This work examines the effects of RNA interference constructs expressed in host cells on target RNAs in Blumeria graminis, an obligate biotrophic fungal pathogen of barley, and finds that RNAs in the host can affect pathogen transcript levels and pathogen development, thereby providing both a useful research tool and a potentially important means for engineering plant disease resistance.
Abstract
Powdery mildew fungi are obligate biotrophic pathogens that only grow on living hosts and cause damage in thousands of plant species. Despite their agronomical importance, little direct functional evidence for genes of pathogenicity and virulence is currently available because mutagenesis and transformation protocols are lacking. Here, we show that the accumulation in barley (Hordeum vulgare) and wheat (Triticum aestivum) of double-stranded or antisense RNA targeting fungal transcripts affects the development of the powdery mildew fungus Blumeria graminis. Proof of concept for host-induced gene silencing was obtained by silencing the effector gene Avra10, which resulted in reduced fungal development in the absence, but not in the presence, of the matching resistance gene Mla10. The fungus could be rescued from the silencing of Avra10 by the transient expression of a synthetic gene that was resistant to RNA interference (RNAi) due to silent point mutations. The results suggest traffic of RNA molecules from host plants into B. graminis and may lead to an RNAi-based crop protection strategy against fungal pathogens.
INTRODUCTION
Obligate biotrophic plant pathogens, such as powdery mildew and rust fungi, represent a large group of organisms that together cause diseases on thousands of plant species (Schulze-Lefert and Vogel, 2000; Zhang et al., 2005). In sharp contrast with their overall importance, little is known about genes and factors of powdery mildew and rust fungi required for pathogenesis and virulence (Zhang et al., 2005). Besides the lack of robust transformation protocols, one of the obstacles to a better understanding of gene function in these pathogens is the fact that any stable transformation event or mutation causing apathogenicity or avirulence will be lost because it thus becomes impossible to maintain them on living plant material. On the other hand, the expected public release of the genome sequence of the barley powdery mildew Blumeria graminis f. sp hordei (B. graminis) will offer new opportunities for a better understanding of the molecular basis of its host interaction provided postgenomic tools and resources will also become available (http://www.blugen.org).
Plants control viral diseases by RNA interference (RNAi) and exhibit enhanced resistance when carrying suitable antisense or hairpin RNAi constructs (Waterhouse and Fusaro, 2006; Sudarshana et al., 2007). Recently, reduced development of root-knot nematodes as well as Lepidoptera and Coleoptera insects feeding on transgenic plants that carry RNAi constructs against target genes in these pests has been reported (Huang et al., 2006; Baum et al., 2007; Mao et al., 2007). The uptake of double-stranded RNA (dsRNA) or small interfering RNA (siRNA) into these animals occurs by sucking or chewing of plant material, followed by resorption in the (mid)gut system. On the other hand, biotrophic fungal pathogens such as B. graminis interact with their corresponding host intimately via a highly specialized cell called a haustorium, which is surrounded by the extrahaustorial matrix bordered by plant and fungal membranes on either side and that represents the interface for signal exchange as well as nutrient uptake (Panstruga, 2003) (see Supplemental Figure 1 online). This close contact of the interaction partners might facilitate not only the exchange of proteins but also of RNA as a carrier of biological information affecting the outcome of the interaction. This hypothesis would have far-reaching implications for our understanding of plant–pathogen interactions and disease control.
As in fungal or oomycete pathogens, parasitic plants also form haustoria as feeding organs during their interaction with host plants. Recently, exchange of mRNA and small noncoding RNAs has been described between host and parasitic plants, such as Cuscuta pentagona or Tryphisaria versicolor (Westwood et al., 2009). These findings lend initial support to the hypothesis of exchange of genetic information between plants and pathogens via exchange of RNA molecules. Here, we tested the possibility that silencing-inducing RNA molecules are exchanged between cereal hosts and the obligate biotrophic fungal pathogen B. graminis by assessing fungal development on plants that express RNAi constructs directed against fungal target transcripts.
RESULTS
Screening for Fungal RNAi Target Genes
To determine whether the accumulation in barley (Hordeum vulgare) cells of dsRNA or siRNA targeting fungal transcripts could affect the outcome of the interaction with B. graminis, we used RNAi in the host cell to target fungal transcripts and examined the progression of the fungal infection. We tested 76 fungal candidate genes that were found to be expressed in planta during the interaction (Zierold et al., 2005). For this aim, we used a transient assay system based on biolistic bombardment of RNAi constructs into single epidermal cells (Douchkov et al., 2005) (see Supplemental Figure 1 online). None of the RNAi constructs was predicted to possess off-targets in the barley transcriptome as determined by the SI-FI software tool (http://labtools.ipk-gatersleben.de/). The 76 candidate mRNAs were targeted by host-induced gene silencing (HIGS) in B. graminis growing either on cv Golden Promise (GP) or on cells of the resistant mutant Ingrid BC7 mlo5 (I mlo5) that were rendered susceptible by transiently expressing the Mlo wild-type allele (Shirasu et al., 1999). The use of I mlo5 also allowed the quantification of hyphal growth rates without interference from neighboring colonies growing out from nontransformed cells. The two screens, in which nonoverlapping sets of genes were tested, revealed 16 out of 76 (21%) target mRNAs of B. graminis that resulted in a significant reduction of the percentage of fungal conidia able to form a haustorium upon introduction of the RNAi construct into the host cell (Tables 1 and 2; see Supplemental Table 1 online for full details). No RNAi construct was identified that significantly altered the rate of secondary hyphae elongation on I mlo5 hosts (data not shown). The regulation of the gene candidates, which had been selected for repeated experiments based on initial screening results, was analyzed using publicly available transcript-profiling data from a B. graminis–barley interaction (Both et al., 2005a; see Supplemental Table 2 online). As shown in Supplemental Figure 2 online, all candidates represented on the array were transiently expressed during the interaction, with no bias for an expression peak of the confirmed candidate genes early or late during the interaction. However, mRNA levels of all confirmed candidates were detectable 4 to 15 h after inoculation, which is in agreement with the observed reduction of haustorium formation occurring within the first 24 h of the interaction.
Table 1.
