This work identifies a semidominant allele of an Arabidopsis conserved receptor-like protein that activates plant immunity and whose signal is transduced through the transcription factor WRKY70.
Abstract
Plant immune receptors belonging to the receptor-like protein (RLP) family contain extracellular leucine-rich repeats (LRRs) and a short cytoplasmic tail linked by a single transmembrane motif. Here, we report the identification of snc2-1D (for suppressor of npr1-1, constitutive 2), a semidominant Arabidopsis thaliana mutant with constitutively activated defense responses. Map-based cloning of snc2-1D showed that it encodes an RLP. The point mutation in snc2-1D leads to substitution of the second Gly for Arg in the conserved GXXXG motif of the transmembrane helix, suggesting that this residue is important for negative regulation of the protein. Epistasis analysis revealed that the snc2-1D mutant phenotype is not affected by mutations in genes known to be required for the nucleotide binding (NB)-LRR Resistance (R) protein signaling. A suppressor screen of snc2-1D was performed, and map-based cloning of one suppressor revealed that mutations in WRKY70 suppress the constitutive defense responses in snc2-1D, suggesting that WRKY70 functions downstream of snc2-1D. The identification of snc2-1D provides us with a unique system for genetic analysis of resistance pathways downstream of RLPs, which may be distinct from those downstream of NB-LRR type R proteins.
INTRODUCTION
Plants have evolved sophisticated defense mechanisms to fend off a vast variety of microbial pathogens in nature. The plant immune system consists of pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (Jones and Dangl, 2006). Recognition of pathogen-associated molecular patterns is facilitated by surface receptors such as FLS2, EFR, and CERK1 (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006; Miya et al., 2007; Wan et al., 2008). By contrast, detection of pathogen effectors involves both intracellular and transmembrane receptors collectively called Resistance (R) proteins. Effector-triggered immunity is usually a stronger immune response compared with PTI, and it is often associated with the hypersensitive response (HR), a form of localized cell death at the site of infection that may help restrict the growth and spread of pathogens.
There are three major classes of R genes in plants (Dangl and Jones, 2001). The largest class encodes nucleotide binding-leucine-rich repeat (NB-LRR)–type R proteins with an NB domain and LRRs. The NB-LRR class of R proteins is predicted to be intracellular. The two remaining classes of R genes encode transmembrane receptor-like kinases (RLKs) and receptor-like proteins (RLPs). Unlike RLKs, RLPs do not have an intracellular kinase domain. The functional R proteins known in RLPs include tomato (Solanum lycopersicum) Cf and Ve proteins and the apple (Malus domestica) HcrVf2 protein, which confer resistance against the fungal pathogens Cladosporium fulvum, Verticillium species, and Venturia inaequalis, respectively (Jones et al., 1994; Kawchuk et al., 2001; Belfanti et al., 2004; Fradin et al., 2009). Interestingly, two RLPs in tomato, Eix1 and Eix2, also function as receptors of the fungal ethylene-inducing xylanase, which is a potent elicitor of plant defense responses (Ron and Avni, 2004).
Reverse genetic analysis of Avr9/Cf-9 Rapidly Elicited genes identified multiple components, including the protein kinase ACIK1, the U-box protein CMPG1, and F-box protein ACIF1 that are required for Cf-mediated HR or resistance against C. fulvum (Rowland et al., 2005; Gonzalez-Lamothe et al., 2006; van den Burg et al., 2008). Another interesting protein found to be required for Cf-mediated resistance responses is NRC1, an NB-LRR–type R protein. Silencing NRC1 in Nicotiana benthamiana compromises HR induced by Eix and multiple R proteins, including Cf-9, Pto, Rx, and Mi (Gabriels et al., 2007).
In Arabidopsis thaliana, there are 57 putative RLP genes (Wang et al., 2008). The functions of these proteins are largely unknown, with the exception of CLAVATA2 (CLV2) and TOO MANY MOUTHS (TMM). CLV2 regulates meristem and organ development (Jeong et al., 1999), whereas TMM regulates proper stomata distribution (Nadeau and Sack, 2002). Arabidopsis RLP52 was implicated in resistance against the powdery mildew pathogen Erysiphe cichoracearum (Ramonell et al., 2005), whereas Arabidopsis RLP30 was suggested to influence nonhost resistance toward Pseudomonas syringae pv phaseolicola (Wang et al., 2008). None of the Arabidopsis RLP proteins have been shown to function as R proteins.
In this study, we report the characterization of snc2-1D (for suppressor of npr1-1, constitutive 2), which was identified in a forward genetic screen to search for components functioning in the NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1)-independent resistance pathways. snc2-1D mutant plants accumulate high levels of endogenous salicylic acid (SA) and exhibit enhanced disease resistance against the virulent oomycete Hyaloperonospora arabidopsidis (H. a.) Noco2. Positional cloning revealed that snc2-1D encodes an RLP with a gain-of-function mutation in the conserved GXXXG motif in the transmembrane domain. A suppressor screen of snc2-1D further revealed that WRKY70 is required for snc2-1D–mediated resistance, suggesting that WRKY70 is an important component of RLP-mediated innate immunity.
