This report describes the genome sequence of Chlorella variabilis NC64A. Surprisingly, given that NC64A has been thought to be asexual and nonmotile, this work identifies homologs of genes involved in meiosis, gamete fusion, and flagella.
Abstract
Chlorella variabilis NC64A, a unicellular photosynthetic green alga (Trebouxiophyceae), is an intracellular photobiont of Paramecium bursaria and a model system for studying virus/algal interactions. We sequenced its 46-Mb nuclear genome, revealing an expansion of protein families that could have participated in adaptation to symbiosis. NC64A exhibits variations in GC content across its genome that correlate with global expression level, average intron size, and codon usage bias. Although Chlorella species have been assumed to be asexual and nonmotile, the NC64A genome encodes all the known meiosis-specific proteins and a subset of proteins found in flagella. We hypothesize that Chlorella might have retained a flagella-derived structure that could be involved in sexual reproduction. Furthermore, a survey of phytohormone pathways in chlorophyte algae identified algal orthologs of Arabidopsis thaliana genes involved in hormone biosynthesis and signaling, suggesting that these functions were established prior to the evolution of land plants. We show that the ability of Chlorella to produce chitinous cell walls likely resulted from the capture of metabolic genes by horizontal gene transfer from algal viruses, prokaryotes, or fungi. Analysis of the NC64A genome substantially advances our understanding of the green lineage evolution, including the genomic interplay with viruses and symbiosis between eukaryotes.
INTRODUCTION
Green algae (phylum Chlorophyta) are a highly diverse group of photosynthetic eukaryotes from which the terrestrial plant lineage emerged >1 billion years ago (Heckman et al., 2001). During the evolutionary history of Earth, they have become major players in global energy/biomass production and biogeochemical recycling (Grossman, 2005). Algae originally included in the genus Chlorella are among the most widely distributed and frequently encountered algae in freshwaters (Fott and Novakova, 1969). They exist in aqueous environments as well as on land. They are typically small (~2 to 10 μm in diameter), unicellular, coccoid, nonmotile, and contain a single chloroplast. Some have a rigid cell wall, and they are reported to lack a sexual cycle (Takeda, 1991). Accessions of Chlorella were extensively used as model systems in early research on photosynthesis (Benson, 2002).
Over a hundred algal isolates were originally assigned to the genus Chlorella, but their taxonomy classification has long remained unreliable because of their lack of conspicuous morphological characters. Recent molecular analyses now separate them into two classes of chlorophytes, the Trebouxiophyceae, which contains the true Chlorella, and the Chlorophyceae (Takeda, 1988; Huss et al., 1999). Here, we report on the genome of Chlorella sp NC64A (NC64A), recently renamed Chlorella variabilis NC64A (Ryo et al., 2010), that is a bona fide member of the true Chlorella genus, belonging to the class Trebouxiophyceae (see Supplemental Figure 1 online). The true Chlorella species, including NC64A, are characterized by glucosamine as a major component of their rigid cell walls (Takeda, 1991; Chuchird et al., 2001). The Trebouxiophyceae contain most of the known green algal endosymbionts (Friedl and Bhattacharya, 2002), living in lichens, unicellular eukaryotes, plants, and animals (e.g., mussels, hydra, etc). Most Chlorella species are naturally free-living; however, NC64A is a hereditary photosynthetic endosymbiont (i.e., photobiont) of the unicellular protozoan Paramecium bursaria (Karakashian and Karakashian, 1965). This symbiosis is facultative in lab conditions since both the paramecium and NC64A can be cultivated separately. NC64A is also a host for a family of large double-stranded DNA viruses that are found in freshwater throughout the world; the genomes of six of these viruses have been sequenced (Wilson et al., 2009). Like other microalgae, there is an increasing interest in using Chlorella in a variety of biotechnological applications, such as biofuels (Schenk et al., 2008), sequestering CO2 (Chelf et al., 1993), producing molecules of high economic value, or removing heavy metals from wastewaters (Rajamani et al., 2007). The sequence of the NC644A genome presented here will help in the optimization of these various processes, while further documenting the evolution of the green lineage.
RESULTS AND DISCUSSION
Global Genome Structure
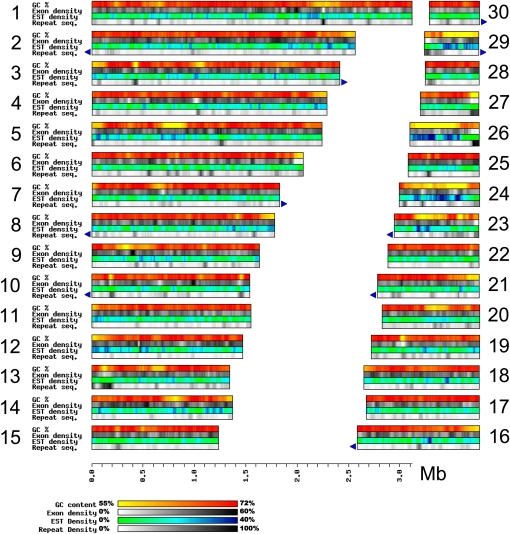
The 46.2-Mb NC64A nuclear genome was sequenced at 9× coverage using the whole-genome shotgun Sanger sequencing approach. The genome size of NC64A is intermediate compared with those of Mamiellale (12.6 to 21.9 Mb) and Chlamydomonas reinhardtii (121 Mb) (Table 1). Sequence assembly yielded 413 scaffolds with lengths >1 kb (see Supplemental Table 1 online). Eighty-nine percent of the genome assembly is contained in 30 scaffolds with lengths ranging from 494 kb to 3.12 Mb (Figure 1). Mapping of 7624 clustered EST sequences onto the genome sequences suggests that the assembly contains >97% of the gene complement. The NC64A karyotype resolved by pulse field gel electrophoresis analysis revealed 12 chromosomes ranging in size from ~1.1 to 8.6 Mb (see Supplemental Figure 2 online).
Table 1.
Comparison of NC64A Genome Statistics to Those of Sequenced Chlorophyte Genomes
| Features | NC64A | C. reinhardtii | Micromonas CCMP1545 | Micromonas RCC299 | O. tauri | O. lucimarinus |
| Nuclear genome size (Mb) | 46.2 | 121 | 21.9 | 20.9 | 12.6 | 13.2 |
| Number of chromosomes | 12a | 17 | 19 | 17 | 20 | 21 |
| GC content total (%) | 67 | 64 | 65 | 64 | 59 | 60 |
| Gene count | 9,791 | 15,143 | 10,575 | 10,056 | 7,892 | 7.651 |
| Avg. protein length (aa) | 456 | 444 | 439 | 473 | 387 | 399 |
| Avg. gene density (kb/gene) | 4.7 | 5.0 | 2.1 | 2.2 | 1.6 | 1.7 |
| Avg. number of exons per gene | 7.3 | 8.3 | 1.9 | 1.6 | 1.6 | 1.3 |
| Avg. exon length (nt) | 170 | 190 | 731 | 958 | 750 | 970 |
| Avg. intron length (nt) | 209 | 373 | 187 | 163 | 126 | 187 |
| Avg. coding sequence (%) | 29 | 17 | 64 | 68 | 73 | 69 |
aa, amino acids; nt, nucleotides.
Estimation based upon pulse field gel electrophoresis analysis.
Figure 1.
General Characteristics of the Chlorella sp NC64A Genome Assembly.
This figure represents the 30 major scaffolds, which contain 89% of the total genome. GC percentage, exon density, EST density, and repeat density were calculated in 40-kb sliding windows with a step of 5 kb. Density was calculated as the percentage of nucleotide in the window covered by the relevant feature (i.e., exon, EST, or repeat sequence). Blue triangles represent telomeric repeat arrays.
