Replication of positive-strand RNA viruses, the largest group of plant viruses, is initiated by viral RNA-dependent RNA polymerase (RdRp). This work shows that Turnip yellow mosaic virus RdRp is a target of the ubiquitin-proteasome system in plant cells during viral infection and supports the idea that proteasomal degradation may constitute another level of regulation of viral replication.
Abstract
Replication of positive-strand RNA viruses, the largest group of plant viruses, is initiated by viral RNA-dependent RNA polymerase (RdRp). Given its essential function in viral replication, understanding the regulation of RdRp is of great importance. Here, we show that Turnip yellow mosaic virus (TYMV) RdRp (termed 66K) is degraded by the proteasome at late time points during viral infection and that the accumulation level of 66K affects viral RNA replication in infected Arabidopsis thaliana cells. We mapped the cis-determinants responsible for 66K degradation within its N-terminal noncatalytic domain, but we conclude that 66K is not a natural N-end rule substrate. Instead, we show that a proposed PEST sequence within 66K functions as a transferable degradation motif. In addition, several Lys residues that constitute target sites for ubiquitylation were mapped; mutation of these Lys residues leads to stabilization of 66K. Altogether, these results demonstrate that TYMV RdRp is a target of the ubiquitin-proteasome system in plant cells and support the idea that proteasomal degradation may constitute yet another fundamental level of regulation of viral replication.
INTRODUCTION
Genome replication of positive-strand RNA [(+)RNA] viruses, the largest class of viruses, which includes significant pathogens of humans, animals, and plants (van Regenmortel et al., 2000), requires assembly of an intricate replication complex comprising both viral and host proteins that forms on intracellular membranes (Ahlquist et al., 2003; Nagy and Pogany, 2009). Successful replication depends to a large extent on the ability of these viruses to generate viral proteins at the required concentration and in a coordinated manner during the various stages of the replication cycle.
Within the replication complex, viral RNA-dependent RNA polymerase (RdRp) plays a pivotal role in the viral infection process, catalyzing synthesis of new viral RNA genomes from the original infecting RNA (Ahlquist, 2002). The importance of maintaining the correct balance of RdRp during viral infection has been highlighted in recent studies, as host membrane rearrangements, RNA replication, and RNA recombination efficiencies are affected by the absolute and relative abundance of viral RdRp (Schwartz et al., 2004; Pantaleo et al., 2004; Jaag et al., 2007).
Many (+)RNA viruses produce their replication proteins as polyprotein precursors that are subsequently cleaved to generate functional viral gene products (Buck, 1996). Equimolar synthesis of proteins as part of the initial polyprotein implies that differences in the concentrations of mature viral proteins are determined by differences in maturation rates and by differences in their subsequent degradation rates.
The degradation of proteins by the ubiquitin-proteasome system (UPS) is recognized as a major process by which many aspects of cell biology are regulated (Glickman and Ciechanover, 2002). Ubiquitin (Ub)-mediated degradation has been widely conserved across the eukaryotic kingdoms, including yeast, plants, and mammals, and as judged from the large number of Arabidopsis thaliana genes involved in Ub-dependent protein turnover, as well as accumulating genetic or biochemical studies, protein degradation by the UPS plays a central role in many processes in plants (Bachmair et al., 2001; Vierstra, 2009).
It has also become increasingly clear in recent years that the UPS is involved in the interactions between plants and pathogens (Zeng et al., 2006; Dreher and Callis, 2007; Citovsky et al., 2009; Craig et al., 2009; Trujillo and Shirasu, 2010). Signaling and regulation of nonhost disease resistance, basal immunity, resistance gene-mediated responses, and systemic acquired resistance appear to involve members of the UPS (Kim and Delaney, 2002; Liu et al., 2002; Peart et al., 2002; Trujillo et al., 2008), and perturbation of the Ub conjugation pathway was also shown to alter plant responses to pathogens (Becker et al., 1993; Dong et al., 2006; Goritschnig et al., 2007).
Interestingly, the UPS is not only used by host cells in immunity and biotic stress responses, but it can also be manipulated and subverted by pathogens, including viruses, for their own use (Isaacson et al., 2009; Spallek et al., 2009; Dielen et al., 2010). Yet, appreciation of its full capability is limited by the extreme paucity of known targets (Zeng et al., 2006; Dreher and Callis, 2007; Citovsky et al., 2009; Vierstra, 2009; Trujillo and Shirasu, 2010).
A better understanding of the involvement of the UPS during (+)RNA virus infection may thus provide deeper insights into plant–pathogens interactions and open new avenues for the development of antiviral strategies. Here, we address this question by studying the stability and degradation pathway of Turnip yellow mosaic virus (TYMV) RdRp in Arabidopsis cells.
TYMV, the type member of the genus Tymovirus, is a plant (+)RNA virus that has proven useful in the study of fundamental aspects of viral multiplication (Dreher, 2004). The 6.3-kb genomic RNA (gRNA) encodes two nonstructural proteins of 69 and 206 kD (206K), the coat protein (CP) being expressed from a subgenomic RNA (sgRNA) produced during viral replication. 206K, the only viral protein required for TYMV replication, shares considerable sequence similarity with replication proteins of other (+)RNA viruses (Buck, 1996) and contains domains indicative of methyltransferase, proteinase, NTPase/helicase, and RdRp activities. Self-cleavage of 206K generates a C-terminal 66K protein encompassing the RdRp domain and an N-terminal 140K protein that is further processed into 98K and 42K proteins (Prod'homme et al., 2001; Jakubiec et al., 2007). Assembly of TYMV replication complexes depends on interactions between the 66K RdRp and the membrane-bound 140K protein (Prod'homme et al., 2003; Jakubiec et al., 2004).
Several lines of evidence suggest that TYMV 66K RdRp is conditionally targeted for degradation: first, we reported that 66K is phosphorylated and ubiquitylated when expressed in insect cells (Héricourt et al., 2000). Two phosphoresidues (Thr-64 and Ser-80) were mapped within an N-terminal PEST sequence, a conditional signal for protein degradation (Rechsteiner and Rogers,1996) that is conserved among tymovirus RdRps (Héricourt et al., 2000). The same phosphoresidues were identified in plant cells, where their phosphorylation status appears to be regulated by the virus-encoded 140K protein (Jakubiec et al., 2006). Mimicking their constitutive phosphorylation led to a severe decrease in the accumulation of 66K in infected cells and appeared strongly deleterious for viral infectivity (Jakubiec et al., 2006; Jakubiec and Jupin, 2007). Time-course experiments demonstrated that 66K RdRp accumulates transiently during viral infection (Prod'homme et al., 2001), while analysis of the stoichiometry within viral replication complexes revealed that the amount of 66K was only ~1/20 of that of its 98K protein counterpart (A. Jakubiec and I. Jupin, unpublished data).
Here, we provide evidence that TYMV RdRp is indeed a target of the UPS in plant cells, with the degradation machinery targeting the PEST squence in the N-terminal domain of 66K. Several Lys residues that act as target sites for ubiquitylation were also mapped. Our findings suggest UPS degradation as yet another fundamental level of regulation of viral replication.