RESULTS
Identification and Characterization of snc2-1D
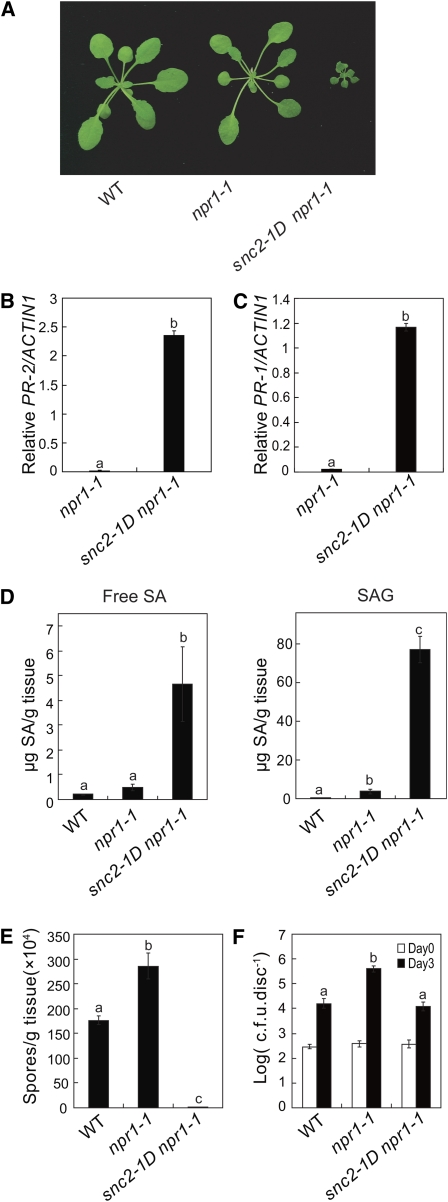
To search for defense signaling components independent of NPR1, we screened for suppressor mutations that caused the activation of defense gene expression in the absence of NPR1. To that end, seeds of npr1-1 were mutagenized with ethane methylsulfonate (EMS), and the M2 seedlings were screened for constitutive expression of a BGL2 (PR2)-GUS reporter gene, which is not expressed in the npr1-1 single mutants. One mutant with strong GUS staining displayed dwarf morphology similar to the previously identified snc1 (Li et al., 2001); therefore, we named the new mutation suppressor of npr1-1, constitutive 2 (snc2) (Figure 1A). When backcrossed with the wild type, the F1 plants exhibited an intermediate morphology, indicating that the mutation is semidominant. Thus, we named this mutant snc2-1D. Macroscopically, like snc1, snc2-1D plants do not exhibit spontaneous lesions. The dwarf morphology of snc2-1D can be partially suppressed by growing the plants at high temperature (see Supplemental Figure 1 online).
Figure 1.
Characterization of the snc2-1D Mutant.
(A) Phenotypes of the wild type, npr1-1, and snc2-1D npr1-1. Plants were grown on soil and photographed 4 weeks after planting.
(B) and (C) PR-2 (B) and PR-1 (C) expression in the wild type, npr1-1, and snc2-1D npr1-1 determined by quantitative RT-PCR. Values were normalized to the expression of ACTIN1. Statistical differences among the samples are labeled with different letters (P < 0.001).
(D) Free SA and SAG in wild-type, npr1-1, and snc2-1D npr1-1 plants. Statistical differences among the samples are labeled with different letters (P < 0.01).
(E) Growth of H. a. Noco2 on the wild type, npr1-1, and snc2-1D npr1-1. Three-week-old seedlings were sprayed with H. a. Noco2 spores (50,000 spores/mL). Infection was scored 7 d after inoculation by counting the number of spores per gram of leaf samples. Error bars represent sd from three measurements. Statistical differences among the samples are labeled with different letters (P < 0.01).
(F) Growth of P. s. t. DC3000 on the wild type, npr1-1, and snc2-1D npr1-1. Leaves of 5-week old plants were infiltrated with a bacterial suspension at OD600 = 0.0002. The values presented are averages of four replicates ± sd. Statistical differences among the samples are labeled with different letters (P < 0.01). c.f.u., colony-forming units.
[See online article for color version of this figure.]
Real-time RT-PCR analysis confirmed that the endogenous defense marker gene PR-2 (BGL2) was constitutively expressed in snc2-1D plants (Figure 1B). The expression level of PR-1 was also elevated, but not to the same level as PR-2 (Figure 1C). Salicylic acid (SA), a key signal molecule of plant defense, was found to accumulate more in snc2-1D npr1-1 plants (Figure 1D). To determine whether pathogen resistance was constitutively activated in snc2-1D npr1-1, the mutant plants were challenged with the virulent oomycete pathogen H. a. Noco2 and the bacterial pathogen Pseudomonas syringae pv tomato (P. s. t.) DC3000. As shown in Figure 1E, H. a. Noco2 growth on the snc2-1D npr1-1 plants is much lower than that on the wild-type plants. Growth of P. s. t. DC3000 on the snc2-1D npr1-1 plants was also lower than on npr1-1 plants (Figure 1F), indicating that snc2-1D confers enhanced pathogen resistance.
Epistasis Analysis
To determine whether snc2-1D activates resistance pathways shared by NB-LRR–type R proteins, snc2-1D npr1-1 was crossed with known defense signaling mutants, and triple mutants were obtained in the F2. These known mutants carry mutations in ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), PHYTOALEXIN DEFICIENT4 (PAD4), and NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1). EDS1 and PAD4 are required generally for resistance mediated by NB-LRR–type R proteins with an N-terminal Toll Interleukin I Receptor (TIR) domain (Glazebrook et al., 1996; Parker et al., 1996; Aarts et al., 1998), whereas NDR1 is required for NB-LRR–type R proteins with an N-terminal coiled coil domain (Century et al., 1995). As shown in Supplemental Figure 2A online, mutations in EDS1, PAD4, and NDR1 had little effect on the morphology of snc2-1D npr1-1. In addition, the mutations in EDS1, PAD4, and NDR1 had no obvious effects on the expression of PR-2 (see Supplemental Figure 2B online) or resistance to H. a. Noco2 (see Supplemental Figure 2C online), suggesting that snc2-1D activates resistance pathways distinct from those downstream of NB-LRR–type R proteins.
Since the snc2-1D mutation lead to the accumulation of high levels of SA, the requirement of SA for the phenotypes of snc2-1D npr1-1 was tested by making the snc2-1D eds5-3 npr1-1 triple mutant. EDS5 encodes a member of the MATE transporter family. Mutations in EDS5 abolish pathogen-induced SA accumulation in Arabidopsis (Nawrath et al., 2002). In snc2-1D eds5-3 npr1-1, SA accumulation was blocked (see Supplemental Figure 2D online). The eds5-3 mutation had little effect on the morphology (see Supplemental Figure 2A online) and resistance to H. a. Noco2 in snc2-1D npr1-1 (see Supplemental Figure 2C online), but the expression of PR-2 was modestly reduced in snc2-1D eds5-3 npr1-1 (see Supplemental Figure 2B online). This suggests that the contribution of the heightened SA levels to the constitutive defense phenotypes in snc2-1D is relatively small.