The nuclear genome sequences have the highest average GC content (67.2% GC) reported so far in sequenced eukaryotic genomes (Table 1). However, several genomic segments present in scaffolds, ranging from 40 to 625 kb, have conspicuously lower GC contents (55 to 65% GC) than the rest of the genome (Figure 1). These low-GC regions represent 15.6% of the total genome size (6.20 Mb). They have a significantly higher frequency of genes with EST support than does the rest of the genome (Kruskal-Wallis test P value = PKWT < 0.0001), suggesting that they correspond to regions of higher transcriptional activity (Figure 2A). In addition, genes located in low-GC regions exhibit significantly shorter introns (Figure 2B) and a less biased codon usage (Figure 2C) relative to the high-GC regions (PKWT < 0.0001). Low-GC regions are also enriched in repeated sequences (most prominent in the 60 to 65% GC range; Figure 2E), but the trend is only marginally significant (PKWT = 0.024). Although the median exon density is slightly smaller for low-GC regions (Figure 2D), the difference from that found in the high-GC regions is not statistically significant (PKWT > 0.05). The majority (1100) of the 1384 NC64A proteins encoded in low-GC regions have their best BLASTP match to homologs in chlorophytes and land plants (see Supplemental Figure 3 online). This suggests that the low-GC regions did not result from an invasion of horizontally transferred foreign DNA sequences.
Figure 2.
Features of Low-GC Regions in Chlorella sp NC64A.
Nonoverlapping 40-kb segments of the NC64A genome assembly were classified into four GC content classes. The distributions of genomic segments in each of the GC content classes are depicted by box plots for the following features: EST density (as defined in the Figure 1 legend) (A), average size of introns supported by EST data (B), mean effective number of codons (ENc) per gene (C), exon density (D), and repeat density (E). (F) shows the distribution of genomic segments as a function of their GC content. The bottom and top of boxes represent the 1st and 3rd quartiles, Q1 and Q3, respectively, and the band near the middle of boxes represents the median. The extremities of the lines appearing below and above the boxes represent the lowest value still within 1.5 IQR (interquartile range = Q3 to Q1) of the lower quartile Q1, and the highest value still within 1.5 IQR of the upper quartile Q3. We applied the Kruskal-Wallis statistical test to each genomic feature to test the null hypothesis of equivalence between the distributions of values in the four GC bins. Distributions of EST density, intron size, and Enc were found to be significantly different between the four GC bins (P < 0.0001), whereas for repeat density, the difference was only marginally significant (P = 0.024). The null hypothesis of equivalence of distributions could not be rejected at α = 0.05 for exon density (P = 0.468).
Low-GC genomic regions also exist in the prasinophytes Micromonas and Ostreococcus, where their origin and function are still unclear (Palenik et al., 2007; Worden et al., 2009). As in NC64A, the Micromonas low-GC chromosomes exhibit higher transcription levels than do the normal-GC chromosomes (Worden et al., 2009). These features common to Micromonas, Ostreococcus, and Chlorella suggest that variation in GC content is a characteristic of many chlorophytes. However, the nature of genes present in the NC64A low-GC regions does not suggest a specific function or a mechanism by which their compositional shift evolved. However, we noticed that the NC64A low-GC regions exhibited a significant underrepresentation of genes involved in transcription, chromatin structure and dynamics, and extracellular structures (see Supplemental Table 2 online).
Repeated Sequences
Only a few algal repetitive sequences are available in public databases. This prevented us from performing an exhaustive search for repetitive sequences based on a set of reference sequences. Therefore, we used a de novo identification approach where repeated sequences are defined as any sequence with more than one copy in the genome (as detected by BLASTN with an E-value < 1e-5), regardless of its size and nature (transposable element, simple repeat, duplicated gene, or low complexity sequences). The cumulative lengths of such repeated sequences represent 5.53 Mb (12%) of the genome (see Supplemental Table 3 online), which makes NC64A relatively repeat-poor compared with land plants (repeat content ranges from 20 to 30% in Arabidopsis thaliana to >90% in large genomes such as wheat [Triticum aestivum]). The content in repeated sequences is probably slightly underestimated because repeats frequently flanked sequence gaps. Half of the repeated sequences (51.6%) have no resemblance to known repeat families (see Supplemental Table 3 online). About 10% (536 kb or 1.2% of the genome) contain open reading frames with deduced protein sequences similar to proteins in public databases (excluding transposable element related proteins) and therefore correspond to highly similar gene duplicates or gene fragments (at the nucleotide level). An additional 40.2% could be classified in known repetitive sequence families based on TBLASTX sequence similarity searches (E-value < 1e-15) against the Repbase database. NC64A has the major classes of known transposable elements (see Supplemental Table 3 online): long terminal repeat (LTR) retrotransposons (Gypsy-like elements and TY1/Copia-like elements), non-LTR retrotransposons (RandI, L1, RTE, and GilM elements form the most prominent families), endogenous-retrovirus-like sequences, and DNA transposons (Novosib-like). The NC64A telomeric repeat unit is identical to that of flowering plants [i.e., (TTTAGGG)n]. Eighteen scaffolds exhibit telomeric repeat arrays at a terminus and represent ends of chromosomes (Figure 1).
Algae- and Land Plant–Specific Protein Families
We predicted and annotated 9791 protein genes in the NC64A genome, a number comparable to that of the Micromonas species (Table 1). Like Chlamydomonas, the NC64A protein genes are intron rich with 7.3 exons per gene on average, but the average NC64A intron length is shorter than in Chlamydomonas. An overview of the NC64A gene repertoire is provided in Supplemental Results, Supplemental Table 1, and Supplemental Figures 4 to 6 online. Comparison of the numbers of PFAM protein domains revealed 27 protein families that are present in all completely sequenced chlorophyte algae (NC64A, C. reinhardtii, Micromonas sp RCC299 and CCMP1545, Ostreococcus lucimarinus, and Ostreococcus tauri) but absent in three representative and completely sequenced land plants (Physcomitrella patens, Arabidopsis, and Oryza sativa) (see Supplemental Table 4 online). Most of these algal genes probably existed in the last common ancestor shared with terrestrial plants since all of them have homologs in other eukaryotes. This would imply that they were subsequently lost in the branch leading to land plants. Many of these protein families are involved in basic metabolism, such as respiration (cytochrome c/c1 heme lyase), amino acid synthesis (asparaginase), carbohydrate metabolism (including ACN9 protein and iron-containing alcohol dehydrogenase), protein synthesis (including the selenocystein aminotransferase and posttranslational modification enzyme PAM), and DNA or RNA metabolism (DNA binding protein HU and helicase family) (see Supplemental Table 4 online). The six chlorophytes contain a 3′5′-cyclic nucleotide phosphodiesterase gene that modulates the levels of the secondary messenger 3′:5′-cyclic nucleotides in signal transduction pathways (Beavo, 1995). Chlorophyte-specific protein families also included the formate/nitrite transporter, type I polyketide synthase, and pyruvate decarboxylase (fatty acid metabolism).
By contrast, 184 protein domain families were present in all three land plants but absent in chlorophytes, including NC64A (see Supplemental Data Set 1 online); 102 of them have homologs in eukaryotes (excluding viridiplantae) and may have existed in the common ancestor with green algae and subsequently been lost in the Chlorophyta lineage. Furthermore, 12 protein domain families are exclusively found in land plants and bacteria or archea. The corresponding genes may have been exchanged by lateral gene transfer between the nuclear genome of land plants and the genomes of prokaryotes or organelles. The remaining 70 protein domains have no recognizable homologs outside of land plants. Many of the 184 land plant protein domain families are involved in development, cell signaling, stress and hormonal response, transcriptional regulation, defense, and polysaccharide and cell wall metabolism (see Supplemental Data Set 1 online). Thus, in addition to the higher number of gene duplications that are characteristic of land plants (Flagel and Wendel, 2009), some of these proteins were probably important in the rise of multicellularity and terrestrial colonization in the Streptophyte lineage. For example, land plant–specific protein families involved in auxin signaling have presumably played a significant role in the emergence of organs, establishment of a complex developmental program, and adaptation to changing environment (Galván-Ampudia and Offringa, 2007). They include the auxin/indole-3-acetic acid transcriptional regulator family, auxin response factor transcription factor family and dormancy-associated and protein products of the ARG7 auxin-responsive gene family. We also found protein families involved in resistance to drought (e.g., dehydrin and Di19 proteins) that are specific to land plants; these families have perhaps been important in the adaptation to water-limiting conditions during the colonization of land (Bateman et al., 1998). Unlike chlorophyte algae, terrestrial plants have proteins involved in polysaccharide metabolism, lignin metabolism (e.g., Phe ammonia lyase and caffeic acid 3-O-methyltransferase) and cell wall metabolism (e.g., pectate lyase and pectinesterase), some of which probably contributed in the stiffening and consolidation of cell walls to withstand the weight of land plants subjected to gravity.