RESULTS
UPS Involvement during Viral Infection
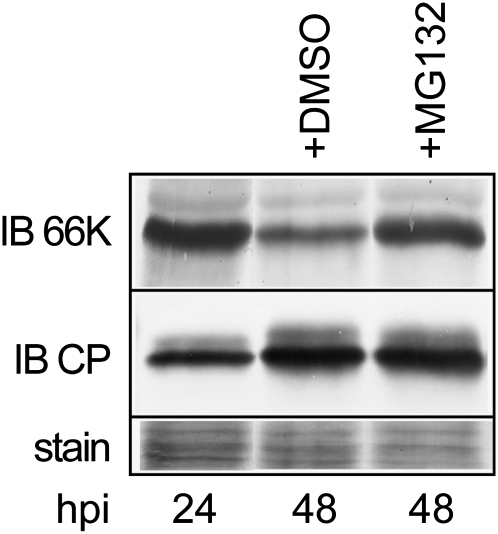
Following infection of Arabidopsis protoplasts with TYMV RNA, the amount of 66K protein is maximal at 24 to 30 h postinfection (hpi) and then decreases gradually until 48 hpi (Prod'homme et al., 2001). To assess the involvement of the 26S proteasome, the major proteolytic degradation system in eukaryotic cells (Glickman and Ciechanover, 2002), during this step of the viral infection process, the proteasome inhibitor MG132 was added at both 28 and 36 hpi (Lee and Goldberg, 1998). As shown in Figure 1, this treatment markedly increased the 66K protein level compared with control DMSO treatment, consistent with the hypothesis that downregulation of 66K at late time points in viral infection is proteasome dependent. No effect was observed on the level of CP, ruling out a general effect of MG132 on the level of protein synthesis.
Figure 1.
Involvement of UPS in the Accumulation of 66K during Viral Infection.
Arabidopsis protoplasts were infected with TYMV RNA and were treated with DMSO or the proteasome inhibitor MG132, as indicated. Cells were collected at 24 or 48 hpi, and equivalent protein amounts were subjected to immunoblot (IB) analysis using anti-66K and anti-CP antibodies. Ponceau staining of the membrane (stain) is shown to indicate protein loading.
These results indicate that the UPS is likely to be involved in 66K accumulation during TYMV infection.
UPS-Mediated Degradation of 66K
To determine whether 66K constitutes a direct target for the UPS and to further characterize the 66K degradation process in plant cells, a combination of two different techniques was used.
First, we measured protein half-life by performing pulse-chase labeling experiments using a stably transformed Arabidopsis suspension cell line that constitutively expresses a 66K-His protein. The encoded protein is biologically active, as previously demonstrated by its ability to trans-complement a defective viral mutant (Prod'homme et al., 2003).
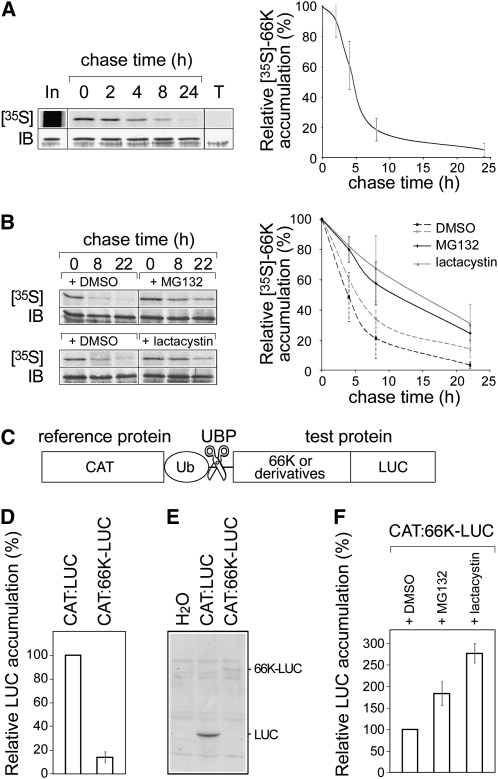
To determine the half-life of 66K, Arabidopsis cells were pulse labeled, followed by incubation in chase medium for varying periods. The cells were lysed and the 66K protein was immunoprecipitated using specific antibodies (Figure 2A, left). After correcting for the amount of labeled protein present in the immunoprecipitates, the in vivo half-life of 66K in Arabidopsis cells was estimated to be ~5 h (Figure 2A, right). Addition of the proteasome inhibitors MG132 and clastolactacystin β-lactone to cells expressing 66K-His led to a stabilization of the protein in both cases (Figure 2B), demonstrating that 66K is targeted for degradation by the 26S proteasome.
Figure 2.
Degradation of 66K Is Mediated by the UPS.
(A) Pulse-chase experiments: Arabidopsis transgenic cells expressing 66K-His were pulse labeled with [35S]Met and [35S]Cys and then chased for the times indicated. The left panel shows a representative experiment. Cell lysates were subjected to immunoprecipitation with anti-66K antibody, and the resulting precipitates were subjected to SDS-PAGE and phosphor imaging ([35S]; top). In, input protein recovered immediately after the labeling period; T, control experiment performed using non transformed cells. The band intensities were quantified, corrected for the total amount of 66K present in the sample as determined by immunoblotting (IB; bottom), and expressed as a percentage of the corresponding value at the beginning of the chase period. The right panel shows the data as mean ± sd of n = 5 replicates.
(B) Pulse-chase experiments as in (A) were performed in the presence of DMSO, MG132 (n = 4), or clastolactacystin β-lactone (n = 2).
(C) Schematic representation of the chimeric protein used in the UPR assay. Reference and test proteins are separated by a ubiquitin moiety (UbK48R) that is cleaved by cellular ubiquitin-specific processing proteases (UBP).
(D) Stability of LUC fusion proteins as measured using the UPR assay. LUC activity was determined and divided by the activity of the CAT internal control. The results are shown as percentages of the control. Data are mean ± sd of n = 14 replicates.
(E) Protein extracts from cells transfected with pΩ-CAT:LUC or pΩ-CAT:LUC-66K were immunodetected using anti-LUC antibody. Samples were adjusted to equal CAT activity and equal total protein by addition of negative control (water) extract to samples with lower protein content.
(F) Stability of LUC fusion proteins was measured using the UPR assay in the presence of DMSO, MG132 (n = 8), or clastolactacystin β-lactone (n = 6).
In a complementary approach, we used the ubiquitin/protein/reference (UPR) technique (Lévy et al., 1996). In this system, a test protein is produced as a translational fusion to a stable reference protein separated by a Ub monomer. Such fusions are rapidly and precisely cleaved at the C terminus of Ub by cellular Ub-specific processing proteases, yielding equimolar amounts of the test and reference proteins (Figure 2C). This technique avoids the need for pulse-chase labeling to study protein degradation signals, as determining the steady state molar ratios of test and reference proteins in cell extracts yields a direct ranking of their metabolic stabilities (Lévy et al., 1996). Different variations of this method have been used successfully in plants (Karsies et al., 2001; Stary et al., 2003), and we adapted it using two stable reporter proteins whose activity can be quantified directly in crude cell lysates. We constructed the plant expression vectors pΩ-CAT:LUC and pΩ-CAT:66K-LUC, in which chloramphenicol acetyl transferase (CAT) served as the internal control and luciferase (LUC), unfused or N-terminally fused to the 66K protein, served as the test protein (Figure 2C). The LUC/CAT activity ratio reflects the instability of the test protein. Fusion of the 66K protein to LUC caused a significant reduction in both the LUC/CAT activity ratio (Figure 2D) and the steady state level of LUC protein as determined by immunoblotting (Figure 2E), thus validating this approach and confirming the instability of the 66K protein in plant cells.