Map-Based Cloning of snc2-1D
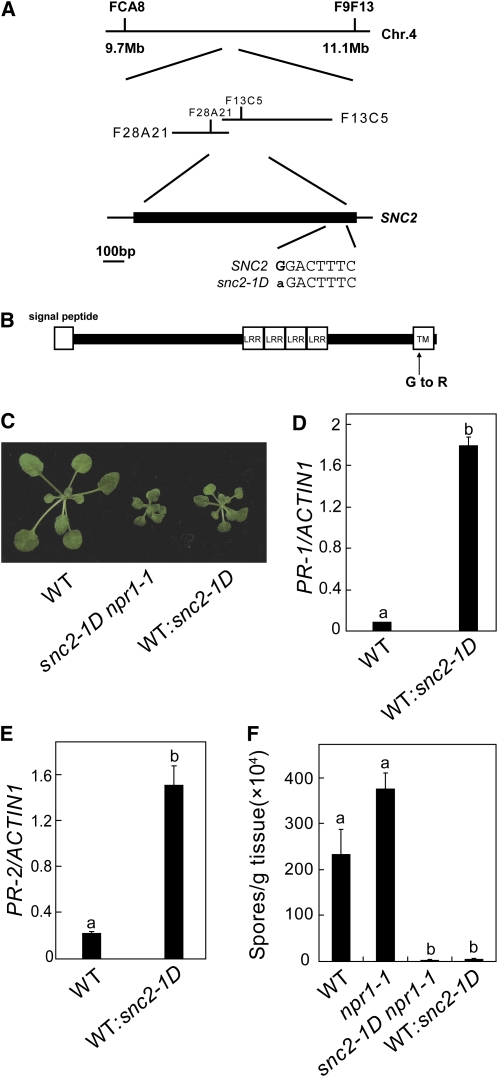
To map the snc2-1D mutation, snc2-1D (in Columbia [Col] background) was crossed with Landsberg erecta (Ler), and F2 plants with snc2-1D morphology were selected for linkage analysis. The snc2-1D mutation was initially flanked between FCA8 and F9F13 on chromosome 4. Further analysis of 1100 F2 plants flanked the mutation between F28A21 and F13C5, a region of ~47 kb (Figure 2A). The coding sequences in this region were amplified from snc2-1D genomic DNA by PCR and sequenced. Comparison of the sequences from snc2-1D with wild-type Arabidopsis genomic sequence revealed a single G-to-A mutation in At4g18760.
Figure 2.
Map-Based Cloning of snc2-1D.
(A) Map position and the mutation in snc2-1D.
(B) Predicted protein structure of SNC2. TM, transmembrane motif.
(C) Morphological phenotypes of 4-week-old wild-type and transgenic plants expressing the snc2-1D mutant gene.
(D) and (E) PR-1 (D) and PR-2 (E) expression in wild-type and transgenic plants expressing the snc2-1D mutant gene under its own promoter determined by quantitative RT-PCR. Values were normalized to the expression of ACTIN1. Statistical differences among the samples are labeled with different letters (P < 0.001).
(F) Growth of H. a. Noco2 on the wild type and transgenic plants expressing the snc2-1D mutant gene. The experiment was performed as in Figure 1E. Statistical differences among the samples are labeled with different letters (P < 0.01).
[See online article for color version of this figure.]
At4g18760 encodes a receptor-like protein, also named Receptor-like protein 51 (RLP 51). It contains a predicted N-terminal signal peptide, four extracellular LRRs, a single transmembrane domain, and a short cytoplasmic tail of four amino acids (Figure 2B). Compared with the Cf-9 class of R proteins previously characterized, the number of LRRs in RLP51 is rather small, and the domain between the signal peptide and the LRRs is atypically long. The transmembrane domain contains a GXXXG motif that is highly conserved in RLPs and many RLKs (Fritz-Laylin et al., 2005). The mutation in snc2-1D resulted in the substitution of the second Gly (Gly-412) in the GXXXG motif for Arg (Arg-412). To confirm that the mutation identified in snc2-1D is responsible for the constitutive defense phenotypes of the mutant, genomic clones containing the wild type or the mutant At4g18760 gene were constructed and transformed into wild-type plants. While transgenic plants with the wild-type transgene all displayed wild-type morphology, about half of the transgenic plants with the mutant transgene exhibited snc2-1D–like morphology (Figure 2C). Two representative lines with snc2-1D morphology were analyzed further. As shown in Figures 2D and 2E, PR-1 and PR-2 were constitutively expressed in both lines. When challenged with H. a. Noco2, the transgenic plants displayed enhanced resistance to H. a. Noco2 (Figure 2F). These results indicate that At4g18760 is SNC2, and the point mutation in snc1-2D is responsible for the autoimmunity phenotypes of the mutant.
SNC2 Is Localized to the Plasma Membrane
To determine the subcellular localization of SNC2, transgenic plants expressing SNC2 with N- or C-terminal green fluorescent protein (GFP) tags under its native promoter were generated. GFP fluorescence was not detected in these transgenic plants, suggesting that the SNC2 protein is expressed at very low levels. As shown in Supplemental Figure 3 online, wild-type plants expressing the snc2-1D-GFP fusion protein displayed snc2-1D–like morphology, suggesting that the fusion protein is functional. Subsequently, we transformed Arabidopsis mesophyll protoplasts with a construct expressing the SNC2-GFP fusion protein under the cauliflower mosaic virus 35S promoter. As shown in Supplemental Figure 4A online, the SNC2-GFP fusion protein was localized to the plasma membrane, suggesting that SNC2 is a plasma membrane protein as predicted. To examine whether the mutation in snc2-1D affects the protein localization, snc2-1D-GFP fusion protein was also expressed in mesophyll protoplasts under 35S promoter. As shown in Supplemental Figure 4A online, snc2-1D-GFP was localized to the plasma membrane, suggesting that the mutation in snc2-1D does not affect the localization of the protein. Membrane localization of SNC2 was further confirmed by fractionation analysis using transgenic plants expressing the SNC2-HA protein (see Supplemental Figure 4B online).