Summary of the Screening for Fungal Target mRNAs Affected by HIGS
| Screening in Barley Genotype |
||
| RNAi Target mRNAs | GP | I mlo5 |
| All tested candidates | 38 | 38 |
| First-round candidates | 13 (16) | 14 |
| Confirmed candidates | 10 | 6 |
RNAi constructs targeting candidate genes of B. graminis were cobombarded with pUbiGUS (for the detection of transformed cells by GUS staining) into barley epidermal cells, followed by inoculation with B. graminis 1 d (I mlo5) or 3 d (GP) later. RNAi constructs that reduced haustorium formation by at least 20% in I mlo5 or 33% in GP were selected for a final number of at least five independent experiments. The number in parentheses in the GP screen includes three additional candidates selected randomly or due to sequence similarity to GTF proteins. The RNAi effects of confirmed candidates differed significantly from the empty vector control (P < 0.05).
Table 2.
Significant Disruption of Fungal Development by RNAi Constructs Targeting Fungal Genes
| Clone IDa | HIGS Screen | Rel. HI (%)b | Min.c | nd | Pe | Short Description (BLASTX) | E-Value |
| HO09I03 | GP | 82.3 ± 3.3 | 75 | 5 | 0.0058 | Hypothetical (Magnaporthe oryzae) | 3 × 10−89 |
| HO07E08 | GP | 76.0 ± 8.0 | 38 | 11 | 0.0135 | Hypothetical (N. crassa) | 4 × 10−43 |
| HO15J13 | GP | 74.9 ± 8.7 | 20 | 20 | 0.0097 | 1,3-β-glucanosyltransferase | 1 × 10−52 |
| HO11N21 | GP | 72.1 ± 7.6 | 47 | 7 | 0.0106 | 1,3-β-glucanosyltransferase | 1 × 10−82 |
| HO08J20 | GP | 71.3 ± 8.5 | 50 | 5 | 0.0285 | No match | |
| HO04M11 | GP | 71.2 ± 6.5 | 50 | 7 | 0.0043 | Hypothetical (G. zeae) | 5 × 10−18 |
| HO15I23 | GP | 65.8 ± 10.2 | 38 | 5 | 0.0286 | ADP/ATP carrier protein | 4 × 10−74 |
| HO14N21 | GP | 64.4 ± 8.0 | 38 | 8 | 0.0030 | Heat shock protein 70 | 1 × 10−133 |
| HO06C14 | GP | 59.6 ± 5.9 | 36 | 11 | <0.0001 | NADH-ubiquinone oxidoreductase | 1 × 10−111 |
| HO06F11 | GP | 59.1 ± 7.6 | 28 | 7 | 0.0017 | Vacuolar Ser protease | 2 × 10−55 |
| HO03F17 | I mlo5 | 82.5 ± 4.4 | 70 | 5 | 0.0170 | Endosomal cargo receptor | 9 × 10−65 |
| HO08A09 | I mlo5 | 79.7 ± 5.0 | 62 | 6 | 0.0096 | Hypothetical (Magnaporthe oryzae) | 1 × 10−22 |
| HO10J21 | I mlo5 | 76.3 ± 6.6 | 63 | 5 | 0.0228 | Hypothetical (Magnaporthe oryzae) | 6 × 10−29 |
| HO12C01 | I mlo5 | 74.9 ± 3.0 | 68 | 5 | 0.0011 | 40S Ribosomal protein | 6 × 10−32 |
| HO06N16 | I mlo5 | 73.6 ± 9.2 | 43 | 5 | 0.0458 | No match | |
| HO10G24 | I mlo5 | 66.4 ± 9.2 | 40 | 5 | 0.0215 | No match |
Clone ID from the CR-EST database (http://pgrc.ipk-gatersleben.de/cr-est/index.php).
Haustorium index relative to the empty vector control set to 100%.
Min, minimum value (in percent) of rel. HI observed among n independent experiments.
Number of independent experiments.
P value for null hypothesis (no deviation from the empty vector control). The fungal origin of all RNAi target sequences was supported by an EST sequence from in vitro–germinated B. graminis, except for HO06C14 that was instead supported by a highly similar sequence in the draft genomic sequence of B. graminis.
Silencing of Fungal Glucanosyltransferase Genes
Two RNAi constructs reducing haustorium formation were derived from the EST clones HO15J13 and HO11N21 (Kunne et al., 2005) and target mRNAs with similarity to 1,3-β-glucanosyltransferase (GTF1 and GTF2). The occurrence of glycosylphosphatidylinositol-anchored GTF proteins appears to be restricted to the fungal kingdom where they act in cell wall elongation and as virulence factors (Caracuel et al., 2005; Mouyna et al., 2005). In addition, GTF1 was found to belong to the penetration-associated cap20 regulon of B. graminis (Both et al., 2005a) (see Supplemental Figure 2 online). The encoded proteins fell into two different phylogenetic clades of the fungal GTF gene family that differ by the absence or presence of a Cys-rich module near the C terminus of the protein (Ragni et al., 2007) (Figure 1). We decided to address the function of GTF1 and GTF2 in a series of additional experiments including virus-induced gene silencing (VIGS) of both genes in wheat and transgenic barley plants that carry an RNAi construct against GTF1. Figure 2 shows that several constructs targeting nonoverlapping sequences of GTF1 or GTF2 significantly reduced haustorium formation, which indicates target specificity of RNAi. These results suggest a role of both GTF proteins in fungal host invasion because a cross-silencing by either RNAi construct was not predicted by the SI-FI tool.
Figure 1.
Unrooted Phylogenetic Tree Indicating Relationships of GTF1 and GTF2 Protein Sequences to GTF Reference Proteins from Different Fungi Inferred by a Neighbor-Joining Analysis.
Bootstrap values (%) are indicated along branches. Common gene names and accession numbers of GTF proteins are shown. The following species abbreviations were used: Asfu, Aspergillus fumigatus; Caal, Candida albicans; Fuox, Fusarium oxysporum; Magr, Magnaporthe grisea; Pabr, Paracoccidioides brasiliensis; Sace, Saccharomyces cerevisiae.
Figure 2.
HIGS of GTF1 and GTF2 Genes in B. graminis Affects Early Fungal Development.
Reduction of haustorium formation induced by RNAi constructs that target different regions of BgGTF mRNAs in the single-cell HIGS assay. Black lines below the mRNA sequences show the length and locations of HIGS target sequences, with exact start/end positions given above the line. Rel. HI (%), haustorium index, relative to the empty vector control set to 100%. White, gray, and black boxes indicate poly(A) tails, noncoding untranslated regions, and coding regions, respectively. Mean values of at least five independent experiments in cv GP are shown, with P values for significant difference from the empty vector control.
