This work systematically examines the expression as well as the actin binding and actin regulatory activities of the Arabidopsis LIM proteins, finding differences in expression and changes in activity, which, for a subset of the LIM proteins, depends on pH and calcium.
Abstract
Recently, a number of two LIM-domain containing proteins (LIMs) have been reported to trigger the formation of actin bundles, a major higher-order cytoskeletal assembly. Here, we analyzed the six Arabidopsis thaliana LIM proteins. Promoter-β-glucuronidase reporter studies revealed that WLIM1, WLIM2a, and WLIM2b are widely expressed, whereas PLIM2a, PLIM2b, and PLIM2c are predominantly expressed in pollen. LIM-green fluorescent protein (GFP) fusions all decorated the actin cytoskeleton and increased actin bundle thickness in transgenic plants and in vitro, although with different affinities and efficiencies. Remarkably, the activities of WLIMs were calcium and pH independent, whereas those of PLIMs were inhibited by high pH and, in the case of PLIM2c, by high [Ca2+]. Domain analysis showed that the C-terminal domain is key for the responsiveness of PLIM2c to pH and calcium. Regulation of LIM by pH was further analyzed in vivo by tracking GFP-WLIM1 and GFP-PLIM2c during intracellular pH modifications. Cytoplasmic alkalinization specifically promoted release of GFP-PLIM2c but not GFP-WLIM1, from filamentous actin. Consistent with these data, GFP-PLIM2c decorated long actin bundles in the pollen tube shank, a region of relatively low pH. Together, our data support a prominent role of Arabidopsis LIM proteins in the regulation of actin cytoskeleton organization and dynamics in sporophytic tissues and pollen.
INTRODUCTION
Actin is one of the most abundant and highly conserved proteins in eukaryotes. In the cytoplasm, actin monomers polymerize into actin filaments (AFs), which constitute the core elements of the actin cytoskeleton, providing mechanical support to the cytoplasm and serving as tracks for myosin-dependent intracellular transport (Hepler et al., 2001; Shimmen, 2007). In animal cells, AF-myosin interactions power cell division, cell contraction, and cell migration. AF polymerization itself is used as a driving force that directs the growth of membrane protrusions and enables cells to alter their shape and to move. In plant cells, AFs are essential for the establishment and maintenance of cell polarity (Vidali and Hepler, 2001) as well as for the formation of plant-specific cytoskeletal structures, such as the phragmoplast and the preprophase band (Schmit, 2000). In addition to its direct functions, the actin cytoskeleton is a key target of many signaling events and acts itself as a transducer of signals in both animal and plant cells (Drobak et al., 2004). To fulfill its various roles, the actin cytoskeleton requires a sophisticated regulatory system to control its organization and dynamics at both spatial and temporal levels. Primary components of this system are the actin binding proteins (ABPs) that directly interact with monomeric and/or polymerized actin to promote AF nucleation, polymerization, depolymerization, stabilization, severing, capping, and cross-linking (Winder and Ayscough, 2005). ABP activities themselves are tightly regulated by many cellular parameters, including Ca2+, pH, phosphoinositides, phosphorylation, and protein–protein interactions. The coordinated regulation of ABP activities ultimately defines AF positioning, turnover, and supraorganization in orthogonal networks or parallel bundles.
ABPs have been categorized into different classes according to the main or the historically first function attributed to them. Each class subdivides into several ABP families, which differ in their domain composition and/or organization and usually comprise several members. However, many ABPs display multiple actin regulatory activities. In addition, all the members belonging to one given ABP family do not necessarily retain the full range of possible activities, neither do they respond similarly to regulatory signals. A typical example is the villin family whose members exhibit distinct features. Indeed, the Arabidopsis thaliana VLIN1 generates actin bundles in an unusual Ca2+/CaM-independent manner and lacks the nucleating, severing, and capping activities that are predicted (although not yet experimentally confirmed) for other Arabidopsis villins (Huang et al., 2005). Other villins, such as the lily (Lilium longiflorum) villin P-135-ABP, have been biochemically demonstrated to bundle and cap AFs as well as to accelerate the rate of AF depolymerization in a Ca2+/CaM -sensitive manner (Yokota et al., 2005). Another functionally heterogeneous ABP family is the formin family. In Arabidopsis, it comprises >20 members that significantly differ in their domain organization, suggesting disparities in activities and modes of regulation (Blanchoin and Staiger, 2008; Grunt et al., 2008). Hence, the biological functions of one given ABP family cannot be fully appreciated by examining a limited number of its members.
Recently, a number of vertebrate LIM domain proteins belonging to the Cys-rich protein (CRP) family and several structurally related plant proteins (hereafter referred as to plant LIMs) have been shown to function as ABPs (Grubinger and Gimona, 2004; Tran et al., 2005; Thomas et al., 2006; Wang et al., 2008a). CRPs and plant LIMs define a subset of relatively short (~200 amino acids) proteins that are characterized by two LIM domains and an unusually long interLIM spacer (40 to 50 amino acids; Weiskirchen and Gunther, 2003). In vitro, both chicken CRP1 and tobacco (Nicotiana tabacum) WLIM1 directly bind to AFs and trigger the formation of thick actin bundles. Importantly, overexpression of CRP1 and WLIM1 proteins was sufficient to significantly increase the bundling of AFs in rat fibroblasts and tobacco cells, respectively (Tran et al., 2005; Thomas et al., 2006, 2008). Similar results have been reported for the pollen-enriched Lilium LIM1 protein (Wang et al., 2008a), suggesting that actin bundling is a prevalent, rather than an accessory, activity among plant LIMs. However, this remains to be confirmed by the analysis of additional plant LIMs.
Interestingly, LIM1 overexpression induced an oscillatory formation of asterisk-shaped AF aggregates in the subapical region of growing pollen tubes (Wang et al., 2008a). In vitro investigations have suggested that the interaction of LIM1 and AFs is regulated by pH and Ca2+, two central regulators of pollen tube oscillatory growth that are assumed to function through the activation/deactivation of several ABPs (Cheung and Wu, 2008; Staiger et al., 2010). Therefore, LIM1 has been proposed, along with pH and Ca2+, to be part of the central oscillatory mechanism that regulates actin cytoskeleton remodeling during pollen tube elongation (Wang et al., 2008a). However, evidence of pH- and/or Ca2+-dependent regulation of Lillium LIM1 or other plant LIMs in the context of live cells is still lacking.
Plants possess multimember LIM gene families, with six members in Arabidopsis and rice (Oryza sativa) and up to 12 members in poplar (Populus spp; Arnaud et al., 2007). Earlier studies have suggested the existence of two main LIM gene subfamilies that differ in their expression patterns (Eliasson et al., 2000). The WLIM subfamily includes genes that exhibit a wide expression pattern throughout the sporophytic plant tissues, whereas the PLIM subfamily includes genes with exclusive or preferential expression in pollen. A more complex classification of LIM genes has been recently proposed based on the phylogenetic analysis of 149 LIMs and the comparison of available expression data (Arnaud et al., 2007). According to this classification, the Arabidopsis LIM gene family comprises three vegetative (WLIM1 and WLIM2a and b) and three reproductive (PLIM2a-c) isoforms, one of which, PLIM2a, has been shown to be expressed in flowers (Alves-Ferreira et al., 2007).
By conducting a detailed analysis of the entire LIM gene/protein family in Arabidopsis, we addressed a number of central issues regarding the functions and modes of action of plant LIMs. Organ- and tissue-specific expression patterns of the six Arabidopsis LIMs were characterized in transgenic plants expressing a β-glucuronidase (GUS) reporter gene under the control of individual LIM regulatory sequences. Our data demonstrate the existence of two differentially expressed LIM subfamilies, although not always with a strict separation between vegetative and reproductive patterns. Both in vitro and in vivo investigations provide evidence that all six Arabidopsis LIMs display actin binding, -stabilizing, and -bundling activities, although with different efficiencies. By contrast, all LIMs did not respond similarly to pH and [Ca2+] variations. Most strikingly, the three pollen-enriched PLIMs were inactivated by relatively high pH values (≥6.8), whereas the three WLIMs remained fully active in the range of conditions tested. PLIM2c exhibited additional responsiveness to calcium. A domain analysis pointed out a central role of the C-terminal domain in the regulation of PLIM activities. Specific pH-dependent regulation of PLIMs was confirmed in live Arabidopsis cells whose pH was artificially modified. Finally, a green fluorescent protein (GFP)-fused PLIM2c fusion protein expressed in pollen interacted with the long actin bundles in the shank of elongating pollen tubes and occasionally decorated a subapical actin fringe-like structure. Together, our data strongly support that plant LIMs define a highly specialized ABP family, which contributes to the regulation of actin bundling in virtually all plant cells. Specific control of PLIM actin regulatory activities by pH is particularly relevant with regard to the potential biological functions of these proteins in tip-growing pollen tubes.
RESULTS
Tissue-Specific Expression of Arabidopsis LIM Genes Reveals Two Differentially Expressed Subfamilies
According to a recent wide-range phylogenetic analysis, the six Arabidopsis LIM genes have been renamed WLIM1 (At1g10200), WLIM2a (At2g39900), WLIM2b (At3g55770), PLIM2a (At2g45800), PLIM2b (At1g01780), and PLIM2c (At3g61230) (Arnaud et al., 2007). The possibility that individual Arabidopsis LIMs have specific functions in specific tissues or cell types due to nonoverlapping expression patterns was examined in Arabidopsis plants expressing the GUS reporter gene under the control of individual LIM gene 5′upstream sequences (ProLIM).
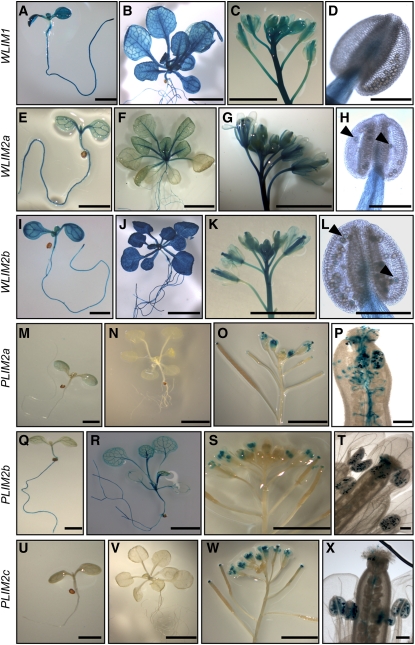
Preliminary RNA gel blot analysis shows expression of all three WLIMs in a wide range of organs, including roots, leaves, stem, flowers, and siliques (see Supplemental Figure 1A online). By contrast, PLIM transcripts were predominantly detected in flowers. GUS histochemical assays confirmed and refined these data (Figure 1). Indeed, ProWLIM1-GUS and ProWLIM2b-GUS expression was high in virtually all organs and tissues, including root, stem, leaf, and apical bud tissues (Figures 1A to 1D and 1I to 1L). WLIM2a promoter activity was also strong in roots and leaf vasculature but resulted in rather weak staining in other leaf tissues (Figures 1E to 1H). Significant expression of the three ProWLIM-GUS fusions was detected in floral tissues, including peduncle, pedicels, pistils, and stamen filaments (Figures 1C, 1D, 1G, 1H, 1K, and 1L). However, no (ProWLIM1) or only a faint signal (ProWLIM2a and ProWLIM2b) could be detected in pollen grains, even after long staining periods (>15 h; Figures 1D, 1H, and 1L). By contrast, the three ProPLIM-GUS fusions exhibited prominent expression in pollen grains (Figures 1O, 1P, 1S, 1T, 1W, and 1X). Particularly high expression levels of PLIMs were indicated by a fast (<30 min) and intense staining. ProPLIM2c-GUS expression exclusively appeared in pollen (Figures 1U to 1X). A weak GUS staining was sometimes (four out of nine lines) observed in leaves for Pro-PLIM2a (Figure 1M), and PLIM2b promoter activity was regularly detected in roots and leaf vasculature (Figures 1Q and 1R). Together, these results are in good agreement with the analysis of publicly available microarray data (see Supplemental Figures 1B and 1C online). Indeed, much higher signal intensities were detected for WLIMs than for PLIMs in vegetative tissues (see Supplemental Figure 1B online), whereas the situation was inverted in pollen (seeSupplemental Figure 1C online). In addition, PLIM2b was the PLIM subfamily member exhibiting the highest expression level in vegetative tissues (see Supplemental Figure 1B online). The strong upregulation of the three PLIMs in tricellular and mature pollen grains suggests that PLIM activity is required during the late stages of pollen development and during pollen germination.