Dynamics of Chlorella Protein Families
Twenty-eight PFAM protein families showed a biased distribution of proteins among the six chlorophyte algae (Figure 3; see Supplemental Table 5 online). Some PFAM domains were specifically overrepresented in NC64A compared with the five other chlorophytes. A subset of those PFAM domains was also found in excess in organisms that have intracellular or symbiotic life styles. We therefore hypothesize that the corresponding proteins in NC64A could also play a role in the mutualistic symbiosis with the protozoan P. bursaria. These PFAM domains include several families of proteins containing protein–protein interaction domains (F-box and MYND) and adhesion domains (fasciclin). Although the functions of domains may differ, proteins containing protein–protein interaction domains generally exist in excess in intracellular bacteria and symbiotic eukaryotes compared with their free-living relatives. For example, in intracellular bacteria, ankyrin proteins and tetratricopeptide repeat proteins are implicated in host–pathogen interactions (Petri et al., 2000), linked to the cytoplasmic incompatibility phenotype of the eukaryotic host (Tram et al., 2003; Iturbe-Ormaetxe et al., 2005) and directly secreted into the host (Wu et al., 2004). Protein families that contain ankyrin and WD40 domains are also prominent in the plant symbiont Laccaria bicolor (Martin et al., 2008), although there is no direct evidence that these proteins are involved in symbiosis.
Figure 3.
Heat Map of PFAM Protein Families with Significantly Biased Distribution among Chlorophyte Algae.
PFAM protein families that have either significantly expanded or shrunk in one or more sequenced chlorophytes (χ2 test, α = 0.05 after Bonferroni correction). Full red and black indicate 100 and 0%, respectively, of the total number of proteins in the PFAM family for the six algae. Real counts and description of PFAM protein families are given in Supplemental Table 5 online. The leftmost graph represents the hierarchical clustering of the PFAM domains by the average linkage methods using correlation coefficients between profiles.
NC64A also has an excess of proteins with Cys-rich GCC2_GCC3 PFAM motifs (Figure 3; see Supplemental Table 5 online), which are found in a wide variety of extracellular proteins. The symbiont L. bicolor secretes Cys-rich proteins (albeit not of the GCC2_GCC3 type) into their host, some of which are upregulated in symbiotic tissues and implicated in the establishment of symbiosis (Martin et al., 2008).
We found a significant increase in the number of amino acid transporters (Aa_trans domain) in NC64A (35 proteins). Fourteen of them have ESTs, indicating they are expressed. Some of these transporters may be expressed when in a symbiotic environment (note: the ESTs in this study were from NC64A cells not engaged in symbiosis). This observation is consistent with previous studies, which suggest that Chlorella symbionts, including NC64A, possess an efficient system for importing amino acids from the P. bursaria host and can use amino acids as a source of nitrogen instead of nitrate (Kato et al., 2006). As a complement to amino acid transporters, NC64A contains many trypsin-like proteases that may be involved in degrading peptides into amino acids.
We also found an increased number of proteins with a class 3 lipase signature (Lipase_3 domain) (Figure 3; see Supplemental Table 5 online). A previous study reported that algal symbionts, such as zooxanthellae, translocate photosynthetic carbon into their animal hosts in the form of intact lipids, glycerol, and fatty acids (Battey and Patton, 1984); this process is mobilized by lipases. The hypothesis that NC64A feeds its P. bursaria host by translocating lipid molecules remains to be confirmed experimentally.
Protein families, such as protein kinases (Pkinase domain), guanylate and adenylate cyclases (Guanylate_cyc), and 3′,5′-cyclic nucleotide phosphodiesterases (PDEase_I domain), are more prevalent in Chlamydomonas compared with Chlorella and the mamiellalean algae, suggesting that cellular signaling is more complex in Chlamydomonas. Chlamydomonas also has a complex set of arylsulfatase-like proteins (sulfatase domain), some of which are secreted in response to sulfur deprivation (Pollock et al., 2005); by contrast, NC64A has only one homolog, and the four Mamiellale species have none, indicating that they adapt differently to low sulfur environments.
Evidence of Sexual Reproduction in Chlorella
Although Chlorella species have long been assumed to be asexual, NC64A encodes all of the known meiosis-specific proteins inventoried by Schurko and Logsdon (2008) and Malik et al. (2008), namely, dosage suppressor of MCk1 DMC1, homologous-pairing proteins HOP1 and HOP2, meiotic recombination protein MER3, meiotic nuclear division protein MND1, and mutS homolog protein MSH4. These genes are also found in most of the other sequenced chlorophyte algal species (see Supplemental Table 6 online). Chlorella species, including symbionts of P. bursaria, have been observed only in the haploid phase (Pickett-Heaps, 1975; Gerashchenko et al., 2001; Kadono et al., 2004), but the presence of meiosis genes suggests that NC64A also has a diploid phase and that its sexual reproductive cycle might have been overlooked, like the cryptic sex recently identified in Ostreococcus species (Grimsley et al., 2010). In addition, we found 19 NC64A homologs of the Chlamydomonas gametolysin proteins that promote the disassembly of the gametic cell walls and allow gamete fusion as well as an NC64A ortholog (id:137637) to the Chlamydomonas GCS1 protein essential for cell fusion (Goodenough et al., 2007). These results suggest that meiosis and sexual reproduction are part of the NC64A life cycle.
Conserved Flagella Proteins in Chlorella
NC64A, O. lucimarinus, and O. tauri are thought to be nonmotile green algae because flagella have never been observed in these organisms. Conversely, Micromonas sp CCMP1545, Micromonas pusilla RCC299, and C. reinhardtii are motile green algae: both Micromonas species have one flagellum and Chlamydomonas has two flagella. A proteomic study (Pazour et al., 2005) identified 360 Chlamydomonas flagellar proteins with high confidence. We used the reciprocal best BLASTP hit (RBH) criterion between the Chlamydomonas proteome and that of other sequenced chlorophytes to identify orthologs to the Chlamydomonas flagella proteins. Unexpectedly, we identified many putative (RBH) orthologs to the Chlamydomonas flagellar proteins in the NC64A genome (103 out of 360 Chlamydomonas flagella proteins = 29%; see Supplemental Data Set 2 online), of which 48% (50/103) and 53% (55/103) were also found in O. tauri and O. lucimarinus, respectively, while 85% (88/103) were also found in both Micromonas sp CCMP1545 and M. pusilla RCC299.
