Copper is an essential micronutrient of plants and also an important element in a number of pesticides for crop protection. A pathogenic strain of bacterial blight, the most devastating bacterial disease of plants worldwide is sensitive to copper. Here, we show that this bacterium overcomes rice by regulating rice genes to remove copper from the site where it multiplies to cause disease.
Abstract
Pathogen effectors are virulence factors causing plant diseases. How the host targets of these effectors facilitate pathogen infection is largely unknown. An effector of Xanthomonas oryzae pv oryzae (Xoo) transcriptionally activates rice (Oryza sativa) susceptibility gene Xa13 to cause bacterial blight disease. Xa13 encodes an indispensable plasma membrane protein of the MtN3/saliva family, which is prevalent in eukaryotes with unknown biochemical function. We show that the XA13 protein cooperates with two other proteins, COPT1 and COPT5, to promote removal of copper from xylem vessels, where Xoo multiplies and spreads to cause disease. Copper, an essential micronutrient of plants and an important element for a number of pesticides in agriculture, suppresses Xoo growth. Xoo strain PXO99 is more sensitive to copper than other strains; its infection of rice is associated with activation of XA13, COPT1, and COPT5, which modulate copper redistribution in rice. The involvement of XA13 in copper redistribution has led us to propose a mechanism of bacterial virulence.
INTRODUCTION
The battle between pathogens and plants is never-ending due to the coevolution of the parasites and their hosts. Pathogenic bacteria interact with plants by secreting proteins into host cells. The proteins, known as effectors, are injected into host cells by the type III secretion system, which is highly conserved in plant and animal pathogens, and these effectors play essential roles in pathogenicity in plants. The type III effectors with known functions have either enzymatic or transcription activator–like (TAL) activities that modify or degrade host proteins or regulate host gene expression (Kay and Bonas, 2009). Mutation of type III effectors is an important mechanism of evolution in pathogenic bacteria that are subjected to the selective pressures of a host defense system (Ma and Guttman, 2008; Stavrinides et al., 2008). In addition, bactericides can also exert selective pressure on pathogens, resulting in the evolution of bactericide-resistant races. For example, copper (Cu) is an important element for a number of pesticides in agriculture. The mechanisms of the antimicrobial activity of Cu are suggested to be associated with denaturation of nucleic acids, alteration and inhibition of protein activity, and changes in plasma membrane permeabilization (Borkow and Gabbay, 2004). Cu-resistant plant pathogenic bacteria have been reported because of the wide application of Cu-containing pesticides in agriculture (Bender et al., 1990; Cooksey, 1990).
Some host plants have evolved sophisticated strategies to counter bacterial effectors and avoid diseases. For example, one strategy uses host disease resistance (R) gene promoters to trap the TAL effectors; mutation of R gene promoters results in induction of dominant R genes by specific effectors and subsequent host defense responses (Gu et al., 2005; Römer et al., 2007, 2009a, 2009b). Another strategy is mutation of a host susceptibility gene promoter to become unresponsive to the TAL effector; this mutation results in a recessive R gene that has lost pathogen-induced expression and subsequent avoidance of disease (Chu et al., 2006b; Yang et al., 2006). Although different pathogen effectors have been characterized, it is largely unknown how the host targets of these effectors act to facilitate pathogen infection.
In addition to being an important element in a number of pesticides, Cu is also an essential micronutrient of plants. There are multiple members of the COPT (copper transporter) protein family that act in Cu homeostasis by Cu uptake in each analyzed plant species (Kampfenkel et al., 1995; Sancenón et al., 2004; Page et al., 2009). These COPTs are the homologs of yeast and human Ctr (copper transporter) proteins (Sancenón et al., 2003; Page et al., 2009). Some Ctrs can interact with themselves or with other Ctr proteins to mediate Cu uptake toward the cytosol (Zhou and Thiele, 2001; Lee et al., 2002; Beaudoin et al., 2006; Nose et al., 2006).
Rice (Oryza sativa) bacterial blight, caused by Xanthomonas oryzae pv oryzae (Xoo), is one of the most devastating rice diseases worldwide (Nino-Liu et al., 2006). The fully recessive R gene xa13 mediates race-specific resistance to Xoo strain PXO99 in a manner different from other characterized R genes (Chu et al., 2006b). Eleven recessive xa13 alleles have been identified. Nine of the 11 alleles encode proteins with one to three amino acid differences from that encoded by their dominant (susceptible) allele Xa13 and another two recessive alleles encode an identical protein to that encoded by dominant Xa13. However, the xa13 and Xa13 alleles have sequence polymorphisms in their promoter regions (Chu et al., 2006b). The expression of dominant Xa13 but not recessive xa13 is induced on PXO99 infection; suppressing dominant Xa13 can result in a similar level of resistance to PXO99 as conferred by xa13 in rice, suggesting that promoter mutations may result in recessive xa13 (Chu et al., 2006b). Further analysis confirmed that transcriptional nonreaction to Xoo infection caused by promoter mutation, not its protein composition, is the key factor for xa13-mediated resistance (Yuan et al., 2009). The dominant Xa13 was also referred to as 8N3, which is transcriptionally activated by the TAL effector PthXo1 of PXO99 by directly interacting with a UTP (upregulated by TAL effector) box in Xa13 promoter, suggesting that the dominant Xa13 is a susceptibility gene used by the bacterium to infect rice in a gene-for-gene manner (Yang et al., 2006; Römer et al., 2010).
XA13 protein belongs to the MtN3/saliva family (Chu et al., 2006a), which contains no known functional domain or motif. To elucidate the biochemical function of XA13 protein in the rice–Xoo interaction, we screened for interacting proteins and identified two XA13-interacting proteins that were homologs of COPT or Ctr proteins in other organisms. We then designed a series of experiments to examine whether XA13 and its interacting proteins were involved in Cu redistribution and whether Xoo was sensitive to Cu both in vitro, in yeast cells, and in planta. A proposed function for XA13 in regulating disease susceptibility is discussed.
