers and types of loci to be evaluated, and the interpretation and quality control of the results. Associated with the standard itself will be the establishment and maintenance of a public STR profile database under the auspices of the National Center for Biotechnology Information. The consensus standard is anticipated to be adopted by granting agencies and scientific journals as appropriate methodology for authenticating human cell lines, stem cells, and tissues.
Keywords: Cell authentication, STR profiling, Consensus standard, Quality control
Introduction
Animal and human primary cell cultures, continuous (immortalized) cell lines, and tissues are of overwhelming importance to the biopharmaceutical industry and to biomedical research as reagents, therapeutic modalities, and as proxy materials for the study of more complex physiological systems. Cell cultures have, from the beginning, been at risk for misidentification due to labeling errors, incorrect classification by pathologists, and cross-contamination with other cell types. Continuous cell lines are potentially jeopardized due to the extended time these are in culture and the frequent manipulations involved in the course of feeding and subculturing. Human stem cell preparations which are propagated in the presence of non-human feeder cell layers are at risk of cross-contamination with the feeder cells. Tumor cells propagated by xenografting onto host animals are at risk of cross-contamination with the host cells.
We know the risks involved in establishing and maintaining cell cultures. We know that periodic identity testing (authentication) is the only way to prove that the cell we are studying is the cell that we believe it to be, and not a contaminating tumor cell line such as HeLa. Why then are many investigators blindly assuming that they are using correctly identified cells? Recent publications appear to indicate that the problem of cell misidentification is not going away. For instance, Berglind et al. (2008) evaluated the p53 status of 1,211 cell lines published between 1989 and 2007 and found discrepancies in the p53 status for 23% of the cell lines. Schweppe et al. (2008) evaluated 40 human thyroid cancer cell lines and found that only 23 of these actually had unique genetic profiles, as determined using short tandem repeat (STR) profiling and single nucleotide polymorphism analysis. Certain of the presumed thyroid cancer cell lines were found to have profiles matching colon cancer or melanoma cells. Another recent revelation was that of Boonstra et al. (2010) indicating that three widely used esophageal cancer cell lines are, in fact, derived from other tumor types. Dittmar et al. (2010) have reported two new cases of misidentification of supposed human cells. Their work clearly demonstrates that phenotypic evaluation alone cannot provide adequate assurance of the authenticity of a cell line. More extensive lists of misidentified cells are available from a number of sources (e.g., ATCC SDO Workgroup ASN-0002 2010; Capes-Davis et al. 2010).
Within the highly regulated biopharmaceutical industry, cell lines used as production substrates must be characterized for identity through phenotypic analysis and confirmation of animal species of origin (US FDA 1993). This, together with implementation of current good manufacturing practices, is believed to have contributed to the relatively low frequency of cell line misidentification reported in this industry (Nims and Herbstritt 2005).
Remediation of the problem of cell line misidentification within the biomedical research community may eventually need to be driven by requirements for authentication from granting agencies and journal editors. An international group of scientists is now preparing a consensus standard which will provide investigators with guidance on the appropriate methodology for authenticating human cells. In this article, we describe the rationale for and the process involved in preparing this standard.
Efforts to Remediate the Problem of Cell Misidentification
The earliest efforts toward tackling the problem of cell misidentification centered on disclosure of the issue through conference presentations and publications. Gartler (e.g., Gartler 1967) and Nelson-Rees (e.g., Nelson-Rees et al. 1974) were among the first and most vocal of those attempting to convince the scientific community of the seriousness of the issue. They hoped that such disclosures would motivate scientists to voluntarily take actions to remediate the problem.
More recently, Roland Nardone championed a series of efforts intended to reemphasize the seriousness of the cell misidentification problem and take any required steps to begin remediating the various causes for the continuing issue. His efforts began with the authoring of a white paper, entitled “Eradication of Cross-Contaminated Cell Lines: A Call for Action” (Nardone 2007). This paper presented recommendations for strict compliance measures in addition to continuing efforts to educate scientists. Nardone believed the time had come for granting agencies to demand cell line authentication as a condition for the receipt of funds and for journals to add a similar requirement to their instructions for authors for manuscripts submitted for publication.