Proteins normally involved in the axonemal outer dynein arm, a structure responsible for movement of flagella, are included among putative Chlorella orthologs (Figure 4; see Supplemental Data Set 2 online): outer dynein arm docking complex proteins (ODA-DC proteins; numbers of proteins in NC64A/Chlamydomonas = 3/3), outer dynein arm heavy chain proteins (ODA-DHC: 3/3), outer dynein arm intermediate chain proteins (OAD-IC: 2/2), and outer dynein arm light chain proteins (ODA-LC: 4/5). Putative orthologs were identified in Micromonas for each of these proteins (except ODA-DC1), but none were found in the Ostreococcus species (except ODA-LC8). In Chlamydomonas, the assembly and maintenance of flagella depend on a process called intraflagellar transport (IFT) (Cole, 2003). The IFT system consists of a motor complex associated with groups of large protein complexes called IFT particles. Chlorella encodes putative orthologs to the proteins ITF52, ITF57, and ITF88 involved in the IFT particle (Figure 4) as well as putative orthologs to the kinesin-2 motor protein FLA8 (Joint Genome Initiative [JGI] 37158) and the kinesin-associated protein KAP (JGI 139946). Surprisingly, two of the proteins identified in Chlorella, namely, ITF57 and ITF88, were until now exclusively found in organisms that have flagella (Wickstead and Gull, 2007) except Plasmodium falciparum that is known to build its flagella throughout an IFT-independent mechanism.
Figure 4.
Distribution of Selected Flagellar Proteins across Chlorophytes.
(A) Cladogram showing the likely evolutionary relationships of sequenced chlorophytes and T. pseudonana based on the 18S phylogenetic tree shown in Supplemental Figure 1 online. The ƒ mark shows organisms known to build motile flagella. Crei, C. reinhardtii; NC64A, Chlorella sp NC64A; Otau, O. tauri; Oluc, O. lucimarinus; M. CCMP, M. pusilla CCMP1545; M. RCC, Micromonas sp RC299.
(B) Presence (dot) or absence (circle) of putative (Reciprocal Best Hit) orthologs to Chlamydomonas outer dynein proteins, inner dynein proteins, radial spoke proteins, central pair proteins, and IFT proteins.
C. reinhardii has 249 flagellar proteins that exhibit no RBH with NC64A. Micromonas spp retained many of them (101/249 [41%] RBHs in both Micromonas species). By contrast, only 18/249 (7%) and 19/249 (8%) putative orthologs were identified in O. tauri and O. lucimarinus, respectively. Overall, 89 proteins were present in all motile sequenced chlorophyte algae but absent in NC64A and the Ostreococcus species. This flagella-specific set includes most proteins known to function in inner-arm dynein complexes (including inner-arm dyneins), the central pair complex, the IFT particle, and all proteins of the Chlamydomonas radial spoke (Figure 4; see Supplemental Data Set 2 online).
The conservation of a substantial subset of the C. reinhardtii flagella proteins in NC64A is intriguing. In particular, our results suggest that NC64A has retained an almost complete set of outer-arm dynein proteins (heavy, intermediate, and light chains and docking complex) that are found only in eukaryotes that exhibit motile cilia/flagella at some point in their life cycle (Wickstead and Gull, 2007). Merchant et al. (2007) identified 195 C. reinhardtii proteins that have homologs in two motile ciliates (Homo sapiens and Phytophthora spp) but not in a group of reference aciliates (Arabidopsis, Neurospora, Cyanidioschyzon, Dictyostelium, eubacteria, and archaea). This protein set, designated the CiliaCut, is thought to contain proteins involved in flagellar function. In agreement with the results obtained above, 63 proteins of the CiliaCut (63/195 = 32%) had putative orthologs (RBH) in NC64A (see Supplemental Figure 7 online). Merchant et al. (2007) further subdivided the CiliaCut on the basis of whether or not a homolog was present in Caenorhabditis elegans, which has only nonmotile sensory cilia, and Thalassiosira pseudonana, which builds unusual motile flagella during gametogenesis. The 62 CiliaCut proteins with homologs in C. elegans were predicted to have structural, sensory, or assembly roles and designated the SSA, whereas the 69 CiliaCut proteins with homologs in T. pseudonana were designated the CentricCut. Interestingly, two-thirds of the CiliaCut proteins with putative orthologs in NC64A (42/63 = 67%) were classified in the CentricCut. This distribution was found to be significantly nonrandom (P value < 2E-7; χ2 test). By contrast, we found no significant association of the NC64A orthologs with the SSA subset. Thus, the pattern of conservation of putative flagellar proteins in NC64A is most similar to that of T. pseudonana, which like NC64A, lacks the genes encoding the radial spoke, central pair, and inner dynein proteins (Figure 4) (Merchant et al., 2007; Wickstead and Gull, 2007).
Altogether, these results lead to two hypotheses that should be verified experimentally: (1) the conserved flagella proteins might have acquired other biological roles when the flagellar apparatus was lost, which allowed the corresponding genes (i.e., encoding the retained flagella proteins) to remain under selective pressure; (2) given that NC64A is probably capable of sexual reproduction as suggested above, we speculate that Chlorella retained the ability to form rudimentary, possibly motile, flagella or flagellum-derived structures, similar to those of T. pseudonana. If true, we hypothesize that this inferred structure might serve in the recognition of the mating partner and initiate cell fusion, producing an as yet unidentified zygote.
Phytohormones in Algae
Phytohormones regulate much of the growth and development in land plants, and they are involved in the plant’s response to infection. Most types of land plant hormones have been biochemically detected in green algae, including chlorophytes (Tarakhovskaya et al., 2007). Some of those hormones appear to play the same roles as in land plants (e.g., cytokinin [Stirk et al., 2002] and auxin [de-Bashan et al., 2008]), but little is known about algal hormone biosynthesis (Bajguz, 2009). Hormone biosynthetic pathways in land plants are associated with plastids. Since chlorophyte algae contain plastids, we anticipated finding orthologs to the enzymes that synthesize hormones in land plants, as well as to their hormone receptors. We did not attempt to compile an exhaustive search of all chlorophyte hormone pathway steps or their receptors. Extensive gene duplication in the Arabidopsis genome used as reference prevented us from identifying clear algal orthologs of some enzymes involved in hormone synthesis. Instead, we looked for the presence of one or more clear orthologous enzymes for some key steps in plant hormone pathways and receptors. Orthology assignation was performed by combining information from reciprocal best hit analysis, phylogenetic tree reconstruction, and protein domain organization (see Supplemental Results online).
We explored the NC64A genome as well as five other chlorophyte genomes and found probable orthologs to Arabidopsis enzymes involved in the synthesis of a variety of plant hormones, including abscisic acid, cytokinin, brassinosteroid, and polyamines (Table 2; see Supplemental Results online). The sequenced chlorophyte algae did not exhibit homologs (BLASTP and TBLASTN analyses E-value cutoff = 1e-5) to Arabidopsis enzymes involved in the gibberellin biosynthetic pathway (gibberellin biosynthetic proteins GA1, GA2, and GA3; gibberellin oxidase proteins GA20OX1, GA2OX1, and GA3OX1) or the ethylene biosynthetic pathway (1-aminocyclopropane-1-carboxylate synthase and 1-aminocyclopropane-1-carboxylate oxidase [ACO]). We did find putative orthologs to some of the known Arabidopsis hormone receptors, including those for abscisic acid (chelatase H subunit [CHLH]), auxin (Auxin Binding Protein1 [ABP1]), and cytokinin (high osmolarity glycerol protein [HOG]) (Table 2). A recent survey of genomic data also reported the existence of orthologs of some of the components of the auxin signaling systems, including ABP1, in chlorophyte algae (Lau et al., 2009). In Arabidopsis, the auxin signaling cascade alternative to ABP1 involves the TIR1/AFB family of F-box proteins, auxin response factor, and auxin/indole-3-acetic acid proteins (Lau et al., 2008). None of these proteins were found to have a significant match with the sequenced chlorophytes, suggesting that this signaling cascade is absent in these organisms (Lau et al., 2009). By contrast, all major components of this pathway were identified in the moss P. patens, which implies that their origin goes back to at least the early evolution of land plants (Rensing et al., 2008).
Table 2.