Figure 1.
Arabidopsis LIM Gene Expression Patterns in Transgenic Plants Expressing Fusions of LIM Regulatory Sequences with the GUS Reporter Gene.
(A) to (D) ProWLIM1-GUS expression.
(E) to (H) ProWLIM2a-GUS expression.
(I) to (L) ProWLIM2b-GUS expression.
(M) to (P) ProPLIM2a-GUS expression.
(Q) to (T) ProPLIM2b-GUS expression.
(U) to (X) ProPLIM2c-GUS expression.
Histological GUS assays were performed on 7-d-old seedlings ([A], [E], [I], [M], [Q], and [U]; bars = 2 mm), 4-week-old plantlets ([B], [F], [J], [N], [R], and [V]; bars = 5 mm), inflorescences ([C], [G], [K], [O], [S], and [W]; bars = 5 mm), anthers ([D], [H], and [L]), pistil with pollen tubes (P), and single flowers ([T] and [X]). Bars in (D), (H), (L), (P), (T), and (X) = 200 μm. Arrows in (H) and (L) indicate faint-blue staining in pollen grains.
In conclusion, our data provide strong evidence for the separation of Arabidopsis LIMs into two subfamilies with different, to some extent complementary, expression patterns. WLIM1, WLIM2a, and WLIM2b are widely expressed in most sporophytic tissues with no or very weak expression in pollen, whereas PLIM2a, PLIM2b, and PLIM2c are predominantly and abundantly expressed in pollen grains.
The Six Arabidopsis LIM Proteins Interact with the Actin Cytoskeleton in Different Cell Types
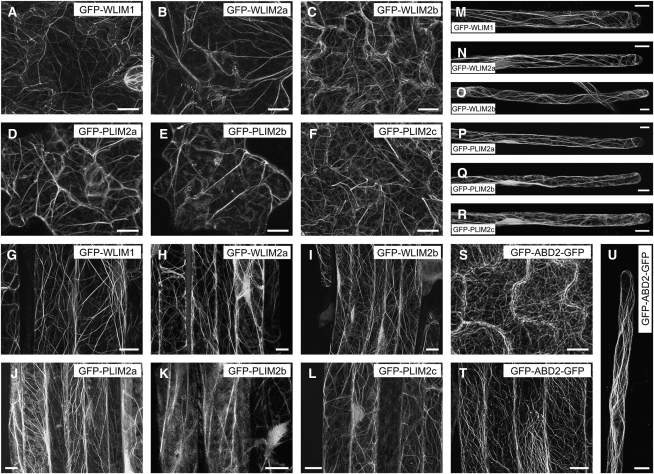
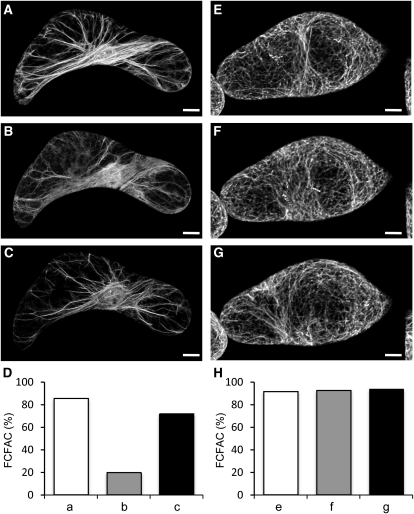
Two tobacco and one lily LIM have been previously reported to interact with the actin cytoskeleton in live cells (Thomas et al., 2006; Cheung et al., 2008; Wang et al., 2008a). The possibility that all six members of the Arabidopsis LIM family display a similar activity was examined in transgenic Arabidopsis plants that constitutively express individual LIMs fused to GFP (GFP-LIMs). Figure 2 shows typical confocal microscopy images obtained for different cell types, including leaf epidermal, root, and root hair cells. All six GFP-LIMs associated with a cytoplasmic filamentous network (Figures 2A to 2R) similar to the one revealed by the fimbrin-derived actin cytoskeleton marker GFP-ABD2-GFP (Wang et al., 2008b; Figures 2S to 2U). This network was confirmed to be the actin cytoskeleton by both rhodamine-phalloidin colabeling and latrunculin B depolymerizing experiments (see Supplemental Figure 2 online). Comparison of confocal images suggested that PLIMs interact less efficiently with the cytoskeleton than WLIMs. Indeed, most of GFP-PLIM–expressing cells exhibited a relatively high level of diffuse cytoplasmic fluorescence, whereas GFP-WLIMs more sharply decorated the cytoskeleton (cf. Figures 2A to 2C to 2D to 2F, 2G to 2I to 2J to 2L, and 2M to 2O to 2P to 2R). Differences in the subcellular distribution of GFP-PLIMs and GFP-WLIMs were further characterized by quantifying the fraction of cytoplasmic fluorescence associated with the cytoskeleton (FCFAC) in GFP-PLIM2c, GFP-WLIM1, and GFP-ABD2-GFP (control) expressing cells (see Supplemental Figure 3A online). In primary root cells, ~53% ± 5% of the fluorescent signal due to GFP-PLIM2c concentrated on the cytoskeleton. In root hairs, the FCFAC dropped to 31% ± 5%, indicating that roughly 70% of the GFP-PLIM2c population was unbound in the cytoplasm. By contrast, GFP-WLIM1 and GFP-ABD2-GFP predominantly associated with the cytoskeleton, as indicated by FCFAC values of roughly 70 and 90% in both types of cells, respectively. Immunoblot analysis performed with an anti-GFP antibody confirmed that GFP-WLIM1 and GFP-PLIM2c were expressed at similar levels in the Arabidopsis lines used in the above analyses, ruling out that results were due to differences in transgene expression levels (see Supplemental Figure 3B online).
Figure 2.
Localization of GFP-Fused LIMs in Different Tissues of Transgenic Arabidopsis Seedlings.
Typical fluorescent patterns observed for GFP-WLIM1 ([A], [G], and [M]), GFP-WLIM2a ([B], [H], and [N]), GFP-WLIM2b ([C], [I], and [O]), GFP-PLIM2a ([D], [J], and [P]), GFP-PLIM2b ([E], [K], and [Q]), GFP-PLIM2c ([F], [L], and [R]), and the actin cytoskeleton marker GFP-ABD2-GFP ([S] to [U]) in epidermal leaf cells ([A] to [F] and [S]), main root cells ([G] to [L] and [T]), and root hairs ([M] to [R] and [U]). Bars = 10 μm.
Remarkably, no obvious developmental or morphological phenotype was noticed in LIM-overexpressing transgenic plants. However, confocal microscopy images revealed substantial modifications of the actin cytoskeleton organization. Indeed, actin bundles were usually thicker and less abundant than in control cells (cf. Figures 2A to 2R to 2S to 2U). These observations are consistent with those previously reported in the case of tobacco WLIM1 overexpression (Thomas et al., 2006, 2008) and support that all six LIMs promote cross-linking of AFs into thick bundles.
Although this study focuses on the cytoplasmic functions of plant LIMs, it is noteworthy that, contrary to GFP-ABD2-GFP, GFP-LIMs also accumulated within the nucleus, suggesting potential nuclear functions for LIMs.
The Six Arabidopsis LIM Proteins Display Actin Binding, -Stabilizing, and -Bundling Activities
To date, actin regulatory activities have been biochemically demonstrated for only two plant LIMs (i.e., tobacco WLIM1 [Thomas et al., 2006, 2007] and lily LIM1 [Wang et al., 2008a]). Here, we assessed the actin binding, -stabilizing, and -bundling activities of the whole Arabidopsis LIM family. Wang et al. (2008a) recently reported that LIM1 preferentially binds AFs under low pH and low [Ca2+]. We therefore initiated biochemical investigations using similar conditions.
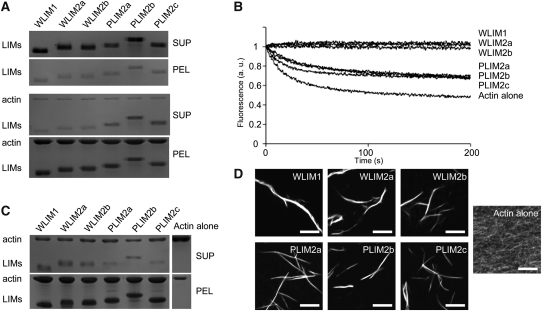
The ability of LIMs to directly bind to AFs was evaluated by high-speed cosedimentation assays. Recombinant LIMs were produced in Escherichia coli and purified by affinity chromatography. AFs (4 μM) were copolymerized with individual LIMs (4 μM) and centrifuged at 100,000g, and the resulting pellet and supernatant fractions were analyzed by SDS-PAGE (Figure 3A). Control experiments showed that recombinant LIMs do not sediment significantly when centrifuged alone (Figure 3A, top gel series). In the presence of AFs, the six LIMs accumulated in the pellet fraction, indicating that they directly interact with AFs (Figure 3A, bottom gel series). However, the relative amount of pelleted LIM was slightly lower in the case of the three PLIMs, suggesting differences in affinity for AFs. This possibility was further investigated by conducting additional high-speed cosedimentation assays with increasing concentrations of LIM proteins and calculating apparent equilibrium dissociation constant (Kd) values as previously described by Thomas et al. (2006, 2007) (Table 1; see Supplemental Figure 4 online). In the conditions used (pH 6.2 and 100 nM Ca2+), the three WLIMs displayed relatively high and similar affinities for AFs as indicated by apparent Kd values of 0.4 ± 0.2 μM (WLIM1), 0.4 ± 0.2 μM (WLIM2a), and 0.5 ± 0.3 (n = 3). Noticeably, these values were significantly lower than those calculated for PLIMs, which ranged from 1.3 ± 0.2 μM (PLIM2a) to 1.7 ± 0.9 μM (PLIM2b).
Figure 3.
Arabidopsis LIMs Bind to, Stabilize, and Bundle AFs.
(A) High-speed cosedimentation assay. After centrifugation at 100,000g, LIMs (4 μM) accumulate in the pellet fraction in the presence (bottom gel panel) but not in the absence of AFs (4 μM; top gel panel).
(B) Depolymerization assay. Time course of AF (4 μM) depolymerization in the absence and in the presence of individual LIMs (6 μM) was monitored by pyrene fluorescence. Initial fluorescence was set to 1. Note that WLIMs stabilize AFs more efficiently than PLIMs.
(C) Low-speed cosedimentation assay. After centrifugation at 12,500g, AFs (4 μM) sediment in the presence but not in the absence of LIMs (6 μM).