RESULTS
Polytopic Membrane Protein XA13 Interacts with COPT1 and COPT5
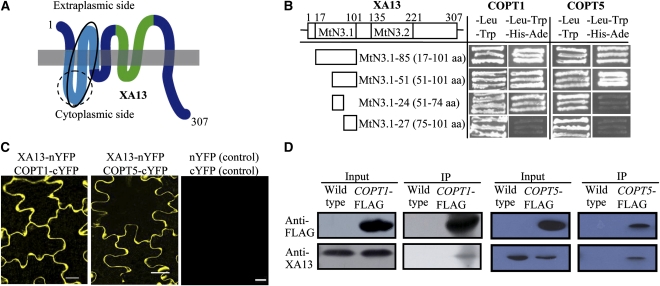
XA13 is a plasma membrane protein (Chu et al., 2006b). Because the arrangement of membrane proteins is an important determinant of their biochemical functions, we first analyzed the topology of XA13. Bioinformatic analyses predicted that XA13 contained six to nine transmembrane helices (see Supplemental Table 1 online), suggesting that it may be a polytopic membrane protein. This hypothesis was confirmed by analyzing a set of truncated XA13 proteins designed based on the prediction using a split-ubiquitin system for integral membrane proteins (Reinders et al., 2002). The growth of yeast cells on selective medium and the expression of reporter protein suggest that XA13 has seven transmembrane regions and an extracellularly located N-terminus and intracellularly located C-terminus (Figure 1A; see Supplemental Figure 1 online).
Figure 1.
Topology and Interaction of XA13 and COPT1 or COPT5 Proteins.
(A) XA13 is a polytopic transmembrane protein. The MtN3.1 and MtN3.2 domains are indicated with light blue and green colors, respectively. The dotted- and solid-lined circles indicate XA13 regions harboring the site interacting with COPT1 and COPT5, respectively.
(B) COPT1 and COPT5 proteins interacted with different fragments of the MtN3.1 domain of XA13 in the yeast two-hybrid assay. The interactions were assessed by growth of yeast cells on selective medium lacking Leu, Trp, His, and Ade.
(C) XA13 interacted with COPT1 and COPT5 in the plasma membrane of tobacco leaf cells as shown by yellow fluorescence signal. Bars = 20 μm.
(D) Coimmunoprecipitation assays verified the interaction between XA13 and COPT1 or COPT5 in rice. Total proteins from wild-type Zhonghua 11 and transgenic plants carrying COPT1-FLAG or COPT5-FLAG were used. Proteins before (input) and after immunoprecipitation (IP) were detected with antibodies, as specified.
XA13 contains two MtN3/saliva domains for which the biochemical function is unknown (Chu et al., 2006a). Based on the topology analysis, the first domain MtN3.1 (17 to 101 amino acids) harbored the N-terminal three transmembrane helices (1 to 3), and the second domain MtN3.2 (135 to 221 amino acids) harbored helices 4 and 5 (Figures 1A and 1B). A cDNA fragment encoding MtN3.1 was used to screen proteins putatively interacting with XA13 by yeast two-hybrid assays. Several candidate MtN3.1-interacting proteins were identified from ~105 colonies screened. Among the putative interacting proteins, a fragment (87 to 161 amino acids) of a putative Cu transporter named COPT1 showed the strongest interaction with MtN3.1 but not with MtN3.2 (see Supplemental Figure 2 online). BLAST analysis showed that there were another six COPT-type genes in the rice genome; we named them COPT2 to COPT7 (see Supplemental Table 2 online). COPT1 shared 51% sequence identity and 66% sequence similarity with Arabidopsis thaliana COPT1, the first reported plant plasma membrane–associated Cu transporter from Arabidopsis (Kampfenkel et al., 1995). We then analyzed the interaction between full-length XA13 and the rice COPT proteins using the split-ubiquitin system. XA13 interacted with COPT1 and COPT5 but not other COPT members (see Supplemental Figure 3 online). COPT1 and COPT5 consist of 161 and 151 amino acids, respectively. Both MtN3.1-51 (51 to 101 amino acids) and MtN3.1-24 (51 to 74 amino acids) fragments of XA13 interacted with COPT1, but only MtN3.1-51 interacted with COPT5 (Figure 1B). These results suggest that COPT1 and COPT5 interacted with different sites of the first MtN3/saliva domain of XA13 (Figure 1A).
Characterized COPT homologs from other species are either plasma membrane or vacuole membrane proteins (Kim et al., 2008). Transient expression of COPT1—green fluorescence protein (GFP) and COPT5-GFP fusion proteins in onion epithelial cells showed that COPT1 and COPT5 are plasma membrane proteins (see Supplemental Figure 4 online). Immunoblot analysis of proteins from transgenic plants carrying COPT1-FLAG or COPT5-FLAG using anti-FLAG antibody also detected these fusion proteins only in the membrane fractions but not soluble protein fractions (see Supplemental Figure 5 online). In addition, the positive interactions of XA13 and COPT1 or COPT5 in the split-ubiquitin system suggest that the C-terminals of COPT1 and COPT5 are localized in the cytoplasmic side (see Supplemental Figure 3 online).
The interactions between XA13 and COPT1 or COPT5 were further confirmed in planta by two approaches. Cotransient expression of XA13-nYFP (N-terminal fragment of yellow fluorescent protein) and COPT1/5-cYFP (C-terminal fragment of YFP) in tobacco (Nicotiana tabacum) leaves using the bimolecular fluorescence complementation (BiFC) system detected the yellow fluorescence signal only in the border of the epithelial cells (Figure 1C), suggesting that XA13, COPT1, and COPT5 colocalize and interact in the plasma membrane. The interactions of XA13 and COPT1 or COPT5 were also confirmed by a coimmunoprecipitation assay using transgenic plants carrying COPT1-FLAG and COPT5-FLAG. After immunoprecipitation of the proteins with anti-FLAG antibody, enrichment of XA13 was detected by anti-XA13 antibody (Figure 1D).