Accession Numbers of Putative Chlorophyte Orthologs to Arabidopsis Proteins Involved in Phytohormone Biosynthesis or Reception
| Arabidopsis Enzyme Namea | Chlorella sp NC64Ab | C. reinhardtii | O. tauri | O. lucimarinus | Micromonas sp RC299 | Micromonas sp CCMP1545 |
| Abscisic acid pathway | ||||||
| Abscisic-aldehyde oxidase (AAO3) NP_180283 | 58208 (30%) | |||||
| 9-cis-epoxycarotenoid dioxygenase (NCED5) NP_174302 | 138368 (37%) | XP_ 001695565 (32%) | ||||
| Zeaxanthin epoxidase (ABA1) NP_201504 | 138731 (57%) | XP_ 001701701 (58%) | CAL 58065 (42%) | XP_ 001421564 (41%) | ACO 64017 (44%) | EEH 54518 (43%) |
| Violaxanthin deepoxidase (NPQ1) NP_172331 | 35609 (42%) | CAL 58064 (46%) | XP_ 001421704 (41%) | ACO 63977 (41%) | EEH 54773 (40%) | |
| Abscisic acid receptor (CHLH) NP_001078578 | 143922 (50%) | XP_ 001700895 (66%) | CAL 51621 (68%) | XP_ 001417229 (68%) | ACO 63109 (68%) | EEH 57631 (67%) |
| Cytokinin pathway | ||||||
| Isopentenyl-transferase 9 (ATIPT9) NP_851043 | 55198 (36%) | CAL 53743 (35%) | XP_ 001418572 (36%) | ACO61527 (42%) | EEH 53639 (41%) | |
| Cytokinin receptor (HOG) BAH19670 | 37522 (81%) | XP_ 001693339 (77%) | CAL 55423 (75%) | XP_ 001419579 (74%) | ACO67241 (73%) | EEH 58817 (69%) |
| Brassinosteroid pathway | ||||||
| lathosterol oxidase (STE1) NP_186907 | 37407 (45%) | XP_ 001701457 (50%) | ||||
| 7-Hydrocholesterol reductase (DWF5) NP_001077693 | ACO 69953 (51%) | EEH 53090 (51%) | ||||
| Steroid reductase (DET2) NP_181340 | 18410 (37%) | XP_ 001696975 (34%) | CAL 52707 (32%) | XP_ 001416556 (33%) | ACO 66602 (34%) | EEH 52455 (34%) |
| Jasmonic acid pathway | ||||||
| 12-Oxophytodienoate reductase (OPR1) NP_177794 | 52565 (48%) | XP_ 001699402 (51%) | ||||
| 3-Hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase (MFP2) NP_187342 | 52565 (54%) | XP_ 001696661 (45%) | CAL 53100 (44%) | XP_ 001417042 (52%) | ACO 65308 (52%) | EEH 60148 (51%) |
| Polyamine (spermidine) pathway | ||||||
| Arg decarboxylase (ADC1) NP_179243 | 25497 (40%) | EEH 59440 (38%) | ||||
| Agmatine iminohydrolase (ATAIH) NP_196434 | 133066 (54%) | |||||
| N-Carbamoyl-putrescine amidohydrolase (NLP1) NP_850101 | 18182 (57%) | XP_ 001692986 (53%) XP_ 001690094 (53%) | ||||
| Spermidine synthase 2 (SPDS2) NP_177188 | 26108 (53%) | XP_ 001702843 | ABO 98745 (56%) | XP_ 001420452 (58%) | ACO 70332 (55%) | EEH 54321 (56%) |
| Orn decarboxylase NP_001063827c | 133981 (50%) | XP_ 001698872 (46%) | CAL 51811 (45%) | XP_ 001417323 (45%) | ACO 63617 (46%) | EEH 58717 (46%) |
| Auxin pathway | ||||||
| Auxin receptor ABP1 NP_192207 | 17596 (48%), 26559 (40%) | |||||
The percentages of sequence identity in the best high-scoring pair (BLASTP) between proteins and their putative orthologous Arabidopsis protein are shown in parentheses.
Arabidopsis accession number of protein used as query in BLAST search.
At the JGI portal site (http://genomeportal.jgi-psf.org/), select Chlorella NC64A.
This enzyme is not found in Arabidopsis; accession number is for O. sativa.
The presence of putative chlorophyte orthologs to Arabidopsis proteins involved in phytohormone biosynthesis and perception does not necessarily imply that these green algae can produce, sense, and respond to hormones through pathways analogous to those in land plants. To our knowledge, some of the identified enzymes have no other role than hormone biosynthesis in land plants (e.g., ATP/ADP isopentenyltransferase AtIPT, Sterol 1 protein STE1, and DWARF5), while others are also involved in the production of molecules with no hormonal function (e.g., abscisic acid 1 protein [ABA1] and nonphotochemical quenching protein [NPQ1] involved both in the xanthophyll cycle and in the synthesis of ABA precursors). However, the presence of putative orthologs to the Arabidopsis auxin receptor ABP1 in NC64A is congruent with earlier studies demonstrating that auxin induces cell division in Chlorella pyrenoidosa (Vance, 1987) and cell enlargement in Chlorella vulgaris (Yin, 1937), two species closely related to NC64A (see Supplemental Figure 1 online). Our analysis suggests that at least some of the genes specifically involved in phytohormone biosynthesis and perception in land plants were established prior to their evolution. Unicellular ancestors of streptophytes and chlorophytes were perhaps able to communicate with each other before the emergence of multicellular land plants. We suggest that the existence of these features likely facilitated the evolution of multicellularity.
Cell Wall Metabolism and Interplay with Chlorella Viruses
With 233 predicted enzymes involved in carbohydrate metabolism, NC64A appears much better equipped for synthesizing and modifying polysaccharides than the other sequenced chlorophytes that have between 92 (O. tauri) and 168 (C. reinhardtii) of such predicted enzymes (see Supplemental Data Set 3 online) (Cantarel et al., 2009). However, we did not find homologs of the Arabidopsis proteins involved in the synthesis of cellulose (cellulose synthase CesA) or hemicellulose (hemicellulose synthase CLS), the major components of the primary cell wall of land plants. Instead, experimental evidence suggests that the cell wall of Chlorella species, including NC64A, contain glucosamine polymers such as chitin and chitosan (Kapaun and Reisser, 1995; Sun et al., 1999). We found two NC64A paralogs for chitin synthase and, remarkably, 25 paralogs for chitin deacetylase, which converts chitin into chitosan. Both NC64A chitin syntase proteins contain conserved amino acids essential for the catalytic activity of the Saccharomyces cerevisiae enzyme (i.e., Asp-441, Asp-562, Gln-601, Arg-604, Trp-605, Asn-797, Asp-800, Trp-803, and Thr-805; Yabe et al., 1998) (see Supplemental Figure 8A online). We also identified putative proteins involved in the degradation of these polysaccharides: two chitinase genes (plant and prokaryotic types [glycosyl hydrolase families GH19 and GH18, respectively]) and four chitosanase genes. The prokaryotic type chitinase protein exhibits protein domains that are homologous to the PF-ChiA chitinase and cellulose binding domains found in the chitinase of archaeon Pyrococcus furiosus. It also exhibits the conserved amino acid sequence (DXDXE motif) that plays an important role in the catalytic mechanism of family 18 chitinases (Watanabe et al., 1994) (see Supplemental Figure 8B online). The four NC64A chitosanases contain the three catalytic residues Glu-36, Asp-40, and Thr-45 of Streptomyces sp N174 chitosanase (Lacombe-Harvey et al., 2009) (see Supplemental Figure 8C online).