As part of his efforts to convince granting agencies of the need for their participation in his overall remediation strategy, Nardone and a group of prominent cell scientists composed and signed an open letter to Michael O. Leavitt, Secretary of Health and Human Resources (Nardone et al. 2007), beseeching the NIH to take appropriate actions. On November 28, 2007, the NIH published an addition to their Guidelines for Research—Notice Regarding Authentication of Cultured Cell Lines (National Institutes of Health 2007) calling for diligence and more careful peer review.
Communications between Nardone (and others) and journal editors have achieved the desired result as slowly and surely, journals are beginning to add the requirement for cell authentication to their instructions for authors (e.g., Cell Biochemistry and Biophysics, In Vitro Cellular & Developmental Biology—Animal, International Journal of Cancer, and the journals of the American Association for Cancer Research).
Attempts to educate scientists in general of the need for cell authentication must go beyond simply raising the level of awareness of the problem. In his white paper, Nardone also stressed the need for training in cell authentication to be added to conference agendas. He recommended that societies sponsor conferences, workshops, and/or training activities to facilitate the adoption of cell line authentication standards (Nardone 2007).
As the requirement for cell authentication is adopted by granting agencies and scientific journals, the need for standardized methods and expectations regarding authentication itself to be defined becomes more critical. Recognizing this, an effort to prepare a consensus standard on authentication of human cells was initiated.
The Concept of the Consensus Standard
The idea of the consensus standard is to allow a greater input from the overall international biomedical community into standards. The derivation of a standard through the consensus process improves the chance of universal voluntary acceptance. In turn, that acceptance will foster reproducibility and comparability of research employing human cells. Such a consensus-driven standard, if universally adopted, should ultimately lead to a marked decrease in the misidentification of human cells used by the biomedical community.
The ATCC® Standards Development Organization
The mission of the ATCC® Standards Development Organization (SDO) is to develop best practices (standards) for use in the life science industry and to promote their global use, using a consensus-driven process that balances the viewpoints of the stakeholder community. Membership is free and open to all stakeholders in the biomedical community, including those involved in the development, production, application, and regulation of life science products. Stakeholders include, but are not limited to, members from academia, government, regulatory, and industry. All members are participants in the consensus review, comment, and voting process.
In 2007, the SDO became the first biological resource organization to become an American National Standards Institute (ANSI)-accredited standards development organization. ANSI accreditation ensures that procedures used by standards developers meet requirements for openness, balance, consensus, and due process.
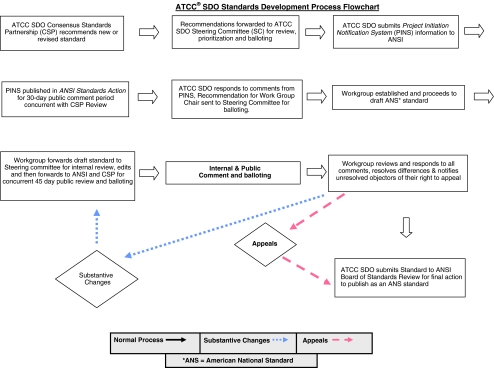
The standard development process employed by the ATCC® SDO is shown in Fig. 1.
Figure 1.
ATCC SDO standards development process flowchart.
ATCC SDO Workgroup ASN-0002
ATCC® SDO workgroup ASN-0002 “Development of a consensus standard for the authentication of human cells: standardization of STR profiling” was formally assembled in early 2009 as a result of a proposal submitted in 2008 by John Masters and Roland Nardone. The workgroup constitutes an international group of concerned and experienced scientists. Chaired by Masters, the workgroup includes individuals with relevant and current experience in DNA profiling technologies, as well as “stakeholders” or representatives from major cell repositories, industry, academia, and government agencies.
Preparation of the Standard.
Working under the auspices of the ATCC SDO, the ASN-0002 workgroup has met monthly since early 2009. The overall effort was divided between two subgroups which have met independently at monthly or more frequent intervals.
The first subgroup is charged with drafting the introduction to the Standard, defining what is meant by “human cell line authentication,” describing the historical aspects, from early discovery of cell line misidentification through to the present efforts encouraging remediation of the problem. The subgroup also is delineating the causes of cell line misidentification, surveying the existing technologies for cell line authentication, and providing the rationale for selection of STR profiling for the Standard. The subgroup is chaired by Raymond Nims.