The proteasome inhibitors MG132 and clastolactacystin β-lactone both had a strong inhibitory effect on protein production in plant cells, consistent with previous reports (Karsies et al., 2001; Deroo and Archer, 2002). However, after correcting for this effect using the control plasmid pΩ-CAT:LUC, both inhibitors led to a stabilization of 66K, compared with DMSO treatment (Figure 2F), again confirming the involvement of the 26S proteasome in degradation of 66K in plant cells.
66K Is Not Degraded by the N-End Rule Pathway
A major determinant of protein half-life is the presence of degradation signals, or degrons (Glickman and Ciechanover, 2002; Ravid and Hochstrasser, 2008; Jadhav and Wooten, 2009). Both modular domains and small peptide sequences have been shown to act as degradation signals. Among them is the N-end rule degron, which relates the in vivo half-life of a protein to the identity of its N-terminal residue (Bachmair et al., 1986; Varshavsky, 1996).
The 66K derives from proteolytic processing of the precursor 206K polyprotein and bears an N-terminal Thr residue, a stabilizing N-terminal residue in yeast, but a destabilizing one in mammals (Gonda et al., 1989). To determine whether the N-end rule is involved in 66K instability in plant cells, we performed site-directed mutagenesis of plasmid pΩ-CAT:66K-LUC, replacing the first residue of 66K with either a stable Met residue or with Trp, Phe, Tyr, Leu, or Arg residues, which are known N-end rule substrates in plants (Worley et al., 1998; Stary et al., 2003; Garzon et al., 2007).
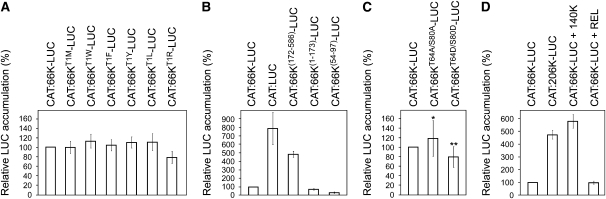
As shown in Figure 3A, no significant difference in 66K stability was seen when the wild-type Thr was compared with Met at the N terminus. Other substitutions also did not affect the 66K turnover, except Arg, which had a modest effect. These results thus suggest that the N-terminal Thr of 66K does not act as an instability determinant, indicating that TYMV 66K is not a natural N-end rule substrate in plant cells.
Figure 3.
Delineation of the 66K Degron and Impact of 66K Phosphorylation on Its Stability.
Stability of LUC fusion proteins was measured using the UPR assay as in Figure 2.
(A) Effects of mutation of the N-terminal Thr to Met, Trp, Phe, Tyr, Leu, or Arg on the stability of 66K-LUC. Data are mean ± sd of n = 10 replicates.
(B) Effects of N- or C-terminal deletions on the stability of 66K-LUC. Data are mean ± sd of n = 8 replicates.
(C) Effects of mutation of phosphorylation target sites on stability of 66K-LUC. Data are mean ± sd of n = 24 replicates. Mann-Whitney rank test was used to test the significance of the results (*P < 0.05; **P < 0.01).
(D) Effects of coexpressing 140K replication protein or control protein (REL, Renilla luciferase) on stability of 66K-LUC. Data are mean ± sd of n = 8 replicates.
Importance of the N-Terminal Domain and the PEST Sequence in 66K Instability
To further characterize domains directing degradation, 66K N- or C-terminal sequences were fused to LUC, and protein stability was analyzed using the UPR system. As shown in Figure 3B, the 66K(172-586)-LUC fusion protein appeared much more stable, whereas the 66K(1-173)-LUC fusion was unstable and accumulated to a steady state level comparable to that of the full-length 66K protein. From these data, we conclude that the major cis-determinants responsible for the instability of 66K lie within the N-terminal domain.
Interestingly, a high turnover rate was also observed when amino acids 54 to 97, corresponding to the proposed PEST sequence within the 66K protein (Héricourt et al., 2000), were fused to LUC. This result indicates that the PEST sequence could function as a transferable degradation motif that is likely to constitute the 66K degron.
Influence of Phosphorylation on 66K Instability
Because PEST sequences are reported to behave as phosphodegrons (i.e., conditional signals for degradation regulated by phosphorylation/dephosphorylation; Hunter, 2007), we next wanted to examine the importance for 66K stability of residues Thr-64 and Ser-80, previously reported phosphorylation target sites within the PEST sequence (Héricourt et al., 2000; Jakubiec et al., 2006). Thus, mutations affecting these residues were introduced in the plasmid pΩ-CAT:66K-LUC: substituting target residues with Ala led to a phosphorylation-deficient mutant, while phosphorylation was mimicked by substituting Ser and Thr with the negatively charged Asp to reproduce the electrostatic and steric effects of phosphorylation (Dean and Koshland, 1990; Sweeney et al., 1994; Waigmann et al., 2000).
Introducing the phosphorylation-deficient mutation T64A/S80A attenuated 66K protein degradation, whereas the phosphomimic T64D/S80D mutation further destabilized the 66K protein (Figure 3C). The differences observed were statistically significant, indicating that degradation of 66K is influenced by the presence of negative charges within the PEST sequence. However, the effects were modest, suggesting that the phosphorylation status of those residues might contribute only moderately to the regulation of metabolic stability of 66K.
Stabilization of 66K by Coexpression of 140K
TYMV 140K protein is involved in the recruitment of 66K to the sites of viral replication, which are at the periphery of chloroplast envelope membranes (Prod'homme et al., 2001, 2003). As 140K was previously reported to interact physically with the N-terminal region of 66K (Jakubiec et al., 2004) and to prevent phosphorylation of the target residues within the PEST sequence (Jakubiec et al., 2006), we next tested whether coexpression of TYMV 140K protein would influence the stability of 66K.
The 140K protein was expressed either directly from the expression vector pΩ-140K or from the plasmid pΩ-CAT:206K-LUC that expresses the 206K polyprotein, which, after self-processing, generates 140K and 66K-LUC proteins. In both cases, coexpression of the 140K protein, but not of a control protein (REL), led to a profound stabilization of 66K-LUC (Figure 3D). Such an effect was not observed on the control unfused LUC protein (data not shown), thus indicating that 140K exerts a protective effect over 66K degradation in plant cells.
Ubiquitylation of 66K Protein in Plant Cells
The great majority of proteins that are degraded by the 26S proteasome are targeted for degradation by ubiquitylation (Pickart and Eddins, 2004). This involves the covalent conjugation of Ub, a highly conserved 76-residue protein, to the target protein via an isopeptide bond between the C terminus of Ub and the −NH2 group of one or more Lys residues of the protein. Stepwise conjugation of additional Ub moieties to the first Ub molecule then generates the poly-Ub chains that are essential for recognition and subsequent degradation of the target protein by the proteasome (Thrower et al., 2000).