Mutations in the GXXXG Motif of SNC3 Constitutively Activate Defense Responses
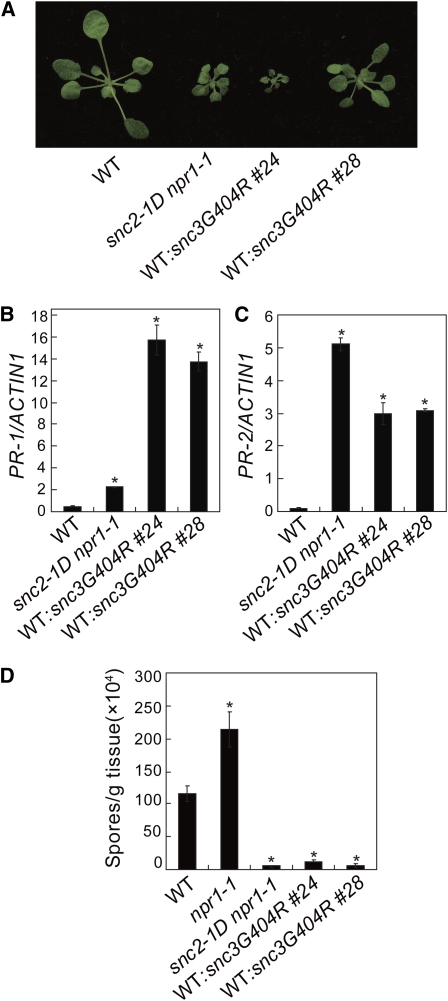
Unlike other R genes, SNC2 and its close homolog At5g45770 in Arabidopsis, which we named SNC3, are quite conserved among different plant species (Fritz-Laylin et al., 2005). SNC3 is 63% identical to SNC2 at amino acid level. Gly-404 in SNC3 is the second Gly in the GXXXG motif that corresponds to Gly-412 in SNC2. To test whether substituting the Gly-404 in SNC3 for Arg results in activation of defense responses, constructs expressing the wild-type SNC3-HA and snc3G404R-HA, both under its native promoter, were made. Upon transformation into wild-type plants, all transgenic plants carrying the SNC3 transgene displayed wild-type morphology. By contrast, about half of the transgenic plants with the snc3G404R mutant transgene exhibited snc2-1D–like morphology (Figure 3A). Analysis of two representative snc3G404R transgenic lines showed that expression of PR-1 and PR-2 is constitutively activated (Figures 3B and 3C). Higher PR-1 expression in the transgenic lines than that in snc2-1D npr1-1 is probably due to the presence of npr1-1 mutation in snc2-1D npr1-1. In addition, these transgenic lines displayed enhanced resistance to H. a. Noco2 (Figure 3D), suggesting that substituting Gly-404 for Arg in SNC3 constitutively activates defense responses.
Figure 3.
Characterization of Transgenic Plants Expressing the snc3G404R Mutant Gene in the Wild-Type Background under Its Own Promoter.
(A) Morphological phenotypes of 4-week-old wild-type and transgenic plants expressing the snc3G404R mutant gene.
(B) and (C) PR-1 (B) and PR-2 (C) expression in wild-type and transgenic plants expressing the snc3G404R mutant gene determined by quantitative RT-PCR. Values were normalized to the expression of ACTIN1. *P < 0.001, significant difference from the wild type.
(D) Growth of H. a. Noco2 on the wild-type and transgenic plants expressing the snc3G404R mutant gene. Error bars represent sd from three measurements. *P < 0.01, significant difference from the wild type.
[See online article for color version of this figure.]
We also tested whether similar mutations in TMM and CLV2 lead to snc2-1D–like phenotypes. We made transgenic plants expressing mutant TMM or CLV2 with the second G or S in the conserved (G/S/T)XXX(G/S/T) motif changed to R and found that all transgenic lines displayed wild-type morphology (see Supplemental Figure 5A online). Real-time RT-PCR analysis showed that TMM and CLV2 are overexpressed in the transgenic lines (see Supplemental Figures 5B and 5C online). These data suggest that the G/S to R mutations do not lead to gain of defense–related functions in these proteins that function in the regulation of plant development.
Identification of Intragenic Suppressors of snc2-1D
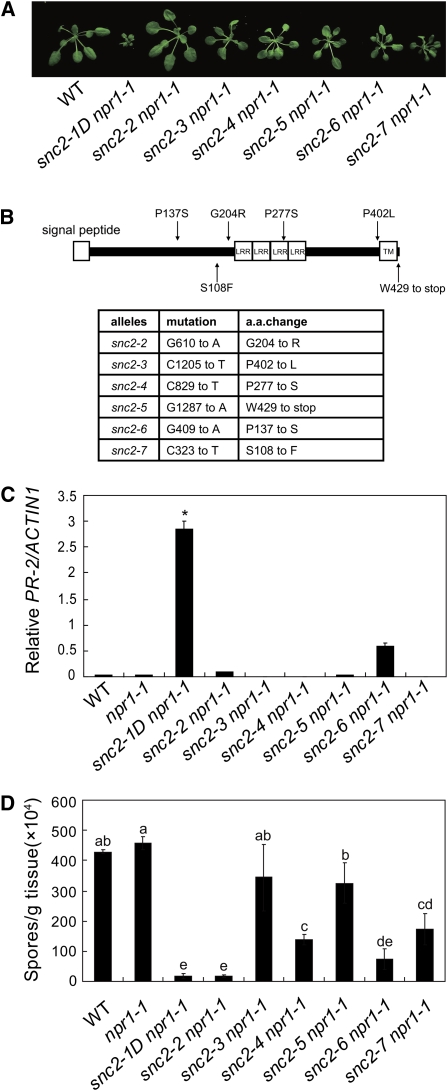
To identify intragenic suppressor mutations of snc2-1D and components functioning downstream of snc2-1D, snc2-1D npr1-1 seeds were mutagenized with EMS, and the M2 population was screened for mutants with wild-type morphology. Six intragenic suppressors with mutations in SNC2 were obtained from the screen (Figure 4A). Five of the mutations are missense mutations of amino acids conserved in SNC2, SNC3, and their homologs in other plants. Among the SNC2 mutations, three were located in the region between the signal peptide and the LRRs, one in the LRRs, and another in the region between the LRRs and the transmembrane helix (Figure 4B). The only nonsense mutation identified was located in the cytoplasmic extension and results in the loss of the last three amino acids of the protein, suggesting that the C-terminal cytoplasmic extension is important for the functions of SNC2. Real-time RT-PCR indicated that PR-2 expression was also suppressed in the snc2-1D revertants (Figure 4C). Further analysis showed that resistance to H. a. Noco2 was suppressed to different extent in the revertants except snc2-2 (Figure 4D).