VIGS mediated by the barley stripe mosaic virus (BSMV), a tripartite sense-strand RNA virus resulting in dsRNA intermediates during its replication, has been established in barley and wheat (Triticum aestivum; Holzberg et al., 2002; Scofield et al., 2005) and was used here to further address the effect of silencing of GTF-encoding genes during the interaction of wheat with the wheat powdery mildew B. graminis f. sp tritici (B. graminis tritici). This allowed addressing HIGS by an independent approach in a related, second pathosystem of high agronomical importance. Due to the irregular, patchy pattern of VIGS in wheat, randomly chosen sites on third and younger leaves of BSMV-symptomatic plants were selected for microscopy assessment of fungal development, which resulted in mixed data from silenced and nonsilenced sectors. B. graminis tritici inoculation of plants infected with BSMV carrying a GTF1 antisense sequence of 774 bp resulted in a highly significant reduction of initial haustorium formation, whereas VIGS of GTF2 induced by an antisense sequence of 468 bp significantly reduced the elongation rate of secondary hyphae of B. graminis tritici (Figure 3). These results suggest that the two GTF genes may be involved in different steps of fungal development that could not be resolved in the single-cell assay.
Figure 3.
VIGS of GTF1 and GTF2 Inhibits Fungal Development in the Wheat–B. graminis tritici Interaction.
Combined data from two to three independent experiments comprising 77 to 105 plants per construct are shown. ***, Significantly different from BSMV wild-type control plants (α = 0.0001; Wilcoxon rank sum test of median); NS, not significant. Mean values ± se are also shown, although not used for statistical analysis because data were not normally distributed. Elongating secondary hyphae were scored short (interaction type II) or long (interaction type III) if their entire length was shorter or longer than 5 times the length of the conidiospore.
To test if HIGS can be used to protect transgenic plants from fungal infection, we transformed barley with the hairpin RNAi construct pIPKb007_BgGTF1 that targets GTF1 under the control of the maize (Zea mays) Ubiquitin-1 promoter (Himmelbach et al., 2007). Several independent transgenic T1 populations were tested for transgene expression and fungal development. As shown in Figure 4, several lines carried both inverted repeats of the RNAi construct. The observed weak and smeary hybridization signals of a GTF1-derived probe on RNA gel blots with RNA from noninoculated plants indicate rapid degradation of the double-stranded transgene RNA. Three T1 lines exhibited significantly reduced B. graminis disease symptoms, whereas transgenic control line E26 that had lost the hairpin RNAi cassette was as susceptible as wild-type control plants.
Figure 4.
Reduced Disease Symptoms by B. graminis on Transgenic Barley Plants Carrying an RNAi Construct against GTF1.
T0 plants were analyzed by genomic PCR for the presence of the selectable marker gene (Hptr) and of both inverted repeats (IR1 and IR2) of the RNAi cassette. The expression of the hairpin RNAi construct in T1 lines was analyzed by RNA gel blotting. 26s rRNA, loading/blotting control. Infection was estimated as percentage of leaf area covered by B. graminis mycelium 7 d after inoculation according to Schweizer et al. (1995) and is show here relative to nontransgenic control plants that were set to 100%. Mean values ± se from five to six independent inoculation experiments using a total of 116 to 170 T1 plants per line are shown. *, Significantly different from control (α = 0.05).
Silencing of Fungal Avra10 Gene
B. graminis appears to deliver the putative effector proteins Avra10 and Avrk1 into host cells where they may support the establishment of disease (Ridout et al., 2006). These proteins are recognized by the matching resistance (R) gene products MLa10 and MLk1 in some barley genotypes, which leads to hypersensitive cell death stopping fungal invasion (Figure 5A). Overexpression of Avra10 and Avrk1 in barley lacking the corresponding resistance genes was found to increase fungal invasion, suggesting indeed a role of the encoded proteins as effectors (Ridout et al., 2006). We took advantage of the predicted functions of Avrk1 and Avra10 inside host cells for a series of proof-of-concept experiments of HIGS. As shown in Table 3, HIGS of either effector gene caused a marked reduction in haustorium formation in a susceptible host, thus supporting the view that in the absence of corresponding R genes, these proteins have effector function already early during the interaction. On the other hand, transient overexpression of a synthetic Avra10 gene induced cell death in the presence of Mla10 (Pallas near-isogenic line P09), which was reflected by a reduced number of epidermal cells accumulating anthocyanin induced by the cobombarded reporter plasmid pBC17 (Schweizer et al., 2000) (Figure 5B). The Mla10 gene induced a rapid hypersensitive cell death reflected by a reduced formation of the first haustorium (Figure 5C) and therefore was a suitable target for assessing an R gene–dependent effect of silencing Avra10 via the host. Figure 5D shows that HIGS of Avra10 reduced haustoria formation in cv Pallas, whereas it had no effect in Pallas P09. The unchanged haustoria formation in Pallas P09 probably reflects the outcome of two opposite HIGS effects in this genotype that neutralized each other: an increase in haustoria formation due to escape from Mla10 recognition and a decrease due to the lack of the Avra10 effector protein. It is important to note that, in the presence of the Avra10 HIGS construct, the Mla10-dependent effect on haustorium formation was eliminated (cf. the second and fourth columns from the left). This is the predicted result because in the absence of Avra10 (due to HIGS), the absence or presence of the matching resistance gene in the host becomes irrelevant for the interaction.
Figure 5.
The Phenotypic Effect of Avra10 Effector Silencing Depends on the R Gene Status of the Host.
(A) The B. graminis isolate used in this study (CH4.8) expresses both Avrk1 and Avra10 effector genes that are recognized by resistance genes Mlk1 and Mla10 in Pallas BC lines P17 and P09, respectively. Effector recognition triggers a defense response, including cell death, which is seen as small dark flecks in the P17 and P09 lines. Lack of Avr recognition produces a susceptible phenotype, seen in the Pallas line as white fungal pustules.
(B) Mla10-dependent cell death induced by transient expression of Avra10 in barley near isogenic line Pallas P09 (Mla10). cv Pallas (mla10) served as negative control. Leaves were cobombarded with reporter plasmid pBC17 leading to anthocyanin accumulation ± the Avra10 overexpression construct pIPKTA9_Avra10, followed by counting of anthocyanin-stained cells 4 d after bombardment. Mean ± se from four biological replicates (two independent experiments). Different letters inside or above columns indicate significant Avra10 effect (analysis of variance).
(C) Mla10 mediates a rapid resistance response in barley epidermis, resulting in a reduction of the formation of the first haustorium that is reflected by a lower haustorial index (HI) of GUS-transformed epidermal cells. Mean ± se from five independent experiments is shown with P values for the comparison of mean values (neighboring columns).
