This study demonstrates that auxin and cytokinin regulate each other's biosynthesis, providing an intrinsic mechanism for optimizing the relative intracellular concentrations of both hormones.
Abstract
Together, auxin and cytokinin regulate many of the processes that are critical to plant growth, development, and environmental responsiveness. We have previously shown that exogenous auxin regulates cytokinin biosynthesis in Arabidopsis thaliana. In this work, we show that, conversely, the application or induced ectopic biosynthesis of cytokinin leads to a rapid increase in auxin biosynthesis in young, developing root and shoot tissues. We also show that reducing endogenous cytokinin levels, either through the induction of CYTOKININ OXIDASE expression or the mutation of one or more of the cytokinin biosynthetic ISOPENTENYLTRANSFERASE genes leads to a reduction in auxin biosynthesis. Cytokinin modifies the abundance of transcripts for several putative auxin biosynthetic genes, suggesting a direct induction of auxin biosynthesis by cytokinin. Our data indicate that cytokinin is essential, not only to maintain basal levels of auxin biosynthesis in developing root and shoot tissues but also for the dynamic regulation of auxin biosynthesis in response to changing developmental or environmental conditions. In combination with our previous work, the data suggest that a homeostatic feedback regulatory loop involving both auxin and cytokinin signaling acts to maintain appropriate auxin and cytokinin concentrations in developing root and shoot tissues.
INTRODUCTION
The relationship between auxin and cytokinin has long been recognized as central to normal plant growth and development (Coenen and Lomax, 1997; Swarup et al., 2002; Rashotte et al., 2005). Fundamental multicellular plant processes, such as embryogenesis (Müller and Sheen, 2008), meristem development and maintenance (Hartig and Beck, 2006; Sablowski 2007; Moubayidin et al., 2009), shoot branching (Ongaro and Leyser, 2008; Shimizu-Sato et al., 2009), and lateral root initiation and development (Casimiro et al., 2003; De Smet et al., 2006; Osmont et al., 2007), rely on both hormones. The mechanisms involved in auxin–cytokinin interactions are not yet fully understood, but discoveries during the last few years have been adding pieces to the puzzle. Both hormones act as growth regulators, both can be biosynthesized in many, if not all, parts of the plant, and both can act as long-distance signaling substances and as paracrine signals during development (Baker, 2000; Ljung et al., 2001, 2005; Nordström et al., 2004; Sakakibara, 2006; Robert and Friml, 2009).
A reliance on both hormones suggests an interrelationship between the auxin and cytokinin response pathways. In fact, auxin and cytokinin interact on many levels. For example, strong evidence has emerged over recent years that they interact at the level of hormone perception and signal transduction (Müller and Sheen, 2008; Moubayidin et al., 2009). It has also been shown that polar auxin transport (PAT) can be regulated via cytokinin modulation of the activity of auxin efflux carriers (Dello Ioio et al., 2008; Ruzicka et al., 2009). At the metabolic level, both auxin and cytokinin are able to modify the expression of the key cytokinin biosynthesis ISOPENTENYLTRANSFERASE (IPT) genes (Miyawaki et al., 2004). Direct cytokinin biosynthesis measurements in Arabidopsis thaliana have shown that auxin can rapidly downregulate cytokinin biosynthesis (Nordström et al., 2004). These authors were the first to demonstrate that cytokinins are biosynthesised in both aerial and root tissues and that young, developing leaves have the highest cytokinin biosynthetic capacity. Tanaka et al. (2006) subsequently showed a similar phenomenon exists in pea (Pisum sativum), in which apical dominance is maintained at least partially by auxin-induced downregulation of cytokinin biosynthesis in the stem.
Interactions at the metabolic level have also been reported between auxin and ethylene (Ruzicka et al., 2007; Stepanova et al., 2007, 2008; Swarup et al., 2007) and auxin and brassinosteroids (Mouchel et al., 2006). In this article, we present evidence that auxin biosynthesis rates are modulated by cytokinin. In experiments combining inducible cytokinin overproduction and exogenous cytokinin application with stable isotope labeling, we show that an elevation of cytokinin levels leads to a rapid increase in auxin biosynthesis rates in young, developing Arabidopsis tissues. We also show that, conversely, a reduction in cytokinin levels leads to lower rates of endogenous auxin biosynthesis, suggesting that cytokinin is an essential regulator of auxin biosynthesis.
Compared with other classic plant hormones, comparatively little is known about the molecular basis of auxin biosynthesis. In part, this may be because of the difficulties arising from the existence of multiple auxin biosynthesis pathways and the high level of redundancy between these pathways (Woodward and Bartel, 2005; Normanly, 2010). Taken together, our data indicate that the regulation of auxin biosynthesis by cytokinin is an important homeostatic control mechanism in developing root and shoot tissues. In combination with previous data that shows that auxin downregulates cytokinin levels (Miyawaki et al., 2004; Nordström et al., 2004; Tanaka et al., 2006; Shimizu-Sato et al., 2009), the data presented here suggest that cellular concentrations of auxin and cytokinin are regulated via an active homeostatic feedback loop, mediated through both auxin and cytokinin signal transduction mechanisms.
RESULTS
Cytokinin Induces de Novo Auxin Biosynthesis in Arabidopsis
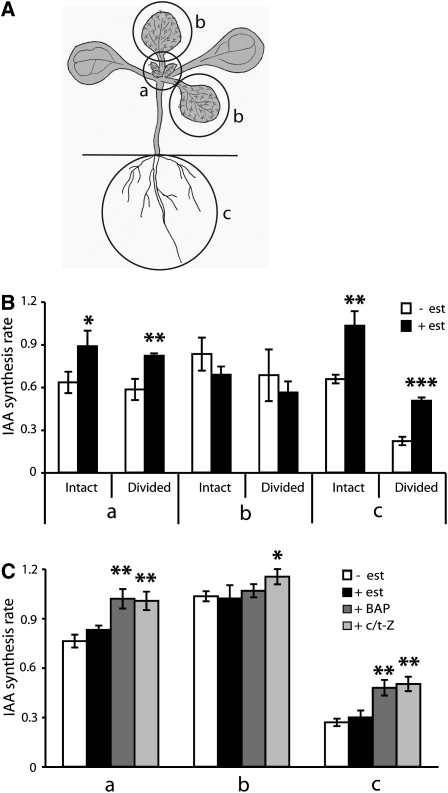
We have previously shown that IAA is synthesized at a high rate in the Arabidopsis root apex (Ljung et al., 2005; Petersson et al., 2009). When we screened different classes of plant hormones for their effects on IAA biosynthesis in the root, we observed that increasing cytokinin levels via the estradiol induction of IPT8 in the transgenic line IPT8/plant growth activator 22 (pga22) (Sun et al., 2003) caused a rapid increase in the rate of indole-3-acetic acid (IAA) biosynthesis and in the levels of IAA. To test if there were tissue-specific differences in the ability of cytokinin to alter the rate of IAA biosynthesis, we grew IPT8/pga22 seedlings on vertical plates in long days (LDs; 16 h light/8 h dark cycle) for 10 d and took measurements of separate tissues. Intact seedlings or seedlings divided into root and shoot were incubated for 12 h in a medium containing 17-β-estradiol to induce IPT8 and cytokinin biosynthesis and then for 24 h in a medium containing 17-β-estradiol and 30% 2H2O (deuterated water). After incubation, the seedlings were divided into (a) apex and young, developing leaves (leaf 3 and smaller), (b) the first two true leaves, and (c) the root system (Figure 1A). De novo IAA biosynthesis was analyzed in these tissues by gas chromatography–multiple reaction monitoring–mass spectrometry (GC-MRM-MS) (Ljung et al., 2005). A significantly higher rate of IAA biosynthesis was observed in the apex and young leaves and in the root system after the estradiol induction of IPT8, both in intact seedlings and in seedlings divided into root and shoot before incubation (Figure 1B, a and c). Interestingly, in contrast with younger tissues, we did not observe an increase in the rate of IAA biosynthesis in older leaves after the estradiol treatment (Figure 1B, b), indicating that elevated cytokinin levels increase the rate of IAA biosynthesis only in young, developing tissues. This may explain the findings of Nordström et al. (2004) who measured the effects of cytokinin on IAA biosynthesis in Arabidopsis on a whole plant basis. In their experiments, an increase in cytokinin levels via the induction of a bacterial IPT transgene led to a reduction in the IAA pool size and in the rate of IAA biosynthesis at 36 to 48 h of induction. The mass of older tissues in these 3-week-old plants may have masked an increase in the rate of IAA biosynthesis in young, developing tissues. It is also possible that longer term developmental and/or homeostatic effects would have predominated at the time scale used in their experiments, rather than the direct effects that can be seen with shorter treatments used in our experiments.