Figure 4.
Analysis of Intragenic Suppressors of snc2-1D.
(A) Morphological phenotypes of the intragenic suppressors of snc2-1D npr1-1. Plants were grown on soil and photographed 4 weeks after planting. All mutants are in the npr1-1 background.
(B) Map of six intragenic SNC2 mutations.
(C) PR-2 expression in the intragenic suppressors of snc2-1D determined by quantitative RT-PCR. Values were normalized to the expression of ACTIN1. *P < 0.001, significant difference from all other genotypes.
(D) Growth of H. a. Noco2 on the intragenic suppressors of snc2-1D. Statistical differences among the samples are labeled with different letters (P < 0.05).
[See online article for color version of this figure.]
SNC2 Is Required for Basal Resistance against P. s. t. DC3000
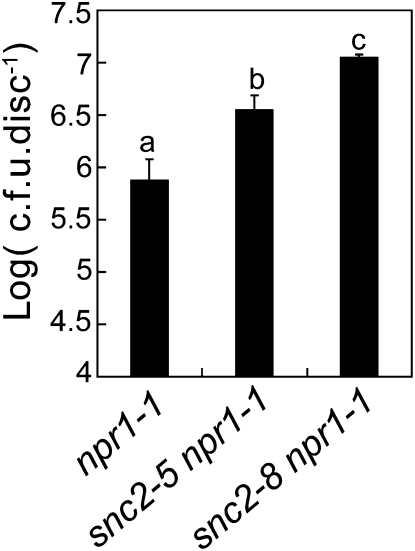
To test whether knocking out SNC2 affects resistance to pathogens, we obtained two T-DNA insertion lines, SALK_143038 and SAIL_740_C06, from the ABRC. We did not find the predicted T-DNA insertion in the SALK_143038 line. The T-DNA insertion in SAIL_740_C06 is located upstream of the 5′-untranslated region (UTR) of SNC2, and we found that the expression of SNC2 was not reduced in the homozygous line for SAIL_740_C06. To identify a knockout mutant for SNC2, we screened for additional intragenic suppressors of snc2-1D. One mutant, snc2-8, was found to revert snc2-1D to wild-type morphology and contain a mutation in SNC2 that changes Gln-52 to a stop codon. Since snc2-8 results in early termination of the protein, it is most likely a null allele of snc2. When spray inoculated with P. s. t. DC3000, snc2-8 npr1-1 supported ~10-fold more bacterial growth than npr1-1 plants (Figure 5). Analysis of another intragenic suppressor of snc2-1D, snc2-5, showed that bacterial growth in snc2-5 npr1-1 is also higher than in npr1-1. These data suggest that SNC2 is required for basal resistance against P. s. t. DC3000. As bacterial growth in snc2-5 npr1-1 is lower than that in snc2-8 npr1-1, the point mutation in snc2-5 probably results in only partial loss of the function of SNC2.
Figure 5.
Growth of P. s. t. DC3000 on npr1-1, snc2-5 npr1-1, and snc2-8 npr1-1.
Leaves were collected 2 d after spray inoculation with a bacterial suspension at OD600 = 0.2. The values presented are averages of five replicates ± sd. Statistical differences among the samples are labeled with different letters (P < 0.01). cfu, colony-forming units.
WRKY70 Functions Downstream of SNC2
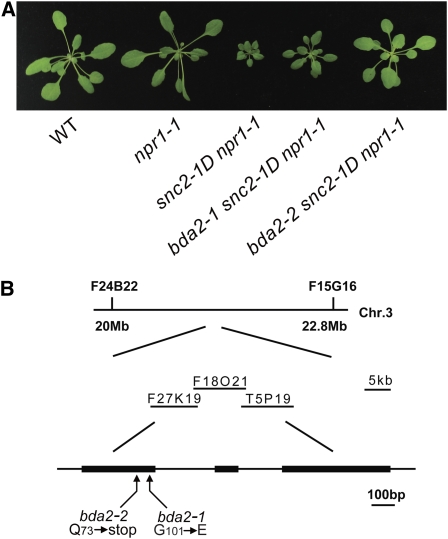
To map second-site suppressor mutants of snc2-1D npr1-1 that do not contain mutations in SNC2, we crossed each mutant with Ler to generate a mapping population. F2 plants homozygous for the snc2-1D mutation with wild-type morphology were selected for linkage analysis. Crude mapping showed that two allelic recessive mutants, bda2-1 (bian da 2-1; bian da means becoming big in Chinese) and bda2-2 (Figure 6A), were located on the lower arm of chromosome 3. Fine mapping of bda2-1 using roughly 2400 F2 plants positioned the mutation in a 220-kb region between marker F27K19 and T5P19. Sequence analysis of genes within this region identified a G-to-A mutation in WRKY70 (Figure 6B). This mutation changes Gly-101 in the protein to Glu-101. Sequence analysis of the WRKY70 locus in the bda2-2 identified a C-to-T mutation that creates an early stop codon in the cDNA, which probably results in complete loss of the function of the protein. These data suggest that mutations in WRKY70 were responsible for the suppression of snc2-1D mutant morphology. Since suppression of snc2-1D morphology by bda2-1 is not as complete as that by bda2-2, the point mutation in bda2-1 probably results in only partial loss of the function of WRKY70. As shown in Supplemental Figure 6 online, bda2-1 and bda2-2 did not affect the expression of SNC2.
Figure 6.
bda2-1 and bda2-2 Contain Mutations in WRKY70.
(A) Morphological phenotypes of 4-week-old bda2-1 snc2-1D npr1-1 and bda2-2 snc2-1D npr1-1.
(B) Map-based cloning of BDA2 (WRKY70).
[See online article for color version of this figure.]