XA13, COPT1, and COPT5 Are Cooperatively Associated with Cu Transport
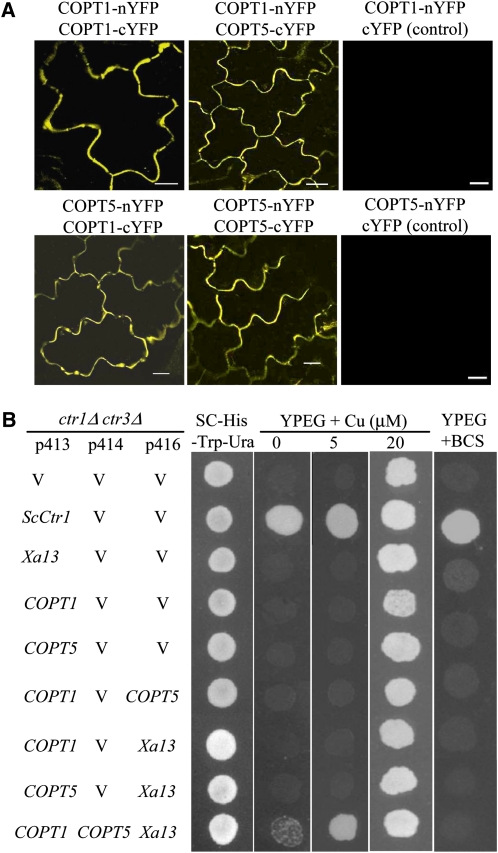
COPT1 and COPT5 interacted with themselves and each other as revealed by both the BiFC and the split-ubiquitin assays (Figure 2A; see Supplemental Figure 6 online). However, XA13 could not interact with itself (see Supplemental Figure 6 online). To ascertain the role of COPT1 and COPT5 in Cu transport, functional complementation analyses were performed using the yeast (Saccharomyces cerevisiae) mutant MPY17, which lacked the functions of Ctr1 and Ctr3 for Cu uptake (Pena et al., 1998). Expression of COPT1, COPT5, or XA13 alone could not complement the phenotype of MPY17 as detection of cell growth in selection medium plus 5 μM Cu or without Cu and neither did the coexpression of any two of the three proteins. Only coexpression of all the three proteins complemented MPY17 phenotype (Figure 2B).
Figure 2.
Interaction and Function of COPT1, COPT5, and XA13 Proteins.
(A) COPT1 and COPT5 interact with themselves and each other in the plasma membrane of tobacco leaf cells as shown by yellow fluorescence signal. Bars = 20 μm.
(B) Functional complementation of S. cerevisiae ctr1Δctr3Δ mutant (MPY17) by coexpression of Xa13, COPT1, and COPT5. Complementation is indicated by growth on the media with 0 or 5 μM Cu. The p413, p414, and p416 are yeast expression vectors. Yeast ScCtr1 and empty vector (V) were used as positive and negative controls, respectively. Transformants were grown in SC-His-Trp-Ura medium to exponential phase and spotted onto SC-His-Trp-Ura, ethanol/glycerol (YPEG), and bathocuproine disulfonic acid (BCS) plates.
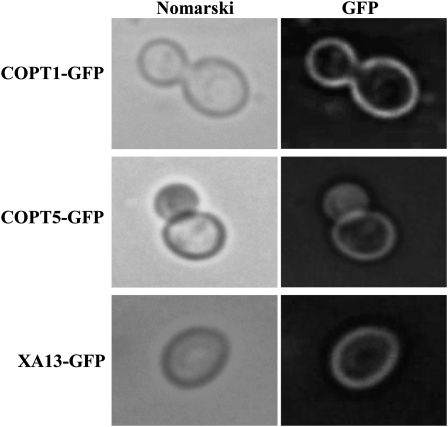
Fission yeast (Schizosaccharomyces pombe) Ctr4 and Ctr5 proteins are interdependent of each other in localization to the plasma membrane for mediating Cu transport (Zhou and Thiele, 2001; Beaudoin et al., 2006). To ascertain whether the requirement of all three proteins to complement MPY17 phenotype was due to one or two of the COPT1, COPT5, or XA13 functioning as cofactors to help protein trafficking to the plasma membrane, we examined the localization of the three proteins in the MPY17 cells. All three proteins localized in the plasma membrane when they were expressed alone in the MPY17 cells (Figure 3). These results suggest that localization of COPT1, COPT5, or XA13 in the plasma membrane does not require the presence of any other two proteins in the MPY17 cells. Thus, XA13, COPT1, and COPT5 appear to be cooperatively required for Cu uptake in the MPY17 cells.
Figure 3.
Localization of COPT1-GFP, COPT5-GFP, and XA13-GFP in Yeast MPY17 Cells.
MPY17 cells were transformed with p413GPD-COPT1-GFP, p413GPD-COPT5-GFP, or p413GPD-Xa13-GFP and grown to log phase on SC-His. The GFP signal and Nomarski optical images were observed using fluorescence microscopy.