Figure 1.
Induction of IAA Biosynthesis by Cytokinin.
(A) The de novo IAA biosynthesis rate was analyzed in specific tissues after incubation of 10 DAG Arabidopsis seedlings in a medium containing 30% 2H2O. Seedlings were divided into (a) apex and young developing leaves (leaf 3 and smaller), (b) the first two true leaves, and (c) the root system after incubation.
(B) IPT8/pga22 seedlings (10 DAG; either intact seedlings or seedlings dissected into root and shoot before incubation) were incubated first for 12 h in a medium containing 5 μM 17-β-estradiol and then for 24 h in a medium containing 30% 2H2O and 5 μM 17-β-estradiol (+ est). As a control, seedlings were incubated without estradiol in the medium (− est). The IAA biosynthesis rate was analyzed in the different tissues described in (A).
(C) Wild-type Ws seedlings 10 DAG were divided into root and shoot and incubated for 24 h in liquid medium containing 30% 2H2O alone or with 5 μM 17-β-estradiol, 10 μM BAP, or 10 μM c/t-Z as indicated. The IAA biosynthesis rate was analyzed in the different tissues described in (A).
Data are mean ± sd of three independent experiments (n = 3). P values in (B) (+ est versus − est) and (C) (+ est, + BAP, or + c/t-Z versus − est) were determined by two-tailed Student’s t test assuming equal variances (* P < 0.05, ** P < 0.01, and *** P < 0.001).
To test our observations, we incubated wild-type seedlings with two different exogenous cytokinins, 6-benzylamino purine (BAP) and zeatin. Ten-day-old wild-type seedlings were divided into shoot and root and incubated for 24 h in a liquid medium containing 30% 2H2O and BAP or cis/trans-zeatin (c/t-Z). We observed a significant increase in the rate of IAA biosynthesis in the shoot apex and young leaves and in the root system (Figure 1C, a and c). No increase in IAA biosynthesis was observed in older leaves after the BAP treatment. However, there was an increase after the c/t-Z treatment (Figure 1C, b). This difference suggests that very high concentrations of exogenous zeatin (two to three orders of magnitude higher than the t-zeatin (t-Z) levels observed in older leaves in the IPT8/pga22 induced seedlings; see Supplemental Table 1 online) are able to overcome the relative insensitivity of older leaves to cytokinins that we observed in the IPT8/pga22-inducible line and in the BAP treatment. Mock treatments with 17-β-estradiol in wild-type plants did not induce IAA biosynthesis in either young or old tissues (Figure 1C).
Direct measurements of cytokinin levels in the IPT8/pga22 line before and after the estradiol treatment showed that the induction of the IPT8 transgene led to a strong induction of iP and Z-type cytokinins in all tissues investigated (see Supplemental Table 1 online). In general, before the estradiol treatment, there was a very low concentration of the free bases, iP and Z, with the highest levels occurring in apex and young, developing leaves. After the induction of the IPT8 transgene, there was a rapid increase in the cytokinin precursors, iPMP, iPA, t-ZMP, and t-ZR, and the free bases iP and t-Z. Interestingly, the induction of iP and its precursors (iPA and iPMP) was stronger in the shoot than in the root system. The opposite was observed for t-Z and its precursors (t-ZR and t-ZMP) and conjugates (Z7G, Z9G, and ZOG) that were more abundant in the root system than in the shoot. This suggests that the hydroxylation mechanisms that convert iP and its precursors to zeatin-type cytokinins (Sakakibara, 2006) are more active in the root than in the shoot. This is consistent with the expression pattern of the genes involved in the process, the cytochrome P450 monoxygenase genes CYP735A1 and CYP735A2, which are more highly expressed in roots than in other organs (Takei et al., 2004). We also observed that the induction of IPT8 led to very high levels of the iP precursor, iPMP, in all tissues. This is in contrast with previous findings in transgenic Arabidopsis plants carrying the dexamethasone-inducible Agrobacterium tumefaciens IPT gene. In this line, only zeatin-type cytokinins were induced (Åstot et al., 2000).
Cytokinin Induction of Auxin Biosynthesis Is Mediated by Cytokinin and Auxin Signal Transduction
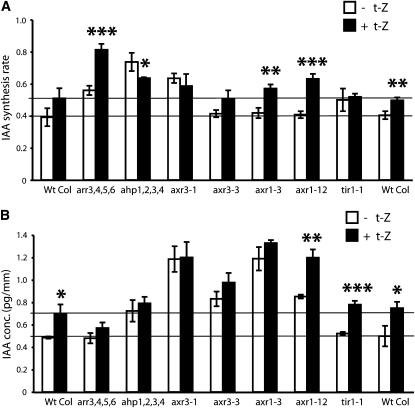
The rapid upregulation of auxin biosynthesis by cytokinin has not been observed previously. Our data indicate that these interactions occur at the metabolic level and are tissue specific. To investigate the mechanisms underlying this cytokinin–auxin interaction, we analyzed root tip–specific cytokinin induction of auxin biosynthesis in different auxin and cytokinin signal perception and signal transduction mutants. Excised roots from 6-d-old wild-type and mutant Arabidopsis seedlings were incubated in media containing 30% 2H2O with or without 5 μM t-Z for 24 h. The IAA concentrations and IAA biosynthetic rates were subsequently measured by GC-MRM-MS in 2-mm-long root apex sections. Two cytokinin signaling mutants were investigated, the cytokinin-hypersensitive ARABIDOPSIS RESPONSE REGULATOR (ARR) quadruple mutant arr3 arr4 arr5 arr6 and the cytokinin-insensitive quadruple ARABIDOPSIS HISTIDINE-PHOSPHOTRANSFER PROTEIN (AHP) mutant ahp1 ahp2 ahp3 ahp4. The arr3 arr4 arr5 arr6 line has mutations in four of the 10 type A ARR genes that are believed to function as negative regulators of cytokinin signaling (To and Kieber, 2008). Mutations in these genes result in plants that are more sensitive to cytokinin. Higher order ARR mutants such as this line have been shown to be considerably cytokinin hypersensitive, and several of the type A ARR genes (including ARR3, ARR4, ARR5, and ARR6) are induced by cytokinin (To et al., 2004). The IAA biosynthetic rate was higher in the arr3 arr4 arr5 arr6 mutant than in the wild type prior to the cytokinin treatment, and the effect of the treatment was exaggerated in the mutant (Figure 2A). Despite the increased biosynthetic rate, we did not observe a significant difference between the steady state IAA concentrations in the root tips of wild-type and arr3 arr4 arr5 arr6 plants (Figure 2B).
Figure 2.
IAA Biosynthesis in Auxin and Cytokinin Signal Transduction Mutants.
De novo IAA biosynthesis rate (A) and IAA concentration (B) were analyzed in root tips of auxin and cytokinin signal transduction mutants after treatment of 6 DAG seedling roots for 24 h with liquid medium containing 30% 2H2O with or without 5 μM t-Z. Data are mean ± sd of three independent experiments (n = 3). P values (+ t-Z versus – t-Z) were determined by two-tailed Student’s t test assuming equal variances (* P < 0.05, ** P < 0.01, and *** P < 0.001).
AHP gene products are believed to be positive elements in cytokinin signal transduction, mediating signaling between the plasma membrane–bound cytokinin receptors and the response regulators within the nucleus (Hutchison et al., 2006). The ahp1 ahp2 ahp3 ahp4 line has mutations in four of the five AHP genes. Surprisingly, similarly to the cytokinin-hypersensitive arr3 arr4 arr5 arr6 line, the cytokinin-insensitive ahp1 ahp2 ahp3 ahp4 line had a higher rate of IAA biosynthesis in the roots than did the wild type prior to cytokinin treatment (Figure 2A). In contrast with both the wild-type and arr3 arr4 arr5 arr6 plants, however, cytokinin treatment led to a significant decrease in the IAA biosynthesis rate in this line, suggesting that the relationship between cytokinin and auxin biosynthesis is fundamentally altered (Figure 2A).