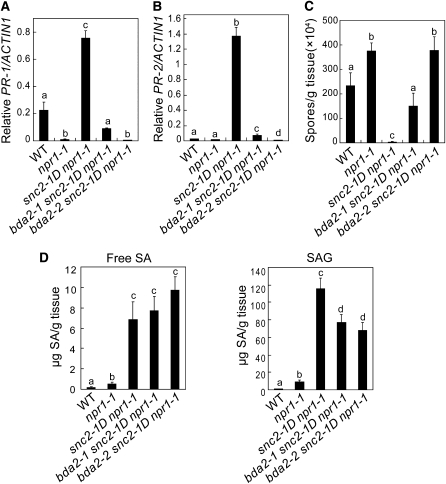
Further analysis of the bda2-1 snc2-1D npr1-1 and bda2-2 snc2-1D npr1-1 mutant plants showed that constitutive expression of PR-1 and PR-2 was reduced in bda2-1 snc2-1D npr1-1 and completely suppressed in bda2-2 snc2-1D npr1-1 (Figures 7A and 7B). In addition, enhanced resistance to H. a. Noco2 was attenuated in bda2-1 snc2-1D npr1-1 and completely suppressed in bda2-2 snc2-1D npr1-1 (Figure 7C), suggesting that snc2-1D activates WRKY70-mediated resistance pathways.
Figure 7.
WRKY70 Is Required for snc2-1D–Mediated Defense Responses.
(A) and (B) PR-1 (A) and PR-2 (B) expression in the wild type, snc2-1D npr1-1, bda2-1 snc2-1D npr1-1, and bda2-2 snc2-1D npr1-1 determined by quantitative RT-PCR. Statistical differences among the samples are labeled with different letters (P < 0.01).
(C) Growth of H. a. Noco2 on the wild type, snc2-1D npr1-1, bda2-1 snc2-1D npr1-1, and bda2-2 snc2-1D npr1-1. Error bars represent sd from three measurements. Statistical differences among the samples are labeled with different letters (P < 0.05).
(D) Free SA and SAG in the wild type, snc2-1D npr1-1, bda2-1 snc2-1D npr1-1, and bda2-2 snc2-1D npr1-1. Statistical differences among the samples are labeled with different letters (P < 0.01).
To test whether SA accumulation in snc2-1D npr1-1 is affected by the wrky70 mutations, SA levels in the bda2-1 snc2-1D npr1-1 and bda2-2 snc2-1D npr1-1 mutant plants were measured. As shown in Figure 7D, free SA levels in bda2-1 snc2-1D npr1-1 and bda2-2 snc2-1D npr1-1 are comparable to those in snc2-1D npr1-1, suggesting that WRKY70 functions in an SA-independent pathway downstream of snc2-1D. Interestingly, accumulation of salicylic acid glucoside (SAG) was modestly reduced in bda2-1 snc2-1D npr1-1 and bda2-2 snc2-1D npr1-1 compared to that in snc2-1D npr1-1, suggesting that WRKY70 may have a role in the regulation of conversion of SA to SAG.
DISCUSSION
Cf-like R genes have been scarcely studied in Arabidopsis. Here, we report the identification and characterization of snc2-1D, a semidominant mutant that exhibits constitutive activation of PR genes and pathogen resistance. The mutant phenotypes of snc2-1D are very similar to those previously reported in the snc1 mutant (Li et al., 2001). Unlike snc1, which is a TIR-NB-LRR R gene mutant (Zhang et al., 2003a), snc2-1D carries a mutation in a receptor-like protein. Since similar mutations in CLV2 and TMM do not result in the same phenotypes as snc2-1D, snc2-1D is probably not a neomorphic mutation. The identification of snc2-1D provides us with a unique system to perform genetic analysis of RLP-mediated immunity in Arabidopsis. It will be interesting to test whether SNC2 and Cf-9-like R proteins identified in other plant species use similar downstream components to activate defense responses.
Unlike the previously characterized RLP-type R proteins, which are generally not well conserved, phylogenetic analysis indicates that SNC2 is highly conserved in plants (Fritz-Laylin et al., 2005). What activates SNC2 remains to be determined. Since loss-of-function mutations in SNC2 cause enhanced susceptibility to P. s. t. DC3000, it is likely that SNC2 functions as a receptor for a conserved molecule from microbial pathogens. Detection of this molecule by SNC2 could activate downstream defense responses. Alternatively, SNC2 could be involved in guarding a conserved component of plant innate immunity. Alteration of this component by microbial pathogens would trigger activation of SNC2.
The conserved GXXXG motif is found in the transmembrane domains of many proteins where it facilitates protein–protein interactions in the plasma membrane (Senes et al., 2004). The mutation in snc2-1D changes the second Gly in the GXXXG motif to Arg. Site-directed mutagenesis of SNC3 showed that changing the corresponding Gly to Arg in SNC3 also results in activation of defense responses, suggesting that this residue may play important roles in preventing autoactivation of these RLPs. Activation of the SNC2 and SNC3 through the Gly to Arg mutations might be the result of disrupting their interaction with an unidentified negative regulator that associates with SNC2 or SNC3 through the GXXXG motif. Interestingly, a similar mutation in Cf-9 (G825R) abolished the function of Cf-9 (Wulff et al., 2004). Whether this mutation causes alteration of the structure or stability of Cf-9 is unknown. It is possible that the GXXXG motif in different RLPs may function through interacting with different proteins.
Epistasis analysis showed that snc2-1D–mediated defense responses do not require NDR1, EDS1, or PAD4, suggesting that it activates resistance pathways distinct from the NB-LRR–type R proteins. Like snc1, snc2-1D also accumulates high levels of SA. Blocking the SA synthesis through the loss of EDS5 function partially blocks the expression of the defense marker gene PR-2 but has limited effects on the snc2-1D mutant morphology, suggesting that snc2-1D activates both SA-dependent and SA-independent resistance pathways.
In a suppressor screen of snc2-1D, we found that the snc2-1D mutant phenotypes were largely suppressed by mutations in WRKY70, indicating that this WRKY transcription factor plays a crucial role in RLP-mediated immunity. In Arabidopsis, there are more than 70 WRKY transcription factors, and many are induced during plant defense (Eulgem and Somssich, 2007). It has been suggested that WRKY transcription factors play important roles in the regulation of plant immune responses (Maleck et al., 2000). Using reverse genetics, a number of WRKY transcription factors have been identified as critical regulators of plant defense responses (Journot-Catalino et al., 2006; Wang et al., 2006; Xu et al., 2006; Zheng et al., 2006; Shen et al., 2007; Kim et al., 2008). However, WRKY transcription factors have rarely been identified in forward genetic screens to search for regulators of plant immunity. One possibility to explain this rarity is that individual WRKY transcription factors may have specialized functions in regulating expression of a subset of defense genes and the mutant phenotypes of these WRKY transcription factors can only be identified in sensitized mutant backgrounds.