The second subgroup, chaired by Yvonne Reid, is fleshing out the procedural details of the general protocol to be recommended for STR profiling. This subgroup is also responsible for determining the format and structure of an associated public database of STR profiles of human cell lines. Such a database is indispensable with regard to the establishment of a set of global reference STR profiles for human cell lines and critical in support of the Standard.
Methodology.
STR profiling was selected as the recommended authentication technology for inclusion in this Standard primarily because it is capable of resolving human cells to the individual level. In contrast, historically important authentication technologies such as karyotyping, isoenzyme analysis, immunotyping, and human leukocyte antigen typing do not have sufficient resolving (discriminating) power to enable unambiguous authentication of human cells to the individual level (see ATCC SDO Workgroup ASN-0002 2010 for a more detailed discussion of the relative discriminating power of these technologies). In addition, the STR profiling technology is commercially available in kit form and is rapid and economical. Masters et al. (2001) demonstrated that the technology can provide a universal reference standard for human cell lines. The STR profiling technology, as normally used, detects only human cells, and therefore additional methods may need to be used to detect contamination with non-human cells. There are a number of different commercial kits now available for STR profiling, and the users will be encouraged to follow the protocol specific to the kit being used and to refer to the Standard for additional methodological information. The most important aspects of the Standard will be the discussions on the numbers and types of loci to be evaluated, quality control of the data, interpretation of the results (matching criteria, loss of alleles, etc.), and implementation of a universal STR database.
Associated database.
Associated with the issuance of the Standard will be the construction of a comprehensive and continuously updatable public database of STR profiles based on results subject to agreed-upon interpretation guidelines and quality control parameters. Comparison of STR profiles generated from individual cell stocks to such a database will help reduce the frequency of misidentification of human cells, enhance confidence in results, assure the user’s ability to compare scientific results between laboratories, and verify that important data originated from intended samples. STR profiles submitted to the database may, at the request of the user, be verified by staff at the National Institutes of Standards and Technology (NIST). The user also will have the option of submitting STR profiles to the database without verification. The database will indicate which profiles have been verified by NIST. The STR database will be established and maintained under the auspices of the National Center for Biotechnology Information and NIST.
Timeline for completion of the Standard.
The Standard, once drafted, will be submitted to the SDO Steering Committee for initial review (Fig. 1). After a nominal 14-d review period, the ASN-0002 workgroup will have a chance to respond to any comments provided by the Steering Committee. At this point, the Standard will be submitted for public review and comment. ANSI notifies the public via its weekly publication “ANSI Standards Action.” Concurrent with this, the Standard will be sent to all ATCC SDO members for review and comment. At the end of the 45-d public review and comment period, the workgroup will review and respond to all negative comments, resolve differences, and notify unresolved objectors of their right to appeal, if necessary.
The final document will be submitted for review and approval as an American National Standard by the ANSI.
Once the consensus standard has been approved and published by ANSI, the workgroup will take appropriate actions to raise awareness throughout the biomedical community of the existence of the new standard.
Anticipated Flow and Impact
For newly developed human primary cell cultures and cell lines, including feeder layer-dependent human stem cells and tumor cells propagated through xenografting, an initial STR profile should be established on the earliest material, for example biopsy, frozen tissue, or formalin-fixed paraffin-embedded tissue. For feeder layer-dependent human stem cell preparations, DNA amplification and barcoding (e.g., Cooper et al. 2007), an isoenzyme analysis assay or an alternative species identification method may need to be performed to demonstrate that there are no cross-contaminating mouse feeder cells in the preparation. Additional testing may also be necessary in the case of tumor cell isolation from xenografts to demonstrate that there are no host cells remaining in the recovered tumor cell line.
For existing human cell lines, investigators will be encouraged to: (1) check the public database to see if the cell line is represented within the STR database; (2) perform an STR profile and compare the results to those within the STR database; and (3) ensure that the STR database indicates that this cell line is not misidentified. The Standard will provide the necessary matching criteria.