We also analyzed the effect of cytokinin on the rate of IAA biosynthesis and steady state IAA concentrations in the auxin perception and signal transduction mutants axr3-1, axr3-3, axr1-3, axr1-12, and tir1 (Figures 2A and 2B). The axr3-1 and axr3-3 mutants have been described as being hypersensitive to auxin, with the axr3-1 mutant showing the more severe phenotype of the two (Leyser et al., 1996). AXR3 is identical to IAA17, a member of the auxin-inducible AUX/IAA gene family of auxin response transcriptional regulators (Rouse et al., 1998). axr3-1 and axr3-3 are both semidominant gain-of-function mutants that increase the stability of the AXR3/IAA17 protein (Chapman and Estelle, 2009). Leyser et al. (1996) reported that axr3 plants show striking differences in phenotypic responses to exogenous cytokinin compared with wild-type plants (e.g., in root elongation and adventitious rooting). Prior to cytokinin treatment, we observed that steady state IAA levels were higher in the root tips of the axr3-1 and axr3-3 mutants compared with wild-type plants (Figure 2B). The axr3-1 mutant also had a higher rate of IAA biosynthesis than the wild-type plants (Figure 2A). Interestingly, the axr3-1 mutant did not respond to exogenous cytokinin. These results suggest that low levels of AXR3/IAA17 are essential for the maintenance of wild-type basal auxin levels and that cytokinin induction of auxin biosynthesis is the result of increased AXR3/IAA17 expression or stabilization of its encoded protein.
We next analyzed affect of cytokinin induction on auxin biosynthesis and levels in the auxin-insensitive, axr1-3, axr1-12, and tir1-1 loss-of-function mutant lines. The axr1 lines were discovered in a screen for auxin-resistant mutants (Lincoln et al., 1990). AXR1 is part of the RUB-activating enzyme required for normal SCF activity and auxin responsiveness (del Pozo et al., 2002; Dharmasiri et al., 2007). The axr1-12 mutant has a more severe phenotype and a stronger resistance to exogenous IAA than does axr1-3. The pretreatment rate of IAA biosynthesis was unchanged from the wild type in both the axr1-3 and axr1-12 lines. However, there was an increased response to cytokinin in both mutant lines compared with the wild type (Figure 2A). There was also a considerable increase in steady state IAA levels in the root tips of the mutants compared with the wild type before and after the cytokinin treatment, particularly in the less severe axr1-3 (Figure 2B). TIR1 codes for an auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). There were no significant differences in IAA levels and biosynthesis rates between the wild type and tir1-1 before or after the cytokinin treatment (Figures 2A and 2B). As a whole, the data from these mutants indicate that both cytokinin and auxin signal transduction mechanisms are implicated in the cytokinin regulation of the level and biosynthesis rate of IAA in the root apex.
Downregulation of Cytokinin Levels Leads to a Decrease in Auxin Biosynthesis Rates
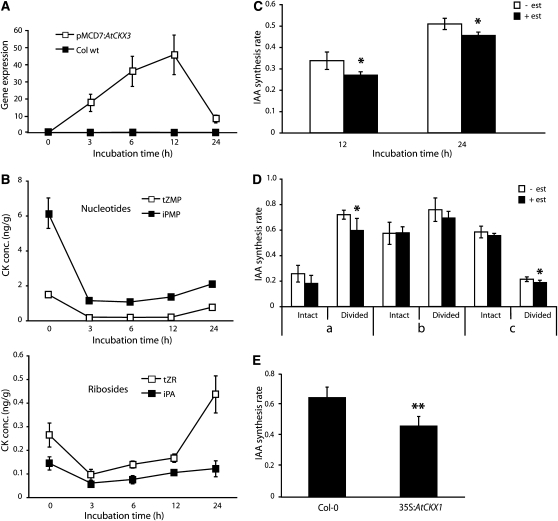
Having observed that elevated levels of cytokinin lead to an increase in auxin biosynthesis, we next asked whether endogenous basal levels of cytokinin play a role in maintaining the metabolic rate of auxin biosynthesis. We constructed an Arabidopsis line carrying an inducible cytokinin oxidase (pMDC7:AtCKX3) gene. Six days after germination (DAG), pMDC7:AtCKX3 and wild-type seedlings were incubated in liquid medium containing 17-β-estradiol, and CKX3 transcript levels were analyzed after 0, 3, 6, 12, and 24 h of incubation. We observed a rapid induction of CKX3 expression in pMDC7:AtCKX3 seedlings, with maximum transcript levels occurring after 12 h of induction (Figure 3A). This coincided with a decrease in cytokinin levels after induction (see Supplemental Table 2 online). Endogenous cytokinin precursor levels were lower after 3 h of induction, and they remained low for 12 h, after which the levels started to rise again (Figure 3B). The concentration of the active cytokinin, iP, was halved between 6 and 24 h of CKX3 induction (see Supplemental Table 2 online). In 6 DAG pMDC7:AtCKX3 seedlings, we observed a significant downregulation of IAA biosynthesis rates 12 and 24 h after the onset of incubation (Figure 3C). Tissue-specific measurements of IAA biosynthesis rates in 10 DAG pMDC7:AtCKX3 seedlings showed that induction of CKX3 led to a significant downregulation of IAA biosynthesis rates in the shoot apex, in young developing leaves, and in the root system of dissected seedlings (Figure 3D). When we analyzed root tip–specific IAA biosynthesis in estradiol-induced pMDC7:AtCKX3 seedlings, we observed a small, but not significant, downregulation of IAA synthesis rates (see Supplemental Figure 1 online). To study if constitutive overexpression of cytokinin oxidase was more effective in downregulating IAA biosynthesis, we used the 35S:AtCKX1 line, which has a CKX1 transgene under the control of the constitutive cauliflower mosaic virus 35S promoter. The levels of cytokinin precursors, active cytokinins, and IAA have already been shown to be reduced in this line (Werner et al., 2003). In the root tips of this line, we observed a significant downregulation of IAA biosynthesis compared with the wild type (Figure 3E), indicating that the constitutive downregulation of cytokinin leads to lower IAA synthesis rates.
Figure 3.
Lowering of Cytokinin Levels Leads to a Downregulation of IAA Biosynthesis.
(A) Analysis of CKX3 transcript levels by qPCR after incubation of 6 DAG Arabidopsis wild-type Columbia and pMDC7:AtCKX3 seedlings with 5 μM 17-β-estradiol (est) for 0, 3, 6, 12, and 24 h. Error bars indicate sd (n = 3).
(B) Quantification of the nucleotide (tZMP and iPMP) and riboside (tZR and iPA) cytokinin precursors after incubation of 6 DAG pMDC7:AtCKX3 seedlings with 5 μM 17-β-estradiol for 0, 3, 6, 12, and 24 h. Error bars indicate sd (n = 3).
(C) IAA biosynthesis rate in 6 DAG pMDC7:AtCKX3 seedlings incubated with medium containing 30% 2H2O with or without 5 μM 17-β-estradiol for 12 and 24 h. Error bars indicate sd (n = 3).
(D) pMDC7:AtCKX3 seedlings 10 DAG were divided into root and shoot and incubated for 12 h in liquid medium containing 30% 2H2O with or without 5 μM 17-β-estradiol. The IAA biosynthesis rate was analyzed in the different tissues described in Figure 1A. Error bars indicate sd (n = 5).
(E) Col-0 and 35S:AtCKX1 seedling roots 6 DAG were incubated for 24 h in liquid medium containing 30% 2H2O. The IAA biosynthesis rate was analyzed in root tips. Error bars indicate sd (n = 5).
Data are mean ± sd of independent experiments. P values in (C) to (E) (+ est versus – est) were determined by two-tailed Student’s t test assuming equal variances (* P < 0.05, ** P < 0.01, and *** P < 0.001).
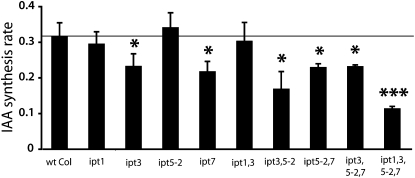
In a second experiment, we analyzed root-specific IAA biosynthesis in different lines carrying mutations in genes encoding IPT proteins. The Arabidopsis genes IPT1, IPT3, IPT5, and IPT7 are all expressed in different cell types of the root tip and are expected to be important in the biosynthesis of cytokinin in those cells (Miyawaki et al., 2004). IPT1 is expressed in xylem precursor cells in the procambium, IPT3 in the vasculature (phloem and pericycle), IPT5 in the columella root cap and in lateral root primordial, and IPT7 in the endodermis of the elongation zone. Arabidopsis wild-type and ipt mutant lines were grown for 6 d in LDs and divided into roots and shoots. Isolated roots were then incubated for 24 h in a medium containing 30% 2H2O. We observed a downregulation of IAA biosynthesis in the root apex of the ipt3 and ipt7 single mutant lines and in the double and triple mutant lines ipt3 ipt5-2, ipt5-2 ipt7, and ipt3 ipt5-2 ipt7 (Figure 4). The quadruple mutant line ipt1 ipt3 ipt5-2 ipt7 showed the strongest reduction in the rate of IAA biosynthesis. These results provide further indication that the downregulation of endogenous cytokinin levels leads to lower auxin biosynthesis rates and that cytokinin is an essential regulator of auxin homeostasis in Arabidopsis.