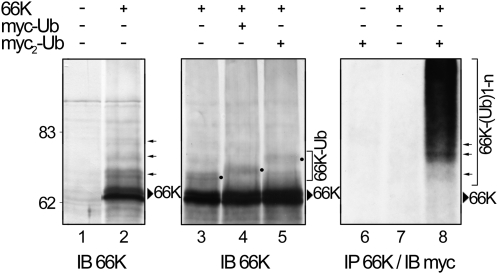
To investigate if 66K-Ub conjugates are generated in plant cells, 66K transiently expressed using the pΩ-66K expression vector was immunodetected using anti-66K antiserum. A characteristic ladder of discrete bands was observed (Figure 4, lane 2, arrows), consistent with 66K polyubiquitylation.
Figure 4.
Ubiquitylation of 66K in Plant Cells.
Arabidopsis protoplasts were transfected with expression vectors encoding 66K, myc-Ub, or myc2-Ub as indicated, and 66K was detected by immunoblotting (IB) using anti-66K antibody (lanes 2 to 5). The 66K-Ub conjugates were identified based on the ~8-kD increase in molecular mass (lane 2, arrows) and the ~2-kD difference between 66K-Ub, 66K-myc-Ub, and 66K-myc2-Ub conjugates (lanes 3 to 5, dots). In lanes 6 to 8, samples were immunoprecipitated (IP) under denaturing conditions using anti-66K antibody and subjected to immunodetection using anti-myc antibody. The position of 66K is indicated by an arrowhead.
To demonstrate that these higher molecular mass products indeed represent ubiquitylated 66K, cells were cotransfected with a plasmid encoding Ub tagged with one or two c-myc epitope tags (myc-Ub and myc2-Ub), which are ~2 and 4 kD larger than Ub, respectively. Upon immunodetection of the 66K protein, slightly slower migrating products were detected in the presence of the c-myc–tagged Ubs (Figure 4, lanes 4 and 5, dots), confirming that the corresponding bands represent ubiquitylated 66K.
To further confirm this result, 66K was immunoprecipitated under denaturing conditions from cells coexpressing myc2-Ub, and immunoblotting via anti-c-myc antibody was used to detect the conjugation of myc2-Ub to 66K. As shown in Figure 4, lane 8, discrete bands and a smear of conjugated species, characteristic of the formation of a heterogeneous mixture of poly-Ub-66K conjugates, were detected. Control samples lacked similar ubiquitylated proteins (Figure 4, lanes 6 and 7), demonstrating the specificity of the observed signals. Altogether, these results demonstrate that 66K is ubiquitylated when expressed in plant cells.
Several Lys Residues Are Ub Acceptor Sites, and Their Mutation Affects 66K Stability
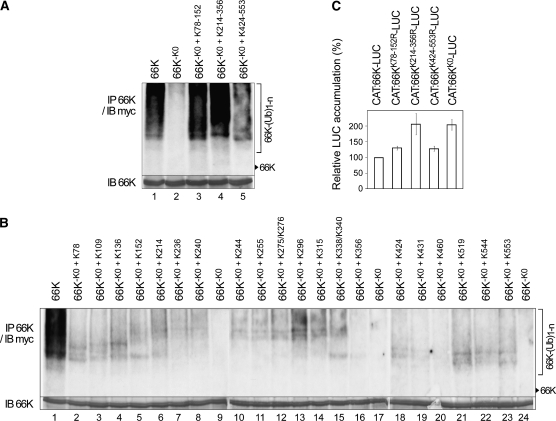
Poly-Ub is generally conjugated with internal Lys residues, although N-terminal Cys, Ser, or Thr residues have sometimes been reported to constitute acceptor sites (Breitschopf et al., 1998; Glickman and Ciechanover, 2002; Cadwell and Coscoy, 2005; Vosper et al., 2009). To define residues critical for 66K ubiquitylation, we performed a mutational scanning analysis, substituting each of the 22 Lys residues in the 66K protein by Arg residues. As preliminary experiments suggested that all single mutants retained susceptibility to ubiquitylation, those mutations were combined altogether to generate the 66K-K0 mutant, in which all Lys residues were substituted. No ubiquitylation of 66K-K0 was detected (Figure 5A, lane 2). Domain swapping experiments between 66K and 66K-K0 were then performed, generating 66K mutants 66K-K0+K78-152, 66K-K0+K214-356, and 66K-K0+K424-553 containing Lys residues exclusively in the N-terminal, central, and C-terminal domains of 66K, respectively. Ubiquitylation was detected in all three cases (Figure 5A, lanes 3 to 5), indicating that Ub acceptor sites must exist throughout the entire 66K.
Figure 5.
Several Lys Residues Are Ub-Acceptor Sites, and Their Mutation Affects 66K Stability.
(A) and (B) Arabidopsis protoplasts were cotransfected with expression vectors encoding myc2-Ub and various 66K mutants as indicated. Formation of poly-Ub-66K conjugates was determined by immunoprecipitation as in Figure 4. The amount of 66K present in the immunoprecipitates was determined by immunoblotting (IB).
(C) Effect of mutation of Lys to Arg on the stability of 66K-LUC, measured using the UPR assay as in Figure 2.
To further define the ubiquitylation sites, single Lys residues were reintroduced individually into the 66-K0 mutant, and in vivo ubiquitylation assays were performed as described above (Figure 5B). Immunodetection of 66K confirmed that all mutants were immunoprecipitated in comparable amounts and that they displayed the electrophoretic mobility shift characteristic of 66K phosphorylation (Jakubiec et al., 2006). It should be noted that the Ub conjugates of these 66K mutants appeared as much fainter bands or smears than in wild-type 66K due to the presence of only a single putative Ub acceptor site. However, when compared with the 66K-K0 protein, which was used as a control in all experiments (Figure 5B, lanes 9, 17, and 24), specific signals are detected. Strikingly, we observed that many Lys residues, spanning the entire 66K protein, had the capacity to be ubiquitylated. However, the efficiency varied substantially between mutants: Lys-296 (lane 13) displayed the highest level of ubiquitylation, whereas Lys-460 (lane 20) was apparently not conjugated. Interestingly, while 66K mutants containing a Lys acceptor residue in the central domain (Lys-214 to Lys-340) generated polyubiquitin conjugates that were >150 kD in size, the other mutants generated chains of lower molecular mass, accumulating predominantly di-, tri-, and tetraubiquitylated 66K.
To further establish the role of each domain in the degradation of 66K, Arg substitutions affecting Lys residues 78 to 152, 214 to 356, and 424 to 553 were introduced into plasmid pΩ-CAT:66K-LUC, generating pΩ-CAT:66KK78-152R-LUC, pΩ-CAT:66KK214-356R-LUC, and pΩ-CAT:66KK424-553R-LUC, respectively, while combining the mutations altogether generated the pΩ-CAT:66KK0-LUC expression vector. The degradation rate of the corresponding mutated 66K was then compared with that of wild-type 66K. We observed that all these substitutions led to a significant stabilization of 66K (Figure 5C), particularly those affecting the central domain.