Chitin is a natural component of fungal cell walls and of the exoskeleton of arthropods but is not normally present in green algae. The origin of chitin and its derivatives in the Chlorella genus has long been an enigma. Except for the plant-type chitinase gene, which is found in land plants (but not in chlorophytes apart from Chlorella), the four gene classes involved in forming and remodeling chitin cell walls (i.e., chitin synthase, chitin deacetylase, chitinase, and chitosanase) are absent in all the other fully sequenced Viridiplantae species. By contrast, homologs for each of these families exist in genomes of Chlorella viruses. The viral genes are presumably involved in degradation of the Chlorella cell wall (chitinase and chitosanase) (Kang et al., 2005) and production of chitinous fibers on the external surface of virus-infected cells (chitin synthase and chitin deacetylase) (Kawasaki et al., 2002). Phylogenetic analysis suggests that the Chlorella ancestor exchanged the bacterial-type chitinase and chitin-deacetylase genes with the chloroviruses (Figure 5). The fact that these genes are absent in the other Viridiplantae species studied to date argues in favor of the capture of the viral genes by Chlorella. Alternatively, capture of the Chlorella genes by chloroviruses would imply that Chlorella genes were vertically inherited from the Viridiplantae ancestor and that these genes were independently lost in many lineages of the Viridiplantae, a very improbable scenario. Another scenario would imply a first capture of the genes by HGT from prokaryotes or fungi to Chlorella, after which a Chlorella virus picked up the two genes from Chlorella. Phylogenetic reconstructions of the chitosanase and chitin synthase proteins indicate that the corresponding Chlorella and Chlorella virus genes are phylogenetically related, but no direct gene exchange occurred between Chlorella and the known Chlorella viruses (see Supplemental Figures 9 and 10 online). Collectively, our results are congruent with the hypothesis that components of the Chlorella chitin metabolism were acquired horizontally from viruses or distantly related chitin-producing cellular organisms rather than from a Viridiplantae ancestor.
Figure 5.
Maximum Likelihood Phylogenetic Tree of the Chitin Deacetylase and Chitinase Proteins.
For both protein families, we used the WAG+I+G model of substitutions. Approximate likelihood ratio test values >50% are indicated beside branches. Phylogenetic trees are midpoint rooted. Alignments used to generate these trees are available as Supplemental Data Sets 4 and 5 online.
(A) Phylogenetic tree of chitin deacetylases. The multiple sequence alignment contained 134 gap-free sites.
(B) Phylogenetic tree of chitinases. The multiple sequence alignment contained 228 gap-free sites.
[See online article for color version of this figure.]
Conclusion
The first sequence of a trebouxiophycean genome unveiled important features of the evolution and genomic organization of the green phylum. For instance, the existence of genomic regions displaying large differences in GC content, correlating with differences in their expression levels, now appear to be a characteristic feature of many chlorophyte genomes. Understanding the role and mechanism by which this compositional shift is established and maintained is one of the next challenges in phycology.
We presented evidence suggesting that Chlorella could have acquired components of its chitin biosynthetic pathway by HGT from a chlorovirus or a microorganism. A similar evolutionary scenario was also evoked for the eukaryotic microalga Emiliania huxleyi that exchanged seven genes of the sphingolipid biosynthesis pathway with its large DNA virus, EhV (Monier et al., 2009), though the direction of gene transfer is unknown. Thus, the large DNA viruses predominantly associated with microalgae and marine protists might have played a much larger role in the evolution of their hosts than previously recognized. Conversely to the traditional view of viruses as gene pickpockets, large DNA viruses might have a propensity to enhance the metabolic capabilities of their host by donating genes (Villarreal, 2004). In the case of Chlorella, the acquisition of a chitinous cell wall may have conferred a protective barrier against other viral and bacterial parasites lacking the chitinase/chitosanase enzymes required to penetrate and/or escape the algal cell. This might have increased the fitness of Chlorella compared with its ancestors unable to synthesize chitin. This HGT might be the key event that promoted the radiation and success of the Chlorella genus (i.e., Chlorella may have achieved a cosmopolitan distribution because most of its previous parasites failed to penetrate its newly acquired chitinous cell wall).
Our results illustrate the role that comparative genomics can play in uncovering unsuspected biological functions; here, the identification of genes involved in meiosis, gamete fusion, and flagella. This led us to hypothesize that Chlorella retained the capability of sexual reproduction despite the fact that no sexual life cycle has been described in this genus. These findings naturally pose the question of the maintenance of sexual reproduction in an organism capable of rapid clonal population growth. In C. reinhardtii, mating between two haploid partners is induced by stress conditions (e.g., lack of nitrogen), producing a zygote resistant to freezing and desiccation (Goodenough et al., 2007). There is some recent evidence that viruses may have played a role in the success of sexual reproduction. Sexual reproduction can confer a selective advantage to the host in the arms race against its parasites (the so-called Red Queen hypothesis) by increasing the efficiency with which selection can fix beneficial mutations that result in virus resistance (Morin, 2008). A more direct viral pressure is illustrated by the haptophyte microalga E. huxleyi escaping infection by the phycodnavirus EhV by switching from its virus-sensitive diploid stage to a morphologically distinct haploid stage immune to the virus (the Cheshire Cat escape strategy) (Frada et al., 2008). Ostreococcus and Chlorella species are normally haploid but contain meiosis-related genes. They are both infected by phycodnaviruses (OsV and Chlorella viruses, respectively) that are phylogenetically related to EhV (Wu et al., 2009). By analogy to the EhV–E. huxleyi model, it is tempting to speculate that these microalgae have a virus-resistant diploid phase that might only become detectable after viruses have decimated the haploid population.
The presence of putative chlorophyte orthologs for land plant proteins functioning in critical hormone metabolic steps and as hormone receptors opens the possibility that phytohormone biosynthesis and perception could also be present in chlorophyte algae, although perhaps in a rudimentary form compared with land plants. Consequently, it has been suggested that green algae would be a model organism for the study of plant hormones (and receptors) because they are unicellular and can be grown axenically in the laboratory (Stirk et al., 2002). A fuller understanding of the role of plant hormone molecules in green algae as well as of their synthesis and perception would possibly lead to the selection and improvement of better algal strains that could benefit agricultural practices in developing countries (Stirk et al., 2002), result in better production of biodiesel, and improve the quality and quantity of nutrient supplements (proteins, vitamins, etc.). While bioinformatics/genomics can provide strong clues, enzyme and receptor functions remain to be experimentally tested to verify these many predictions.
METHODS
A detailed description of methods is provided in Supplemental Methods online.
Genome Sequencing and Assembly
The NC64A genome was sequenced using the whole-genome sequencing strategy. The data were assembled using release 2.10.11 of Jazz, a whole-genome sequencing assembler developed at the JGI (Aparicio et al., 2002). After excluding redundant and short scaffolds from the initial assembly, there was 46.4 Mb of scaffold sequence, of which 4.0 Mb (8.5%) were gaps. The filtered assembly contained 431 scaffolds, with a scaffold N/L50 of 12/1.5 Mb (the number of scaffolds/length of the shortest scaffold, respectively, such that the sum of scaffolds of equal length or longer is at least 50% of the total length of all scaffolds), and a contig N/L50 of 441/27.6 kb. The sequence depth derived from the assembly was 8.95 ± 0.15.
Pulse Field Gel Electrophoresis
Pulse field gel electrophoresis studies were performed according to Agarkova et al. (2006). Chromosomal DNAs were separated in a CHEF-DR II (Bio-Rad) unit in a 0.8% agarose gel. Electrophoresis conditions and running buffer were selected to resolve the target chromosome sizes. The exact conditions are described in the figure legends.
EST Sequencing and Assembly
Chlorella sp NC64A cells were grown to log phase (1.5 × 107 cells/mL). NC64A poly(A)+ RNA was isolated from total RNA using the Absolutely mRNA Purification kit (Stratagene). One to two micrograms of poly(A)+ RNA, reverse transcriptase SuperScript II (Invitrogen), and oligo(dT)-NotI primer were used to synthesize first-strand cDNA. Second-strand synthesis was performed with Escherichia coli DNA ligase, polymerase I, and RNaseH followed by end repair using T4 DNA polymerase. The cDNA inserts were directionally ligated into the SalI- and NotI-digested vector pCMVsport6 (Invitrogen). Subcloned inserts were then sequenced with Big Dye terminator chemistry (Applied Biosystems). A total of 38,400 ESTs were generated. The ESTs were processed through the JGI EST pipeline. A total of 23,828 ESTs remained after trimming vector sequences and removing short sequences. EST clusters were assembled using CAP3 (Huang and Madan, 1999) to form consensus sequences. Clustering and assembly of all 23,828 ESTs resulted in 7499 consensus sequences.