(D) Direct visualization of actin bundles induced by LIMs. After polymerization in the absence (actin alone) or in the presence of individual LIMs (6 μM), AFs (4 μM) were labeled with rhodamine-phalloidin and examined by light fluorescence microscopy. Bars = 5 μm. All above assays were conducted at pH 6.2 and in low [Ca2+] (~5 nM free Ca2+). SUP, supernatant fraction; PEL, pellet fraction.
Table 1.
Affinities of the Six Arabidopsis LIM Proteins for AFs
| Protein | Kd (μM) | Bmax |
| WLIM1 | 0.4 ± 0.2 | 1.4 ± 0.2 |
| WLIM2a | 0.4 ± 0.2 | 1.1 ± 0.2 |
| WLIM2b | 0.5 ± 0.3 | 1.5 ± 0.2 |
| PLIM2a | 1.3 ± 0.2 | 1.5 ± 0.1 |
| PLIM2b | 1.7 ± 0.9 | 1.5 ± 0.1 |
| PLIM2c | 1.5 ± 1.1 | 1.8 ± 0.2 |
Apparent equilibrium dissociation constants (Kd) values (±sd) were calculated from three independent high-speed cosedimentation assay experiments after fitting the data (bound protein plotted against free protein; see Supplemental Figure 4 online) with a hyperbolic function.
The effect of LIM binding on AF stability was examined in depolymerization assays. Pyrene-labeled AFs (4 μM) were copolymerized with individual LIMs (6 μM) and subsequently subjected to depolymerization by diluting the samples to an actin concentration below the critical concentration (i.e., 0.2 μM). Depolymerization kinetics were recorded by monitoring fluorescence intensity over time (Figure 3B). In the absence of LIMs (actin alone), AFs promptly depolymerized, as shown by the rapid decline of fluorescence. By contrast, in the presence of individual LIMs, the AF depolymerization rate was markedly decreased. Consistent with the Kd values calculated above, the two LIM subfamilies exhibited different stabilization capabilities. Whereas WLIMs fully stabilized AFs, as indicated by stable fluorescence curves, PLIMs only reduced the AF depolymerization rate. Remarkably, the three members of each LIM subfamily displayed roughly identical stabilizing capabilities.
Finally, the ability of each LIM to cross-link AFs was assessed using low-speed cosedimentation assays. AFs (4 μM) were copolymerized with individual LIMs (6 μM) and centrifuged at 12,500g, and the resulting pellet and supernatant fractions were analyzed by SDS-PAGE (Figure 3C). In the absence of LIM (actin alone), most of the actin was detected in the supernatant fraction. By contrast, in the presence of individual LIMs, actin massively sedimented, indicating the presence of high-order actin structures. The latter were directly examined by fluorescence light microscopy after rhodamine-phalloidin labeling and identified as actin bundles (Figure 3D).
In conclusion, all six Arabidopsis LIMs function as true ABPs (i.e., they directly interact with AFs). In addition, they display autonomous actin-stabilizing and -bundling activities. However, differences are also pointed out by data (the three WLIMs exhibiting a higher affinity for AFs than the three PLIMs).
WLIM1 and PLIM2c Activities Are Differently Regulated by pH and Ca2+
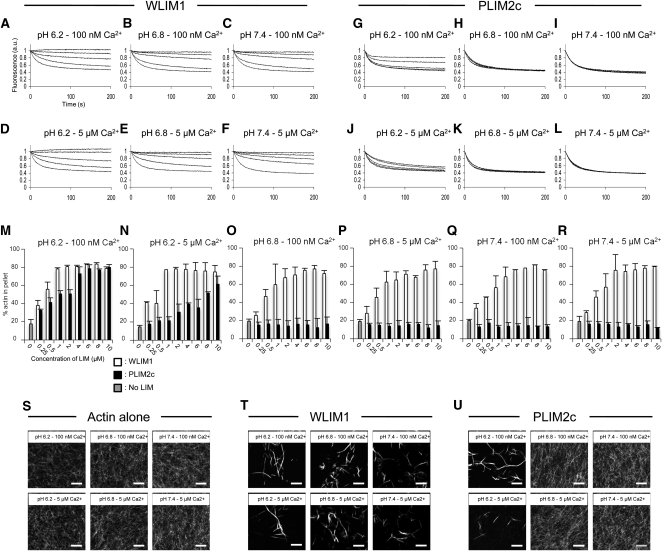
The potential regulation of LIM activities by pH and Ca2+ were assessed by focusing on one member of each LIM subfamily, namely, WLIM1 and PLIM2c. Both proteins were first subjected to a series of actin depolymerization assays conducted at three different pH conditions, 6.2, 6.8, and 7.4, and in the presence of low or high amounts of free Ca2+, corresponding to ~100 nM and ~5 μM, respectively. For each pH and [Ca2+] condition, different concentrations of WLIM1 and PLIM2c (ranging from 1 to 10 μM, concentrations before sample dilution) have been tested, whereas the concentration of actin before dilution was set at 4 μM (Figures 4A to 4L). Under low pH and low [Ca2+], both WLIM1 and PLIM2c stabilized AFs in a concentration-dependent manner (Figures 4A and 4G). Data confirmed the higher stabilizing capability of WLIM1. Indeed, 1 μM of WLIM1 was sufficient to slow down the AF depolymerization rate, whereas 3 μM of PLIM2c were required to produce a significant effect. In addition, full stabilization of AFs was achieved for WLIM1 concentrations ≥6 μM, whereas it required >10 μM of PLIM2c.
Figure 4.
Detailed Comparison of WLIM1 and PLIM2c Actin Regulatory Activities in Different Combinations of pH and [Ca2+].
(A) to (L) Depolymerization assays. Pyrene-labeled AFs (4 μM) were copolymerized in the presence of increasing concentrations of WLIM1 or PLIM2c (0, 1, 3, 6, and 10 μM from bottom to top curves) and were induced to depolymerize by dilution below the critical concentration (i.e., 0.2 μM). Initial fluorescence was set to 1.
(M) to (R) Low-speed cosedimentation assays. AFs (4 μM) were copolymerized in the presence of increasing concentrations of WLIM1 or PLIM2c (0 to 10 μM) and centrifuged at 12,500g. The amount of actin in the pellet and supernatant fractions was quantified. Results are expressed as the percentage of total actin in the pellet as a function of LIM concentration (n = 3; error bars indicate sd).
(S) to (U) Direct visualization of AFs (4 μM) that were polymerized alone (left panel) or in the presence of WLIM1 (6 μM; middle panel) or PLIM2c (6 μM; left panel) using rhodamine-phalloidin labeling. Bars = 5 μm.
Increasing pH from 6.2 to 6.8 or 7.4 caused the loss of PLIM2c stabilizing activity, as indicated by depolymerization curves similar to the controls (AFs alone; Figures 4H and 4I). At higher pH, even the highest PLIM2c concentrations failed to slow down AF depolymerization, indicating a strong pH-dependent inhibition. By contrast, WLIM1 preserved prominent activity at both intermediate and high pH values (Figures 4B and 4C). Furthermore, its stabilizing efficiency appeared unmodified by pH increase since, in all pH conditions, maximal AF stabilization was observed for WLIM1 concentrations ≥6 μM. To check whether high pH values can also inhibit the actin-stabilizing activity of AF-bound PLIM2c, AFs (4 μM) were first copolymerized with PLIM2c (8 μM) in optimal conditions (i.e., pH 6.0 and 100 nM of free Ca2+), and the pH was subsequently shifted by the addition of alkalizing buffers of increasing strength (final pH 6.0, 6.2, 6.4, 6.8, and 7.4). Supplemental Figure 5A online shows that the rate of depolymerization increased proportionally to the shift of pH applied, indicating that pH can inactivate PLIM2c when the latter is associated with AFs. By contrast, the ability of WLIM1 to stabilize AFs was preserved whatever the shift of pH applied (see Supplemental Figure 5B online).
Additional depolymerization experiments were performed at high [Ca2+] levels (Figures 4D to 4F and 4J to 4L). At pH 6.2, the high [Ca2+] significantly reduced the actin-stabilizing capability of PLIM2c (Figure 4J). However, this inhibition was less strong than that of pH, as indicated by the weak but significant levels of stabilization observed for the highest concentrations of PLIM2c (Figure 4J versus 4G). In higher pH conditions (i.e., pH 6.8 and 7.4), high [Ca2+] showed no visible effect, since PLIM2c activity remained turned off (Figures 4K and 4L). The depolymerization curves obtained for WLIM1 were similar to those obtained at low [Ca2+], indicating that WLIM1 was not responsive to Ca2+ (Figures 4D to 4F versus 4A to 4C).
LIM-induced stabilization most likely results from the cross-linking of AFs. Thus, from the above data, one might anticipate that PLIM2c bundling activity is negatively regulated by high pH and/or high [Ca2+], whereas the one of WLIM1 is not. To confirm these assumptions, low-speed (12,500g) cosedimentation assays were conducted in different pH and [Ca2+] conditions using a fixed concentration of actin (4 μM) and increasing concentrations of LIM (0.5 to 10 μM; Figures 4M to 4R). To make comparisons easier, SDS-PAGE gels were scanned after staining, and the respective amounts of actin in the pellet and in supernatant fractions were quantified using ImageJ software. As expected, WLIM1 induced actin sedimentation in a concentration-dependent manner in all tested pH and [Ca2+] combinations (Figures 4M to 4R, white bars). Lack of responsiveness to pH and Ca2+ was further demonstrated by the fact that maximal sedimentation (75 to 80% of total actin in the pellet fraction) was invariably achieved for WLIM1 concentrations ≥2 μM. PLIM2c also induced efficient cross-linking of AFs under low pH and [Ca2+] conditions (Figures 4M to 4R, black bars). However, maximal sedimentation required ~6 μM of PLIM2c, confirming its weaker activity compared with WLIM1 (Figure 4M, black versus white bars). Higher pH values (i.e., 6.8 and 7.4) fully inhibited PLIM2c cross-linking activity, as shown by the relative amounts of actin in the pellet similar to those measured in controls (actin alone, ~20%; Figures 4O to 4R, black bars). Consistent with the depolymerization data, high [Ca2+] partially inhibited PLIM2c activity at pH 6.2, as shown by the reduced but significant amounts of actin sedimented in the presence of high PLIM2c concentrations (Figure 4N, black bars).
Direct observation of AFs polymerized alone (Figure 4S) or in the presence of WLIM1 (Figure 4T) or PLIM2c (Figure 4U) confirmed the above data. Indeed, WLIM1 triggered the formation of actin bundles in all pH and [Ca2+] combinations tested (Figure 4S versus 4T), whereas PLIM2c only induced similar structures under both low pH and low [Ca2+] (Figure 4S versus 4U). As expected, PLIM2c bundling activity was partially inhibited by high [Ca2+], as shown by the rare bundles observed at pH 6.2 and 5 μM Ca2+ (Figure 4U, bottom left image).
In conclusion, WLIM1 and PLIM2c respond differently to pH and [Ca2+] in vitro. WLIM1 actin regulatory activities are not regulated by pH and [Ca2+], whereas those of PLIM2c are inhibited by pH values ≥ 6.8 and/or high [Ca2+].