The Accumulation Level of 66K Affects Viral RNA Replication
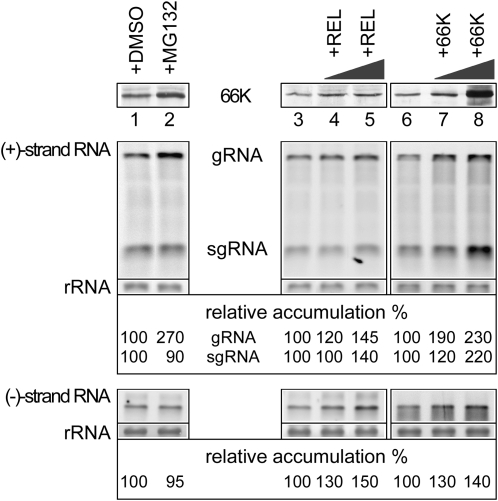
To test if the accumulation level of 66K is important for viral replication, we analyzed the levels of viral RNA accumulating in infected cells treated with MG132 at late time points of viral infection as this treatment has been shown to increase the 66K protein level (Figure 1). As shown in Figure 6 (lanes 1 and 2), RNA gel blotting analysis revealed that the level of viral gRNA was increased ~2.5-fold by MG132 treatment compared with the control DMSO treatment, whereas accumulation of sgRNA or (−)-strand RNA was unaffected.
Figure 6.
The Accumulation Level of 66K Affects Viral RNA Replication.
Arabidopsis protoplasts were infected with TYMV RNA and were either treated with DMSO or MG132 (lanes 1 and 2) or cotransfected with water (lanes 3 and 6) or with 1 or 10 μg of the expression vector encoding 66K (lanes 7 and 8) or control protein (REL, Renilla luciferase; lanes 4 and 5) as indicated. Cells were collected at 48 hpi. Equivalent protein amounts were subjected to immunoblot analysis using anti-66K antibody, and equivalent RNA amounts were subjected to strand-specific RNA gel blot analysis to detect (−)-strand and (+)-strand gRNA and sgRNA TYMV RNAs. Methylene blue staining of the membrane (rRNA) is shown to indicate RNA loading.
As MG132 is likely to stabilize many proteins other than 66K, including host factors that may contribute directly or indirectly to viral replication, we next tested whether overexpressing 66K in infected cells might also affect viral replication. To this end, Arabidopsis protoplasts were transfected with viral RNA transcripts, together with increasing amounts of the pΩ-66K expression vector or pΩ-REL as a negative control. As shown in Figure 6, pΩ-REL caused a slight increase in viral accumulation (lanes 3 to 5) that might be due to improved transfection efficiency due to a carrier DNA effect, as similar results were also obtained upon cotransfection of the empty expression vector pΩ (data not shown). However, the amounts of viral gRNA and sgRNA synthesized were further increased upon expression of 66K (lanes 6 to 8), thus indicating that a higher level of 66K can stimulate TYMV replication.
DISCUSSION
66K Is Degraded by UPS during Viral Infection
As judged from the large number of genes involved in Ub-dependent protein turnover, as well as accumulating genetic or biochemical studies, protein degradation by the UPS is important in many plant processes, and either continuous or regulated turnover is essential in the function of many proteins occupying key positions in regulatory networks (Bachmair et al., 2001; Vierstra, 2009).
Although it has become increasingly clear in recent years that the UPS plays a major role in the battle between plants and pathogens, only few protein targets have been identified so far (Zeng et al., 2006; Dreher and Callis, 2007; Citovsky et al., 2009; Vierstra, 2009; Trujillo and Shirasu, 2010). The results reported here show that UPS-mediated degradation of a viral RdRp occurs in plant cells during viral infection (Figure 1). This result is consistent with the observed transient accumulation of 66K during infection (Prod'homme et al., 2001) and with its stoichiometry within viral replication complexes being only 1:20 of that of 98K protein (A. Jakubiec and I. Jupin, unpublished data). Two independent experimental systems demonstrated 66K instability and confirmed the contribution of the UPS to the in vivo degradation of 66K independently of other viral proteins (Figure 2).
The 66K Degron Maps to the N-Terminal PEST Sequence
Degradation signals usually comprise two essential and separable determinants: an amino acid or conformational primary determinant that functions as a binding site for a substrate recognition factor and one or more internal Lys residue that is the site of ubiquitylation (Glickman and Ciechanover, 2002).
The major primary determinant for 66K degradation was mapped within its N-terminal noncatalytic domain (Figure 3B). In contrast with Sindbis polymerase nsp4 (De Groot et al., 1991), a viral polymerase also generated by polyprotein cleavage, 66K does not appear to be a natural N-end rule substrate (Figure 3A). Instead, based on its ability to function as a minimal transferable instability determinant, we conclude that the 66K degron maps to the PEST sequence (Figure 3B). Notably, PEST sequences are invariably identified in this particular region within different tymoviral RdRp proteins (Héricourt et al., 2000), suggesting that RdRp degradation may constitute a conserved feature among tymoviruses.
Phosphorylation is one of the major signals regulating degradation of proteins by the UPS (Karin and Ben-Neriah, 2000; Hunter, 2007). We previously proposed that phosphorylation of Thr-64 and Ser-80 within the PEST sequence may serve to control the metabolic stability of the protein (Héricourt et al., 2000; Jakubiec et al., 2006). Indeed, we observed that dephosphorylation of these residues slowed down degradation, whereas phosphomimic mutations further destabilized 66K (Figure 3C). However, the differences in 66K stability were modest, suggesting that phosphorylation of these residues is likely to have only a moderate impact on the control of 66K turnover. We cannot rule out the possibility that other, as yet uncharacterized, phosphorylation sites exist within the N-terminal region of 66K and contribute to its degradation.
Alternatively, the PEST sequence may also constitute a conditional degradation signal regulated by other means than phosphorylation. In a number of cases, PEST sequences were reported to become activated by conformational changes occurring upon ligand binding or protein–protein interactions (Rechsteiner and Rogers, 1996; García-Alai et al., 2006). In that respect, TYMV 140K was shown previously to interact with 66K, promoting its recruitment to chloroplast envelope membranes (Prod'homme et al., 2003; Jakubiec et al., 2004) and preventing phosphorylation of the target residues within the PEST sequence (Jakubiec et al., 2006). Because coexpression of 140K led to a more efficient stabilization of 66K than mutations affecting the Thr-64/Ser-80 phosphoresidues (Figures 3C and 3D), it is likely that 140K plays additional roles in that process than just inhibiting phosphorylation. As the domain of 66K that controls its stability is also part of the region to which 140K binds (Jakubiec et al., 2004), formation of a 66K/140K complex may interfere with recognition of 66K by the UPS by masking the degradation signal. It is also possible that subsequent compartmentalization, association with other proteins, or other events sequesters the 66K polymerase from the degradation machinery.
In vivo ubiquitylation assays using 66K mutant proteins containing a single Lys residue revealed that many residues, spanning the entire protein, can be involved in Ub conjugation (Figures 5A and 5B). Naturally unstable proteins are frequently modified on multiple Lys residues, but the mechanisms that control mono- or polyubiquitylation, chain topology, and Lys selection remain poorly understood (Catic et al., 2004; Jadhav and Wooten, 2009; Sadowski et al., 2010). As the ubiquitylation apparatus shows remarkable flexibility (Vosper et al., 2009), it is possible that some of the identified Lys residues may serve as acceptor sites only in the absence of other available residues. In that respect, the observation that 66KK0-LUC is not fully stabilized (Figure 5C) suggests that noncanonical conjugation events might possibly occur when all Lys residues are mutated. Cys, Ser, or Thr residues can constitute alternative Ub conjugation sites, as shown in a few examples (Cadwell and Coscoy, 2005; Wang et al., 2007; Vosper et al., 2009). Such noncanonical conjugation events could have been overlooked in the immunoprecipitation experiments shown in Figure 5A, as the experimental conditions involved high-temperature and reducing agents, conditions that will break these noncanonical linkages (Vosper et al., 2009). More direct approaches, such as mass spectrometry (Peng et al., 2003), may help clarify this point, but the low level of 66K expressed during viral infection has so far precluded such analyses.