(D) HIGS of Avra10 eliminates the Mla10-mediated difference in initial haustorium formation by selectively reducing HI in Pallas lacking the R gene. Mean values ± se from five independent experiments are shown with P values for the comparison of mean values (neighboring columns).
Table 3.
Reduction of Fungal Development by RNAi Constructs Targeting Fungal Effector Genes
| Construct | Rel. HI (%)a | Mina | na | Pb |
| pIPKTA30N | 100 | – | 8 | |
| pIPKTA30N_Avrk1 | 53.6 ± 10.0 | 18.0 | 8 | 0.0024 |
| pIPKTA30N_Avra10 | 57.9 ± 16.3 | 7.8 | 8 | 0.0368 |
For abbreviations, see Table 2.
P value for null hypothesis (no deviation from the empty vector control).
The reduction of target mRNA abundance has to be postulated if RNAi is the underlying mechanism leading to a phenotypic effect. However, to demonstrate this in the obligate biotrophic fungus B. graminis is difficult because no reporter strains and almost no information about gene function exist in this organism due to its recalcitrance to genetic transformation. Therefore, no known nondetrimental gene knockout phenotypes are available for phenocopying by RNAi, which means that silencing of any gene may compromise fungal development or pathogenicity. This may result in a selection against efficiently silenced sporelings during host interactions and, consequently, in difficulty to observe target mRNA reduction in the surviving fungal cells. To circumvent this problem, we analyzed Avra10 mRNA abundance in fungal hausoria interacting with epidermal cells that expressed both Avra10_RNAi and the matching Mla10 gene, which should create a selection pressure for efficient silencing of Avra10 in order to escape Mla10 detection. Furthermore, only fungal haustoria interacting with transformed epidermal cells must be analyzed, which was achieved by coexpressing wild-type Mlo in the resistant barley line I mlo5 resulting in cell-autonomous complementation of basal susceptibility (Shirasu et al., 1999). Assuming that the Avra10 effector is also expressed in young haustoria and not only at the prehaustorial stage, which has to be postulated due to the HIGS effect on haustoria formation and early (prehaustorial) recognition by the MLa10 protein (Figure 5), RNA was extracted from bombarded and challenge-inoculated leaves 24 h after inoculation following mechanical removal of epiphytic fungal mycelium. This allowed for the selective extraction of fungal transcripts for TaqMan real-time RT-PCR analysis from young haustoria, which are a likely target tissue of HIGS due to their intimate interaction with host cells. Our assumption that Avra10 is expressed in young haustoria is supported by an expression time course that showed low expression in conidia that just landed on the leaf surface, followed by rapid induction within 6 h and high expression until 24 h after inoculation, with a drop thereafter (see Supplemental Figure 3 online). Transient expression of the fast-acting Mla10 allele resulted in a strong, though not complete, reduction of the formation of the first haustorium (Table 4), thereby creating a strong selection pressure for Avra10 silencing that will counteract the selection against silencing of this gene due to loss of effector functionality. As a result, at least a partial reduction of Avra10 mRNA should become visible assuming that the strength of HIGS is not equal among a large population of haustoria but follows a normal distribution. This assumption is supported by our observation that with none of the tested fungal target genes a complete elimination of fungal development occurred. Indeed, as shown in Figure 6, the relative abundance of Avra10 transcript was significantly reduced by the Avra10 HIGS construct in the presence of the cobombarded Mla10 overexpression construct strongly suggesting target gene silencing inside the fungus. The undetectable target transcript reduction in the absence of coexpressed Mla10 probably reflects inefficient Avra10 silencing in those surviving haustoria that actually yielded the RNA for transcript quantification. The nondetectable and partial RNAi effect in the absence and presence of Mla10, respectively, suggests indeed a virulence penalty resulting from Avra10 silencing. At first sight, this is not in line with the frequent observation that effectors can be easily mutated or deleted by pathogens without a significant loss of virulence. However, Avra10 was found to belong to a complex gene family in B. graminis with at least six clades (unisequences) differing by at least 1% in DNA sequence, and the RNAi construct used here is predicted by the SI-FI software to efficiently silence five out of these six unisequences, which could explain the strong induced phenotype (see Supplemental Table 3 online; Sacristan et al., 2009). Taken together, our data provide direct evidence for HIGS of the Avra10 effector family inside young B. graminis haustoria.
Table 4.
Resistance against B. graminis Isolate CH4.8 Mediated by Transient Expression of Mla10
| Constructs | SIa | n | Pb |
| pUbiGUS + pU-Mlo | 0.402 ± 0.063 | 4 | – |
| pUbiGUS + pU-Mlo + pENTR-(ubi)-Mla10-(c) | 0.112 ± 0.015 | 4 | 0.0042 |
Susceptibility index.
P value for null hypothesis (no deviation from absence of Mla10).
Figure 6.
Target Transcript Reduction by HIGS in B. graminis.
Transcript abundance of Avra10 in young fungal haustoria interacting with transformed epidermal cells was quantified by TaqMan real-time RT-PCR 24 h after bombardment with the Avra10 HIGS construct (pIPKTA30_Avra10) in the absence or presence of coexpressed Mla10 [construct pENTR-(ubi)-Mla10-(c), here abbreviated as pMla10]. Basic host compatibility of transformed cells of resistant line Ingrid BC mlo5 was established by coexpressing wild-type Mlo (construct pU-Mlo). Twenty-four hours after B. graminis inoculation, poly(A)+ RNA was extracted, reverse transcribed, and used for real-time PCR. Data represent mean values ± se from three independent quantitative PCR runs using cDNA from two independent poly(A)+ preparations of one bombardment experiment. Similar data were obtained from a second independent biological replicate. The ratio of Avra10 transcript relative to the monoglyceride lipase reference transcript is shown (Both et al., 2005b). Statistical significance of the HIGS effect was determined by Student’s t test (one-tailed; Avra10 HIGS construct versus empty vector control in the absence or presence of Mla10 coexpression).