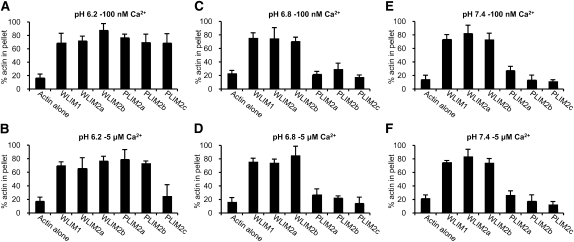
LIM Subfamily-Specific Modes of pH- and Ca2+-Dependent Regulation
To test whether responsiveness to pH and Ca2+ is a specific property of Arabidopsis PLIM subfamily members, additional low-speed cosedimentation assays were performed using 4 μM actin and 6 μM each Arabidopsis LIM (Figure 5). Similarly to WLIM1, WLIM2a and WLIM2b induced actin sedimentation in a pH- and Ca2+-independent manner. Indeed, ~80% of actin were pelleted in all the combinations of pH and [Ca2+] tested (Figures 5A to 5F). Similarly to PLIM2c, PLIM2a and PLIM2b were unable to promote actin sedimentation at relatively high pH values (i.e., pH 6.8 and pH 7.4) (Figures 5C to 5F). However, both PLIM2a and PLIM2b efficiently cross-linked AFs at low pH (6.2) whatever the [Ca2+], indicating that they are not responsive to Ca2+ (Figures 5A and 5B).
Figure 5.
Actin Cross-Linking Activity of the Six Arabidopsis LIMs in Different Combinations of pH and [Ca2+].
Low-speed (12,500g) cosedimentation assays were performed after copolymerization of AFs (4 μM) with individual LIM proteins (6 μM) at pH 6.2 ([A] and [B]), pH 6.8 ([C] and [D]), or pH 7.4 ([E] and [F]) and in the presence of low [Ca2+]free (~100 nM; [A], [C], and [E]) or high [Ca2+]free (5 μM; [B], [D], and [F]). The relative amount of actin in the pellet and supernatant fractions was quantified, and results are expressed as the percentage of total actin in the pellet for each of the Arabidopsis LIM tested (n = 3; error bars indicate sd).
In summary, the three WLIM subfamily members cross-link AFs in a pH- and Ca2+-independent manner. By contrast, the cross-linking activity of the three PLIM subfamily members is inhibited by pH values ≥6.8. Finally, PLIM2c is the only Arabidopsis LIM to clearly respond to Ca2+, its activity being downregulated by high [Ca2+].
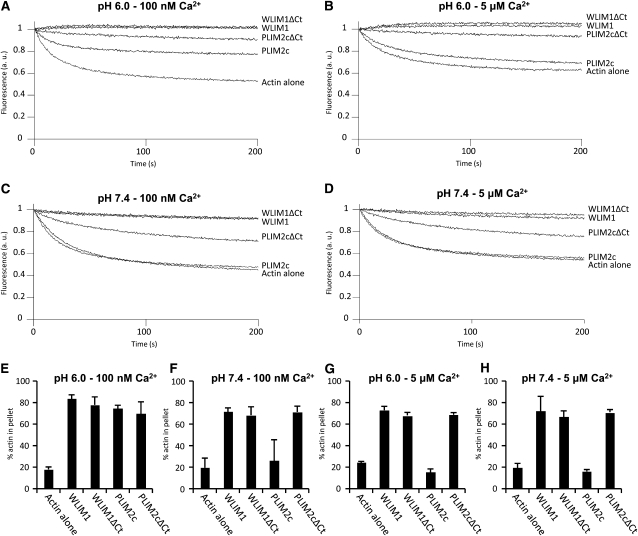
Deletion of the C-Terminal Domain Abolishes PLIM2c Responsiveness to pH and Calcium
The most divergent domain in size and in amino acid sequence between WLIM and PLIM family members is the C-terminal domain (see Supplemental Figure 6 online). Noticeably, this domain is longer and contains a significantly higher number of acidic residues in PLIMs than in WLIMs. To test whether the C-terminal domain is involved in the specific responsiveness of PLIMs to pH (and calcium in the case of PLIM2c), we generated C-terminal deleted versions of PLIM2c and WLIM1 (control), namely, PLIM2cΔCt and WLIM1ΔCt, and studied their actin-stabilizing and -bundling activities in different conditions of pH (6.2 and 7.4) and Ca2+ (100 nM and 5 μM). Our data indicate that, in contrast with PLIM2c, whose activities are inhibited by high pH and/or high [Ca2+], PLIM2cΔCt efficiently stabilized and cross-linked AFs in all the conditions tested (Figure 6). Indeed, the deletion of the C-terminal domain is sufficient to abolish the ability of PLIM2c to respond to pH and calcium variations. In addition, WLIM1 and WLIM1ΔCt displayed strong and nearly identical activities whatever the pH and [Ca2+], supporting that the deletion of the C-terminal domain is not detrimental to the basal activity of WLIM proteins.
Figure 6.
Comparison of the Actin-Stabilizing and -Bundling Activities of WLIM1, PLIM2c, and Their Corresponding C-Terminal Deleted Versions WLIM1ΔCt and PLIM2cΔCt in Different pH and [Ca2+] Conditions.
(A) to (D) Depolymerization assays. Pyrene-labeled AFs (4 μM) were copolymerized with PLIM2c, WLIM1, PLIM2cΔCt, or WLIM1ΔCt (10 μM) in different combinations of pH and [Ca2+] and were induced to depolymerize by dilution. Initial fluorescence was set to 1.
(E) to (H) Low-speed cosedimentation assays. AFs (4 μM) were copolymerized with PLIM2c, WLIM1, PLIM2cΔCt, or WLIM1ΔCt (6 μM) and centrifuged at 12,500g. The relative amount of actin in the pellet and supernatant fractions was quantified and results are expressed as the percentage of total actin in the pellet as a function of the LIM or LIM variant tested (n ≥ 4; error bars indicate sd).
In Vivo pH-Dependent Regulation Arabidopsis LIM Proteins
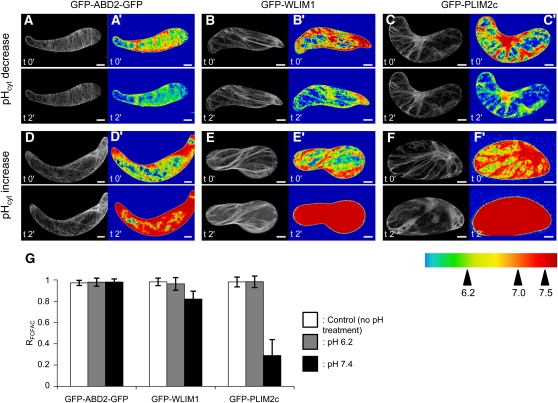
The above biochemical data strongly suggest that cytoplasmic pH acts as a regulator of PLIM activities. This hypothesis is consistent with the existence of intracellular pH gradients in growing pollen tubes and the concept that pH variations regulate the actin cytoskeleton organization and dynamics through the in/activation of different classes of ABPs (Feijo et al., 1999; Hepler et al., 2006; Lovy-Wheeler et al., 2006). However, pH-specific monitoring of PLIM actin regulatory activities in the context of live cells requires direct demonstration. In that attempt, we artificially modulated the cytoplasmic pH of Arabidopsis cells and analyzed the resulting effect of such manipulations on the ability of GFP-WLIM1 and GFP-PLIM2c fusion proteins to interact with the actin cytoskeleton.
Briefly, cell culture lines were produced from transgenic Arabidopsis plants expressing GFP-WLIM1, GFP-PLIM2c, and the actin cytoskeleton marker GFP-ABD2-GFP (control). Prior to pH treatment, cells were immobilized on polylysine-coated cover slips and incubated in standard culture medium supplemented with the ratiometric pH-sensitive dye SNARF-5F. Both concentration and incubation time of the dye were optimized for Arabidopsis cells to 5 μM and 30 min, respectively. The pH dye most frequently diffused in a relatively homogenous manner into the cytoplasm and the nucleus (Figures 7A to 7F). The average cytoplasmic pH was estimated to be 7.06 ± 0.06 (n = 15). Occasionally, the dye also penetrated the central vacuole. As expected, the vacuole was found to be the most acidic cellular compartment, with pH values ≤ 6.0 (see Supplemental Figure 7 online). To decrease or increase the cytoplasmic pH, cells were incubated in buffers containing the cell-permeant weak acid sodium propionate (pH adjusted to 6.2) or the cell-permeant weak base ammonium chloride (pH adjusted to 7.4; Parton et al., 1997). Confocal acquisitions of the pH-sensitive dye and of the GFP-fused proteins were collected just before and 2 min after the application of pH buffers. It should be noted that, after longer incubation times in pH buffers, the evolution of pH frequently reversed, although the recovery was much slower than the initial shift (see Supplemental Figure 8 online). Figure 7 presents typical results obtained with the three cell lines of interest. Successful modifications of pH were confirmed by pseudocolored ratio images, and the cytoplasmic pH was found to reach values close to those of pH buffers (i.e., 6.2 ± 0.1 or 7.4 ± 0.1). The effect of pH modifications on the ability of each GFP fusion protein to interact with the actin cytoskeleton was carefully analyzed by quantifying the FCFAC and by comparing this value before and after pH treatment (RFCFAC = FCFAC t2'/FCFACt0'). Lowering the pH had no significant effects on the actin binding activity of GFP-WLIM1, GFP-PLIM2c, and GFP-ABD2-GFP, as indicated by direct comparison of confocal images (Figures 7A to 7C) and RFCFAC values close to 1 (Figure 7G). By contrast, pH increase dramatically weakened the binding of GFP-PLIM2c to the cytoskeleton, as shown by prominent diffuse fluorescent signal (Figure 7F). Massive release of GFP-PLIM2c from the actin network upon pH elevation is supported by a calculated RFCFAC value of 0.29 ± 0.15, indicating that ~70% of the fluorescence initially associated with the cytoskeleton has been displaced toward the cytoplasmic diffuse pool. Contrary to GFP-PLIM2c, GFP-ABD2-GFP and GFP-WLIM1 remained largely associated with the actin cytoskeleton upon cytoplasmic pH elevation (Figures 7D and 7E). However, quantitative analyses revealed a slight modification in the GFP-WLIM1 subcellular distribution (Figure 7G). Indeed, we calculated a RFCFAC value of 0.82 ± 0.075, indicating an 18% decrease of the cytoskeleton-bound GFP-WLIM1 population upon pH elevation. By contrast, the subcellular distribution of GFP-ABD2-GFP was unaffected, as indicated by RFCFAC = 0.98 ± 0.029. However, considering the relatively weak effect of high pH on WLIM1 subcellular localization, its biological significance remains uncertain. Importantly, rhodamine-phalloidin colabeling experiments confirmed the persistence of a filamentous actin cytoskeleton after increase of cytoplasmic pH in GFP-PLIM2–expressing cells (see Supplemental Figure 9 online). The reversibility of the process was assessed by successive treatments of cells with the alkalizing and acidifying buffers. As shown in Figures 8A to 8D, GFP-PLIM2c efficiently dissociated and reassociated with the cytoskeleton, indicating that its actin binding ability was not irreversibly damaged by elevated pH conditions. A control experiment conducted with GFP-ABD2-GFP–expressing cells confirmed that the predominant cytoskeletal localization of GFP-ABD2-GFP is not significantly affected during similar pH treatment (Figures 8E to 8H).
Figure 7.
Increase of Cytoplasmic pH Specifically Impairs PLIM2c Interaction with the Actin Cytoskeleton.
(A) to (F’) Arabidopsis cells expressing GFP-ABD2-GFP ([A], [A'], [D], and [D']), GFP-WLIM1 ([B], [B'], [E], and [E']), and GFP-PLIM2c ([C], [C'], [F], and [F']) were treated with acidifying ([A] to [C] and [A'] to [C']) or alkalizing buffers ([D] to [F] and [D'] to [F']). Typical confocal images showing the localization of GFP fusion proteins before (t0') and 2 min after pH treatment (t2') are presented on the left of each image panel ([A] to [F]). Modifications of cytoplasmic pH were controlled using the ratiometric SNARF-5F dye, and rainbow pH images are presented at the right of each image panel ([A'] to [F']). Note the prominent diffuse cytoplasmic localization of GFP-PLIM2c after increase of cytoplasmic pH ([F] and [F']).