Interestingly, we observed that the pattern of ubiquitylation varied among mutants as Lys within the central domain appeared to form longer polyubiquitin chains. Ubiquitin ligases for 66K have yet to be identified, and at present it is not clear whether the same enzyme catalyzes ubiquitylation of all target residues. Based on the stabilization of 66K observed upon their substitution (Figure 5C), it is likely that Lys residues spanning the entire 66K protein contribute substantially to its degradation, although central Lys residues appear to be more important in that process.
The Role of UPS Degradation in Viral Replication
UPS degradation of viral or cellular proteins is a major mechanism regulating viral infection (Glickman and Ciechanover, 2002; Isaacson et al., 2009). For (+)RNA viruses, ubiquitylation of viral proteins or requirement for proteasomal activity during viral infection has been documented in members of the Flaviviradae (Gao et al., 2003; Gilfoy et al., 2009), Picornaviridae (Luo et al., 2003), Coronaviridae (Yu and Lai, 2005; Raaben et al., 2010), and Tombusviridae (Li et al., 2008; Barajas et al., 2009; Barajas and Nagy, 2010) families. To date, the roles played by the UPS in replication and infection are still unclear. As discussed by Citovsky et al. (2009), proteasomal degradation pathways may play a dual, ambivalent role in the infection process: being either a host’s defense against the pathogen, a pathogen strategy to enhance its infectivity, or possibly both due to the close coevolution of the host and its pathogens.
The fact that accumulation of TYMV gRNA is improved when 66K is stabilized or overexpressed in infected cells (Figure 6) suggests that the amount of RdRp available at late infection times may be a limiting factor for viral replication and that UPS degradation might contribute to a host cell defense pathway. This idea is consistent with several recent reports linking the UPS to biotic defense responses in plants (Zeng et al., 2006; Dreher and Callis, 2007; Citovsky et al., 2009; Trujillo and Shirasu, 2010).
We observed that overexpression of 66K from an expression vector affected the accumulation of both gRNA and sgRNA, whereas treatment with MG132 affected mostly the accumulation of gRNA. The basis for this difference is unclear but may reflect a difference between cis- and trans-complementation, to which TYMV replication was previously shown to be sensitive (Weiland and Dreher, 1993). It is also conceivable that MG132 treatment led to the stabilization of protein(s) other than 66K that may contribute directly or indirectly to the viral replication process.
In any case, these results suggest that RdRp degradation might also constitute a regulatory step in the viral multiplication cycle, consistent with previous findings reporting the importance of maintaining a correct balance of RdRp during viral infection (Pantaleo et al., 2004; Schwartz et al., 2004; Jaag et al., 2007) and in agreement with the evolutionary conservation of the PEST sequence among tymovirus RdRps (Héricourt et al., 2000).
These results raise some interesting questions on the role that RdRp degradation may play during viral infection, in particular at late time points of the replication cycle (i.e., 24 to 48 hpi). One possibility is that decreasing the amount of RdRp may provide a means to temporally regulate the synthesis of the various RNA species [(−)-strand versus (+)-strand genomic or sgRNAs] in the course of viral replication. Although detailed kinetics of TYMV (+)- and (−)-strand RNA accumulation are not available, it has been shown in the case of Cucumber mosaic virus that switching of RNA replication from (−)-strand to (+)-strand synthesis occurs around 36 hpi (Seo et al., 2009). This switch has been proposed to correlate with changes in protein–protein interactions among the viral replication complexes, possibly leading to their dissociation and to the release of free RdRp. Whether such a model would also apply to TYMV and whether degradation of the RdRp is the cause, or the consequence, of such a switch remains to be determined.
Another interesting possibility is the idea that keeping low levels of polymerase may constitute a viral strategy to maintain the integrity of the viral genome, as high levels of RdRp may promote viral genomes rearrangements. In support of this hypothesis is the observation by Jaag et al. (2007) that high polymerase levels had a pronounced effect on the frequency of viral RNA recombination events due to template switching.
It can also be envisaged that low levels of RdRp at a late stage of cell infection may be used as a means to prevent the formation of new viral replication complexes or induce disassembly of the existing complexes, therefore allowing the pool of viral RNAs to be involved in other processes besides replication, such as packaging or cell-to-cell movement.
Finally, the down-modulation of viral replication consecutive to RdRp degradation may also be advantageous for the virus to escape from the host’s defense mechanisms, such as RNA silencing in plants or type I interferon in mammals, which have both been reported to be triggered by the double-stranded RNA intermediates produced during viral replication (Voinnet, 2005; Gantier and Williams, 2007).
In this respect, it is notable that many (+)RNA viruses have developed multiple strategies to downregulate the expression and ratio of their RdRp. These include translation from separate genomes, inhibition of translation initiation, ribosomal readthrough, frameshift, and/or differential processing of polyproteins (Buck, 1996; Pe’ery and Mathews, 2000). Targeting for degradation by the UPS may thus represent a functionally related method of gene regulation that can be added to the list of strategies by which (+)RNA viruses regulate levels of mature cleavage products. This possibility opens new avenues for future research that may reveal yet more levels of control of viral replication with even greater levels of complexity.
METHODS
Plasmids
All DNA manipulations were performed using standard techniques. Cloning details and primer sequences are indicated in Supplemental Methods online. Plant expression vectors are derived from pΩ-66K, pΩ-66K-His, pΩ-140K, and pΩ-206K (Prod'homme et al., 2001, 2003). Infectious transcripts were derived from the full-length TYMV cDNA clone E17 (Drugeon and Jupin, 2002). Genes encoding CAT, LUC, REL, Ub, and UbK48R were subcloned from plasmids pBLCAT6 (Boshart et al., 1992), pGL3 and pRL-CMV (Promega), and Yep96 and Yep110 (Hochstrasser et al., 1991), respectively. The overall structures of all plasmids were confirmed by restriction analysis and the sequences of PCR-generated DNA fragments were confirmed by DNA sequencing.
Generation of a Stably Transformed Arabidopsis thaliana Suspension Cell Line
The 66K-His open reading frame was cloned in the binary vector pBin19 (Bevan, 1984). Recombinant C58C1 agrobacteria were used to transform an Arabidopsis suspension cell culture by cocultivation as described (An, 1985). Transformed cells were selected on kanamycin, and resistant calli expressing 66K-His were transferred to Gamborg liquid medium to establish a suspension cell line as described (Camborde et al., 2007).
Pulse-Chase, Immunoprecipitation, and Immunoblotting Experiments
In vivo metabolic labeling of proteins was performed by adding 1.85 MBq/mL of l-[35S]Met and l-[35S]Cys (Pro-Mix; Amersham) to the Arabidopsis suspension cell line. After 2 h, chase was performed by addition of 5 mM Met and Cys. 66K-His was immunoprecipitated using anti-66K antiserum, subjected to SDS-PAGE, and blotted to nitrocellulose membranes as described (Prod'homme et al., 2001; Jakubiec et al., 2004). Radioactivity was detected by phosphor imaging prior to immunodetection. Quantitation of [35S] incorporation was performed using ImageQuant software (Molecular Dynamics) on at least three independent replicate experiments and was correlated with the amount of 66K-His determined by NIH Image after scanning of the immunoblots.