XA13, COPT1, and COPT5 Are Associated with Cu Redistribution in Rice
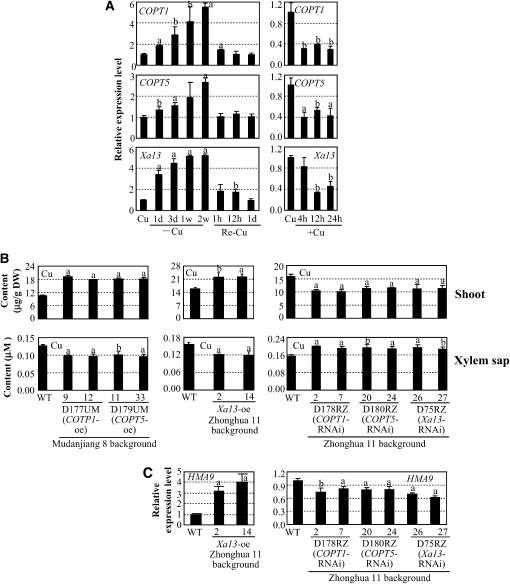
To examine whether the three proteins were also involved in Cu uptake in rice, we first analyzed the expression patterns of Xa13, COPT1, and COPT5 in response to Cu treatment. The three genes showed a similar expression pattern in the shoot and root of rice plants with Cu treatments (Figure 4A; see Supplemental Figure 7 online). The expression of the three genes was induced by Cu deficiency and returned to a normal level when the plants were returned to a normal culture containing 0.2 μM Cu. Furthermore, 50 μM Cu suppressed the expression of the three genes. The coinduction or cosuppression of the three genes in Cu deficiency or excess supports the concept that XA13, COPT1, and COPT5 may cooperate in Cu homeostasis in rice.
Figure 4.
Xa13, COPT1, and COPT5 in Cu Transport in Rice.
Gene expression was analyzed by qRT-PCR. Each data point represents mean (three replicates) ± sd; the “a” or “b” indicates a significant difference was detected between a treatment or a transgenic line and the control or wild-type plants at P < 0.01 or P < 0.05 level, respectively.
(A) Expression of Xa13, COPT1, and COPT5 in shoot is influenced by Cu levels. Rice variety Zhonghua 11 at the four-leaf stage grown in hydroponic culture was used for qRT-PCR analysis. Cu, standard physiologic Cu (0.2 μM); –Cu, Cu deficiency; Re-Cu, retransferring plants to normal culture (containing 0.2 μM Cu); +Cu, 50 μM Cu.
(B) Modulating COPT1, COPT5, and Xa13 expression influenced Cu accumulation in shoots and xylem sap in rice at booting stage. Xa13-suppressing plants were in T0 generation, and other transgenic plants were in T2 generation. DW, dry weight; oe, overexpression, RNAi, RNA interference (suppression).
(C) Modulating COPT1, COPT5, and Xa13 expression influenced the expression of HMA9. The samples were from the same plants described in (B).
We then modified the expression of COPT1 and COPT5 in rice varieties Mudanjiang 8 and Zhonghua 11 carrying Xa13. Sixteen independent transgenic plants carrying the COPT1- and COPT5-overexpressing construct, respectively, and 11 and 10 independent transgenic plants carrying the COPT1- and COPT5-suppressing construct, respectively, were obtained (see Supplemental Figure 8 online). Plants overexpressing COPT1 or COPT5 contained 71 to 83% and 43 to 52% more Cu in shoots and roots, respectively, compared with wild-type Mudanjiang 8 (Figure 4B; see Supplemental Figure 9 online). Xa13-overexpressing lines (Yuan et al., 2009) also contained ~40% and 19 to 22% more Cu in shoots and roots, respectively, compared with wild-type Zhonghua 11 (Figure 4B; see Supplemental Figure 9 online). By contrast, COPT1- or COPT5-suppressing lines showed 27 to 36% and 30 to 40% less Cu in shoots and roots, respectively, compared with wild-type Zhonghua 11; Xa13-suppressing plants also showed ~30 and 33% less Cu in shoots and roots, respectively, compared with wild-type Zhonghua 11 (Figure 4B; see Supplemental Figure 9 online). However, the contents of iron (Fe), manganese (Mn), and zinc (Zn) in the COPT1, COPT5, and Xa13 transgenic plants showed no obvious difference from their corresponding wild-type plants (see Supplemental Figure 10 online). These results suggest that XA13, COPT1, and COPT5 may be specially associated with Cu uptake in rice.
Plants take up Cu from roots and store excess Cu in the root cells; only a small amount of Cu is translocated to the shoot through xylem, one of the two components of the vascular bundles, in vascular plants (Fernandes and Henriques, 1991). In the shoot, Cu is transpired from xylem into the phloem, another component of the vascular bundles, to reach sink tissues such as developing leaves and flowers (Burkhead et al., 2009). To ascertain whether the XA13, COPT1, and COPT5 were associated with Cu redistribution, the same plants used for quantifying Cu contents in shoots were used for quantifying Cu content in xylem sap. The Cu contents in the xylem sap showed opposite patterns as those in shoot tissues in the transgenic plants compared with wild-type plants (Figure 4B). The COPT1-, COPT5-, and Xa13-overexpressing lines had 20 to 25% less Cu in the xylem sap compared with their corresponding wild-type plants, whereas the COPT1-, COPT5-, and Xa13-suppressing plants had 19 to 28% more Cu in the xylem sap compared with wild-type plants. These results suggest that XA13, COPT1, and COPT5 are involved in removing Cu from xylem.
To ascertain whether the removal of Cu from xylem was associated with increased Cu movement from the extracellular spaces into cells, we used expression levels of rice HMA9 as a reporter for intracellular Cu levels in rice. The expression of HMA9, encoding a metal efflux protein, was slightly induced in plants grown in liquid medium containing 10 μM Cu and markedly induced in the shoots of plants grown in medium containing 100 μM Cu (Lee et al., 2007). COPT1-, COPT5- or Xa13-overexpressing plants, which had increased Cu content in shoot tissues, all showed markedly increased levels of HMA9 transcripts (Figure 4C; see Supplemental Figure 11 online), whereas COPT1-, COPT5-, or Xa13-suppressing plants, which had reduced Cu content in tissues, all showed slightly reduced levels of HMA9 transcripts (Figure 4C). The expression of Xa13, COPT1, and COPT5 was also regulated by Cu concentration (Figure 4A). Overexpressing one of COPT1, COPT5, or Xa13 slightly suppressed the expression of the other two genes, while suppressing Xa13 markedly induced COPT1 and COPT5 (see Supplemental Figure 11 online). Likewise, suppressing COPT1 or COPT5 markedly induced Xa13 but cosuppressed COPT5 or COPT1. Consistent with this result, suppressing COPT1 and COPT5 also increased accumulation of XA13 protein in the transgenic plants (see Supplemental Figure 12 online). On the other hand, the expression of the Fe deficiency-induced IRT1, encoding an Fe transporter (Bughio et al., 2002), and the Zn-responsive ZIP4, encoding a Zn transporter (Ishimaru et al., 2005), was not influenced in these transgenic plants (see Supplemental Figure 11 online). These results suggest that XA13, COPT1, and COPT5 may all be involved in promoting Cu influx into rice cells.