(G) Quantitative analyses of the above experiments. The FCFAC was quantified for each GFP fusion protein before (FCFACt0') and after pH treatment (FCFACt2'), and ratios were calculated (RFCFAC = FCFACt2'/ FCFACt0'). Gray and black bars indicate the RFCFAC values calculated after a decrease and increase of cytoplasmic pH respectively (n ≥ 10; errors bars indicate sd). White bars indicate the RFCFAC values calculated for control experiments in which cells have not been submitted to any pH treatment (n ≥ 10; errors bars indicate sd). Note the low RFCFAC value (0.29 ± 0.15) calculated for GFP-PLIM2c upon increase of cytoplasmic pH indicating that GFP-PLIM2c has massively detached from the actin cytoskeleton. Bars = 10 μm.
Figure 8.
Specific Dissociation and Reassociation of GFP-PLIM2c with the Actin Cytoskeleton Induced by Successive Increase and Decrease of Cytoplasmic pH.
(A) to (C) Subcellular localization of GFP-PLIM2c in a transgenic Arabidopsis cell before pH treatment (A), after 2 min of incubation in the alkalizing buffer (B), and after 2 min of subsequent incubation in the acidifying buffer (C).
(D) Quantitative analysis of the FCFAC in (A) (white bar), (B) (gray bar), and (C) (black bar).
(E) to (H) Corresponding control experiment conducted with the GFP-ABD2-GFP–expressing cell line. Note that GFP-ABD2-GFP is predominantly associated with the actin cytoskeleton before pH treatment ([E] and [H], white bar), after 2 min of incubation in the alkalizing buffer ([F] and [H], gray bar), and after 2 min of subsequent incubation in the acidifying buffer ([G] and [H], black bar). Bars = 10 μm.
GFP-PLIM2c Predominantly Associates with Long and Dynamic Actin Bundles in the Shank of Growing Pollen Tubes
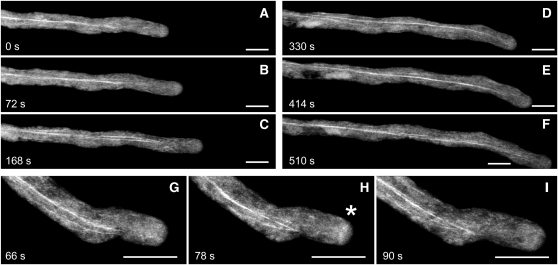
To examine the cellular distribution of PLIM2c in live growing pollen tubes of Arabidopsis, we produced transgenic plants expressing a GFP-PLIM2c fusion protein under the control of the PLIM2c promoter. Analyses performed on heterozygous plants revealed that transgenic and wild-type nonfluorescent pollen grains germinate and elongate with very similar rates (i.e., 3.56 ± 0.65 μm·s−1 and 3.41 ± 0.76 μm·s−1, respectively; n ≥ 10), indicating that the recombinant protein does not significantly disturb pollen tube physiology. Figure 9A shows that GFP-PLIM2c interacted with a population of long actin bundles running along the pollen tube shank. Time-lapse imaging revealed that these bundles are highly dynamic and do not penetrate subapical and apical regions (see Supplemental Movie 1 online). Interestingly, GFP-PLIM2c occasionally decorates a structure in the subapical region resembling the cortical actin fringe, which has been described by several studies (e.g., Kost et al., 1998; Sheahan et al., 2004; Lovy-Wheeler et al., 2005; Cheung et al., 2008; Vidali et al., 2009; Figure 9B; see Supplemental Movie 2 online).
Figure 9.
Localization of GFP-PLIM2c in Growing Pollen Tubes.
(A) to (F) Selected confocal stack images in a time series of a GFP-PLIM2c–expressing pollen tube.
A movie for this time series (see Supplemental Movie 1 online) shows that the PLIM2c-decorated actin bundles are dynamic.
(G) to (I) Selected confocal stack images in a time series of a GFP-PLIM2c–expressing pollen tube showing the decoration of an actin fringe-like structure in the subapical region (indicated by an asterisk). A movie for this time series (see Supplemental Movie 2 online) shows the morphological changes of this structure during growth. Bars = 10 μM.
DISCUSSION
Actin bundles are key structural components in eukaryotes (Bartles, 2000; Thomas et al., 2009). In plant cells, they are required to stabilize the strands that cross the vacuole to connect distant cytoplasmic regions (Shimmen et al., 1995; Tominaga et al., 2000). In addition, actin bundles represent the main long-distance tracks for (myosin-dependent) vesicle and organelle transport and are therefore particularly important for cytoplasmic streaming and during tip growth processes, such as pollen tube and root hair growth (Tominaga et al., 2000; Vidali and Hepler, 2001). Four families of ABPs are commonly assumed to be involved in the formation and/or the maintenance of actin bundles in plants: the villin (Vidali et al., 1999; Tominaga et al., 2000; Huang et al., 2005), the fimbrin (McCurdy and Kim, 1998; Kovar et al., 2000; Klein et al., 2004), the formin (Cheung and Wu, 2004; Michelot et al., 2005, 2006; Ye et al., 2009), and the two LIM domain–containing protein (LIM) families (Thomas et al., 2006, 2007; Wang et al., 2008a). However, their exact contribution in actin bundling remains difficult to appreciate because only a few of their members have been closely studied so far.
Two plant LIMs, namely, tobacco WLIM1 and lily LIM1, were reported to bind to AFs and trigger the formation of actin bundles (Thomas et al., 2006, 2007; Wang et al., 2008a). Our data provide evidence that the six Arabidopsis LIMs display direct actin binding activity, indicating that most and possibly all plant LIMs function as ABPs. Interestingly, the mammalian counterparts of plant LIMs, namely, the CRPs, were initially suggested to interact only indirectly with AFs via ABPs, such as zyxin and α-actinin (Sadler et al., 1992; Louis et al., 1997; Pomies et al., 1997). However, recently, two out of the three mammalian CRPs have been reported to bind AFs autonomously (Grubinger and Gimona, 2004; Tran et al., 2005; Jang and Greenwood, 2009). This strongly suggests that direct actin binding activity is a common feature to all CRPs and CRP-like proteins, such as the plant LIMs.
Remarkably, the six Arabidopsis LIMs not only bind to AFs but also cross-link them into bundles, suggesting that actin bundling is a major actin regulatory function of LIMs rather than an accessory function displayed by only few family members. Fluorimetric data ruled out a cofilin-like severing activity, which would have been identified by faster actin depolymerization rates in the presence of LIMs. Additional evidence that actin bundling is a predominant function of LIMs is provided by the increase of actin bundle thickness and the concurrent decrease of the number of bundles observed in most LIM overexpressing cells (Figure 2; Thomas et al., 2006, 2008). Interestingly, such cytoskeletal rearrangements differ from those resulting from the overexpression of Arabidopsis FH1, an Arabidopsis formin that has been reported to bundle AFs in vitro (Michelot et al., 2005, 2006). Indeed, FH1 overexpression increases the number of actin bundles in pollen tubes (Cheung and Wu, 2004). Opposite effects on bundle population induced by the two classes of actin-bundling proteins may be explained by the fact that only FH1 displays actin-nucleating activity. Mechanistic studies have suggested that FH1 functions as a nonprocessive nucleating factor that detaches from the barbed end after nucleation and moves to the side of the growing filament to promote the assembly of a novel filament, thereby facilitating the formation of actin bundles (Michelot et al., 2006; Blanchoin and Staiger, 2008). Therefore, FH1, in addition to its bundling activity, promotes de novo formation of actin bundles, whereas LIMs only cross-link existing filaments, thereby reducing the number of individual filaments and small bundles. A recent loss-of-function study has provided evidence of the central role played by another formin, namely, AFH3, in the nucleation of longitudinal actin bundles in Arabidopsis pollen tubes (Ye et al., 2009). However, in vitro analyses failed to reveal autonomous cross-linking activity, suggesting that, in vivo, the bundling of AFH3-nucleated AFs requires the action of other ABPs (e.g., villins or LIMs). Noticeably, the supernumerary actin bundles induced by AFH3 overexpression were abnormally thin (Ye et al., 2009). This possibly results from insufficient levels of actin-bundling proteins to assemble bundles of normal thickness.
The expression pattern of a number of plant LIM genes has been previously examined to some extent (Eliasson et al., 2000; Mundel et al., 2000; Arnaud et al., 2007). Here, we show that, like actin and most ABP genes, including ADFs and profilins (McDowell et al., 1996; Hussey et al., 2002; Kandasamy et al., 2002; Ruzicka et al., 2007), Arabidopsis LIM genes can be categorized into two major groups according to their expression pattern. Indeed, WLIM1, WLIM2a, and WLIM2b are widely expressed throughout sporophytic tissues but are not or only very weakly expressed in pollen. By contrast, PLIM2a, PLIM2b, and PLIM2c are predominantly and abundantly expressed in pollen. As already reported for cytoskeletal gene families (e.g., Ruzicka et al., 2007), the separation between vegetative and reproductive patterns is not always clear. Most noteworthy are the relatively high expression levels of PLIM2b observed in vasculature and roots. The expression of the various LIM gene family members considerably overlaps in plant tissues with at least two, usually three, members coexpressed at significant levels. On the one hand, this supports the high degree of functional redundancy suggested by the lack of clear phenotypes in single insertion mutants (M. Dieterle, unpublished data). On the other hand, the expression of multiple actin or ABP isovariants in the same cells has been proposed to be a key element of the extreme flexibility in dynamic behavior of the cytoskeleton (Meagher et al., 1999). Interestingly, the wide expression of LIMs in plant tissues contrasts with the more restricted expression patterns of mammalian CRPs. Indeed, CRPs are predominantly expressed in muscle tissues (Louis et al., 1997; Yet et al., 1998; Henderson et al., 1999) where they are assumed to participate in the organization of contractile fibers (Arber et al., 1997; Kim-Kaneyama et al., 2005; Tran et al., 2005; Sagave et al., 2008). Similarly, villins, another important class of actin-bundling proteins, exhibit wide expression patterns in plants, whereas mammalian villin expression is limited to microvilli of brush border cells, indicating a higher degree of functional specialization (Klahre et al., 2000). The high overall level of actin bundling in plant cells (Thomas et al., 2009) supports the idea that actin bundles and associated bundling proteins are involved in ubiquitous plant-specific processes (e.g., the formation and/or the maintenance of the transvacuolar cytoplasmic strand network) (Shimmen et al., 1995; Tominaga et al., 2000; Yokota et al., 2005).