Total protein extraction, SDS-PAGE, and anti-66K immunoblotting were performed as described using NBT/BCIP (Sigma-Aldrich) as a substrate (Prod'homme et al., 2001). For detection of Ub conjugates, N-ethylmaleimide (45 μM) was included in all buffers used to immunoprecipitate 66K. Samples were subjected to SDS-PAGE and blotted to polyvinylidene difluoride membranes. Membranes were treated with Pierce protein gel blot signal enhancer and incubated with anti-myc monoclonal antibody (9E10) (1/3000), followed by chemoluminescent detection using CDP-Star as a substrate (Roche). Membranes were exposed to Super RX films (Fujifilm). For detection of LUC derivatives, protein samples were subjected to SDS-PAGE and blotted to nitrocellulose. Membranes were incubated with anti-LUC antibody (Santa-Cruz) (1/100), followed by detection with NBT/BCIP.
Preparation and Transfection of Arabidopsis Protoplasts
Protoplasts of Arabidopsis were prepared and transfected with 5 to 15 μg of plasmids, viral RNAs, or in vitro transcripts as described (Boudsocq et al., 2004; Camborde et al., 2007).
Reporter Assays
Protoplasts collected 48 h posttransfection (hpt) were lysed in Cell Culture Lysis reagent (Promega), and luciferase activity was measured on a Berthold microplate luminometer using the Luciferase assay system (Promega). The amount of CAT enzyme present in the same protein sample was assessed using ELISA immunoassay (Roche).
Inhibitor Treatments
MG132 and clastolactacystin β-lactone (Calbiochem) were dissolved in DMSO and used at a concentration of 100 and 25 μM, respectively. Stably transformed cells were treated during metabolic labeling, and further addition was performed at 4 and 10 h of chase. Transfected protoplasts were treated 44 hpt for reporter gene assays and immunoprecipitation of Ub conjugates experiments or 28 and 36 hpt for viral infectivity assays.
RNA Extraction and RNA Gel Blot Hybridization
Total RNA extraction from protoplasts, electrophoresis, and blotting was performed as previously described (Jakubiec et al., 2006). The RNA gel blot was hybridized with a digoxygenin-labeled RNA probe as described (Prod'homme et al., 2003) and was revealed with CDP-Star as a substrate (Roche). Signal was acquired using the Fujifilm Image Analyzer LAS-3000, and quantitation of viral RNAs was performed using NIH Image and serial dilutions of RNA extracts to ensure signals were in the linear range.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: NC_004063 (TYMV genome), NP_733818 (TYMV 140K), and NP_733819 (TYMV 66K).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Nucleotide Sequences of Primers Used in This Study.
Supplemental Methods. Plasmid Constructions.
Acknowledgments
This work was supported in part by grants from the Centre National de la Recherche Scientifique and Agence Nationale de la Recherche (contract ANR-06-BLAN-0062). A.J. was the recipient of a fellowship from the Ministère de l’Education Nationale de la Recherche et de la Technologie. We thank A. Rousseau for constructing some of the plasmids used in this study, M. Mantovan for help with immunoprecipitation experiments, M. Brault for kind advice regarding protoplasts, C. Dargemont and M. Glickman for useful discussions, and H. Rothnie for careful editing of the manuscript.
References
- Ahlquist P. (2002). RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273 [DOI] [PubMed] [Google Scholar]
- Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. (2003). Host factors in positive-strand RNA virus genome replication. J. Virol. 77: 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. (1985). High efficiency transformation of cultured tobacco cells. Plant Physiol. 79: 568–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. (1986). In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Bachmair A., Novatchkova M., Potuschak T., Eisenhaber F. (2001). Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci. 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Barajas D., Li Z., Nagy P.D. (2009). The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 83: 11751–11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Nagy P.D. (2010). Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 397: 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker F., Buschfeld E., Schell J., Bachmair A. (1993). Altered response to viral infection by tobacco plants perturbed in ubiquitin system. Plant J. 3: 875–881 [Google Scholar]
- Bevan M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M., Klüppel M., Schmidt A., Schütz G., Luckow B. (1992). Reporter constructs with low background activity utilizing the CAT gene. Gene 110: 129–130 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279: 41758–41766 [DOI] [PubMed] [Google Scholar]
- Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. (1998). A novel site for ubiquitination: The N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17: 5964–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K.W. (1996). Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47: 159–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Coscoy L. (2005). Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309: 127–130 [DOI] [PubMed] [Google Scholar]
- Camborde L., Tournier V., Noizet M., Jupin I. (2007). A turnip yellow mosaic virus infection system in Arabidopsis suspension cell culture. FEBS Lett. 581: 337–341 [DOI] [PubMed] [Google Scholar]
- Catic A., Collins C., Church G.M., Ploegh H.L. (2004). Preferred in vivo ubiquitination sites. Bioinformatics 20: 3302–3307 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Zaltsman A., Kozlovsky S.V., Gafni Y., Krichevsky A. (2009). Proteasomal degradation in plant-pathogen interactions. Semin. Cell Dev. Biol. 20: 1048–1054 [DOI] [PubMed] [Google Scholar]
- Craig A., Ewan R., Mesmar J., Gudipati V., Sadanandom A. (2009). E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot. 60: 1123–1132 [DOI] [PubMed] [Google Scholar]
- Dean A.M., Koshland D.E., Jr (1990). Electrostatic and steric contributions to regulation at the active site of isocitrate dehydrogenase. Science 249: 1044–1046 [DOI] [PubMed] [Google Scholar]
- De Groot R.J., Rumenapf T., Kuhn R.J., Strauss E.G., Strauss J.H. (1991). Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 88: 8967–8971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroo B.J., Archer T.K. (2002). Proteasome inhibitors reduce luciferase and beta-galactosidase activity in tissue culture cells. J. Biol. Chem. 277: 20120–20123 [DOI] [PubMed] [Google Scholar]
- Dielen A.S., Badaoui S., Candresse T., German-Retana S. (2010). The ubiquitin/26S proteasome system in plant–pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 11: 293–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Nowara D., Schweizer P. (2006). Protein polyubiquitination plays a role in basal host resistance of barley. Plant Cell 18: 3321–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T.W. (2004). Turnip yellow mosaic virus: Transfer RNA mimicry, chloroplasts and a C-rich genome. Mol. Plant Pathol. 5: 367–375 [DOI] [PubMed] [Google Scholar]
- Drugeon G., Jupin I. (2002). Stability in vitro of the 69K movement protein of turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83: 3187–3197 [DOI] [PubMed] [Google Scholar]
- Gantier M.P., Williams B.R. (2007). The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 18: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Tu H., Shi S.T., Lee K.J., Asanaka M., Hwang S.B., Lai M.M.C. (2003). Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77: 4149–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Alai M.M., Gallo M., Salame M., Wetzler D.E., McBride A.A., Paci M., Cicero D.O., de Prat-Gay G. (2006). Molecular basis for phosphorylation-dependent, PEST-mediated protein turnover. Structure 14: 309–319 [DOI] [PubMed] [Google Scholar]
- Garzon M., Eifler K., Faust A., Scheel H., Hofmann K., Koncz C., Yephremov A., Bachmair A. (2007). PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581: 3189–3196 [DOI] [PubMed] [Google Scholar]
- Gilfoy F., Fayzulin R., Mason P.W. (2009). West Nile virus genome amplification requires the functional activities of the proteasome. Virology 385: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Ciechanover A. (2002). The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Gonda D., Bachmair A., Wünning I., Tobias J.W., Lane W.S., Varshavsky A. (1989). Universality and structure of the Nend rule. J. Biol. Chem. 264: 16700–16712 [PubMed] [Google Scholar]