Cu Suppresses Xoo Spread in Rice
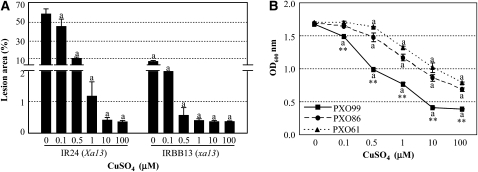
To explore the effect of Cu on Xoo strain PXO99, which causes rice disease by specifically inducing Xa13 expression (Chu et al., 2006b; Yang et al., 2006; Yuan et al., 2009), we analyzed the responses of rice plants and PXO99 to Cu treatment. IR24 carrying dominant susceptible Xa13 and IRBB13 carrying recessive resistant allele xa13 are near-isogenic rice lines in the same genetic background. Adding 0.1 μM CuSO4 to the bacterial inoculum right before inoculation of rice significantly reduced disease lesions caused by PXO99 infection in both susceptible IR24 and resistant IRBB13 plants (Figure 5A); increasing the concentration of CuSO4 to 0.5 to 1 μM in the bacterial inoculum further reduced disease symptom. Furthermore, growth of PXO99 was significantly (P < 0.01) suppressed in liquid culture medium containing 0.1 μM CuSO4, but the growth of other Xoo strains (PXO61 and PXO86), which cause disease on xa13-carrying plants (Yuan et al., 2009), was not significantly (P < 0.01) suppressed until the CuSO4 concentration in the media reached 0.5 μM (Figure 5B). In addition, the cell density of PXO99 in the culture medium was 13 to 60% and 10 to 52% lower compared with that of PXO61 and PXO86 after Cu treatment, respectively (Figure 5B). These results suggest that PXO99 is more sensitive to Cu than other Xoo strains, and Cu can greatly reduce disease symptoms caused by PXO99.
Figure 5.
Cu in Rice–Xoo Interaction.
Each data point represents the mean (four replicates for lesion area and three replicates for bacterial growth) ± sd. The “a” indicates a significant difference was detected between a treatment and the control at P < 0.01 level.
(A) Pretreatment of Xoo strain PXO99 with CuSO4 reduced disease symptoms caused by PXO99 infection. Near-isogenic rice lines IR24 and IRBB13 were inoculated with PXO99 at booting stage.
(B) Addition of CuSO4 into liquid culture medium influenced the growth of different Xoo strains PXO99, PXO61, and PXO86. Bacterial cell densities were measured at 12 h after culture. Two asterisks indicate that a significant difference between cell densities of PXO99 and PXO61 or PXO86 in the same treatment was detected at P < 0.01 level.
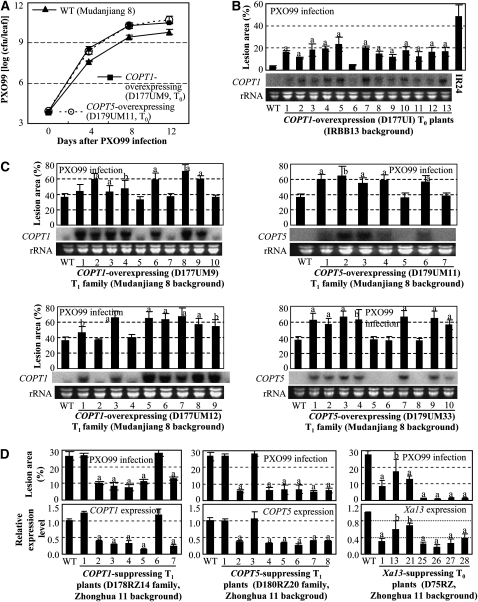
Xoo invades rice plants through hydathodes or wounds, multiplies in the intercellular spaces, then enters xylem vessels, and spreads in the plant through the vessels to cause disease (Nino-Liu et al., 2006). Although more Cu accumulated in COPT1- or COPT5-overexpressing plants, we still expected that COPT1- and COPT5-overexpressing plants would favor disease development because the Cu content in xylem decreased (Figure 4B). To examine this hypothesis, T0 transgenic plants were inoculated with PXO99. The COPT1- or COPT5-overexpressing plants showed increased susceptibility to PXO99 with lesion areas ranging from 50 to 72% compared with 44% for susceptible wild-type Mudanjiang 8 (see Supplemental Figure 8A online). The PXO99 growth rates in COPT1- or COPT5-overexpressing plants were significantly (P < 0.01; 4.3- and 8.5-fold) higher than in wild-type plants on day 12 after infection (Figure 6A). The COPT1-overexpressing plants carrying the recessive xa13 (IRBB13 background) also showed increased susceptibility to PXO99 with lesion areas ranging from 11 to 20% compared with 3% for wild-type plants and 49% for susceptible control IR24 (Figure 6B). T1 families from each of two T0 plants overexpressing COPT1 or COPT5 were further analyzed for susceptibility by inoculation with PXO99 and also measured for COPT1 or COPT5 expression levels. The increased susceptibility was associated with overexpressing COPT1 or COPT5 (Figure 6C).
Figure 6.