To further confirm target specificity of the observed effect of Avra10 silencing, we performed an RNAi rescue experiment (Figure 7) as described for the silencing of barley ubiquitin genes (Dong et al., 2006). In this type of experiment, an RNAi construct is coexpressed with a synthetic gene that encodes the target protein but is insensitive to the RNAi effect due to a high density of silent point mutations at codon wobble positions. The expected result of this experiment will be full complementation of the RNAi phenotype by the RNAi rescue construct if the RNAi phenotype is the result of specific silencing of one target mRNA or of highly similar, functionally redundant target mRNAs. Here, the RNAi construct pIPKTA30_Avra10 was cobombarded together with a synthetic Avra10 gene (pIPKTA9_Avra10_wobble) that is saturated in silent point mutations by replacing B. graminis with barley codons (Figure 8A). Figure 8B shows that pIPKTA30_Avra10 caused a significant reduction of fungal haustorium formation, whereas the rescue construct pIPKTA9_Avra10_wobble alone was without effect. This suggests that the level of Avra10 protein was not limiting during the interaction between the used fungal isolate CH4.8 and barley GP. Importantly, the cobombardment of pIPKTA30_Avra10 with pIPKTA9_Avra10_wobble eliminated the RNAi effect. The Avra10 RNAi rescue construct had no effect on the efficiency of cobombarded RNAi construct pPKTA36 that targets the barley Mlo gene and phenocopies recessive mlo resistance alleles (Schweizer et al., 2000). We conclude that the effects of the Avra10_RNAi as well as the synthetic rescue constructs were gene specific and demonstrate silencing of Avra10 in B. graminis.
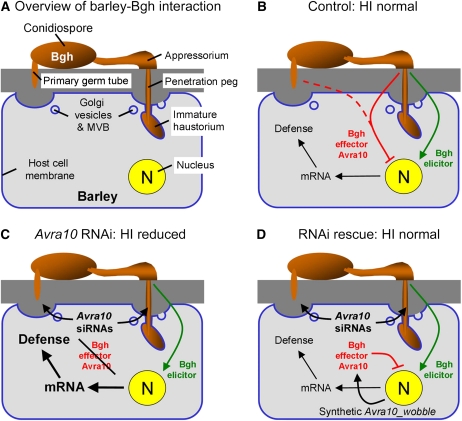
Figure 7.
Model of HIGS and RNAi Rescue of Avra10 in the Barley–B. graminis Interaction.
(A) Overview of interaction-related cellular structures ~16 h after inoculation. Please note that cell wall penetration by the primary germ tube and targeted host secretion leading to a cell wall apposition are occurring ~10 h prior to appressorial penetration. Bgh, B. graminis; MVB, multivesicular bodies.
(B) to (D) Model of silencing and RNAi rescue of Avra10.
Figure 8.
Rescue from Silencing of Avra10 by a Synthetic Gene Restores Fungal Haustorium Formation.
(A) Alignment of Avra10 wild-type (top) and synthetic Avra10_wobble (bottom) DNA sequences. Mismatches are highlighted by white boxes.
(B) Barley leaf segments were cobombarded with plasmid combinations followed by inoculation with B. graminis, and haustorium formation was assessed microscopically. Rel. HI, haustorial index relative to the empty vector control (cobombardment of pIPKTA9 and pIPKTA30) set to 100%. Mean values ± se from eight independent experiments are shown with P values for the null hypothesis. Construct pIPKTA340_Avra10 was used for Avra10 silencing in B. graminis, construct pIPKTA30_Mlo for silencing of barley Mlo was used as positive RNAi control, and construct pIPKTA9_Avra10_wobble was used for RNAi rescue. For a schematic representation of constructs, see Supplemental Figure 4 online.
DISCUSSION
Based on the presented data, we postulate the transfer of dsRNA or siRNA from host plant cells into powdery mildew fungi; these RNAs can disturb the host–pathogen interaction by inducing silencing of fungal housekeeping genes or genes required for development or virulence. RNAs have been found to move systemically in plants, including, for example, the movement of viruses via plasmodesmata and phloem (Voinnet, 2005). However, no such structures have been observed across powdery mildew haustorial walls (Hippe, 1985). Instead, intensive vesicle fusion/budding at plant and fungal membranes of the haustorial complex has been described, which is preceded by anterograde and probably retrograde vesicle traffic in epidermal cells at sites of fungal penetration (Hippe, 1985; An et al., 2006). In this respect, exosomes are of special interest because they have been described to accumulate in multivesicular bodies at plant-powdery mildew contact sites and to be released into cell wall or extrahaustorial matrix (Meyer et al., 2009). It is also important to note that multivesicular bodies and exosomes from monocytes and mast cell lines have been described to contain small noncoding RNAs plus components of the silencing machinery, such as GW182, and this is being discussed with respect to microRNA-mediated intracellular gene silencing and as a mechanism of cell-to-cell exchange of genetic information (Valadi et al., 2007; Gibbings et al., 2009).
Theoretically, the observed phenotypes could also have been caused by degradation in epidermal cells of mRNA that was secreted from the fungus into the host. However, in the case of seven target genes, including GTFI and II, silencing is likely to have occurred inside B. graminis because the encoded proteins clearly fulfill specific functions in fungal cells (Table 2). In the case of Avra10, a role of the encoded signal-peptide free protein inside host cells has been proposed, which could theoretically also be explained by secretion of the corresponding fungal transcript into the host (Shen et al., 2007). However, secretion of fungal transcripts into host cells has not been reported so far, in contrast with the secretion of signal-peptide free proteins through a novel pathway independent from the conventional endoplasmic reticulum–Golgi secretory pathway (Nickel and Rabouille, 2009). In summary, our data are compatible with a model of secreted dsRNA or siRNA trafficking from barley and wheat into B. graminis probably via an exosomal pathway and silencing of target genes inside the fungus.
The rate of ~20% of tested HIGS constructs that induced a reduction in fungal haustorium formation appears relatively high although the corresponding target genes had been preselected for a likely role during host infection based on their expression and, in many cases, upregulation in planta. One possible explanation for the observed efficiency of the HIGS screening procedure comes from the fact that plant multivesicular bodies/exosomes are also observed at sites of primary germ tube penetration, which occurs as early as 2 to 3 h after inoculation (Hückelhoven et al., 2000). This means that the time interval for the kicking-in of HIGS leading to a reduction of haustoria formation may be in the order of 12 to 18 h, which has been shown to be sufficient for transient-induced gene silencing of the Mlo gene in barley epidermal cells (Douchkov et al., 2005). Moreover, several of the effective HIGS constructs target in planta–expressed B. graminis genes with central housekeeping functions whose silencing is predicted to be detrimental to the organism.