- Goritschnig S., Zhang Y., Li X. (2007). The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 49: 540–551 [DOI] [PubMed] [Google Scholar]
- Héricourt F., Blanc S., Redeker V., Jupin I. (2000). Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J. 349: 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M., Ellison M.J., Chau V., Varshavsky A. (1991). The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA 88: 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007). The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Isaacson M.K., Hidde L., Ploegh H.L. (2009). Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag H.M., Stork J., Nagy P.D. (2007). Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368: 388–404 [DOI] [PubMed] [Google Scholar]
- Jadhav T., Wooten M.W. (2009). Defining an embedded code for protein ubiquitination. J. Proteomics Bioinform. 2: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A., Drugeon G., Camborde L., Jupin I. (2007). Proteolytic processing of turnip yellow mosaic virus replication proteins and functional impact on infectivity. J. Virol. 81: 11402–11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A., Jupin I. (2007). Regulation of positive-strand RNA virus replication: The emerging role of phosphorylation. Virus Res. 129: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A., Notaise J., Tournier V., Héricourt F., Block M.A., Drugeon G., Van Aelst L., Jupin I. (2004). Assembly of turnip yellow mosaic virus replication complexes: Interaction between the proteinase and polymerase domains of the replication proteins. J. Virol. 78: 7945–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec A., Tournier V., Drugeon G., Pflieger S., Camborde L., Vinh J., Héricourt F., Redecker V., Jupin I. (2006). Phosphorylation of viral RNA-dependent RNA polymerase and its role in replication of a plus-strand RNA virus. J. Biol. Chem. 281: 21236–21249 [DOI] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. (2000). Phosphorylation meets ubiquitination: The control of NF-κB activity. Annu. Rev. Immunol. 18: 621–663 [DOI] [PubMed] [Google Scholar]
- Karsies A., Hohn T., Leclerc D. (2001). Degradation signals within both terminal domains of the cauliflower mosaic virus capsid protein precursor. Plant J. 27: 335–343 [DOI] [PubMed] [Google Scholar]
- Kim H.S., Delaney T.P. (2002). Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. (1998). Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Lévy F., Johnsson N., Rümenapf T., Varshavsky A. (1996). Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. USA 93: 4907–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Barajas D., Panavas T., Herbst D.A., Nagy P.D. (2008). Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82: 6911–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Serino G., Deng X.W., Dinesh-Kumar S.P. (2002). Role of SCF ubiquitin ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Zhang J., Cheung C., Suarez A., McManus B.M., Yang D. (2003). Proteasome inhibition reduces coxsackievirus B3 replication in murine cardiomyocytes. Am. J. Pathol. 163: 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. (2009). Host factors promoting viral RNA replication. Viral Genome Replication, Cameron C.E., Götte M., Raney K.D., eds (Philadelphia: Springer; ), pp. 267–295 [Google Scholar]
- Pantaleo V., Rubino L., Russo M. (2004). The p36 and p95 replicase proteins of carnation italian ringspot virus cooperate in stabilizing defective interfering RNA. J. Gen. Virol. 85: 2429–2433 [DOI] [PubMed] [Google Scholar]
- Peart J., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’ery T., Mathews M.B. (2000). Viral translational strategies and host defense mechanisms. Translational Control of Gene Expression, Sonenberg N., Hershey J.W.B., Mathews M.B., eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ), pp. 371–424 [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. (2003). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart C.M., Eddins M.J. (2004). Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 1695: 55–72 [DOI] [PubMed] [Google Scholar]
- Prod'homme D., Jakubiec A., Tournier V., Drugeon G., Jupin I. (2003). Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J. Virol. 77: 9124–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod'homme D., Le Panse S., Drugeon G., Jupin I. (2001). Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology 281: 88–101 [DOI] [PubMed] [Google Scholar]
- Raaben M., Posthuma C.C., Verheije M.H., te Lintelo E.G., Kikkert M., Drijfhout J.W., Snijder E.J., Rottier P.J.M., de Haan C.A.M. (2010). The ubiquitin-proteasome system plays an important role during various stages in the coronavirus infection cycle. J. Virol. 84: 7869–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T., Hochstrasser M. (2008). Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S.W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21: 267–271 [PubMed] [Google Scholar]
- Sadowski M., Suryadinata R., Lai X., Heierhorst J., Sarcevic B. (2010). Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol. Cell. Biol. 30: 2316–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Chen J., Lee W.M., Janda M., Ahlquist P. (2004). Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. USA 101: 11263–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.K., Kwon S.J., Choi H.S., Kim K.H. (2009). Evidence for alternate states of Cucumber mosaic virus replicase assembly in positive- and negative-strand RNA synthesis. Virology 383: 248–260 [DOI] [PubMed] [Google Scholar]
- Spallek T., Robatzek S., Gohre V. (2009). How microbes utilize host ubiquitination. Cell. Microbiol. 11: 1425–1434 [DOI] [PubMed] [Google Scholar]
- Stary S., Yin X.J., Potuschak T., Schlögelhofer P., Nizhynska V., Bachmair A. (2003). PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 133: 1360–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney H.L., Yang Z., Zhi G., Stull J.T., Trybus K.M. (1994). Charge replacement near the phosphorylatable serine of the myosin regulatory light chain mimics aspects of phosphorylation. Proc. Natl. Acad. Sci. USA 91: 1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman L., Rechsteiner M., Pickart C.M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Trujillo M., Shirasu K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13: 1–7 [DOI] [PubMed] [Google Scholar]
- van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., McGeoch D.J., Maniloff J., Mayo M.A., Pringle C.R., Wickner R.B. (2000). Virus taxonomy: The classification and nomenclature of viruses. The 7th Report of the International Committee on Taxonomy of Viruses, (San Diego, CA: Academic Press; ). [Google Scholar]
- Varshavsky A. (1996). The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6: 206–220 [DOI] [PubMed] [Google Scholar]
- Vosper J.M., McDowell G.S., Hindley C.J., Fiore-Heriche C.S., Kucerova R., Horan I., Philpott A. (2009). Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 284: 15458–15468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Chen M.H., Bachmaier R., Ghoshroy S., Citovsky V. (2000). Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19: 4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Herr R.A., Chua W.J., Lybarger L., Wiertz E.J., Hansen T.H. (2007). Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland J.J., Dreher T.W. (1993). Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc. Natl. Acad. Sci. USA 90: 6095–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley C.K., Ling R., Callis J. (1998). Engineering in vivo instability of firefly luciferase and Escherichia coli beta-glucuronidase in higher plants using recognition elements from the ubiquitin pathway. Plant Mol. Biol. 37: 337–347 [DOI] [PubMed] [Google Scholar]
- Yu G.Y., Lai M.M.C. (2005). The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J. Virol. 79: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.R., Vega-Sanchez M.E., Zhu T., Wang G.L. (2006). Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 16: 413–426 [DOI] [PubMed] [Google Scholar]