Modulating COPT1, COPT5, or Xa13 Expression Influenced Rice Responses to Xoo Infection.
Each data point represents mean (five replicates for lesion and three replicates for others) ± sd. Gene expression was analyzed by RNA gel blot analysis ([B] and [C]) or qRT-PCR (D). The “a” or “b” indicates a significant difference was detected between a transgenic plant and its corresponding wild-type plants at P < 0.01 or P < 0.05 level, respectively.
(A) COPT1- and COPT5-overexpressing plants showed increased bacterial growth at booting stage. Each T0 plant was divided into three plants in the tillering stage. cfu, colony-forming unit.
(B) COPT1-overexpressing plants showed increased disease symptoms in IRBB13 (xa13) background. IR24, a susceptible near-isogenic line of IRBB13 that carried dominant Xa13.
(C) Increased susceptibility of the transgenic plants to PXO99 was associated with overexpression of COPT1 or COPT5.
(D) Enhanced resistance of the transgenic plants was associated with reduced expression of COPT1, COPT5, or Xa13.
By contrast, COPT1- or COPT5-suppressing plants showed enhanced resistance to PXO99 with lesion areas ranging from 6 to 13% compared with 27% for susceptible wild-type Zhonghua 11 or 2 to 5% compared with 6% for resistant wild-type IRBB13 (Figure 6D; see Supplemental Figure 13 online). In Zhonghua 11 background, Xa13-suppressing plants had lesion areas caused by PXO99 infection ranging from 1 to 17% compared with 27% for wild-type plants (Figure 6D). Xa13-suppressing plants showed on average a greater reduction in lesion areas than did COPT1- or COPT5-suppressing plants. Suppression of Xa13 in other genetic backgrounds also generated highly resistant plants (Chu et al., 2006b). These results suggest that Xa13 may be more important than COPT1 and COPT5 for disease development caused by PXO99. These results also suggest that removing Cu from xylem vessels by activation of XA13, COPT1, and COPT5 may be the basis of Xa13-facilited PXO99 infection.
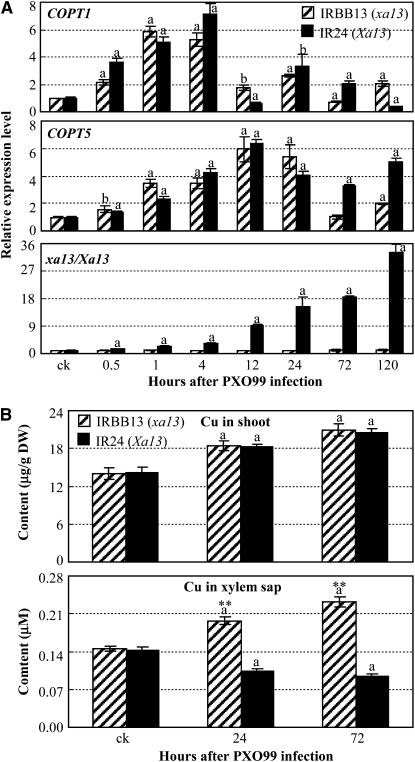
PXO99 Infection Changes Cu Distribution in Rice
The hypothesis that Xa13 may be more important than the COPTs for disease development is supported by the expression patterns of Xa13, COPT1, and COPT5 in a pair of near-isogenic lines, susceptible IR24 (Xa13) and resistant IRBB13 (xa13), after Xoo infection. PXO99 infection specifically induced the expression of dominant Xa13 but not recessive R gene xa13 (Chu et al., 2006b) and expression of xa13 using the Xa13 promoter in xa13-carrying IRBB13 resulted in susceptibility (Yuan et al., 2009). Furthermore, expression of COPT1 and COPT5 was also induced after PXO99 infection in both IR24 and IRBB13 (Figure 7A). The initial induction of COPT1 and COPT5 was accompanied by the induction of Xa13 after Xoo infection (Figure 7A). However, the maximal induction level of Xa13 was 4.6- and 5.2-fold higher than the maximal induction levels of COPT1 and COPT5 in IR24, respectively. These results suggest that activation of COPT1 and COPT5 cannot induce disease, and coactivation of Xa13, COPT1, and COPT5 appears to be critical for disease development in physiologic conditions. Because the three proteins are involved in Cu redistribution, these results also suggest that PXO99 infection may change Cu distribution in rice tissues.
Figure 7.
PXO99 Infection Influenced the Expression of COPT1, COPT5, and Xa13 and the Distribution of Cu in Both Susceptible Rice Line IR24 and Resistant Line IRBB13.
Bars represent mean (three replicates) ± sd. The “a” or “b” indicates a significant difference was detected between noninfected plants and PXO99-infected plants at P < 0.01 or P < 0.05 level, respectively. Two asterisks indicate that a significant difference between IR24 and IRBB13 of the same treatment was detected at P < 0.01 level.
(A) The expression of COPT1, COPT5, and Xa13 analyzed by qRT-PCR.
(B) Cu contents in shoot and xylem sap. DW, dry weight.
This inference is supported by the dynamic distribution of Cu in rice lines with or without Xoo infection. PXO99 infection significantly induced Cu accumulation in shoot tissues of both IR24 and IRBB13; Cu contents in shoots of the two rice lines showed no significant difference either before or after Xoo infection (Figure 7B). However, Cu contents in xylem sap of the same plants used for quantifying Cu contents in shoots were significantly reduced in IR24 and significantly increased in IRBB13 after infection (Figure 7B). The Cu content in the xylem sap of IRBB13 was 2.0- and 2.6-fold higher than that of IR24 at 24 and 72 h after infection. To ascertain whether the different level of Cu in the xylem sap of IR24 was associated with disease development, we analyzed PXO99 growth in xylem sap collected at 72 h after infection. PXO99 growth rates in the xylem sap of IRBB13, which contained 0.23 μM Cu (Figure 7B), were 47, 18, 37, and 20% lower than those in the xylem sap of IR24, which contained 0.09 μM Cu (Figure 7B) at 2, 4, 8, and 12 h after culture, respectively (see Supplemental Figure 14 online). PXO99 growth rates in the xylem sap of IRBB13 and IR24 were similar as those in culturing media supplemented with 0.2 and 0.1 μM CuSO4, respectively. Furthermore, PXO99 growth rates in modified xylem sap of IR24 that was supplemented with CuSO4 to a final concentration of 0.23 μM Cu were similar as those in the xylem sap of IRBB13 (see Supplemental Figure 14 online). These results suggest that PXO99 infection changes Cu distribution in rice leaf tissues and that removing Cu from xylem is associated with development of disease.