HIGS of the Avrk1 and Avra10 effector genes caused a reduction in haustorium formation, which might be at odds with the previously proposed release of such effectors by mature haustoria (Panstruga and Dodds, 2009; Godfrey et al., 2010). Blumeria species form not only the appressorial but also a primary germ tube that has been shown to punch a hole into host cell walls very rapidly after landing on its surface and suggested to be important for water uptake and host recognition (Zhang et al., 2005). Recently, a conditioning by the primary germ tube of B. graminis of barley epidermal cells for enhanced susceptibility to subsequent appressorial penetration and haustorium establishment in the same host cell has been reported (Yamaoka et al., 2007). This indicates a very early release of effectors by the primary germ tube and suggests a role of effector proteins not only in maintenance of haustorium functionality and prevention of hypersensitive death of invaded cells, but also in the inhibition of very early defense responses, either at the prepenetration phase or immediately after appressorial penetration pegs have breached cell walls. Further support for this hypothesis comes from the absence of Avra10 and Avrk1 transcripts from isolated mature haustoria at 7 d after inoculation (Godfrey et al., 2010).
In summary, we propose HIGS as a novel tool to address gene function in obligate biotrophic powdery mildew fungi that also bears the potential of disease control by the use of transgenic plants that express antifungal RNAi constructs. HIGS might be used to control multiple diseases of a given crop because constructs can be designed such as containing multiple stacked RNAi target sequences. Alternatively, multiple transgenic events can be stacked by crossing of corresponding lines. No side effects of stacked events on host plants would be expected provided careful selection of target RNAi sequences without off-targets in the plant. We also propose to address in future research the question of naturally occurring siRNA or miRNA molecules trafficking between plants and fungal pathogens as mediators of disease or resistance.
METHODS
Plants and Fungi
The barley (Hordeum vulgare) line I mlo5 was grown in pots containing soil of the IPK nursery without fertilizers in the greenhouse at ~20°C and a photoperiod of 16 h by supplemental light from sodium halogen lamps. Barley cv Golden Promise (GP), Pallas, and Pallas backcross lines P09 (Mla10) and P17 (Mlk1) were grown in pots containing soil of the IPK nursery without fertilizers in a growth chamber (16 h of light from metal halogen lamps; 8 h of darkness; temperature 20°C; relative humidity 60% on average). The first leaves of 7-d-old GP or I mlo5 plants were used for the particle bombardments. The pathogens Blumeria graminis f. sp hordei (B. graminis, isolate CH4.8) and B. graminis f. sp tritici (B. graminis tritici, isolate FAP93151) were cultivated on susceptible barley and wheat (Triticum aestivum) plants, respectively, in separate climate chambers at 22°C, 70% relative humidity, and a photoperiod of 16 h. Six to seven days after inoculation, B. graminis or B. graminis tritici conidiospores were used for the inoculation of detached leaves or experimental plants by shaking diseased plants in an inoculation tower.
HIGS Single-Cell Assay
The RNAi constructs were designed using pIPKTA38 as entry vector and pIPKTA30N as Gateway destination vector, and the single-cell assay based on microprojectile bombardment was performed using first leaves of 7-d-old barley plants, as described (Douchkov et al., 2005) (see Supplemental Table 4 and Supplemental Figure 4 online). Approximately 400 to 500 bp of each inverted repeat sequence was contained by the final constructs. Barley leaf segments of GP and I mlo5 were inoculated with B. graminis 3 and 1 d after bombardment, respectively. Forty hours after inoculation, the leaves were stained for the β-glucuronidase (GUS) reporter gene and used for microscopy assessment of haustorium formation (see Supplemental Figure 1 online). The haustorial index of transformed cells expressing the cobombarded GUS reporter gene was calculated by dividing the number of counted haustoria by the number of blue-stained cells per bombardment.
RNAi Rescue
The RNAi rescue construct pIPKTA9_Avra10_wobble was made by inserting a synthetic gene of 885 bp (Eurofins MWG Operon) into the XbaI and XhoI sites of the transient expression vector pIPKTA9. The synthetic Avra10 cDNA was designed such as to be mutated on the third wobble position of each redundant codon by replacing the original B. graminis codon usage by the most or second most common barley codon. In silico testing by Si-Fi software of the synthetic gene for silencing by the Avra10 HIGS construct pIPKTA30_Avra10 did not reveal any 21mer oligonucleotide as potential target. Therefore, we assume this rescue construct to be fully immune against Avra10 silencing.
VIGS
cDNA Clones in pBluescript of a coat protein–deleted mutant of the BSMV isolate ND18 were used for the construction of recombinant versions of the γ-genome containing antisense sequences of target genes and for in vitro transcription of capped viral RNA, as described (Bruun-Rasmussen et al., 2007). Inserts for constructs pBSMV_BgGTF1(γ-T7) and pBSMV_BgGTF2(γ-T7) were PCR amplified using B. graminis tritici DNA and the EST-clone HO09P14 as template, respectively. See Supplemental Table 4 online for primer sequences. The fragments were introduced into the BamHI and XmaI sites of pBSMV_MCS(γ-T7)ND18 (Bruun-Rasmussen et al., 2007) in antisense orientation. Five-day-old wheat plants (cv Kanzler) were used for virus inoculation by rubbing the first leaf with 20 μL of 5% bentonite solution until dark green stripes appeared on the leaf. Six microliters of an equimolar mixture of the three different transcripts of the BSMV genome were then rubbed onto the wounded leaf, followed by spraying briefly with water and incubation in a growth chamber (16 h light at 25°C and 8 h darkness at 20°C, 60% relative humidity). Seventeen to nineteen days after BSMV inoculation, all leaves expect the first and second were harvested from plants showing BSMV symptoms. Leaf segments (~6 cm long) were placed onto 1% phytoagar (Duchefa) containing 0.002% benzimidazol. The leaf segments were inoculated with B. graminis tritici at a density of 10 to 15 conidia mm−2 and incubated for 40 h at 20°C in a climatized room with indirect daylight. Epiphytic fungal structures were stained with Coomassie Blue R 250 as described (Seiffert and Schweizer, 2005).
Transgenic Plants
The binary RNAi construct pIPKb007_BgGTF1 was generated using the RNAi vector pIPKb007 (Himmelbach et al., 2007; see Supplemental Figure 4 online). The LR reaction with entry clone pIPKTA38_BgGTF1 that contained 833 bp of BgGTF1 cDNA sequence was performed as described (Douchkov et al., 2005). Immature barley embryos (GP) were transformed with the binary RNAi vector described above using the Agrobacterium tumefaciens strain AGL1 as described (Hensel et al., 2008). The resulting plantlets were selected on medium containing hygromycin (50 mg L−1). Approximately 15 individual T1 plants per selected primary transgenic line were used for one inoculation experiment in 54-well multipot trays, together with GP wild-type plants. Transgenic lines were arranged in rows, with regularly interspersed wild-type control plants. Sixteen-day-old seedlings were inoculated with B. graminis by blowing conidiospores from four infected leaves into an inoculation tower. Disease was scored 7 d after inoculation, as described (Schweizer et al., 1995).