Figure 4.
IAA Biosynthesis Is Downregulated in the Root Apex of ipt Mutant Lines.
The de novo IAA biosynthesis rate was analyzed in root tips of ipt mutant lines after incubation of 6 DAG seedling roots for 24 h with liquid medium containing 30% 2H2O. Data are mean ± sd of three independent experiments (n = 3). P values for the mutant lines versus the wild type were determined by two-tailed Student’s t test assuming equal variances (* P < 0.05, ** P < 0.01, and *** P < 0.001).
Cell Type–Specific Analysis of Auxin Biosynthesis in the Root Apex after Cytokinin Treatment
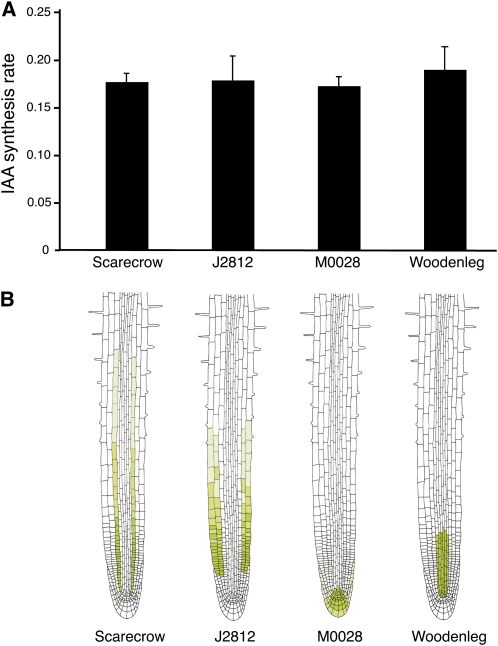
We then analyzed the rates of IAA biosynthesis in Arabidopsis lines expressing green fluorescent protein (GFP) in different root cell types to determine if cytokinin affected auxin biosynthesis more in some cell types than in others. Four different GFP-expressing cell lines were chosen so that all cell types in the root apex were sampled (Figure 5B). We have previously shown that the cell types represented in these lines have a high relative IAA synthesis rate compared with the rest of the root (Petersson et al., 2009). Seedlings were grown on vertical plates as described (Petersson et al., 2009), and intact roots were harvested and incubated for 16 h in a liquid medium containing 30% 2H2O, 10 μM t-Z, and 10 μM naphthylphthalamic acid (NPA; to block transport of newly biosynthesized IAA between cells). Protoplast isolation, cell sorting, and IAA biosynthesis measurements were performed as described in Methods. There were no significant differences in IAA biosynthesis rates between the four lines, indicating that all responded similarly in terms of the response to an exogenous cytokinin treatment (Figure 5A).
Figure 5.
IAA Biosynthesis in Specific Cell Types after Cytokinin Treatment.
(A) Intact roots from 8 DAG Arabidopsis seedlings expressing GFP in specific cell types of the root apex were incubated for 16 h in liquid medium (30% 2H2O, 30 mM sucrose, 4.4 g MS, and 2.6 mM MES) containing 10 μM NPA and 10 μM t-Z. IAA biosynthesis rates were analyzed in isolated cells from the roots after enzymatic protoplasting and fluorescence-activated cell sorting. A general increase of IAA biosynthesis was observed in all cell types examined, but there were no significant differences between the four GFP-expressing lines. The data were processed in a single factor analysis of variance, which gave a P value of 0.38. Data are mean ± sd, and the number of biological and technical replicates was between 2 and 4, and 1 and 3, respectively.
(B) GFP expression pattern in the Arabidopsis lines used in this study.
Transcript Profiling of the Root Apex after Cytokinin Induction of Auxin Biosynthesis
A global transcript profiling of IPT8/pga22 root tips, comparing untreated with estradiol-induced or cytokinin treated seedlings, was conducted to investigate further the mechanisms behind cytokinin regulation of auxin biosynthesis. The profiling was performed on root tip material as used in our previous IAA measurements (Figures 2, 3E, 4, and 5). A time-course experiment was first conducted to identify any differences in the timing of changes in IAA biosynthesis between the estradiol-induced IPT8/pga22 plants and exogenous t-Z–treated plants. Whole roots of 7 DAG IPT8/pga22 seedlings were incubated in a medium containing 30% 2H2O and either 5 μM 17-β-estradiol or 5 μM t-Z for 3, 12, and 24 h, and the rate of IAA biosynthesis was analyzed by GC-MRM-MS in the root apex. A significant increase in the rate of IAA biosynthesis was observed in the t-Z–treated roots at 3 h (see Supplemental Figure 2 online). By contrast, the estradiol-induced roots did not show a significant increase in the rate of IAA biosynthesis until 24 h after incubation. Based on this time course, transcript profiling was performed on IPT8/pga22 seedlings that were first grown for 7 d in LDs and then incubated for 12 and 24 h with medium containing either 0 or 5 μM 17-β-estradiol or 0 or 5 μM t-Z. After incubation, root tips were collected from the seedlings and transcript profiling was performed using the ATH1 Affymetrix array.
As expected, there was a very strong upregulation of IPT8 in the IPT8/pga22 line after estradiol treatment but not after the t-Z treatment (see Supplemental Data Set 1 online). This and the upregulation of a number of genes known to be involved in cytokinin signal transduction, including AHK4/WOL and several cytokinin response regulator genes (ARRs), indicated that the treatments were effective. There was also a strong induction of the cytokinin oxidases CKX3, CKX4, and CKX5, suggesting that cytokinin homeostasis is autoregulatory in root tips. Given our previous observations, it was not surprising that the data indicated that several of the genes believed to be involved in IAA metabolism were also affected. The data showed that transcript levels for the Trp biosynthetic genes ASA1/WEI2, PAT/TRP1, and IGPS were upregulated, as were genes believed to be involved in Trp-dependent IAA biosynthesis, such as CYP79B2, CYP79B3, YUCCA6, and NIT3. The data also indicated that transcripts for some putative auxin biosynthesis genes, such as AMI1, YUCCA5, and a YUCCA5-like gene, were downregulated by the treatments (see Supplemental Data Set 1 online). Increased transcript levels were indicated for the IAA conjugating (GH3.17 and GH3.9) and deconjugating (ILL6) enzymes, suggesting that a range of IAA homeostatic mechanisms are induced by an increase in cytokinin. The data also indicated upregulation of two genes, CYP83B1/SUR2 and SUR1, that are believed to be involved in indole glucosinolate (IG) biosynthesis. Interestingly, a MYB transcription factor (ATR1/MYB34) that has been shown to coregulate several genes in the IAA and IG biosynthetic pathways, including CYP79B2, CYP79B3, and CYP83B1/SUR2 (Celenza et al., 2005), was also induced by cytokinin.
The data also indicated that cytokinin regulates the level of transcripts for several genes known to be involved in auxin signal transduction (ABP1, AXR1, IAA13, IAA17/AXR3, IAA29, IAA30, IAA32, and IAA33) and IAA transport (PIN2, PIN5, PIN6, and PIN7), indicating a multilayered interaction between the two hormones. Gene Ontology (GO) analysis was performed for genes with a >1.5-fold change between treated tissues and control untreated tissues using the GO browser function in Genespring. A range of GO terms was significantly overrepresented (ranked by P value), including cytokinin-mediated signaling pathway, response to cytokinin stimulus, and glucosinolate metabolic process (see Supplemental Data Set 2 online).