This study shows a remarkable correlation between the expression pattern of Arabidopsis LIM genes and the pH responsiveness of the corresponding proteins. Indeed, the in vitro activities of the three PLIMs are virtually turned off by pH values above 6.8, whereas those of the three WLIMs remain optimal in all pH conditions tested. Consistent with these data, the pollen LIM1 protein from Lilium, which belongs to another phylogenetical subgroup than Arabidopsis PLIMs (Arnaud et al., 2007), has been shown to preferentially bind to AFs under low pH conditions (Wang et al., 2008a). Therefore, pH responsiveness appears to be a feature common to pollen LIMs. Importantly, in vivo pH-dependent regulation of pollen LIM activities is strongly supported by our live-cell investigations showing that an increase in cytoplasmic pH specifically disrupts the interaction between PLIM2c and the actin cytoskeleton. Interestingly, the pH threshold above which PLIM2c activities are inhibited is apparently higher in vivo than in vitro. Indeed, when fused to GFP, PLIM2c efficiently decorates the actin cytoskeleton in cells whose average cytoplasmic pH is estimated to be close to 7.0 (Figures 2 and 7), whereas it is already deactivated at pH 6.8 in in vitro biochemical assays. This suggests that other factors than pH might regulate LIM protein activities in the context of a live cell. In contrast with PLIM2c, WLIM1 remains predominantly associated with AFs in both low and high intracellular pH conditions. However, a moderate reduction (~18%) in the WLIM1 cytoskeletal fraction was noticed after an increase in cytoplasmic pH. On the one hand, the biological significance of this response is questionable considering its relative weakness. On the other hand, an indirect regulation of WLIM1 activities by pH through de/activation of pH-dependent factors, such as pH-dependent kinases or phosphatases, cannot be ruled out. It is worth noting that several putative phosphorylation sites have been predicted in both WLIM and PLIM sequences (Arnaud et al., 2007) and that an animal CRP is phosphorylated in vivo (Huber et al., 2000). Possible regulation of plant LIM functions by phosphorylation is therefore an important issue to address in future work. PLIM2c is the only Arabidopsis LIM to obviously respond to Ca2+ in our in vitro assays, its activities being downregulated by high, physiologically relevant, [Ca2+]. This corroborates the previous observation that optimal binding of lily LIM1 to F-actin requires both low pH and [Ca2+] conditions (Wang et al., 2008a). However, inactivation of PLIM2c and lily LIM1 by high cytoplasmic [Ca2+] remains to be experimentally confirmed in vivo. Interestingly, a PLIM2c variant from which the C-terminal domain has been deleted is fully active but is no longer able to respond to variations of pH and/or [Ca2+]. Therefore, the C terminus appears as a candidate of choice for a regulatory domain of PLIMs. Whether this domain is involved in the regulation of WLIMs by other factors than pH and calcium has to be investigated.
Under favorable conditions (i.e., pH 6.2 and 100 nM Ca2+), PLIMs exhibited a lower affinity for AFs than WLIMs, as indicated by 2 to 3 times higher apparent Kd values. Consistent with these data, maximal actin stabilization and sedimentation required significantly higher amounts of PLIM2c than WLIM1 (Figures 4A, 4G, and 4M). Moreover, the three members of each subfamily exhibit similar affinities for AFs as well as similar stabilizing and bundling efficiencies (e.g., Figure 3B). These observations suggest that WLIMs and PLIMs have coevolved with vegetative and reproductive actin isoforms, respectively, so that they exhibit dissimilar, subfamily-specific affinities for a given source of actin. The hypothesis regarding functional specificity and class-specific interaction of actin and ABP isoforms is strongly supported by elegant studies showing that the toxic effect of a misexpressed reproductive actin in vegetative tissues can be neutralized only by coexpression of a reproductive but not a vegetative profilin or ADF isoform (Kandasamy et al., 2007).
Many lines of evidence indicate that pH gradients are present in growing pollen tubes (Messerli and Robinson, 1998; Feijo et al., 2001; Certal et al., 2008). A so-called alkaline band has been characterized in the subapical region of lily pollen tubes (Feijo et al., 1999; Lovy-Wheeler et al., 2006). Within this band, the pH oscillates relative to the oscillatory growth and reaches values above 7.5. Strikingly, the alkaline band is located in the vicinity of the clear zone, a region where the actin cytoskeleton is subjected to profound remodeling. Indeed, in the subapical region, a dense collar of cortical AFs, also referred to as a cortical fringe, replaces the prominent longitudinal bundles that run along the pollen tube shank (e.g., Kost et al., 1998; Sheahan et al., 2004; Lovy-Wheeler et al., 2005; Cheung et al., 2008; Vidali et al., 2009). A number of studies have described the direct implication of this structure in pollen tube elongation and highlighted its high rate of turnover (Gibbon et al., 1999; Fu et al., 2001; Vidali et al., 2001). Therefore, our data regarding the regulation of PLIM activities by pH are consistent with pH and actin cytoskeleton patterns in growing pollen tubes. The relatively low pH in the shank of the tube would activate PLIMs, which in turn would enhance the formation of long actin bundles. By contrast, the cyclic increases of pH in the alkaline band would downregulate PLIM activity, thereby maintaining the actin cytoskeleton in a highly dynamic state. However, it is not excluded that PLIMs transiently protect the cytoskeletal structures forming in the subapical region. Indeed, when the alkaline band reaches its lowest pH values, down to 6.8 (Lovy-Wheeler et al., 2006), PLIMs may reactivate and stabilize the short actin bundles of the cortical fringe until the next pH increase. Our observations conducted in growing pollen strongly support this view. Indeed, as fused to GFP, PLIM2c predominantly associates with long and dynamic actin bundles in the pollen tube shank. Careful analysis revealed that it also occasionally faintly labels a subapical structure resembling the actin cortical fringe. It should be noticed that the latter is expected to be rather difficult to image in live growing pollen, as it is highly dynamic.
Another ABP that has been proposed to play a central role in the regulation of the actin cytoskeleton dynamics in the subapical region of pollen tubes is the actin depolymerization factor (ADF). Remarkably, ADF concentrates in the same area as the cortical actin fringe (Lovy-Wheeler et al., 2006), and its F-actin severing activity is stimulated by alkaline pH conditions (Chen et al., 2002). Therefore, through their antagonist actin regulatory activities, ADF and PLIMs may orchestrate, in a pH-controlled manner and with other players, the successive cycles of disassembly and reassembly of the cortical fringe. Recently, the human CRP3 (or Muscle LIM Protein), a counterpart of plant LIMs, has been reported to directly interact with ADF/cofilin 2 in a pH-dependent manner (Papalouka et al., 2009). The possibility of a similar interaction between plant LIMs and ADFs as well as its potential effects on respective actin regulatory activities are important issues to be addressed by future work.
Calcium is another factor assumed to play crucial roles in the regulation of actin dynamics during pollen tube elongation. Noticeably, a tip-high oscillatory cytosolic Ca2+ gradient (Holdaway-Clarke et al., 1997; Messerli and Robinson, 1997; Pierson et al., 1996; Iwano et al., 2009) is assumed to locally increase the rate of AF turnover through the activation of Ca2+-dependent ABPs, such as profilins and villins/gelsolins (Fan et al., 2004; Huang et al., 2004; Yokota et al., 2005; Xiang et al., 2007; Wang et al., 2008a). Therefore, the downregulation of PLIM2c activity by high [Ca2+] suggested by in vitro data might also contribute to maintain the cytoskeleton in a highly dynamic state in the apex of growing pollen tubes.
In conclusion, we propose that plant LIMs represent a highly specialized family of actin-bundling proteins that is present in virtually all plant cells. Interestingly, the two differentially expressed WLIM and PLIM subsets exhibit similar but not identical activities, suggesting that they have been optimized for vegetative and pollen tissues, respectively. We provide clear evidence for the in vivo regulation of PLIM subfamily members by pH as well as for the involvement of the C-terminal domain in this process. Importantly, our data are highly consistent with the cellular distribution of a GFP-PLIM2c fusion protein in elongating pollen tubes and support a central role of PLIM proteins in the regulation of AF organization and dynamics during pollen tube growth. An aspect that has not been covered here is the nuclear function of plant LIMs. Arabidopsis LIMs accumulate in the nucleus, and this has been found not to be a consequence of passive diffusion (C. Thomas, unpublished data). Therefore, they represent attractive candidates as signal integrating factors connecting the nucleus and the actin cytoskeleton.
METHODS
Plant Material and Generation of Transgenic Lines
Transgenic Arabidopsis thaliana lines described in this study were produced in Landsberg erecta for ProPLIM2c-eGFP-PLIM2c and in Columbia-0 ecotype for all other lines. Plants were greenhouse cultivated and propagated on soil under 16-h-light/8-h-dark cycles. Transformations were performed with Agrobacterium tumefaciens strain GV3101 using the floral dip method (Clough, 2005). For selection, seeds were surface sterilized and plated onto half-strength Murashige and Skoog agar medium (MS255; Duchefa), pH 5.8, supplemented with 1% sucrose and with 50 mg/L of kanamycin or 15 mg/L of hygromycin. After a 3-d stratification at 4°C in the dark, seeds were incubated in a growth chamber at 22°C under 12-h-light/12-h-dark cycles. Arabidopsis cell suspension cultures were generated by transferring 10-d-old seedlings onto a callus-inducing medium (Murashige and Skoog agar medium, 200 mg/L KH2PO4, 1 mg/L thiamine, 0.05 g/L myo-inositol, 20 g/L sucrose, and 1 mg/L 2,4-D, pH 5.7). Plates were wrapped in foil and kept in the dark at 22°C for 4 to 6 weeks. Calli were transferred to 40 mL liquid callus-inducing medium and maintained in the dark at 22°C with gentle agitation (100 rpm). Cells were propagated once a week by transferring 30 mL of 7-d-old suspension to 60 mL of fresh medium.
Arabidopsis LIM coding sequences (cds) were amplified from clones obtained from the ABRC (pUNI clones U18145 for At3g55770 and U50754 forAt2g45800; Yamada et al., 2003) and Institut National de la Recherche Agronomique–Centre National de Ressources Génomiques Végétales (BX817923 for At1g01780 and BX825681 for At3g61230) or from a homemade seedling cDNA library (At1g10200 and At2g39900) and cloned in the pENTR vector. Binary vectors harboring pro35S-GFP-LIM fusions were constructed by transferring LIM cds from pENTR-LIM vectors by Gateway cloning (Invitrogen) into pMDC43 (Curtis and Grossniklaus, 2003).
For each Arabidopsis LIM gene, a promoter region consisting of a minimal sequence of 900 bp upstream the translational start site was amplified from Arabidopsis genomic DNA and subsequently cloned into the binary vector pGPTVII.Kan (Walter et al., 2004) to obtain proLIM-GUS. LIM coding sequences were cloned into pMDC43 (Curtis and Grossniklaus, 2003) to obtain proCaMV35S-GFP-LIM. A ProPLIM2c-eGFP-PLIM2c-TermNos cassette was assembled in pUC18 and transferred in the binary vector pGPTVII.bar (Walter et al., 2004). Detailed cloning procedures and primer sequences can be found in Supplemental Methods online and Table 1, respectively.
The F-actin reporter line, expressing the actin binding domain 2 of Arabidopsis fimbrin 1 fused to GFP at both C and N termini with GFP (GFP-ABD2-GFP; Wang et al., 2008b), was kindly provided by the group of Elison Blancaflor (The Samuel Roberts Noble Foundation).
GUS Histochemical Analyses
GUS histochemical analyses of each reporter gene were performed on at least nine independent transgenic lines. Staining for GUS activity was performed for 12 to 15 h according to Marrocco et al. (2003) and on samples of various stages of development, including 7-d-old seedlings and 4-week-old seedlings and inflorescences. In the case of inflorescences, additional shorter staining periods (30 min to 1 h) were applied to confirm the high expression level of PLIM genes. After staining, samples were dehydrated by a series of ethanol washes and stored in 70% ethanol until observation at the binocular and light microscope (Leica DMI 6000B).
Confocal Microscopy and Imaging
Plants and cells expressing the GFP fusion proteins were imaged using a Zeiss LSM510 META confocal laser scanning microscope equipped with a ×40 Plan-NeoFluar oil immersion objective (numerical aperture 1.3). GFP was detected by exciting samples at a wavelength of 488 nm and using a 505- to 530-nm band-pass emission filter. For rhodamine-phalloidin labeling experiments, a 543-nm excitation wavelength and a 560- to 615-nm band-pass emission filter were used. Confocal images were deconvolved using Huygens Essential image processing software package (Scientific Volume Imaging) and are shown as stacks of neighboring sections reconstructed in ImageJ (National Institutes of Health).