Continuous human cell lines which are manipulated within laboratories employing non-human cell lines may need to be monitored periodically for non-human cell cross-contamination using one of the cell species identification methods mentioned above (isoenzyme analysis or DNA amplification and barcoding).
To the degree to which the Standard is adopted and complied with, issuance of the Standard will have a significant beneficial impact on the quality and validity of research based upon the use of human cells.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- ATCC SDO Workgroup ASN-0002 Cell line misidentification: the beginning of the end. Nature Rev. Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- Berglind H, Pawitan Y, Kato S, Ishioka C, Soussi T. Analysis of p53 mutation status in human cancer cell lines. Cancer Biol. Ther. 2008;7:701–710. doi: 10.4161/cbt.7.5.5712. [DOI] [PubMed] [Google Scholar]
- Boonstra JJ, van Marion R, Beer DG, Lin L, Chaves P, Ribeiro C, Pereira AD, Roque L, Darnton SJ, Altorki NK, Schrump DS, Klimstra DS, Tang LH, Eshleman JR, Alvarez H, Shimada Y, van Dekken H, Tilanus HW, Dinjens WNM. Verification and unmasking of widely used human esophageal adenocarcinoma cell lines. J. Natl. Cancer Inst. 2010;102:1–4. doi: 10.1093/jnci/djp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RAF, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer. 2010;127:1–8. doi: 10.1002/ijc.25379. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P. Species identification in cell culture: a two-pronged molecular approach. In Vitro Cell. Dev. Biol. Anim. 2007;43:344–351. doi: 10.1007/s11626-007-9060-2. [DOI] [PubMed] [Google Scholar]
- Dittmar KEJ, Simann M, Zghoul N, Schön O, Meyring W, Hannig H, Macke L, Dirks WG, Miller K, Garritsen HSP, Lindenmaier W. Quality of cell products: authenticity, identity, genomic stability and status of differentiation. Transfus. Med. Hemother. 2010;37:57–64. doi: 10.1159/000284401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM. Genetic markers as tracers in cell culture. Natl. Cancer Inst. Monogr. 1967;26:167–195. [PubMed] [Google Scholar]
- Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, Toji LH, Ohno T, Tanabe H, Arlett CF, Kelland LR, Harrison M, Virmani A, Ward TH, Ayres KL, Debenham PG. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl. Acad. Sci. USA. 2001;98:8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone RM. Eradication of cross-contaminated cell lines: a call for action. Cell Biol. Toxicol. 2007;23:367–72. doi: 10.1007/s10565-007-9019-9. [DOI] [PubMed] [Google Scholar]
- Nardone R. M.; Masters J. R. W.; Bradlaw J. A; Jacobsen L. B.; Nims R. W.; Price P. J.; Lewis D.; Stacey G.; McCormick J. J.; Gartler S. M.; Pathak S.; Butler J. M.; Buehring G. C.; Massaro E. J.; Steuer A. F.; Gold M.; Freshney R. I.; Krause D.; O’Brien S. J. An open letter regarding the misidentification and cross-contamination of cell lines: significance and recommendations for correction. July 11, 2007. http://cellbank.nibio.go.jp/cellbank/qualitycontrol/OL7-11-07.pdf.
- National Institutes of Health. Notice regarding authentication of cultured cell lines. Nov. 28, 2007. http://grants.nih.gov/grants/guide/notice-files/NOT-OD-08-017.html.
- Nelson-Rees WA, Flandermeyer RR, Hawthorne PK. Banded marker chromosomes as indicators of intraspecies cellular contamination. Science. 1974;184:1093–1096. doi: 10.1126/science.184.4141.1093. [DOI] [PubMed] [Google Scholar]
- Nims RW, Herbstritt CJ. Cell line authentication using isoenzyme analysis: strategies for accurate speciation and case studies for detection of cell line cross-contamination using a commercial kit. BioPharm Int. 2005;18:76–82. [Google Scholar]
- Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland JA, Smallridge RC, Haugen BR. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J. Clin. Endocrinol. Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US FDA. Points to consider in the characterization of cell lines used to produce biologicals. CBER, 1993. http://www.fda.gov/downloads/BiologicsBloodVaccines/SafetyAvailability/UCM162863.pdf.