Analyses of Auxin-Related Genes by Quantitative PCR
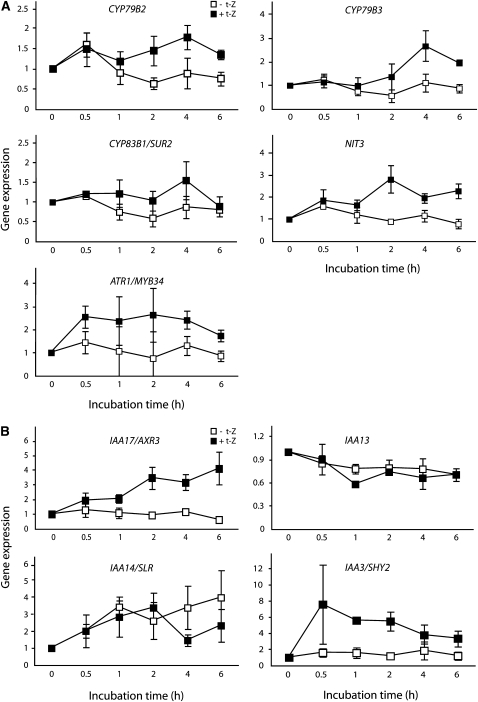
Genes related to auxin biosynthesis, transport, and signaling indicated in the microarray screen as being responsive to cytokinin were analyzed by quantitative PCR (qPCR). To identify genes involved in the primary response to cytokinin, samples were taken earlier after the onset of the treatment than with the microarrays. This shorter time course indicated a rapid response to cytokinin for a number of auxin biosynthesis–related genes. Root tips from 7 DAG wild-type Arabidopsis roots were sampled at 0, 0.5, 1, 2, 4, and 6 h after incubation in liquid medium with and without t-Z. CYP735A2 was used as a positive control, as transcript levels for this gene have previously been shown to increase rapidly in response to elevated cytokinin levels (Takei et al., 2004; see Supplemental Figure 3C online). Transcript levels for the auxin biosynthesis–related genes CYP79B2, CYP79B3, CYP83B1/SUR2, and NIT3 were elevated by 2 h after the onset of the cytokinin treatment (Figure 6A). The transcript level of ATR1/MYB34 was induced by cytokinin treatment even at the earliest sampling, 0.5 h after the onset of the treatment. This supports previous findings (Celenza et al., 2005) and suggests a potential mechanism for the coregulation of genes in the IAA and IG biosynthesis pathways. We also analyzed several other genes believed to be involved in IAA biosynthesis (ASA1/WEI2, PAT1/TRP1, IGPS, TSA1, TAA1, AMI1, YUCCA3, YUCCA5, YUCCA6, YUCCA7, YUCCA8, YUCCA9, NIT1, NIT2, NIT4, and AAO1 and the TAA1-like genes At4g33070, At5g54960, At5g01320, and At5g01330) (see Supplemental Figures 3A and 3B online). Members of the YUCCA and TAA1-like gene families were selected for qPCR analysis based on the microarray results and whether or not they were previously shown to be expressed in the roots (Genevestigator). Transcript levels for some genes increased after t-Z treatment (IGPS, NIT1, and NIT4), while others were reduced (TAA1, YUCCA3, YUCCA5, YUCCA9, and most of the TAA1-like genes).
Figure 6.
qPCR Analysis of Auxin-Related Genes.
Wild-type Arabidopsis (Columbia) seedlings were grown in LD for 7 DAG. The seedlings were then incubated for 0.5, 1, 2, 4, or 6 h in liquid medium with or without 5 μM t-Z. The root tips were harvested and genes related to auxin biosynthesis (A) or auxin signaling (B) were analyzed by qPCR. Error bars indicate se (n = 3).
A rapid increase in transcript abundance was also observed for AXR3/IAA17 (Figure 6B). Transcript levels for IAA13, another Aux/IAA indicated by the microarray to be significantly upregulated by cytokinin, did not show this early response (Figure 6B). Transcript abundance for IAA14/SLR was downregulated after 4 h. IAA14/SLR is involved in lateral root development and has been reported to function as a negative regulator of the DNA binding auxin response element ARF7/NPH4 (Fukaki et al., 2005). There was no observable early response in ARF7/NPH4, TIR1, or IAA30 transcript levels (see Supplemental Figure 3C online). IAA3/SHY2 and ARR1 have recently been shown to play a role in root development (Dello Ioio et al., 2008; Moubayidin et al., 2009). We observed a strong and rapid increase in IAA3/SHY2 transcript levels in response to cytokinin (Figure 6B), but no early response in the ARR1 transcript levels (see Supplemental Figure 3C online), although the array results indicated that this gene is downregulated at later time points (see Supplemental Data Set 1 online). Finally, we analyzed the transcript levels for three genes believed to be involved in auxin transport that were indicated by the microarray results to be cytokinin responsive. One of these, PIN2, was transiently upregulated, and another, a putative PIN gene (At2g17500), was shown to be significantly induced in the microarray experiment and rapidly upregulated by cytokinin in the qPCR results, but the third (PIN5) did not respond over the time course of the qPCR experiment (see Supplemental Figure 3C online). These data further indicate that auxin transport in the Arabidopsis root apex is regulated by cytokinin through the regulation of specific PIN genes.
DISCUSSION
Critical plant growth and developmental processes are driven largely by defined changes in hormone levels and responsiveness. Environmental integration is also mediated largely through the biosynthesis and action of hormones. Although an individual hormone may play a predominant role in a particular process, in all processes, elements of the response to one hormone are likely to overlap with elements integral to the response to others. Evidence of interrelationships between hormones has been accumulating over many years. One of the most thoroughly investigated of these is the interplay between auxin and cytokinin (Coenen and Lomax, 1997; Swarup et al., 2002; Moubayidin et al., 2009). Both hormones are key regulators of critical processes, such as organogenesis, embryogenesis, and root and shoot meristem function. We have previously shown that one way that auxin and cytokinin levels are regulated is through the auxin-induced downregulation of cytokinin biosynthesis (Nordström et al., 2004). We show here that, conversely, cytokinin induces auxin biosynthesis and increases steady state auxin levels in young, developing tissues in the root and shoot of Arabidopsis. Combined, the data suggest that a feedback homeostasis loop exists to regulate the relative levels of auxin and cytokinin in plants.
Both auxin and cytokinin are involved in the regulation of root growth in response to environmental variables (Osmont et al., 2007). Clearly, mechanisms need to exist to alter the levels and/or responsiveness to the hormones to elicit the appropriate environmental response. Although auxin levels and concentration maxima are maintained partially via the active transport of the hormone (Robert and Friml, 2009), high levels of auxin are produced directly in the root and shoot apices (Ljung et al., 2001, 2005; Petersson et al., 2009). IPT gene expression patterns suggest that cytokinin is also biosynthesized throughout the root apex (Miyawaki et al., 2004).
In this study, we have shown that the induction of a transgene encoding the cytokinin biosynthesis protein, IPT8, leads to a rapid upregulation of IAA biosynthesis in the shoot apex, young leaves, and the root system. Older, fully developed leaves did not show a strong cytokinin induction of IAA, except in leaves treated with very high levels of active cytokinins. It remains to be determined if these high levels are physiologically relevant. However, the data suggest that the ability of cytokinin to induce the biosynthesis and increase the levels of IAA is particularly important in actively growing tissues (Figures 1B and 1C). In the cytokinin hypersensitive arr3 arr4 arr5 arr6 line, the steady state IAA concentration in the root tip remained unchanged from the wild type. However, the rate of IAA biosynthesis was higher in this line than in the wild type prior to the cytokinin treatment, and it had an augmented response to the hormone compared with the wild type (Figure 2). This suggests that a hypersensitivity to both endogenous and exogenous cytokinin increases the ability of cytokinin to induce auxin biosynthesis. Whereas ahp1 ahp2 ahp3 ahp4 also had an increased rate of auxin biosynthesis compared with the wild type prior to cytokinin treatment, in contrast with the wild type and the arr3 arr4 arr5 arr6 line, the rate of IAA biosynthesis declined in response to cytokinin in the ahp1 ahp2 ahp3 ahp4 line. This suggests that the relationship between cytokinin and auxin is fundamentally altered in this mutant. The data indicate clearly that the regulation of auxin biosynthesis is intimately dependent on normal cytokinin responsiveness in root tips.