DISCUSSION
The changed Cu contents in the transgenic plants described here (Figure 4B; see Supplemental Figure 9 online) suggest that Xa13, COPT1, and COPT5 are involved in Cu redistribution in rice. Specifically, they are associated with removal of Cu from xylem. Cu absorbed from plant roots is transported from xylem to intercellular spaces and then into cells (Fernandes and Henriques, 1991; Burkhead et al., 2009). Thus, activation of XA13, COPT1, or COPT5, which was associated with reduced Cu content in xylem sap (Figure 4B), is expected to be accompanied by increased intracellular Cu content. Although direct measurement of rice intracellular Cu content will help elucidate the role of the three proteins in Cu uptake, this expectation is supported by the following indirect evidence. First, the three proteins complemented the S. cerevisiae ctr1Δctr3Δ mutant phenotype by transporting Cu into cells (Figure 2B). Second, the expression of HMA9, a reporter for intracellular excess Cu, was induced in Xa13-, COPT1- or COPT5-overexpressing plants, which had increased Cu content in the shoot but decreased Cu content in xylem sap (Figure 4C; see Supplemental Figure 11 online). However, the putative intracellular accumulation of Cu may not be solely due to the functions of XA13, COPT1, and COPT5. Decrease of Cu in xylem may indirectly affect the expression of other transporters involved in Cu uptake. This conjecture is supported by the indirect evidence that pathogen infection increased Cu content in the shoot (Figure 7B); however, further study is required to determine which Cu transporters are involved in the Cu accumulation.
Xoo strain PXO99 causes rice disease, in part through activation of Xa13 (Chu et al., 2006b; Yuan et al., 2009). Cu could reduce disease level caused by PXO99 (Figure 5A). These results suggest that redistribution of Cu is associated with the development of disease symptoms specially caused by PXO99. This hypothesis is supported by the following evidence. First, the increased susceptibility of COPT1- or COPT5-overexpressing plants to PXO99 was accompanied by reduced Cu in rice xylem where Xoo grows and spreads, whereas reduced susceptibility of COPT1-, COPT5-, or Xa13-suppressing plants was accompanied by increased Cu in xylem (Figures 4B and 6). However, COPT1 or COPT5 transgenic plants all showed the same levels of susceptibility to Xoo strain PXO61 as wild-type plants. Xa13-suppressing plants also showed susceptibility to Xoo strains PXO61 and PXO86 like wild-type plants (Chu et al., 2006b). Furthermore, PXO61 and PXO86 could not induce Xa13 (Chu et al., 2006b; Yuan et al., 2009). Xa13-overexpressing plants did not show significantly (P > 0.05) increased susceptibility to PXO99 (Yuan et al., 2009). One explanation is that the sensitivity of PXO99 to Cu appears to be dosage dependent (Figure 5B) and the Cu content in the xylem sap of Xa13-overexpressing plants was ~20% higher than that in COPT1- or COPT5-overexpressing plants (Figure 4B). Second, disease development and bacterial growth in the susceptible rice line was accompanied by the reduction of Cu content in xylem (Figure 7B; see Supplemental Figure 14 online). The effect of Cu redistribution on disease development specially caused by PXO99 may be because PXO99 was more sensitive to Cu compared with other Xoo strains (Figure 5B). Another explanation is that excess Cu is toxic to rice cells, which may interfere with plant defense. If this is the case, we may expect the Xa13, COPT1, or COPT5 transgenic plants to have a modified response to other Xoo strains. However, this change has not been observed. Although our results cannot answer whether the tolerance of these Xoo strains other than PXO99 to the in vivo level of Cu is due to mutation, a plasmid mutation causing Cu resistance in Xanthomonas has been reported (Bender et al., 1990).
XA13, COPT1, and COPT5 proteins appear to be cooperatively involved in Cu redistribution. This hypothesis is supported by the following evidence. First, the three proteins interacted with each other in the plasma membrane in planta (Figures 1C and 2A). Second, the three genes showed a similar expression pattern in response to the change of Cu concentration (Figure 4A; see Supplemental Figure 7 online). Third, only coexpression of Xa13, COPT1, and COPT5 complemented the phenotype of S. cerevisiae ctr1Δctr3Δ mutant (Figure 2B). Last, only pathogen-induced coactivation of Xa13, COPT1, and COPT5 reduced Cu content in xylem (Figure 7). However, suppressing any one of the three genes influenced Cu content in transgenic plants. One explanation is that the interaction of XA13, COPT1, and COPT5 may be dosage dependent and decreasing the level of any one of the three proteins may obstruct their interaction. This explanation is supported by the evidence that the expression levels of COPT1 and COPT5 are low (Figures 6B and 6C) and the expression level of Xa13 is also low (Yuan et al., 2009) in wild-type plants; thus, suppressing Xa13 even accompanied by compensative induction of COPT1 and COPT5 or suppressing COPT1 and COPT5 even accompanied by compensative induction of Xa13 (see Supplemental Figure 11 online) significantly increased Cu content in xylem. Overexpressing COPT1 or COPT5 reduced Cu content in xylem and increased susceptibility to PXO99. These transgenic plants are likely presenting amplified phenotypes because the induction of only COPT1 and COPT5 in resistant rice line could not promote disease and removal of Cu from xylem in physiologic conditions (Figure 7), although the complementary phenotypes of these plants with their corresponding suppressing plants helped us understand the role of the two proteins in Cu redistribution.