Previous studies have shown that WRKY70 plays complex roles in modulating defense responses and senescence (Li et al., 2004; Knoth et al., 2007; Ulker et al., 2007). Based on epistasis analysis, WRKY70 was suggested to function downstream of NPR1 in an SA-dependent signal pathway where it activates SA-inducible PR gene expression and represses jasmonate-inducible PDF1.2 expression. WRKY70 was proposed to be a determinant of the balance between SA and jasmonate signaling (Li et al., 2004). In addition, knocking out WRKY70 causes modest reduction of resistance to H. a. Emoy2 mediated by the TIR-NB-LRR–type R protein, RPP4 (Knoth et al., 2007). Our data suggest that WRKY70 functions independent of SA and NPR1 in the regulation of snc2-1D–mediated defense responses.
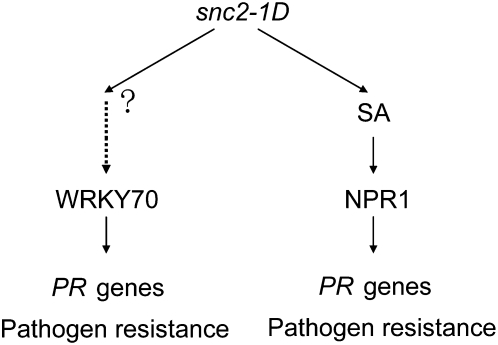
A working model is proposed in Figure 8. Accumulation of high levels of SA in snc2-1D suggests that the SA-dependent resistance pathway is activated in the mutant. Since blocking SA synthesis in snc2-1D does not completely block PR gene expression and pathogen resistance, snc2-1D also activates an SA-independent defense pathway. WRKY70 probably functions as a critical regulator of the SA-independent pathway because it blocks the defense responses but not SA synthesis in snc2-1D npr1-1 plants. It remains to be determined how the signal is transduced from the plasma membrane–localized SNC2 to nucleus to activate WRKY70. Previous studies have shown that recognition of Avr4 by Cf-4 or Avr9 by Cf-9 leads to rapid activation of downstream mitogen-activated protein kinases in tomato and tobacco (Romeis et al., 1999; Stulemeijer et al., 2007). Studies of RLP TMM in stomata development also suggest that a mitogen-activated protein kinase cascade links the surface receptors to its downstream target SPEECHLESS, a transcription factor required for stomata formation. Future research will reveal whether there is also a mitogen-activated protein kinase cascade functioning downstream of snc2-1D to regulate WRKY70-mediated activation of defense responses.
Figure 8.
A Working Model for snc2-1D–Induced Resistance.
The snc2-1D mutation constitutively activates both SA-dependent and SA-independent resistance pathways downstream. SA-dependent resistance pathway requires NPR1. The SA-independent resistance responses are mediated by WRKY70.
METHODS
Mutant Screening and Characterization
The snc2-1D npr1-1 mutant was identified from an EMS-mutagenized npr1-1 mutant population by GUS staining (Gao et al., 2008). To identify the suppressor mutants of snc2-1D, the snc2-1D npr1-1 mutant seeds were treated with EMS. About 40,000 M2 plants representing ~2000 M1 families were grown on soil and screened for loss of the snc2-1D morphological phenotype. All plants were grown under 16 h light at 23°C and 8 h dark at 20°C.
SA was extracted from 0.1 g of leaf tissue using a previously described protocol and measured by HPLC (Li et al., 1999). To analyze resistance to H. a. Noco2, 2-week-old seedlings were sprayed with H. a. Noco2 spores at a concentration of 50,000 spores per mL of water. Plants were grown for another 7 d at 18°C in 12-h-light/12-h-dark cycles with 95% humidity before infections were scored by counting the number of spores with a hemocytometer. P. s. t. DC3000 infections were performed by infiltrating or spraying leaves of 5-week-old plants grown at 22°C under 12-h-light/12-h-dark cycles with bacterial suspensions at different concentrations.
Quantitative Real-Time RT-PCR
For gene expression analysis, RNA was extracted from the 2-week-old seedlings grown on half Murashige and Skoog medium using the RNAiso reagent (Takara). Reverse transcription was performed using the M-MLV reverse transcriptase (Takara). Quantitative real-time RT-PCR was performed using the SYBR Premix Ex Taq II kit (Takara). PCR reactions were performed in triplicate with three independent RNA samples. Statistical analyses were performed with one-way analysis of variance by StatsDirect statistical software (StatsDirect). Primers used for amplification of PR-1, PR-2, and Actin1 were described previously (Zhang et al., 2003b).
Epistasis Analysis
The pad4-1, ndr1-1, and eds5-3 mutants used for epistasis were described previously (Century et al., 1995; Glazebrook et al., 1996; Nawrath and Metraux, 1999). The eds1-2 mutant (Parker et al., 1996) backcrossed into Col background was kindly provided by Jane Parker. To generate various mutant combinations, the snc2-1D npr1-1 plants were crossed with pad4-1, eds1-2, ndr1-1, and eds5-3. Triple mutant plants were identified in the F2 progeny by PCR (Li et al., 2001). To check for npr1-1 homozygosity, mutant lines were plated on Murashige and Skoog plates with 0.2 mM SA because npr1-1 mutants bleach to death on high concentrations of SA.
Map-Based Cloning
The markers used for mapping were designed based on the Monsanto Arabidopsis thaliana polymorphism and Ler sequence collections (Jander et al., 2002). For mapping the snc2-1D and bda2-1 mutations, the phenotypes of the recombinants were determined by assessing the segregation of snc2-1D morphological phenotype in their progeny. Primer sequences of markers used for mapping are shown in Supplemental Table 1 online.