Genome Annotation and Sequence Analysis
The genome assembly v1.0 of NC64A was annotated using the JGI annotation pipeline, which combines several gene predictors and filtering steps (see Supplemental Methods online). Phylogenetic analyses were performed on the phylogeny.fr web tool (Dereeper et al., 2008). De novo identification of repeated sequences was performed by aligning the genome against itself using the BLASTN program (E-value < 1e-15). Individual repeat elements were organized into families with the RECON program using default settings (Bao and Eddy, 2002). RECON constructed 2980 repetitive sequence families from 10,723 individual repeat elements. Second, identification of known repetitive sequences was performed by aligning the prototypic sequences contained in Repbase v12.10 (Jurka et al., 2005) using TBLASTX. The results of the two methods were combined.
Accession Numbers
Assembly and annotations of Chlorella sp NC64A are available from JGI Genome Portal at http://genome.jgi-psf.org/NC64A and can also be found in the GenBank/EMBL data libraries under accession number ADIC00000000.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Position of Chlorella variabilis NC64A among Chlorophyte Algae.
Supplemental Figure 2. Pulsed Field Gel Electrophoresis of Chlorella sp NC64A Chromosomes.
Supplemental Figure 3. Taxonomic Distribution of Best Matches for Proteins Encoded in Low-GC Regions.
Supplemental Figure 4. Gene Duplication in Selected Viridiplantae.
Supplemental Figure 5. Sequence Motifs at Intron Splice Sites.
Supplemental Figure 6. Taxonomic Distribution of Best Matches for Representative Chlorophyte Algae.
Supplemental Figure 7. NC64A Putative Orthologs to Chlamydomonas CiliaCut Proteins.
Supplemental Figure 8. Alignment of NC64A Proteins with Their Reference Proteins Involved in Chitin Metabolism.
Supplemental Figure 9. Maximum Likelihood Phylogenetic Tree of Chitosanase Proteins.
Supplemental Figure 10. Maximum Likelihood Phylogenetic Tree of Chitin Synthase Proteins.
Supplemental Table 1. NC64A Nuclear Genome Assembly Statistics.
Supplemental Table 2. Eukaryotic Ortholog Groups (KOG) Functional Categories among Low-GC and Normal-GC Regions.
Supplemental Table 3. Repeated Sequences in the NC64A Genome.
Supplemental Table 4. Chlorophyte Algae–Specific PFAM Protein Domains.
Supplemental Table 5. PFAM Domains with Biased Distribution in Chlorophyte Green Algae.
Supplemental Table 6. Meiosis-Specific Protein GenBank Identification (gi) Numbers and Percentage of Protein Sequence Identity with Reference Arabidopsis Proteins.
Supplemental Data Set 1. Land Plant–Specific PFAM Protein Domains.
Supplemental Data Set 2. Putative Orthologs to Chlamydomonas Flagellar Proteins in Sequenced Chlorophytes.
Supplemental Data Set 3. Carbohydrate-Active (CAZy) Enzymes in Chlorophyte Green Algae.
Supplemental Data Set 4. Multiple Sequence Alignment of Chitin Deacetylase Proteins Used in Figure 5A.
Supplemental Data Set 5. Multiple Sequence Alignment of Chitinase Proteins Used in Figure 5B.
Supplemental Data Set 6. Multiple Sequence Alignment of 18S Genes Used in Supplemental Figure 1.
Supplemental Data Set 7. Multiple Sequence Alignment of Chitosanase Proteins used in Supplemental Figure 9.
Supplemental Data Set 8. Multiple Sequence Alignment of Chitin Synthase Proteins Used in Supplemental Figure 10.
Acknowledgments
We thank Marek Elias for helpful discussion on flagella proteins. We also thank Magali Lescot, Dmitry Brogun, Timo Greiner, Ming Kang, Gentry L. Lewis, Suzanne Rose, Eliza Wiech, and Giane M. Yanai-Balser for their contribution in the annotation. Genome sequencing conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02-05CH11231. This work was partially supported by Marseille-Nice Génopole, the PACA-Bioinfo platform, the French fund “Infrastructures en Biologie Santé et Agronomie,” and Public Health Service Grant GM32441 from the National Institute of General Medical Sciences (to J.L.V.E.). G.D. was partially funded by National Institutes of Health Grant P20 RR016469.
References
- Agarkova I.V., Dunigan D.D., Van Etten J.L. (2006). Virion-associated restriction endonucleases of chloroviruses. J. Virol. 80: 8114–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S., et al. (2002). Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310 [DOI] [PubMed] [Google Scholar]
- Bajguz A. (2009). Brassinosteroid enhanced the level of abscisic acid in Chlorella vulgaris subjected to short-term heat stress. J. Plant Physiol. 166: 882–886 [DOI] [PubMed] [Google Scholar]
- Bao Z., Eddy S.R. (2002). Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12: 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman R.M., Crane P.R., DiMichele W.A., Kenrick P.R., Rowe N.P., Speck T., Stein W.E. (1998). Early evolution of land plants: phylogeny, physiology, and ecology of the primary terrestrial radiation. Annu. Rev. Ecol. Syst. 29: 263–292 [Google Scholar]
- Battey J.F., Patton J.S. (1984). A reevaluation of the role of glycerol in carbon translocation in zooxanthellae-coelenterate symbiosis. Mar. Biol. 79: 27–38 [Google Scholar]
- Beavo J.A. (1995). Cyclic nucleotide phosphodiesterases: Functional implications of multiple isoforms. Physiol. Rev. 75: 725–748 [DOI] [PubMed] [Google Scholar]
- Benson A.A. (2002). Following the path of carbon in photosynthesis: A personal story. Photosynth. Res. 73: 29–49 [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 37 (Database issue): D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caturegli P., Asanovich K.M., Walls J.J., Bakken J.S., Madigan J.E., Popov V.L., Dumler J.S., Dumler J.S. (2000). ankA: An Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68: 5277–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelf P., Brown L.M., Wyman C.E. (1993). Aquatic biomass resources and carbon dioxide trapping. Biomass Bioenergy 4: 175–183 [Google Scholar]
- Chuchird N., Hiramatsu S., Sugimoto I., Fujie M., Usami S., Yamada T. (2001). Digestion of chlorella cells by chlorovirus-encoded polysaccharide degrading enzymes. Microbes Environ. 16: 206–212 [Google Scholar]
- Cole D.G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4: 435–442 [DOI] [PubMed] [Google Scholar]
- de-Bashan L.E., Antoun H., Bashan Y. (2008). Involvement of indole-3-acetic acid produced by the growth-promoting bacterium Azospirillum spp. in promoting growth of Chlorella vulgaris. J. Phycol. 44: 938–947 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 (Web Server issue): W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564 [DOI] [PubMed] [Google Scholar]
- Fott B., Novakova M. (1969). A monograph of the genus Chlorella. The fresh water species. Stud. Phycol. 11: 1–73 [Google Scholar]
- Frada M., Probert I., Allen M.J., Wilson W.H., de Vargas C. (2008). The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl. Acad. Sci. USA 105: 15944–15949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl T., Bhattacharya D. (2002). Origin and evolution of green lichen algae. Symbiosis: Mechanisms and Model Systems, Seckbach J., ed (Dordrecht, The Netherlands: Springer; ), pp. 341–357 [Google Scholar]
- Galván-Ampudia C.S., Offringa R. (2007). Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 12: 541–547 [DOI] [PubMed] [Google Scholar]
