Abstract
Whole-genome sequencing of an Irish person reveals hundreds of thousands of novel genomic variants. Imputation using previous known information improves the accuracy of low-read-depth sequencing.
See research article: http://genomebiology.com/2010/11/9/R91
Research highlight
In the past 10 years, numerous human genomic variants have been discovered and catalogued, mostly through the efforts of the International HapMap project and personal genome studies [1]. Information on human genomic variants may serve as a valuable resource for developing personalized medicine because some of these variants could potentially predispose humans to complex diseases. The 2009 version (version 130) of the dbSNP database included approximately 13.9 million (13.9 M) single nucleotide polymorphisms (SNPs) and 4.5 M small insertions and deletions (indels). However, many issues need to be addressed before personalized medicine becomes a reality. These include an understanding of: the kind and number of variants that exist in the entire human genome; the number of populations and individuals needed to detect most, if not all, human genomic variants with efficiency and accuracy; the frequency of common and rare variants in an individual genome; and finally the number of variants that influence human diseases.
Recent advancements in next-generation sequencing technology have dramatically decreased both the cost and the time required for sequencing [1]. Consequently, in the past few years, sequencing of individual genomes (personal genomes) has gained in popularity. Currently, whole-genome sequencing is thought to be the best way to detect human genomic variations because it could detect novel variants [2]. Excluding cancer genomes, so far at least 15 personal genomes have been sequenced and analyzed using various platforms (Figure 1). In this issue of Genome Biology, Tong and colleagues [3] present data on the first whole-genome sequence of an Irish person using the Illumina Genome Analyzer platform. As the authors [3] suggest, the Irish population could be a good candidate for genomic studies as it is isolated and located in the western fringes of Europe, and thus may possess many polymorphisms unique to this population.
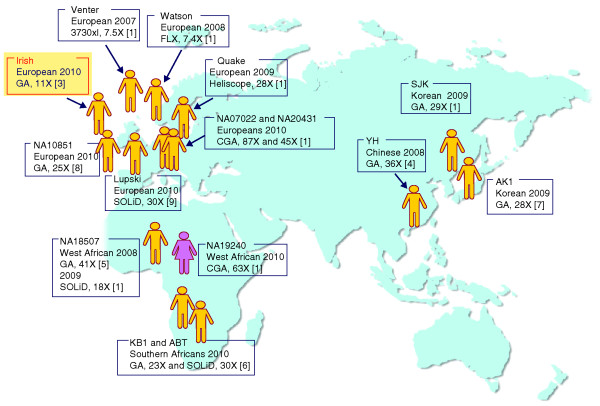
Figure 1.
Personal genomes sequenced so far. Each male (yellow) or female (pink) sequenced is shown in the approximate geographical position of their ethnic origin. The name or codename and ethnicity of the individual sequenced, the date of publication, the sequencing platform used, the coverage and the reference are given for each genome. Platforms: 3730xl (Applied Biosystems by Life Technologies, Carlsbad, CA, USA); Complete Genomics Analysis Platform (CGA) Complete Genomics, Mountain View, CA, USA); FLX (Roche, Basel, Switzerland); Genome Analyzer (GA) (Illumina, San Diego, CA, USA); Heliscope (He6licos BioScience, Cambridge, MA, USA); SOLiD (Applied Biosystems by Life Technologies).
The authors [3] generated 440 M short reads from the Irish genome and obtained 11X sequencing coverage genome-wide. Despite the lower read-depth compared with other personal genomes (Figure 1), they discovered more than 3 M SNPs. Approximately 13% of these SNPs (0.4 M, approximately 3% of the total number of SNPs catalogued in dbSNP version 130) may be designated as new variants, as they were not previously deposited in the SNP database. They also found more than 20,000 potentially disease-related new SNPs. For example, they have identified a new non-synonymous SNP in the Macrophage-stimulating 1 (MST1) gene, which may have a functional role in inflammatory bowel disease. In addition, the authors detected about 200,000 short indel polymorphisms, half of which have not been reported before. Their results [3] clearly suggest that the human genome still harbors a tremendous number of undetected and often population-specific variants, and they provide justification for more personal genome sequencing studies from worldwide populations.