Fungal Transcript Analysis
Primary leaves of barley Ingrid BC mlo5 were cobombarded with a mixture of plasmids containing pU-Mlo (Shirasu et al., 1999), empty RNAi vector pIPKTA30N, or the HIGS construct pIPKTA30_Avra10, plus optionally pENTR-(ubi)-Mla10-(c) (Seeholzer et al., 2010). Twenty-four hours after bombardment, leaves were inoculated with B. graminis, and epiphytic fungal mycelium was wiped off by wet cotton pads prior to RNA extraction 24 h after inoculation. The RNA from leaves and fungal haustoria inside transformed epidermal cells with restored Mlo susceptibility was isolated with the RNeasy kit (Qiagen). Poly(A)+ RNA was isolated using magnetic Dynabeads (Invitrogen Dynal), DNase treated by DNA-free kit (Ambion), and reverse-transcribed by oligo(dT) and random priming into cDNA using the iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was performed in a volume of 15 μL using Maxima Probe qPCR Mastermix (Fermentas) and an ABI 7900HT fast real-time PCR system (Applied Biosystems). Forty cycles (15 s × 94°C, 30 s × 56°C, 30 s × 72°C, preceded by standard denaturation steps) were conducted. Data were analyzed by the ΔCt method using the SDS 2.2.1 software (Applied Biosystems). For this purpose, standard curves were included for each gene, with fivefold dilutions and three technical replicates per cDNA sample. The reverse PCR primer and TaqMan probe of Avra10 was placed outside the inverted repeat region of HIGS construct pIPKTA30_Avra10 to prevent amplification of dsRNA. This was confirmed by the absence of amplification products from RNA of leaf segments bombarded with the Avra10 HIGS construct without subsequent B. graminis inoculation (data not shown). For PCR primers and TaqMan probe sequences, see Supplemental Table 4 online.
Phylogenetic Analysis of GTF Proteins
Amino acid sequences of GTF1 and 2 together with other GTF proteins were aligned with ClustalX (Larkin et al., 2007) using the Slow-Accurate algorithm with the BLOSUM series protein weight matrix and a gap opening penalty of 10 together with a gap extension penalty of 0.1. Afterwards, the alignment was checked and manually adjusted. Phylogenetic analyses using (1) neighbor-joining clustering based on pairwise mean character differences and (2) maximum parsimony with the heuristic search algorithm and TBR branch swapping were conducted in PAUP* 4b10 (Swofford, 1993). Statistical support of branches was assessed by bootstrap analyses using 1000 resamples of the data matrix. As the outcomes of both analyses were nearly identical, only the unrooted neighbor-joining tree is shown. The protein sequence alignment used for tree construction is shown in Supplemental Data Set 1 online.
Statistical Analysis of HIGS Effects
Because the absolute susceptibility levels of empty vector or wild-type controls varied between independent experiments, effects of RNAi or antisense constructs were normalized to these internal controls (set to 100%) in each experiment. Deviations from hypothetical value 100 were tested by one-sample t test or Wilcoxon rank sum test, depending on normal distribution of data. All tests were performed two-sided. Unless otherwise specified, α was set to 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GTF1 of B. graminis f. sp hordei, EU646133; GTF1 of B. graminis f. sp tritici, FJ422119; GTF2 of B. graminis f. sp hordei, HQ234876; and synthetic gene Avra10_wobble, FJ422120.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Examples of Interaction Types in the HIGS Single-Cell Assay of Barley and in the VIGS Assay of Wheat.
Supplemental Figure 2. Regulation in Planta of Tested HIGS Target Genes of B. graminis.
Supplemental Figure 3. Time Course of Avra10 Transcript Levels in B. graminis.
Supplemental Figure 4. Constructs Used for HIGS in the Single-Cell Assay, by VIGS, and in Stable Transgenic Plants.
Supplemental Table 1. Results from the Initial HIGS-Based Screening of All 76 Tested Candidate Genes and from Repeated Experiments of Selected Candidates.
Supplemental Table 2. BLASTN-Based Linking of cDNA Clones Used in This Study to Clones Spotted onto a B. graminis cDNA Array (13), and Log2-Transformed Average Signal Intensities of Selected Transcripts.
Supplemental Table 3. Prediction of Avra10-Like Off-Target Transcripts in B. graminis
Supplemental Table 4. PCR Primers Used for Construct Generation and RT-PCR.
Supplemental Data Set 1. Fasta File of Alignment Used to Generate the Phylogenetic Tree in Figure 1.
Acknowledgments
The technical assistance of Sonja Gentz, Stefanie Lück, and Cornelia Marthe (Leibniz-Institute of Plant Genetics and Crop Plant Research [IPK]) is acknowledged as well as support of the VIGS work by Ingo Hein (Scottish Crop Research Institute). We thank Merete Albrechtsen (Aarhus University, Denmark) for supplying BSMV vectors, Tina Jordan (Zurich University) for the kind gift of the Mla10 expression plasmid, Frank Blattner IPK for calculating phylogenetic relationship of GTF proteins, and I. Schubert IPK for improving the manuscript. This work was supported by the Leibniz-Institute of Plant Genetics and Crop Plant Research (to P.S.) and by EU-FP6 BIOEXPLOIT (to C.R.).