Many plant hormone signaling components have been shown to be regulated by more than one hormone. To determine whether cytokinin led directly to an increase in the auxin pool size and rate of biosynthesis or achieved this through auxin signaling components, we analyzed IAA biosynthesis and levels in auxin perception and signal transduction mutants. In the axr3-1 mutant, both the IAA pool size and auxin biosynthetic rates were increased in comparison to the wild type prior to cytokinin treatment (Figure 2). This suggests that, just as in the cytokinin response mutants, auxin homeostasis is altered in this mutant. As the normal auxin-stimulated degradation of AXR3/IAA17 is perturbed in the axr3-1 mutant, it is possible that this protein plays an important role in a feedback loop mechanism regulating IAA levels and biosynthesis rates. Interestingly, cytokinin did not alter the pool size or the rate of auxin biosynthesis in the axr3-1 mutant, suggesting that an increase in the rate of IAA biosynthesis requires the stabilization and/or increased level of AXR3/IAA17 expression. Stabilization of the protein in the axr3-1 mutant may constitutively activate the molecular response mechanism required for cytokinin induction of auxin biosynthesis. Both the microarray and qPCR analyses showed that cytokinin increased the level of AXR3/IAA17 expression. In contrast with the axr3-1 mutant, the rate of IAA biosynthesis in the auxin-insensitive mutants axr1-3 and axr1-12 was the same as the wild type prior to the cytokinin treatment. However, both had an increased response to cytokinin compared with the wild type. This augmentation of cytokinin-responsive auxin biosynthesis suggests that in the wild type, there is component of feedback inhibition of elevated auxin biosynthesis.
The initial data showing that auxin biosynthesis rapidly increased in response to elevated ectopic or exogenous cytokinin suggested the possibility that endogenous levels of the hormones are similarly interrelated in rapidly growing tissues. In the seedlings with inducible CKX3 expression, treatment with 17-β-estradiol led to a reduction in cytokinin levels between 3 and 12 h after the onset of the treatment (Figure 3B). This correlated with a downregulation of IAA biosynthesis rates after 12 and 24 h of estradiol incubation (Figures 3C and 3D), providing a first indication of the involvement of endogenous cytokinin in the regulation of endogenous auxin biosynthesis in young tissues. Further evidence for an interrelationship between endogenous cytokinin and auxin biosynthesis was provided by the observation of lower auxin biosynthesis rates in the root apex of the constitutive CKX1overexpressor line (Figure 3E). A third line of evidence was provided by the analysis of IAA levels and biosynthesis rates in single and multiple gene mutants for cytokinin biosynthesis IPT genes. All showed reduced rates of endogenous auxin biosynthesis in the root tip (Figure 4) with the reduction varying with the different mutant combinations. As the individual IPT genes are expressed in different cell types, we tested whether the different levels of reduction in auxin biosynthesis in the various ipt mutant combinations was related to a cell specificity for the cytokinin induction of auxin biosynthesis. An analysis of cell-specific responses revealed that cytokinin had a similar capacity to induce auxin biosynthesis in all cell types tested (Figure 5).
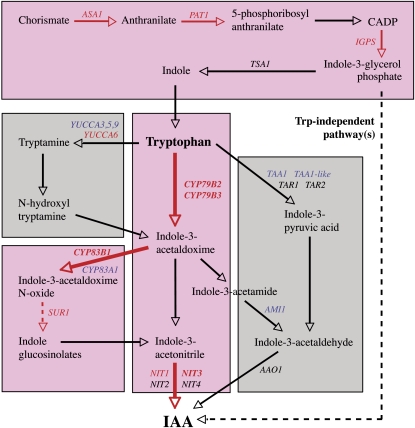
The putative pathways of IAA biosynthesis in Arabidopsis (Figure 7) include many steps for which the enzymes responsible are still unknown (Woodward and Bartel, 2005; Normanly, 2010). Our results have shown that several of those that have been clearly identified are regulated by cytokinin. The data suggest that cytokinin is capable of inducing a rapid upregulation of the Trp biosynthesis pathway through an increase in the precursor for Trp-dependent IAA biosynthesis and an upregulation of the pathway from Trp via indole-3-acetaldoxime and indole-3-acetonitrile to IAA (Figure 7). The data also suggest that cytokinin leads to an induction of the related IG pathway. The MYB-type transcriptional regulator ATR1/MYB34 has been shown to coregulate a number of genes involved in Trp-dependent IAA and IG biosynthesis (Celenza et al., 2005). ATR1/MYB34 transcript levels were increased at the first time point examined, 0.5 h after the onset of the cytokinin treatment, suggesting that the encoded protein regulates the other IAA and IG biosynthesis genes, CYP79B2, CYP79B3, CYP83B1/SUR2, and NIT3, that were also rapidly upregulated by the hormone (Figures 6A and 7).
Figure 7.
IAA Biosynthesis Pathways in Arabidopsis Regulated by Cytokinin.
Putative IAA biosynthesis pathways in Arabidopsis, with genes shown to be differentially regulated in the microarray and/or qPCR analysis indicated (up,red; down, blue). Genes and pathways that are more strongly upregulated are indicated with red arrows and a pink background, respectively. A dotted arrow indicates that there is more than one enzymatic step involved in the pathway.
We provided evidence that cytokinin regulates auxin responses by altering the rate of biosynthesis and steady state levels of auxin. Transcript levels of several auxin-signaling genes were affected by cytokinin treatment. For example, there were strong increases in the levels of IAA17/AXR3 and IAA3/SHY2 transcripts in response to cytokinin, and transcripts for both these genes were shown to be increased after only 30-min incubations (Figure 6B). IAA17/AXR3 and IAA3/SHY2 are both members of the auxin/IAA family of auxin response regulators that usually work as auxin response repressors (Chapman and Estelle, 2009). Our data show that both auxin and cytokinin are able to induce the expression of IAA17/AXR3 and IAA3/SHY2. Cytokinin induction of both auxin biosynthesis and the expression of putative repressors of the auxin response indicate a complex homeostatic interrelationship between the two hormones. Moubayidin et al. (2009) recently suggested that IAA3/SHY2 is involved in auxin–cytokinin interactions in the root apical meristem. Our data and that of other groups (Moubayidin et al., 2009; Ruzicka et al., 2009) have shown that cytokinin can also modulate auxin transport in the root via the regulation of PIN gene expression. There is strong evidence that this is achieved through the regulation of IAA3/SHY2 expression (Dello Ioio et al., 2008).
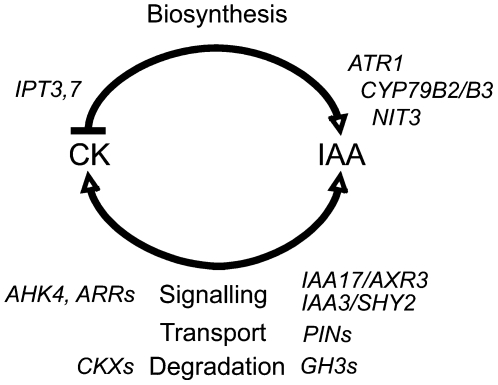
As might be expected for two hormones with a strong capacity to affect critical plant growth and developmental processes, it is clear from the data presented here that complex mechanisms have evolved for regulating the levels of and the responsiveness to the two hormones. We have shown that the rate of auxin biosynthesis and auxin steady state levels are regulated by cytokinin and that these regulatory mechanisms work through both auxin and cytokinin signaling. This regulation is likely to have evolved to ensure that hormone levels are optimal for cell- and tissue-specific developmental and environmental response processes. We propose a model for auxin/cytokinin interactions in the root apex in which cytokinin acts as a positive regulator of auxin biosynthesis and auxin as a negative regulator of cytokinin biosynthesis (Figure 8). This regulatory loop is mediated via cytokinin and auxin signal transduction and through the transcriptional control of specific cytokinin and auxin biosynthesis genes. Our data have shown that other homeostatic mechanisms, such as catabolism/conjugation and hormone transport, are also likely to participate in the crosstalk that modulates the essential intracellular levels of and relative responses to auxin and cytokinin.
Figure 8.
A Model of the Homeostatic Feedback Loop Regulating IAA and Cytokinin Concentrations in the Arabidopsis Root Apex.
Cytokinin (CK) acts as a positive regulator of auxin (IAA) biosynthesis in the root apex, possibly by ATR1 coinduction of the CYP79B2/B3 and NIT3 genes. On the other hand, auxin can act as a repressor of cytokinin biosynthesis via downregulation of specific IPT genes. Both auxin and cytokinin signaling is important for these interactions, and our data suggest that specific auxin/IAA genes (IAA17/AXR3 and IAA3/SHY2) and cytokinin signaling components (AHK4 and ARRs) are involved. Changes in auxin transport (PIN efflux carriers) and in the degradation of auxin (GH3s) and cytokinin (CKXs) may also play important roles in the modulation of the response.