Sequencing read-depth: improving accuracy by imputation
Despite these interesting results from the Irish genome analysis [3], its low read-depth of sequencing coverage (11X) must be examined in some detail. With the exception of the first two personal genomes sequenced by relatively longer reads, most of the other human whole-genome analyses were carried out using more than 20X sequencing coverage [1]. Low coverage may dramatically reduce the accuracy of genome sequencing because it risks misclassification of heterozygous variants as wild type (missing the variant; this is called undercalling) or misclassification of heterozygous variants as homozygous ones (missing the wild-type allele; overcalling). Consequently, in low-depth sequencing, both the detection of sensitivity and the positive predictive value of genomic variants are compromised [4].
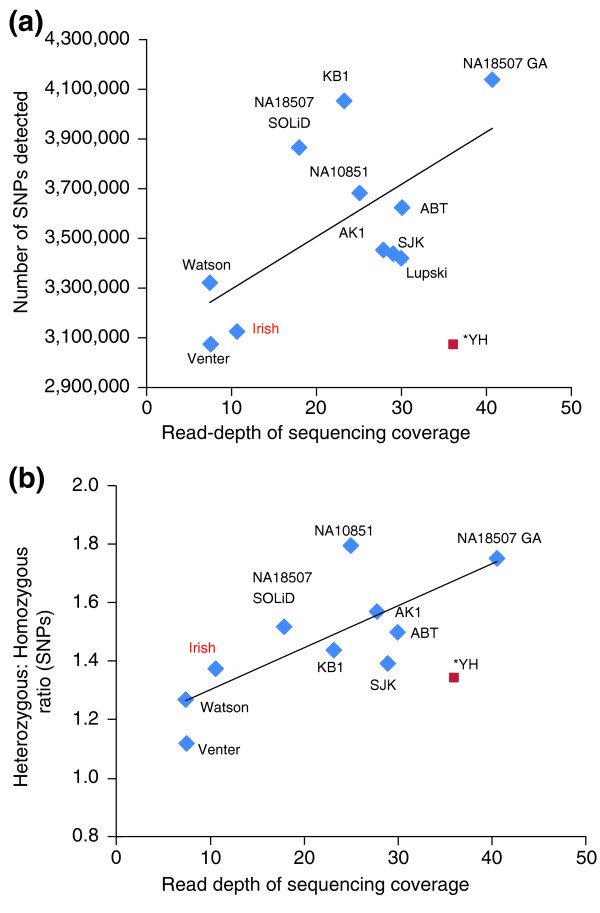
When we look at the data from individual genomes analyzed by high-depth sequencing using the Illumina platform, most of the personal genomes have more than 3.4 M SNPs, approximately 0.3 M more than the number for the Irish (Figure 2a) [4-7]. Recently published personal genomes of European origin also have more than 3.4 M SNPs (NA10851 and Lupski) [8,9]. From this perspective, if the Irish genome was sequenced at higher depth, it could potentially reveal an additional 0.3 M SNPs (Figure 2a). Many of the additional variants might be heterozygous and novel (Figure 2b). Furthermore, higher-depth sequencing would not only increase the total number of genome variants, but would also bring the false discovery rate down to less than 0.1% from the current 1.4%.
Figure 2.
The influence of read-depth on discovering personal genomic variants. Sequences included in our analysis are represented by blue squares, with names or codenames as shown in Figure 1. The lines indicate correlations; the YH genome (red square) was not included in the correlation analysis because it is thought to be an outlier, possibly owing to its higher percentage of single-end-read sequencing. (a) Higher read-depth sequencing can reveal more SNPs, as shown by the positive correlation between read-depth and number of SNPs detected. (b) The ratio of heterozygous to homozygous SNPs shows a positive relationship with read-depth of sequencing, suggesting that high-depth sequencing can detect heterozygous SNPs better.
We could also consider the relative merits of personal genome sequencing from another perspective. Do we want all personal genome sequencing to exceed 99.9% accuracy? If personal genome data are not used for diagnostic purposes, why should we invest a lot of resources, time and effort in doing additional 10X to 20X sequencing to boost the accuracy from 99% to 99.9%? With limited resources, precise estimation of an individual's genetic variation is in direct conflict with analyzing as many individual genomes as possible to obtain a broader picture of the genomic architecture of a given population. For instance, if one is not interested in understanding the detailed genomic architecture of a specific person, but only in gaining a broader understanding of genomic characteristics of an ethnic group, then it would be more prudent to sequence many individuals with lower depth than a limited number with high depth. One of the attractive features of the study by Tong and colleagues [3] is that they have suggested ways to improve the precision of low-coverage sequencing without investing additional resources. The authors [3] have demonstrated that the accuracy of known SNPs in low-depth sequencing can be dramatically improved by integrating the previously known genotype or haplotype data assembled for European populations by the HapMap and 1000 Genomes projects into low coverage sequencing projects. The authors have shown that over 99% accuracy can be achieved using imputation methods using these other datasets, with only 5X sequencing coverage. What is even more interesting is that just 2X sequencing can provide genotype calls with over 95% accuracy.