- Gerashchenko B.I., Kosaka T., Hosoya H. (2001). Growth kinetics of algal populations exsymbiotic from Paramecium bursaria by flow cytometry measurements. Cytometry 44: 257–263 [DOI] [PubMed] [Google Scholar]
- Goodenough U., Lin H., Lee J.H. (2007). Sex determination in Chlamydomonas. Semin. Cell Dev. Biol. 18: 350–361 [DOI] [PubMed] [Google Scholar]
- Grimsley N., Péquin B., Bachy C., Moreau H., Piganeau G. (2010). Cryptic sex in the smallest eukaryotic marine green alga. Mol. Biol. Evol. 27: 47–54 [DOI] [PubMed] [Google Scholar]
- Grossman A.R. (2005). Paths toward algal genomics. Plant Physiol. 137: 410–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman D.S., Geiser D.M., Eidell B.R., Stauffer R.L., Kardos N.L., Hedges S.B. (2001). Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133 [DOI] [PubMed] [Google Scholar]
- Huang X., Madan A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss V.A.R., Frank C., Hartmann E.C., Hirmer M., Kloboucek A., Seidel B.M., Wenzeler P., Kessler E. (1999). Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta). J. Phycol. 35: 587–598 [Google Scholar]
- Iturbe-Ormaetxe I., Burke G.R., Riegler M., O’Neill S.L. (2005). Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J. Bacteriol. 187: 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Kadono T., Kawano T., Hosoya H., Kosaka T. (2004). Flow cytometric studies of the host-regulated cell cycle in algae symbiotic with green paramecium. Protoplasma 223: 133–141 [DOI] [PubMed] [Google Scholar]
- Kang M., Dunigan D.D., VAN Etten J.L. (2005). Chlorovirus: A genus of Phycodnaviridae that infects certain chlorella-like green algae. Mol. Plant Pathol. 6: 213–224 [DOI] [PubMed] [Google Scholar]
- Kapaun E., Reisser W. (1995). A chitin-like glycan in the cell wall of a Chlorella sp. (Chlorococcales, Chlorophyceae). Planta 197: 577–582 [Google Scholar]
- Karakashian S.J., Karakashian M.W. (1965). Evolution and symbiosis in the genus Chlorella and related algae. Evolution 19: 368–377 [Google Scholar]
- Kato Y., Ueno S., Imamura N. (2006). Studies on the nitrogen utilization of endosymbiotic algae isolated from Japanese Paramecium bursaria. Plant Sci. 170: 481–486 [Google Scholar]
- Kawasaki T., Tanaka M., Fujie M., Usami S., Sakai K., Yamada T. (2002). Chitin synthesis in chlorovirus CVK2-infected chlorella cells. Virology 302: 123–131 [DOI] [PubMed] [Google Scholar]
- Lacombe-Harvey M.E., Fukamizo T., Gagnon J., Ghinet M.G., Dennhart N., Letzel T., Brzezinski R. (2009). Accessory active site residues of Streptomyces sp. N174 chitosanase: Variations on a common theme in the lysozyme superfamily. FEBS J. 276: 857–869 [DOI] [PubMed] [Google Scholar]
- Lau S., Jürgens G., De Smet I. (2008). The evolving complexity of the auxin pathway. Plant Cell 20: 1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., Shao N., Bock R., Jürgens G., De Smet I. (2009). Auxin signaling in algal lineages: Fact or myth? Trends Plant Sci. 14: 182–188 [DOI] [PubMed] [Google Scholar]
- Malik S.B., Pightling A.W., Stefaniak L.M., Schurko A.M., Logsdon J.M., Jr (2008). An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS ONE 3: e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., et al. (2008). The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452: 88–92 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A., Pagarete A., de Vargas C., Allen M.J., Read B., Claverie J.-M., Ogata H. (2009). Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 19: 1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P.J. (2008). Sex as an algal antiviral strategy. Proc. Natl. Acad. Sci. USA 105: 15639–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 104: 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J.D. (1975). Green Algae: Structure, Reproduction, and Evolution in Selected Genera. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Pollock S.V., Pootakham W., Shibagaki N., Moseley J.L., Grossman A.R. (2005). Insights into the acclimation of Chlamydomonas reinhardtii to sulfur deprivation. Photosynth. Res. 86: 475–489 [DOI] [PubMed] [Google Scholar]
- Rajamani S., Siripornadulsil S., Falcao V., Torres M., Colepicolo P., Sayre R. (2007). Phycoremediation of heavy metals using transgenic microalgae. Adv. Exp. Med. Biol. 616: 99–109 [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Ryo H., Mitsunori I., Nobutaka I. (2010). Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycological Res. 58: 188–201 [Google Scholar]
- Schenk P.M., Thomas-Hall S.R., Stephens E., Marx U.C., Mussgnug J.H., Posten C., Kruse O., Hankamer B. (2008). Second generation biofuels: High-efficiency microalgae for biodiesel production. Bioenergy Res. 1: 20–43 [Google Scholar]
- Schurko A.M., Logsdon J.M., Jr (2008). Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays 30: 579–589 [DOI] [PubMed] [Google Scholar]
- Stirk W.A., Ördög V., Van Staden J., Jäger K. (2002). Cytokinin and auxin-like activity in Cyanophyta and microalgae. J. Appl. Phycol. 14: 215–221 [Google Scholar]
- Sun L., Adams B., Gurnon J.R., Ye Y., Van Etten J.L. (1999). Characterization of two chitinase genes and one chitosanase gene encoded by Chlorella virus PBCV-1. Virology 263: 376–387 [DOI] [PubMed] [Google Scholar]
- Takeda H. (1988). Classification of Chlorella strains by cell wall sugar composition. Phytochemistry 27: 3823–3826 [Google Scholar]
- Takeda H. (1991). Sugar composition of the cell wall and the taxonomy of Chlorella (Chlorophyceae). J. Phycol. 27: 224–232 [Google Scholar]
- Tarakhovskaya E.R., Maslov Y.I., Shishova M.F. (2007). Phytohormones in algae. Russ. J. Plant Physiol. 54: 163–170 [Google Scholar]
- Tram U., Ferree P.M., Sullivan W. (2003). Identification of Wolbachia—Host interacting factors through cytological analysis. Microbes Infect. 5: 999–1011 [DOI] [PubMed] [Google Scholar]
- Vance B.D. (1987). Phytohormone effects on cell division in Chlorella pyrenoidosa chick (TX-7-11–05) (chlorellaceae). J. Plant Growth Regul. 5: 169–173 [Google Scholar]
- Villarreal L.P.P. (2004). Viruses and the Evolution of Life. (Washington DC: ASM Press; ). [Google Scholar]
- Watanabe T., Uchida M., Kobori K., Tanaka H. (1994). Site-directed mutagenesis of the Asp-197 and Asp-202 residues in chitinase A1 of Bacillus circulans WL-12. Biosci. Biotechnol. Biochem. 58: 2283–2285 [DOI] [PubMed] [Google Scholar]
- Wickstead B., Gull K. (2007). Dyneins across eukaryotes: A comparative genomic analysis. Traffic 8: 1708–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W.H., Van Etten J.L., Allen M.J. (2009). The Phycodnaviridae: The story of how tiny giants rule the world. Lesser Known Large dsDNA Viruses, Van Etten J.L., eds (Berlin-Heidelberg, Germany: Springer-Verlag; ), pp. 1–42 [Google Scholar]
- Worden A.Z., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Wu G.A., Jun S.R., Sims G.E., Kim S.H. (2009). Whole-proteome phylogeny of large dsDNA virus families by an alignment-free method. Proc. Natl. Acad. Sci. USA 106: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T., Yamada-Okabe T., Nakajima T., Sudoh M., Arisawa M., Yamada-Okabe H. (1998). Mutational analysis of chitin synthase 2 of Saccharomyces cerevisiae. Identification of additional amino acid residues involved in its catalytic activity. Eur. J. Biochem. 258: 941–947 [DOI] [PubMed] [Google Scholar]
- Yin H.C. (1937). Effect of auxin on Chlorella vulgaris. Proc. Natl. Acad. Sci. USA 23: 174–176 [DOI] [PMC free article] [PubMed] [Google Scholar]