To test whether the mutation in At4g18760 is responsible for the mutant phenotypes in the snc2-1D mutant, a 3.3-kb genomic fragment of At4g18760 (SNC2) containing its own promoter but without the stop codon and 3′-UTR was amplified from snc2-1D or wild-type genomic DNA using primers 5′-CGGGGTACCGATCATCATGTTCATCGACC-3′ and 5′-CGCGGATCCACCACACCATTTGGCGAGAAG-3′ and cloned into modified pCAMBIA1305 vectors to obtain pCAMBIA1305-snc2-1D (or SNC2)-HA and pCAMBIA1305-snc2-1D (or SNC2)-GFP to express the wild type or mutant snc2-1D fusion proteins under its own promoter. The resulting plasmids were used to transform wild-type plants by the floral dip method (Clough and Bent, 1998).
Mutation Analysis of SNC3
A genomic fragment of At5g45770 (SNC3) without the stop codon and 3′-UTR was amplified from wild-type genomic DNA using primers 5′-CCGGAATTCTAGAGGTGATGGAATTGTC-3′ and 5′-CGCGGATCCTCAAATCAAACGACACCTTTTAG-3′and cloned into a modified pCAMBIA1305 vector to obtain pCAMBIA1305-SNC3-HA to express the wild-type SNC3 fusion protein under its own promoter. The snc3G404R mutation was introduced into pCAMBIA1305-SNC3-HA by site-directed mutagenesis using a PCR-based linker scanning method (Li and Shapiro, 1993). The resulting plasmids were transformed into wild-type plants using the floral dip method (Clough and Bent, 1998).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g18760 (SNC2), At5g45770 (SNC3), AT3G56400 (WRKY70), At2g14610 (PR1), At3g57260 (PR2), and At2g37620 (Actin1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphological Phenotypes of Wild Type, npr1-1, and snc2-1D npr1-1 Plants Grown on Soil at 23 or 28°C.
Supplemental Figure 2. Analysis of snc2-1D pad4-1 npr1-1, snc2-1D eds1-2 npr1-1, snc2-1D ndr1-1 npr1-1, and snc2-1D eds5-3 npr1-1 Mutant Plants.
Supplemental Figure 3. Morphological Phenotypes of Wild-Type, npr1-1, snc2-1D npr1-1, and Transgenic Plants Expressing the snc2-1D-GFP Fusion Protein under Its Own Promoter in the Wild-Type Background.
Supplemental Figure 4. SNC2-GFP and snc2-1D-GFP Are Localized to the Plasma Membrane.
Supplemental Figure 5. Analysis of Transgenic Plants Expressing clv2S697R or tmmG475R under the 35S Promoter in the Wild-Type Background.
Supplemental Figure 6. SNC2 Expression in npr1-1, snc2-1D npr1-1, bda2-1 snc2-1D npr1-1, and bda2-2 snc2-1D npr1-1 Determined by Quantitative RT-PCR.
Supplemental Table 1. Markers Used for Map-Based Cloning.
Acknowledgments
We thank Dongling Bi, Yan Huang, and Yi-Ti Cheng for their help with confocal microscopy and SA measurement. We also thank Youjun Zhang, Shaoning Chen, and Pei Wen for their help on the mutant screening and mapping. Jane Parker is thanked for seeds of eds1-2 in Col. We are grateful for financial support from the Chinese Ministry of Science and Technology to Y.Z.
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfanti E., Silfverberg-Dilworth E., Tartarini S., Patocchi A., Barbieri M., Zhu J., Vinatzer B.A., Gianfranceschi L., Gessler C., Sansavini S. (2004). The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc. Natl. Acad. Sci. USA 101: 886–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K.S., Holub E.B., Staskawicz B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Fradin E.F., Zhang Z., Juarez Ayala J.C., Castroverde C.D., Nazar R.N., Robb J., Liu C.M., Thomma B.P. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin L.K., Krishnamurthy N., Tör M., Sjölander K.V., Jones J.D. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls S.H., Vossen J.H., Ekengren S.K., van Ooijen G., Abd-El-Haliem A.M., van den Berg G.C., Rainey D.Y., Martin G.B., Takken F.L., de Wit P.J., Joosten M.H. (2007). An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 50: 14–28 [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R., Tsitsigiannis D.I., Ludwig A.A., Panicot M., Shirasu K., Jones J.D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.A., Thomas C.M., Hammond-Kosack K.E., Balint-Kurti P.J., Jones J.D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I.E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk L.M., Hachey J., Lynch D.R., Kulcsar F., van Rooijen G., Waterer D.R., Robertson A., Kokko E., Byers R., Howard R.J., Fischer R., Prufer D. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth C., Ringler J., Dangl J.L., Eulgem T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 20: 120–128 [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Clarke J.D., Zhang Y., Dong X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Y., Clarke J.D., Li Y., Dong X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339 [DOI] [PubMed] [Google Scholar]
- Li X.M., Shapiro L.J. (1993). Three-step PCR mutagenesis for ‘linker scanning’. Nucleic Acids Res. 21: 3745–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K.A., Dangl J.L., Dietrich R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Métraux J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Métraux J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.E., Holub E.B., Frost L.N., Falk A., Gunn N.D., Daniels M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell K., Berrocal-Lobo M., Koh S., Wan J., Edwards H., Stacey G., Somerville S. (2005). Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 138: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Piedras P., Zhang S., Klessig D.F., Hirt H., Jones J.D. (1999). Rapid Avr9- and Cf-9 -dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O., Ludwig A.A., Merrick C.J., Baillieul F., Tracy F.E., Durrant W.E., Fritz-Laylin L., Nekrasov V., Sjölander K., Yoshioka H., Jones J.D. (2005). Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17: 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senes A., Engel D.E., DeGrado W.F. (2004). Folding of helical membrane proteins: The role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14: 465–479 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Stulemeijer I.J., Stratmann J.W., Joosten M.H. (2007). Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 144: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B., Shahid Mukhtar M., Somssich I.E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Tsitsigiannis D.I., Rowland O., Lo J., Rallapalli G., Maclean D., Takken F.L., Jones J.D. (2008). The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B.B., Thomas C.M., Parniske M., Jones J.D. (2004). Genetic variation at the tomato Cf-4/Cf-9 locus induced by EMS mutagenesis and intralocus recombination. Genetics 167: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X. (2003a). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M.J., Lassner M., Li X. (2003b). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]