METHODS
Plant Material and Growth Conditions
Seeds of all lines used in this study (wild-type Arabidopsis thaliana ecotypes Columbia and Wassilewskija [Ws]; the European Arabidopsis Stock Centre [NASC]; the lines Woodenleg:GFP, Scarecrow:GFP [Philip Benfey], J2812:GFP, M0028:GFP [NASC], and 35S:AtCKX1 [Thomas Schmülling]; the mutant lines arr3 arr4 arr5 arr6, ahp1 ahp2 ahp3 ahp4 [Joe Kieber], axr3-1, axr3-3, axr1-3, and axr1-12 [Ottoline Leyser], tir1-1 [Mark Estelle], ipt1, ipt3, ipt5-2, ipt7, ipt1 ipt3, ipt3 ipt5-2, ipt5-2 ipt7, ipt3 ipt5-2 ipt7, and ipt1 ipt3 ipt5-2 ipt7 [Tatsuo Kakimoto], and the inducible lines IPT8/pga22 [Nam-Hai Chua] and pMDC7:At-CKX3) were surface sterilized, plated on solid medium (1× Murashige and Skoog [MS] medium [Duchefa], 1% sucrose, and 1% agar [Merck], pH 5.7), and vernalized at 4°C for 3 d prior to germination. The mutant and transgenic lines are all in the Columbia background except for IPT8/pga22 (Ws). The GFP lines used in this study were as described by Petersson et al. (2009). For collection of larger amounts of root material for cell sorting, transcript profiling, and qPCR analysis, seeds were sown at high density (~500 seeds per row) on nylon mesh (Sefar Nitex 03-110/47) on solid MS medium. After vernalization, all plates were placed vertically under 150 μE light in LD conditions (16 h light/8 h darkness) at 22°C.
Construction of an Inducible Cytokinin Oxidase Line
For the inducible cytokinin oxidase line, the coding sequence of CKX3 was amplified with the primers CKX3-F1 (5′-CACCAATGGCGAGTTATAATCTTCGTTC-3′) and CKX3-R1 (5′-CTACTAACTCGAGTTTATTTTTTGA-3′). The PCR product was cloned into the estradiol-inducible binary vector pMDC7 (http://www.unizh.ch/botinst/Devo_website/curtisvector/7cassetteB.pdf) via pTOPO TA GATEWAY (Invitrogen). Arabidopsis plants were transformed by Agrobacterium tumifaciens–mediated floral dip transformation. Analyses of transcript levels by qPCR analysis and quantification of cytokinins were performed on 6 DAG pMDC7:AtCKX3 and wild-type Columbia seedlings as described. The seedlings were incubated for 0, 3, 6, 12, and 24 h in liquid medium with or without 5 μM 17-β-estradiol (Sigma-Aldrich). For each sample, tissues from 10 to 20 seedlings were pooled, frozen in liquid nitrogen, and stored at −80°C until extraction and purification. Samples were collected in triplicates. The pMDC7:AtCKX3 line has been deposited at the NASC (http://Arabidopsis.info/) under the name N9911.
IAA Biosynthesis Measurements
IPT8/pga22 seedlings were grown on vertical plates in LD for 10 d as earlier described. Intact seedlings or seedlings divided into root and shoot were transferred to Petri dishes and incubated first for 12 h (LD with gentle shaking) in liquid medium (1× MS and 1% sucrose, pH 5.7) containing 5 μM 17-β-estradiol to induce cytokinin biosynthesis and then for 24 h in medium containing 5 μM 17-β-estradiol and 30% 2H2O (Cambridge Isotope Laboratories). As a control, IPT8/pga22 seedlings were incubated without estradiol in the medium.
Arabidopsis wild-type Ws seedlings 10 DAG were divided into root and shoot and incubated for 24 h in liquid medium containing 30% 2H2O with or without 5 μM 17-β-estradiol, 10 μM BAP, or 10 μM c/t-Z added to the medium. pMDC7:AtCKX3 seedlings 6 and 10 DAG were incubated for 12 and 24 h in liquid medium containing 30% 2H2O with or without 5 μM 17-β-estradiol. After incubation, the 10 DAG seedlings were rinsed in distilled water and divided into (a) apex and young, developing leaves (leaf 3 and smaller), (b) the first two true leaves, and (c) the root system (Figure 1A). For each sample, tissues from 10 seedlings were pooled, frozen in liquid nitrogen, and stored at −80°C until extraction and purification.
Arabidopsis wild-type, 35S:AtCKX1, and ipt mutant lines were grown for 6 d in LDs and divided into root and shoot. Isolated roots were then incubated for 24 h in medium containing 30% 2H2O. Arabidopsis wild-type and auxin and cytokinin signal transduction mutants were grown for 6 d in LDs and divided into root and shoot. Isolated roots were then incubated for 24 h in medium containing 30% 2H2O with or without 5 μM t-Z. IPT8/pga22 seedlings were grown for 7 DAG in LD and divided into root and shoot. Isolated roots were incubated for 3, 12, and 24 h in medium containing 30% 2H2O with or without 5 μM 17-β-estradiol or 5 μM t-Z. After incubation, the roots were rinsed in distilled water, and 2 mm of the root apex was collected under the stereomicroscope. Fifty root sections were pooled for each sample, frozen in liquid nitrogen, and stored at −80°C until extraction and purification. Samples were purified according to Andersen et al. (2008) and analyzed by GC-MRM-MS as described (Ljung et al., 2005). The IAA biosynthesis rate was calculated from the measured incorporation of one to three deuterium atoms into the IAA molecule, and corrections were made for the contribution of natural isotopic abundances to a mass-to-charge ratio (m/z) of 203 to 205. The amount of endogenous IAA (labeled + unlabeled) was calculated by combining the corrected areas of the ions from deuterium labeled IAA (m/z 203 to 205) with that of unlabeled IAA (m/z 202).
Cell Type–Specific IAA Biosynthesis Measurements
The Arabidopsis lines Woodenleg:GFP, Scarecrow:GFP, J2812:GFP, and M0028:GFP were grown on vertical plates in LDs for 8 d as described by Petersson et al. (2009). Seedling roots were then excised ~10 mm from the root/shoot junction and incubated for 16 h in liquid medium containing 30 mM sucrose, 4.4 g MS, 2.6 mM MES, 10 μM NPA, and 10 μM t-Z in 30% 2H2O. After incubation, the root tissue was treated with cell wall–degrading enzymes, and protoplasts were isolated as described (Petersson et al., 2009). Cell sorting was performed on a FACSAria equipped with a 100-μm nozzle (Becton Dickinson) as described (Birnbaum et al., 2005; Petersson et al., 2009). Intact root protoplasts were identified based on forward and side scatter, and GFP positive and negative cells were then separated according to their GFP emission and auto fluorescence, after excitation by the 488-nm laser. Data were processed using the FACSDiVa 5.0 software. The samples were kept at 4°C during the whole sorting procedure, and 150,000 to 300,000 protoplasts were collected for each technical replicate. The protoplasts were suspended in BD FACSFlow buffer (Becton Dickinson) prior to extraction. 13C6-IAA internal standard (125 pg) was added, and the sample was frozen to rupture the plasma membrane before purification by solid phase extraction, and IAA biosynthesis measurements were performed by GC-MRM-MS as described (Ljung et al., 2005; Petersson et al., 2009). The numbers of biological and technical replicates analyzed were as follows: Woodenleg:GFP (3;3,3,2), Scarecrow:GFP (2;3,3), J2812:GFP (2;2,3), and M0028:GFP. (4;1,1,2,2).
Cytokinin Quantification
IPT8/pga22 seedlings were grown on vertical plates in LDs for 10 d. Intact seedlings were transferred to Petri dishes and incubated for 24 h (LD with gentle shaking) in liquid medium (1× MS and 1% sucrose, pH 5.7) containing 5 μM 17-β-estradiol to induce cytokinin biosynthesis. As a control, IPT8/pga22 seedlings were incubated without estradiol in the medium. After incubation, the seedlings were rinsed in distilled water and divided into (a) apex and young, developing leaves, (b) the first two true leaves, and (c) the root system (Figure 1A). For each sample, tissues from 20 seedlings were pooled, frozen in liquid nitrogen, and stored at −80°C until extraction and purification. Samples were purified, derivatized by propyonylation, and quantified by liquid chromatography–mass spectrometry analysis in the MRM mode on a Micromass Quattro Ultima mass spectrometer according to Nordström et al. (2004). Chromatographic separation was performed on a reversed phase analytical column (150 × 1 mm BetaMax Neutral, 5-μm particle size; Thermo Hypersil). All data were processed by Masslynx software (version 4.0; Waters).