SNARF-5F ratio imaging was performed in the META channel using a 543-nm excitation wavelength and a laser power of 7%. Emitted light was simultaneously collected with a dual channel of 560- to 600-nm (Ch1) and 625- to 665-nm (Ch2) bandwidth. To optimize signal detection, the photomultiplier gain was set over the range 60 to 70%. Ratio images were generated by dividing Ch2/Ch1 using the physiology module of LSM510 acquisition software. A 3 × 3 pixel median filter was applied to improve image quality, and final images were displayed with a rainbow look-up table.
Quantification of the FCFAC in GFP-LIM– and GFP-ABD2-GFP–expressing cells was performed using Metamorph software (Molecular Devices) by adjusting the image threshold to eliminate most of the diffuse cytoplasmic fluorescence. The threshold has to be adjusted for each cell (in the range of ±10%) because the actin cytoskeleton organization and the associated intensity of fluorescence can slightly vary from one cell to another. The remaining integrated fluorescent signal was quantified and expressed as a percentage of the total cellular fluorescence (the nucleus was excluded). To avoid under- or overexposure problem during acquisition, the image intensity histogram was systematically checked, and acquisition settings were adjusted so that the fluorescent signal distributes within the 0 to 255 range (8-bit grayscale image). To estimate how artificial modifications of cytoplasmic pH influence GFP-LIM and GFP-ABD2-GFP interaction with the actin cytoskeleton, FCFAC values were measured before (t0’) and after 2 min (t2’) pH treatment, and FCFAC ratios (RFCFAC = FCFACt2’/FCFACt0’) were calculated.
In the in vivo actin depolymerization experiments, Arabidopsis cells were mounted in an open observation chamber and imaged before and 1 h after addition of 100 μM cytoskeletal inhibitor latrunculin B (Sigma-Aldrich). Rhodamine-phalloidin labeling was performed in PME buffer (50 mM PIPES, 20 mM MgCl2, and 50 mM EGTA).
For confocal analyses conducted with pollen, pollen grains were placed on microscope slides by dipping freshly dehiscent anthers onto pollen germination medium solidified with 0.5% (w/v) low-melting agarose (Duchefa). Pollen germination medium was modified from Li et al. (1999) and consisted of 0.01% boric acid, 5 mM CaCl2, 5 mM Ca(NO3)2, 5 mM MgSO4, 5 mM KCl, and 18% (w/v) sucrose, pH 6.8 to 7.0. Pollen of T1 plants were used for pollen germination studies.
pH Treatments
Approximately 200 μL of Arabidopsis cell suspension culture was plated on a poly-l-lysine–coated cover slip. After removing the culture medium, cells were incubated in 1 mL of diluted ratiometric pH-sensitive dye SNARF-5F 5-(and-6)-carboxylic acid, acetoxymethyl ester, acetate (5 mM; Invitrogen) for 30 min in the dark at 22°C. Modification of the intracellular pH was achieved by incubating cells in 1 mL of callus-inducing medium supplemented with 30 mM ammonium chloride and 10 mM HEPES, pH 7.4 (alkalizing buffer), or with 30 mM sodium propionate and 10 mM MES, pH 6.2 (acidifying buffer). Confocal microscopy observations were performed just before pH buffer application and after 2, 4, and 6 min of treatment.
In situ calibration was performed as recommended by Feijo et al. (1999) using a combination of nigericin (5 μM) and valinomycin (2 μM) and in the presence of a high concentration of potassium ions. Reference pH buffers (half-strength Murashige and Skoog medium, 150 mM KCl, 30 mM NaCl, 25 mM HEPES, and 25 mM MES) were adjusted to a final pH of 6.2, 6.8, 7.0, 7.2, and 7.5. Cells were incubated 15 min in each pH buffer before imaging the SNARF-5F probe. A calibration curve was generated by calculating ratio values in cytoplasmic volumes of 50 μm3 using Metamorph Software (Molecular Devices). This curve was used to estimate the pH in nontreated cells and to control pH modifications induced by the alkalizing and acidifying buffers (n ≥ 10).
Expression and Purification of Recombinant Arabidopsis LIMs
Arabidopsis LIM cds were subcloned into the bacterial expression vector pQE-60 (Qiagen). Due to relatively low levels of expression in bacteria, Escherichia coli codon-optimized sequences have been generated for the three PLIMs (DNA2.0). This did not result in any modification of the predicted amino acid sequences for PLIMs. The deletion of the C-terminal domain of WLIM1 and PLIM2c for production of WLIM1ΔCt and PLIM2cΔCt recombinant proteins has been achieved by PCR using the specific primers CTAT1F2 + CT101 (WLIM1ΔCt) and CT65 + CT80 (PLIM2cΔCt) (see Supplemental Table 1 online). His6-tagged LIMs were expressed in M15[pREP4] bacteria and purified using a Ni-NTA resin following procedures described by the manufacturer (Qiagen). Purified proteins were concentrated in a centrifugal filter (Amicon), buffer exchanged (10 mM Tris-Cl, 50 mM NaCl, 1 mM DTT, 50 μM ZnCl2, and 2 M urea, pH 6.9) using a 7 K molecular weight cutoff dialysis cassette (Pierce), and stored on ice. Prior to an experiment, proteins were preclarified at 150 000g and checked for correct molecular weight by SDS-PAGE analysis, and their concentration was determined by Bradford assay (Bio-Rad) using BSA as standard.
High- and Low-Speed Cosedimentation Assays
High- and low-speed cosedimentation assays were used to assess the actin binding and -crosslinking activities of Arabidopsis LIMs, respectively. In both cases, rabbit muscle actin (Cytoskeleton; concentration indicated in figures) was copolymerized with various concentrations of individual LIMs for 1 h in 50 mM KCl, 2 mM MgCl2, 1 mM ATP, and 0.5 mM DTT. Depending on the pH and [Ca2+] conditions tested, the reaction medium was buffered with either MES and PIPES, pH 6.2, or PIPES and Tris, pH 6.8 and 7.4, and was supplemented with either EGTA (low [Ca2+] conditions) or CaCl2 (high [Ca2+] conditions). Supplemental Table 2 online indicates the concentration of MES, PIPES, Tris, EGTA, and CaCl2 for each of the copolymerization conditions used.
In high-speed experiments, samples were centrifuged at 100,000g for 30 min to pellet AFs. The presence of LIM in the resulting supernatants (F-actin unbound fraction) and pellets (F-actin bound fraction) was analyzed by SDS-PAGE and Coomassie Brillant Blue R (Sigma-Aldrich) staining.
In low-speed experiments, samples were centrifuged at 12,500g for 30 min in a microcentrifuge to pellet high-order F-actin structures. The presence of actin in the resulting supernatants (noncross-linked AFs) and pellets (cross-linked AFs) was analyzed by SDS-PAGE and Coomassie Brillant Blue R (Sigma-Aldrich) staining. In some experiments, the respective amounts of actin in pellet and supernatant fractions were quantified using ImageJ software. The presence of actin bundles in samples was checked by direct visualization using fluorescence microscopy. An aliquot of the copolymerized actin samples was labeled with 4 μM rhodamine-phalloidin (Sigma-Aldrich). One micromolar of sample was diluted in one drop of cityfluor (Agar Scientific) and applied to a cover slip coated with poly-l-lysine (0.01%). Images were recorded via confocal microscope using a pinhole set to produce thick (~2 μm) optical sections.
F-Actin Depolymerization Assay
Pyrene-labeled actin (4 μM, 30% pyrene-labeled; Cytoskeleton) was copolymerized with individual Arabidopsis LIMs in the same conditions as in cosedimentation assays. Depolymerization was induced by diluting samples to a final actin concentration of 0.2 μM. The decrease in pyrene fluorescence accompanying actin depolymerization was recorded over 200 s using a PTI QM-4 QuantaMaster fluorimeter.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or Arabidopsis Genome Initiative databases under the following accession numbers: WLIM1 NM_100894.3 (At1g10200), WLIM2a NM_129548.3 (At2g39900), WLIM2b NM_001035791.1 (At3g55770), PLIM2a NM_001036468.2 (At2g45800), PLIM2b NM_100061.3 (At1g01780), and PLIM2c NM_115987.3 (At3g61230).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RNA Gel Blot and Microarray Analyses of LIM Gene Expression in Arabidopsis.
Supplemental Figure 2. Arabidopsis LIMs Interact with the Actin Cytoskeleton in Arabidopsis Cells.
Supplemental Figure 3. Fraction of Cytoplasmic Fluorescence Associated with the Actin Cytoskeleton in GFP-ABD2-GFP–, GFP-WLIM1–, and GFP-PLIM2c–Expressing Root and Root Hair Cells.
Supplemental Figure 4. Examples of Data Used to Calculate the Apparent Equilibrium Dissociation Constant (Kd) Values Shown in Table 1.
Supplemental Figure 5. Inactivation by pH of the Stabilizing Activity of AF-Bound PLIM2c.
Supplemental Figure 6. Alignment of Arabidopsis LIM Protein Sequences.
Supplemental Figure 7. Imaging of SNARF-5F in a GFP-PLIM2c–Expressing Arabidopsis Cell.
Supplemental Figure 8. Cytoplasmic pH Recovery after Artificially Induced Acidification.
Supplemental Figure 9. Persistence of a Prominent Actin Cytoskeleton after Increase of Cytoplasmic pH in GFP-PLIM2c–Expressing Cells.
Supplemental Table 1. Oligonucleotides Used in the Study.
Supplemental Table 2. Concentrations of MES, PIPES, Tris, EGTA, and CaCl2 Used in the Different in Vitro Assays.
Supplemental Movie 1. Time-Lapse Confocal Scanning Microscopy of a Growing Arabidopsis Pollen Tube Expressing GFP-PILM2c under the Control of the PLIM2c Promoter.
Supplemental Movie 2. Time-Lapse Confocal Scanning Microscopy of a Growing Pollen Tube Showing the Interaction of GFP-PLIM2c with a Subapical Actin Fringe-Like Structure.
Supplemental Methods. Detailed Cloning Procedures.
Acknowledgments
We thank Elison B. Blancaflor (The Samuel Roberts Noble Foundation, OK) who kindly provided Arabidopsis GFP-ABD2-GFP–expressing seeds. We also thank the gardeners of the Institut de Biologie Moléculaire des Plantes (Centre National de la Recherche Scientifique and University of Strasbourg, France) for excellent plant care and Esther Lechner and Katia Marocco for logistic support of plant-related experiments. This work was supported by the Ministry of Culture, Higher Education, and Research and by the National Research Fund (Luxembourg).