References
- An Q.L., Ehlers K., Kogel K.H., van Bel A.J.E., Hückelhoven R. (2006). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172: 563–576 [DOI] [PubMed] [Google Scholar]
- Baum J.A., Bogaert T., Clinton W., Heck G.R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M., Vaughn T., Roberts J. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25: 1322–1326 [DOI] [PubMed] [Google Scholar]
- Both M., Csukai M., Stumpf M.P.H., Spanu P.D. (2005b). Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell 17: 2107–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both M., Eckert S.E., Csukai M., Müller E., Dimopoulos G., Spanu P.D. (2005a). Transcript profiles of Blumeria graminis development during infection reveal a cluster of genes that are potential virulence determinants. Mol. Plant Microbe Interact. 18: 125–133 [DOI] [PubMed] [Google Scholar]
- Bruun-Rasmussen M., Madsen C.T., Jessing S., Albrechtsen M. (2007). Stability of Barley stripe mosaic virus-induced gene silencing in barley. Mol. Plant Microbe Interact. 20: 1323–1331 [DOI] [PubMed] [Google Scholar]
- Caracuel Z., Martínez-Rocha A.L., Di Pietro A., Madrid M.P., Roncero M.I. (2005). Fusarium oxysporum gas1 encodes a putative β-1,3-glucanosyltransferase required for virulence on tomato plants. Mol. Plant Microbe Interact. 18: 1140–1147 [DOI] [PubMed] [Google Scholar]
- Dong W.B., Nowara D., Schweizer P. (2006). Protein polyubiquitination plays a role in basal host resistance of barley. Plant Cell 18: 3321–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D., Nowara D., Zierold U., Schweizer P. (2005). A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol. Plant Microbe Interact. 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O. (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Godfrey D., Böhlenius H., Pedersen C., Zhang Z., Emmersen J., Thordal-Christensen H. (2010). Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel G., Valkov V., Middlefell-Williams J., Kumlehn J. (2008). Efficient generation of transgenic barley: The way forward to modulate plant-microbe interactions. J. Plant Physiol. 165: 71–82 [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Zierold U., Hensel G., Riechen J., Douchkov D., Schweizer P., Kumlehn J. (2007). A set of modular binary vectors for transformation of cereals. Plant Physiol. 145: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippe S. (1985). Ultrastructure of haustoria of Erysiphe graminis f. sp hordei preserved by freeze substitution. Protoplasma 129: 52–61 [Google Scholar]
- Holzberg S., Brosio P., Gross C., Pogue G.P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Huang G.Z., Allen R., Davis E.L., Baum T.J., Hussey R.S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 103: 14302–14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R., Trujillo M., Kogel K.-H. (2000). Mutations in Ror1 and Ror2 genes cause modification of hydrogen peroxide accumulation in mlo-barley under attack from the powdery mildew fungus. Mol. Plant Pathol. 1: 287–292 [DOI] [PubMed] [Google Scholar]
- Künne C., Lange M., Funke T., Miehe H., Thiel T., Grosse I., Scholz U. (2005). CR-EST: A resource for crop ESTs. Nucleic Acids Res. 33 (Database issue): D619–D621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Mao Y.B., Cai W.J., Wang J.W., Hong G.J., Tao X.Y., Wang L.J., Huang Y.P., Chen X.Y. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25: 1307–1313 [DOI] [PubMed] [Google Scholar]
- Meyer D., Pajonk S., Micali C., O’Connell R., Schulze-Lefert P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Mouyna I., Morelle W., Vai M., Monod M., Léchenne B., Fontaine T., Beauvais A., Sarfati J., Prévost M.C., Henry C., Latgé J.P. (2005). Deletion of GEL2 encoding for a beta(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 56: 1675–1688 [DOI] [PubMed] [Google Scholar]
- Nickel W., Rabouille C. (2009). Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10: 148–155 [DOI] [PubMed] [Google Scholar]
- Panstruga R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6: 320–326 [DOI] [PubMed] [Google Scholar]
- Panstruga R., Dodds P.N. (2009). Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni E., Fontaine T., Gissi C., Latgè J.P., Popolo L. (2007). The Gas family of proteins of Saccharomyces cerevisiae: Characterization and evolutionary analysis. Yeast 24: 297–308 [DOI] [PubMed] [Google Scholar]
- Ridout C.J., Skamnioti P., Porritt O., Sacristan S., Jones J.D.G., Brown J.K.M. (2006). Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18: 2402–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristán S., Vigouroux M., Pedersen C., Skamnioti P., Thordal-Christensen H., Micali C., Brown J.K.M., Ridout C.J. (2009). Coevolution between a family of parasite virulence effectors and a class of LINE-1 retrotransposons. PLoS ONE 4: e7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Vogel J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5: 343–348 [DOI] [PubMed] [Google Scholar]
- Schweizer P., Pokorny J., Schulze-Lefert P., Dudler R. (2000). Technical advance. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 24: 895–903 [DOI] [PubMed] [Google Scholar]
- Schweizer P., Vallelian-Bindschedler L., Mosinger E. (1995). Heat-induced resistance in barley to the powdery mildew fungus Erysiphe graminis f.sp. hordei. Physiol. Mol. Plant Pathol. 47: 51–66 [Google Scholar]
- Scofield S.R., Huang L., Brandt A.S., Gill B.S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138: 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer S., Tsuchimatsu T., Jordan T., Bieri S., Pajonk S., Yang W.X., Jahoor A., Shimizu K.K., Keller B., Schulze-Lefert P. (2010). Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact. 23: 497–509 [DOI] [PubMed] [Google Scholar]
- Seiffert U., Schweizer P. (2005). A pattern recognition tool for quantitative analysis of in planta hyphal growth of powdery mildew fungi. Mol. Plant Microbe Interact. 18: 906–912 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shirasu K., Nielsen K., Piffanelli P., Oliver R., Schulze-Lefert P. (1999). Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17: 293–299 [Google Scholar]
- Sudarshana M.R., Roy G., Falk B.W. (2007). Methods for engineering resistance to plant viruses. Methods Mol. Biol. 354: 183–195 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. (1993). Paup - A computer program for phylogenetic inference using maximum parsimony. J. Gen. Physiol. 102: A9–A9 [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9: 654–659 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2005). Non-cell autonomous RNA silencing. FEBS Lett. 579: 5858–5871 [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Fusaro A.F. (2006). Plant science. Viruses face a double defense by plant small RNAs. Science 313: 54–55 [DOI] [PubMed] [Google Scholar]
- Westwood J.H., Roney J.K., Khatibi P.A., Stromberg V.K. (2009). RNA translocation between parasitic plants and their hosts. Pest Manag. Sci. 65: 533–539 [DOI] [PubMed] [Google Scholar]
- Yamaoka N., Ohta T., Danno N., Taniguchi S., Matsumoto I., Nishiguchi M. (2007). The role of primary germ tubes in the life cycle of Blumeria graminis: The primary germ tube is responsible for the suppression of resistance induction of a host plant cell. Physiol. Mol. Plant Pathol. 71: 184–191 [Google Scholar]
- Zhang Z., Henderson C., Perfect E., Carver T.L.W., Thomas B.J., Skamnioti P., Gurr S.J. (2005). Of genes and genomes, needles and haystacks: Blumeria graminis and functionality. Mol. Plant Pathol. 6: 561–575 [DOI] [PubMed] [Google Scholar]
- Zierold U., Scholz U., Schweizer P. (2005). Transcriptome analysis of mlo-mediated resistance in the epidermis of barley. Mol. Plant Pathol. 6: 139–151 [DOI] [PubMed] [Google Scholar]