These tantalizing observations suggest that even low-depth sequencing can be effective with prior detailed information on related genomes. In addition, with accurate genomic data on Irish genomes, the power of imputation methods could be even better, and this would also be the case for other populations. These predictions further emphasize the need for additional personal whole-genome sequencing of a large number of individuals from diverse ethnic groups.
Evidence for positive selection
In the past, many investigators have reported signatures of selection in the human genome. Tong and colleagues [3] have used an interesting approach to study positive selection in the human genome using the Irish genome and the available sequence data on nine personal genomes from previous studies. Despite the small sample size and varied sequencing methods used in previous studies, this attempt can be considered as an initial step toward developing an 'official' whole-genome population genetics study. Thus, this study may give a taste of future insights into population genetics research and of some of the challenges specific to whole human genome data. This study has shown evidence for balancing selection at the sites related to olfactory and taste receptors, mostly confirming the previous results from genome-wide SNP studies. Also, their analysis of ten genomes reveals elevated positive selection in fairly recently duplicated genes. Taken together [3], these results clearly show that whole-genome analysis can shed new light on the field of human evolutionary genetics.
Some population statistics based on haplotype patterns would benefit from complete sequence data, since relatively accurate haplotype phase can be inferred. Recently, Higasa and colleagues [10] showed that errors of population-based haplotype inference affected the results of some statistics for positive selection more than others. Currently available haplotype inference software may have limitations depending on the data size and availability of external information. Development of accurate haplotyping and haplotype inference methods suitable for genome sequence data will be a key to successful population genetics study using haplotype information.
Ten years have passed since the first drafts of human genome sequences were published, and we now have at least 15 individual whole-genome sequences, thanks to the dramatic progress in sequencing technology. However, there remain many unsolved questions on human genome diversity. To expand our understanding, we need more personal genome data from worldwide populations. With limited resources, quality (accuracy) and quantity (number of individuals) are always difficult to balance. The study by Tong and colleagues [3] is the first attempt to tackle this question. This approach is the first of its kind and will likely be improved on in the near future as other researchers see the potential of such an approach, however, this method and the findings, will help to open new prospect in the field of human genome research.
Acknowledgements
We thank DR Govindaraju for his valuable advice.
References
- Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes Dev. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Response: Why it is time to sequence. Cell. 2010;142:353–355. doi: 10.1016/j.cell.2010.07.027. [DOI] [Google Scholar]
- Tong P, Prendergast J, Lohan AJ, Farrington SM, Cronin S, Friel N, Bradley D, Evans A, Wilson JF, Loftus B. Sequencing and analysis of an Irish human genome. Genome Biol. 2010;11:R91. doi: 10.1186/gb-2010-11-9-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L. et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, Alkan C, Kidd JM, Sun Y, Drautz DI, Bouffard P, Muzny DM, Reid JG, Nazareth LV, Wang Q, Burhans R, Riemer C, Wittekindt NE, Moorjani P, Tindall EA, Danko CG, Teo WS, Buboltz AM, Zhang Z, Ma Q, Oosthuysen A. et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Lee S, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Park H, Kim JY, Gokcumen O, Mills RE, Zaranek AW. et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Hong D, Kim S, Park S, Kim S, Lee S, Park H, Kim J, Seo J. Reference-unbiased copy number variant analysis using CGH microarrays. Nucleic Acids Res. 2010. in press . [DOI] [PMC free article] [PubMed]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higasa K, Kukita Y, Kato K, Wake N, Tahira T, Hayashi K. Evaluation of haplotype inference using definitive haplotype data obtained from complete hydatidiform moles, and its significance for the analyses of positively selected regions. PLoS Genet. 2009;5:e1000468. doi: 10.1371/journal.pgen.1000468. [DOI] [PMC free article] [PubMed] [Google Scholar]