References
- Alves-Ferreira M., Wellmer F., Banhara A., Kumar V., Riechmann J.L., Meyerowitz E.M. (2007). Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S., Hunter J.J., Ross J., Jr, Hongo M., Sansig G., Borg J., Perriard J.C., Chien K.R., Caroni P. (1997). MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88: 393–403 [DOI] [PubMed] [Google Scholar]
- Arnaud D., Dejardin A., Leple J.C., Lesage-Descauses M.C., Pilate G. (2007). Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 14: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles J.R. (2000). Parallel actin bundles and their multiple actin-bundling proteins. Curr. Opin. Cell Biol. 12: 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Staiger C.J. (2008). Plant formins: Diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta 1803: 201–206 [DOI] [PubMed] [Google Scholar]
- Certal A.C., Almeida R.B., Carvalho L.M., Wong E., Moreno N., Michard E., Carneiro J., Rodriguez-Leon J., Wu H.M., Cheung A.Y., Feijo J.A. (2008). Exclusion of a proton ATPase from the apical membrane is associated with cell polarity and tip growth in Nicotiana tabacum pollen tubes. Plant Cell 20: 614–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Wong E.I., Vidali L., Estavillo A., Hepler P.K., Wu H.M., Cheung A.Y. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Duan Q.H., Costa S.S., de Graaf B.H., Di Stilio V.S., Feijo J., Wu H.M. (2008). The dynamic pollen tube cytoskeleton: live cell studies using actin-binding and microtubule-binding reporter proteins. Mol. Plant 1: 686–702 [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen ubes. Annu. Rev. Plant Biol. 59: 547–572 [DOI] [PubMed] [Google Scholar]
- Clough S.J. (2005). Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286: 91–102 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobak B.K., Franklin-Tong V.E., Staiger C.J. (2004). The role of the actin cytoskeleton in plant cell signaling. New Phytol. 163: 13–30 [DOI] [PubMed] [Google Scholar]
- Eliasson A., Gass N., Mundel C., Baltz R., Kräuter R., Evrard J.L., Steinmetz A. (2000). Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol. Gen. Genet. 264: 257–267 [DOI] [PubMed] [Google Scholar]
- Fan X., Hou J., Chen X., Chaudhry F., Staiger C.J., Ren H. (2004). Identification and characterization of a Ca2+-dependent actin filament-severing protein from lily pollen. Plant Physiol. 136: 3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijo J.A., Sainhas J., Hackett G.R., Kunkel J.G., Hepler P.K. (1999). Growing pollen tubes possess a constitutive alkaline band in the clear zone and a growth-dependent acidic tip. J. Cell Biol. 144: 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijo J.A., Sainhas J., Holdaway-Clarke T., Cordeiro M.S., Kunkel J.G., Hepler P.K. (2001). Cellular oscillations and the regulation of growth: the pollen tube paradigm. Bioessays 23: 86–94 [DOI] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubinger M., Gimona M. (2004). CRP2 is an autonomous actin-binding protein. FEBS Lett. 557: 88–92 [DOI] [PubMed] [Google Scholar]
- Grunt M., Zarsky V., Cvrckova F. (2008). Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J.R., Macalma T., Brown D., Richardson J.A., Olson E.N., Beckerle M.C. (1999). The LIM protein, CRP1, is a smooth muscle marker. Dev. Dyn. 214: 229–238 [DOI] [PubMed] [Google Scholar]
- Hepler P.K., Lovy-Wheeler A., McKenna S.T., Kunkel J.G. (2006). Ions and pollen tube growth. Plant Cell Monogr. 3: 47–69 [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke T.L., Feijo J.A., Hackett G.R., Kunkel J.G., Hepler P.K. (1997). Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Chaudhry F., Franklin-Tong V.E., Staiger C.J. (2004). A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 279: 23364–23375 [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., Neuhuber W.L., Klugbauer N., Ruth P., Allescher H.D. (2000). Cysteine-rich protein 2, a novel substrate for cGMP kinase I in enteric neurons and intestinal smooth muscle. J. Biol. Chem. 275: 5504–5511 [DOI] [PubMed] [Google Scholar]
- Hussey P.J., Allwood E.G., Smertenko A.P. (2002). Actin-binding proteins in the Arabidopsis genome database: Properties of functionally distinct plant actin-depolymerizing factors/cofilins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M., Entani T., Shiba H., Kakita M., Nagai T., Mizuno H., Miyawaki A., Shoji T., Kubo K., Isogai A., Takayama S. (2009). Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 150: 1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.S., Greenwood J.A. (2009). Glycine-rich region regulates cysteine-rich protein 1 binding to actin cytoskeleton. Biochem. Biophys. Res. Commun. 380: 484–488 [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Burgos-Rivera B., McKinney E.C., Ruzicka D.R., Meagher R.B. (2007). Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19: 3111–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (2002). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Kaneyama J.R., Suzuki W., Ichikawa K., Ohki T., Kohno Y., Sata M., Nose K., Shibanuma M. (2005). Uni-axial stretching regulates intracellular localization of Hic-5 expressed in smooth-muscle cells in vivo. J. Cell Sci. 118: 937–949 [DOI] [PubMed] [Google Scholar]
- Klahre U., Friederich E., Kost B., Louvard D., Chua N.H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 122: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M.G., Shi W., Ramagopal U., Tseng Y., Wirtz D., Kovar D.R., Staiger C.J., Almo S.C. (2004). Structure of the actin crosslinking core of fimbrin. Structure 12: 999–1013 [DOI] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Staiger C.J., Weaver E.A., McCurdy D.W. (2000). AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis H.A., Pino J.D., Schmeichel K.L., Pomies P., Beckerle M.C. (1997). Comparison of three members of the cysteine-rich protein family reveals functional conservation and divergent patterns of gene expression. J. Biol. Chem. 272: 27484–27491 [DOI] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Kunkel J.G., Allwood E.G., Hussey P.J., Hepler P.K. (2006). Oscillatory increases in alkalinity anticipate growth and may regulate actin dynamics in pollen tubes of lily. Plant Cell 18: 2182–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Wilsen K.L., Baskin T.I., Hepler P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221: 95–104 [DOI] [PubMed] [Google Scholar]
- Marrocco K., Lecureuil A., Nicolas P., Guerche P. (2003). The Arabidopsis SKP1-like genes present a spectrum of expression profiles. Plant Mol. Biol. 52: 715–727 [DOI] [PubMed] [Google Scholar]
- McCurdy D.W., Kim M. (1998). Molecular cloning of a novel fimbrin-like cDNA from Arabidopsis thaliana. Plant Mol. Biol. 36: 23–31 [DOI] [PubMed] [Google Scholar]
- McDowell J.M., Huang S., McKinney E.C., An Y.Q., Meagher R.B. (1996). Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142: 587–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.B., McKinney E.C., Kandasamy M.K. (1999). Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell 11: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli M., Robinson K.R. (1997). Tip localized Ca2+ pulses are coincident with peak pulsatile growth rates in pollen tubes of Lilium longiflorum. J. Cell Sci. 110: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Messerli M., Robinson K.R. (1998). Cytoplasmic acidification and current influx follow growth pulses of Lilium longiflorum pollen tubes. Plant J. 16: 87–91 [Google Scholar]
- Michelot A., Derivery E., Paterski-Boujemaa R., Guerin C., Huang S., Parcy F., Staiger C.J., Blanchoin L. (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr. Biol. 16: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guerin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundel C., Baltz R., Eliasson A., Bronner R., Grass N., Krauter R., Evrard J.L., Steinmetz A. (2000). A LIM-domain protein from sunflower is localized to the cytoplasm and/or nucleus in a wide variety of tissues and is associated with the phragmoplast in dividing cells. Plant Mol. Biol. 42: 291–302 [DOI] [PubMed] [Google Scholar]
- Papalouka V., Arvanitis D.A., Vafiadaki E., Mavroidis M., Papadodima S.A., Spiliopoulou C.A., Kremastinos D.T., Kranias E.G., Sanoudou D. (2009). Muscle Lim Protein interacts with Cofilin 2 and regulates F-actin dynamics in cardiac and skeletal muscle. Mol. Cell. Biol. 29: 6046–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.M., Fischer S., Malhó R., Papasouliotis O., Jelitto T.C., Leonard T., Read N.D. (1997). Pronounced cytoplasmic pH gradients are not required for tip growth in plant and fungal cells. J. Cell Sci. 110: 1187–1198 [DOI] [PubMed] [Google Scholar]
- Pierson E.S., Miller D.D., Callaham D.A., van Aken J., Hackett G., Hepler P.K. (1996). Tip-localized calcium entry fluctuates during pollen tube growth. Dev. Biol. 174: 160–173 [DOI] [PubMed] [Google Scholar]
- Pomies P., Louis H.A., Beckerle M.C. (1997). CRP1, a LIM domain protein implicated in muscle differentiation, interacts with alpha-actinin. J. Cell Biol. 139: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka D.R., Kandasamy M.K., McKinney E.C., Burgos-Rivera B., Meagher R.B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52: 460–472 [DOI] [PubMed] [Google Scholar]
- Sadler I., Crawford A.W., Michelsen J.W., Beckerle M.C. (1992). Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J. Cell Biol. 119: 1573–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagave J.F., Moser M., Ehler E., Weiskirchen S., Stoll D., Gunther K., Buttner R., Weiskirchen R. (2008). Targeted disruption of the mouse Csrp2 gene encoding the cysteine- and glycine-rich LIM domain protein CRP2 result in subtle alteration of cardiac ultrastructure. BMC Dev. Biol. 8: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit A.C. (2000). Actin during mitosis and cytokinesis. Actin: A Dynamic Framework for Multiple Plant Cell Functions, Staiger C.J., Baluska F., Volkmann D., Barlow P.W., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 437–456 [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T. (2007). The sliding theory of cytoplasmic streaming: Fifty years of progress. J. Plant Res. 120: 31–43 [DOI] [PubMed] [Google Scholar]
- Shimmen T., Hamatani M., Saito S., Yokota E., Mimura T., Fusetani N., Karaki H. (1995). Roles of actin filaments in cytoplasmic streaming and organization of transvacuolar strands in root hair cells of Hydrocharis. Protoplasma 185: 188–193 [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Thomas C., Dieterle M., Gatti S., Hoffmann C., Moreau F., Papuga J., Steinmetz A. (2008). Actin bundling via LIM domains. Plant Signal. Behav. 3: 320–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18: 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Moreau F., Dieterle M., Hoffmann C., Gatti S., Hofmann C., Van Troys M., Ampe C., Steinmetz A. (2007). The LIM domains of WLIM1 define a new class of actin bundling modules. J. Biol. Chem. 282: 33599–33608 [DOI] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton 66: 940–957 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P.K., Shimmen T. (2000). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta 210: 836–843 [DOI] [PubMed] [Google Scholar]
- Tran T.C., Singleton C., Fraley T.S., Greenwood J.A. (2005). Cysteine-rich protein 1 (CRP1) regulates actin filament bundling. BMC Cell Biol. 6: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Hepler P.K. (2001). Actin and pollen tube growth. Protoplasma 215: 64–76 [DOI] [PubMed] [Google Scholar]
- Vidali L., McKenna S.T., Hepler P.K. (2001). Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12: 2534–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Yokota E., Cheung A.Y., Shimmen T., Hepler P.K. (1999). The 135-kDa actin-bundling protein from Lilium longiflorum pollen is the plant homolog of villin. Protoplasma 209: 283–291 [Google Scholar]
- Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H.J., Wan A.R., Jauh G.Y. (2008a). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147: 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.S., Yoo C.M., Blancaflor E.B. (2008b). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Weiskirchen R., Gunther K. (2003). The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. Bioessays 25: 152–162 [DOI] [PubMed] [Google Scholar]
- Winder S.J., Ayscough K.R. (2005). Actin-binding proteins. J. Cell Sci. 118: 651–654 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis Formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yet S.F., Folta S.C., Jain M.K., Hsieh C.M., Maemura K., Layne M.D., Zhang D., Marria P.B., Yoshizumi M., Chin M.T., Perrella M.A., Lee M.E. (1998). Molecular cloning, characterization, and promoter analysis of the mouse Crp2/SmLim gene. Preferential expression of its promoter in the vascular smooth muscle cells of transgenic mice. J. Biol. Chem. 273: 10530–10537 [DOI] [PubMed] [Google Scholar]
- Yokota E., Tominaga M., Mabuchi I., Tsuji Y., Staiger C.J., Oiwa K., Shimmen T. (2005). Plant villin, lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant Cell Physiol. 46: 1690–1703 [DOI] [PubMed] [Google Scholar]