Transcript Profiling
IPT8/pga22 seedlings were grown on vertical plates in LDs for 7 d. The seedlings were then incubated directly on the plate for 12 or 24 h (LD with gentle shaking) in liquid medium (1× MS and 1% sucrose, pH 5.7) containing either 5 μM 17-β-estradiol or 5 μM t-Z. Control samples were incubated for 12 or 24 h without cytokinins in the medium. Root tips (~5 mm long, 10 mg root tissue/sample collected in triplicates) were harvested by cutting the roots with a scalpel on the nylon mesh. Each sample was immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. The methods and statistical analysis are described in detail in Supplemental Methods online.
qPCR Analysis
To quantify the expression of genes related to auxin and cytokinin biosynthesis and signaling in root tips of 7 DAG wild-type Arabidopsis (Columbia) seedlings incubated with or without 5 μM t-Z, qPCR analysis was performed with the SYBR-Green fluorochrome technique. We chose TIP41-like as the best reference gene for our conditions using GeNorm (Vandesompele et al., 2002). The methods and primer sequences are described in detail in Supplemental Methods and Supplemental Table 3 online. The expression data presented in the figures are means ± se of three biological replicates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ARR3 (At1G59940), ARR4 (At1G10470), ARR5 (At3G48100), ARR6 (At5G62920), AHP1 (At3G21510), AHP2 (At3G29350), AHP3 (At5G39340), AHP4 (At3G16360), AXR1 (At1G05180), AXR3 (At1G04250), TIR1 (At3G62980), CKX1 (At2G41510), CKX3 (At5G56970), IPT1 (At1G68460), IPT3 (At3G63110), IPT5 (At5G19040), IPT7 (At3G23630), IPT8 (At3G19160), and TIP4-like (At4G34270). The pMDC7:AtCKX3 line has been deposited at the NASC (http://Arabidopsis.info/) under the name N9911. The NASCArrays experiment reference number is NASCARRAYS-364, and the data have been donated to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/) with the GEO accession number GSE20231. Accession numbers for the auxin- and cytokinin-related genes identified in the array data and discussed in the text can be found in Supplemental Data Set 1 online. Accession numbers for genes analyzed by qPCR can be found in Supplemental Table 3 online.
Author Contributions
K.L. and G.S. conceived the project. K.L., S.A.G., N.G., and S.M. designed, performed,and analyzed the microarray experiment. B.J., K.L., and S.A.G. designed, performed, and analyzed the qPCR experiments. B.J. constructed the inducible CKX3 line. K.L. and S.V.P designed, performed, and analyzed the IAA biosynthesis measurements. P.T. and K.D. performed the cytokinin analyses. K.L. and B.J. wrote the manuscript. All authors contributed to the interpretation of results and edited the manuscript.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Root Tip–Specific IAA Biosynthesis after Induction of the CKX3 Gene.
Supplemental Figure 2. Timing in the Induction of IAA Biosynthesis by Cytokinins.
Supplemental Figure 3. qPCR Analysis of Genes Related to Auxin and Cytokinin Biosynthesis, Signaling, and Transport after Cytokinin Treatment.
Supplemental Table 1. Cytokinin Concentration in Uninduced and Induced IPT8/pga22 Seedlings.
Supplemental Table 2. Cytokinin Concentration in Uninduced and Induced pMCD7:AtCKX3 Seedlings.
Supplemental Table 3. Oligonucleotide Primers for Genes Analyzed by qPCR.
Supplemental Data Set 1. Expression of Auxin- and Cytokinin-Related Genes in the ATH1 Affymetrix Microarray.
Supplemental Data Set 2. Gene Ontology Analysis.
Supplemental Methods. Microarray Analysis and qPCR Analysis.
Acknowledgments
The mutant lines used in this study were generous gifts from Tatsuo Kakimoto, Ottoline Leyser, Mark Estelle, Nam-Hai Chua, Joe Kieber, and Thomas Schmülling. The GFP lines were obtained from Philip Benfey and from NASC. We thank Roger Granbom, Gun Lövdahl, Gaia Castelain, Liping Ke, and Yuqiang Sun for excellent technical assistance and Henrik Antti and Rishikesh Bhalerao for helpful comments on the manuscript. The work was supported by the Swedish Research Council (Vetenskapsrådet), the Swedish Foundation for Strategic Research centre “Developmental Biology of Plants,” and Kempestiftelserna.
References
- Andersen S.U., Buechel S., Zhao Z., Ljung K., Novák O., Busch W., Schuster C., Lohmann J.U. (2008). Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åstot C., Dolezal K., Nordström A., Wang Q., Kunkel T., Moritz T., Chua N.-H., Sandberg G. (2000). An alternative cytokinin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 97: 14778–14783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.A. (2000). Long-distance vascular transport of endogenous hormones in plants and their role in source:sink regulation. Isr. J. Plant Sci. 48: 199–203 [Google Scholar]
- Birnbaum K., Jung J.W., Wang J.Y., Lambert G.M., Hirst J.A., Galbraith D.W., Benfey P.N. (2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2: 615–619 [DOI] [PubMed] [Google Scholar]
- Casimiro I., Beeckman T., Graham N., Bhalerao R., Zhang H., Casero P., Sandberg G., Bennett M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Coenen C., Lomax T.M. (1997). Auxin-cytokinin interactions in higher plants: Old problems and new tools. Trends Plant Sci. 2: 351–356 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- del Pozo J.C., Dharmasiri S., Hellmann H., Walker L., Gray W.M., Estelle M. (2002). AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inzé D., Beeckman T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60: 871–887 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Karunarathna N., Jurgens G., Estelle M. (2007). AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J. 52: 114–123 [DOI] [PubMed] [Google Scholar]
- Fukaki H., Nakao Y., Okushima Y., Theologis A., Tasaka M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44: 382–395 [DOI] [PubMed] [Google Scholar]
- Hartig K., Beck E. (2006). Crosstalk between auxin, cytokinins, and sugars in the plant cell cycle. Plant Biol. 8: 389–396 [DOI] [PubMed] [Google Scholar]
- Hutchison C.E., Li J., Argueso C., Gonzalez M., Lee E., Lewis M.W., Maxwell B.B., Perdue T.D., Schaller G.E., Alonso J.M., Ecker J.R., Kieber J.J. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Leyser H.M.O., Pickett F.B., Dharmasiri S., Estelle M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K., Bhalerao R.P., Sandberg G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto K. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specifcity and regulation by auxin, cytokinin, and nitrate. Plant J. 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Di Mambro R., Sabatini S. (2009). Cytokinin-auxin crosstalk. Trends Plant Sci. 14: 557–562 [DOI] [PubMed] [Google Scholar]
- Mouchel C.F., Osmont K.S., Hardtke C.S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443: 458–461 [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A., Tarkowski P., Tarkowska D., Norbaek R., Åstot C., Dolezal K., Sandberg G. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (2010). Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V., Leyser O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Osmont K.S., Sibout R., Hardtke C.S. (2007). Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Petersson S.V., Johansson A.I., Kowalczyk K., Makoveychuk A., Wang J.Y., Moritz T., Grebe M., Benfey P.N., Sandberg G., Ljung K. (2009). An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte A.M., Chae H.S., Maxwell B.M., Kieber J.J. (2005). The interaction of cytokinin with other signals. Physiol. Plant. 123: 184–194 [Google Scholar]
- Robert H.S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Rouse D., Mackay P., Stirnberg P., Estelle M., Leyser O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373 [DOI] [PubMed] [Google Scholar]
- Ruzicka K., Ljung K., Vanneste S., Podhorska R., Beeckman T., Friml J., Benkova E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K., Simásková M., Duclercq J., Petrásek J., Zazímalová E., Simon S., Friml J., Van Montagu M.C., Benková E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sablowski R. (2007). The dynamic plant stem cell niches. Curr. Opin. Plant Biol. 10: 639–644 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S., Tanaka M., Mori H. (2009). Auxin-cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69: 429–435 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Niu Q.-W., Tarkowski P., Zheng B., Tarkowska D., Sandberg G., Chua N.-H., Zuo J. (2003). The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol. 131: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Parry G., Graham N., Allen T., Bennett M. (2002). Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol. Biol. 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K., Yamaya T., Sakakibara H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 279: 41866–41872 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Takei K., Kojima M., Sakakibara H., Mori H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- To J.P., Kieber J.J. (2008). Cytokinin signaling: Two components and more. Trends Plant Sci. 13: 85–92 [DOI] [PubMed] [Google Scholar]
- To J.P.C., Haberer G., Ferreira F.J., Deruère J., Mason M.G., Schaller G.E., Alonso J.M., Ecker J.R., Kieber J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]