Previous studies have reported that a single Ctr- or COPT-type plasma membrane–localized protein from human (hCtr1; Zhou and Gitschier, 1997), yeast (Sc Ctr3; Pena et al., 2000), mouse (mCtr1; Lee et al., 2000), Arabidopsis (COPT1, 2, 3, and 5; Kampfenkel et al., 1995; Sancenón et al., 2004), lizard (Ctr1; Riggio et al., 2002), Drosophilia melanogaster (Ctr1A, B, and C; Zhou et al., 2003), or Chlamydomonas reinhardtii (CTR1 and 2; Page et al., 2009) can complement the phenotype of S. cerevisiae ctr1Δctr3Δ mutant. Rice COPT1 and COPT5, which harbor the conserved motifs and residues required for Cu transport (Puig et al., 2007), have 21 to 51% sequence identity and 35 to 68% sequence similarity with these Ctr or COPT proteins (see Supplemental Table 3 online). Furthermore, XA13 appears to be more important than COPT1 and COPT5 in affecting the rice response to PXO99 (Figure 6D). Thus, our results also raise the interesting questions of why three proteins are required for complementation of the S. cerevisiae ctr mutant phenotype and what is the molecular role of XA13 when it interacts with COPT1 and COPT5. Limited data suggest that some proteins interacting with plasma membrane–localized transporter function as chaperones. For example, coexpression of fission yeast SpCtr4 and SpCtr5 is required for complementation of S. cerevisiae ctr mutant or S. pombe ctr mutant because the two proteins are interdependent for trafficking to the plasma membrane and form a heteromeric complex for mediating a high-affinity Cu transport; S. pombe Ctr5 is also required for Ctr4 folding (Zhou and Thiele, 2001; Beaudoin et al., 2006). The rat plasma membrane–localized ion channel protein TRPV2 interacts with the endoplasmic reticulum/Golgi apparatus–localized RGA protein; RGA plays role in trafficking of TRPV2 to plasma membrane (Barnhill et al., 2004; Stokes et al., 2005). The RGA is suggested to have four transmembrane regions and is distantly related to the MtN3/saliva-type proteins (Stokes et al., 2005). XA13 with seven transmembrane regions is also an MtN3/saliva-type protein (Figure 1A), and it has 28% sequence identity and 49% sequence similarity with rat RGA (NP_061333). However, our results suggest that localization of COPT1, COPT5, or XA13 in the plasma membrane does not require the presence of any other two proteins in S. cerevisiae ctr mutant (Figure 3). Further studies are needed to elucidate the kinetics of the interaction of XA13, COPT1, and COPT5 and to determine whether XA13 is associated with the structural stabilization of COPT1 and COPT5. Analysis of the interaction of the three proteins at the three-dimensional level may provide insights into the roles of the three proteins in Cu transport.
The pathogen-induced expression of Xa13 is via the activity of the TAL effector PthXo1 of PXO99 (Yang et al., 2006; Römer et al., 2010). A number of effectors with TAL activity have been characterized (Kay and Bonas, 2009). However, the molecular mechanisms of how the host targets of these effectors help bacterial infection are poorly understood. Only recently, a TAL effector AvrBs3 from Xanthomonas campestris pv Vesicatoria (Xcv), which causes bacterial spot disease on pepper (Capsicum annuum) and tomato (Solanum lycopersicum) plants, was identified to activate upa20 encoding a transcription factor; Upa20 is a key regulator of pepper cell hypertrophy (enlargement) that may play a role in bacterial dispersal (Kay et al., 2007). Upa20 activates the expression of upa7, encoding a putative α-expansin in susceptible pepper plants (Kay et al., 2007). Expansins are wall-loosening proteins of the plant cell wall that play an important role in basal resistance against pathogens; activation of expansin genes results in increased susceptibility to Xoo in rice (Ding et al., 2008). Thus, it is interesting to see if upa20, the target of AvrBs3, helps disease development caused by Xcv via weakening host basal resistance. Xcv and Xoo all belong to the Xanthomonas genus. PXO99 uses a different host mechanism to facilitate its infection, although both pathogens use their TAL effectors to transcriptionally manipulate their host targets. These results suggest that the TAL effectors of bacterial plant pathogens may use various host mechanisms for infection.
The mechanistic understanding of XA13 in rice–Xoo interactions also presents a notable picture of the coevolution of rice and Xoo. Rice bacterial blight disease was first identified in 1884. Chemical control of this disease in rice fields began in the 1950s with the preventive application of Bordeaux mixture (Nino-Liu et al., 2006), which was the world’s first commercially successful fungicide and bactericide, a simple mixture of CuSO4 and hydrated lime that now remains a popular weapon for gardeners and farmers fighting a range of foliar diseases (Ayres, 2004). Some other pesticides also contain Cu as one of the major elements. PXO99 is sensitive to Cu. Xa13 is essential for reproductive development and is also involved in vegetative development (Chu et al., 2006b; Yuan et al., 2009). To survive in rice, this bacterium evolved the specific PthXo1 effector to transcriptionally activate Xa13 (Yang et al., 2006; Römer et al., 2010). Although our data cannot answer whether the evolution of PthXo1 is due to the wide application of Cu-containing pesticides, the bacterium carrying PthXo1 uses the indispensable XA13 to overcome rice defenses by removing toxic Cu from the place where the pathogen grows. To fight this pathogen, rice evolved the recessive R gene xa13, which is the promoter mutant of dominant Xa13 (Chu et al., 2006b). This mutation leaves xa13 unresponsive to PXO99 infection and thus suppresses PXO99 growth in rice (Yuan et al., 2009). PXO99 was identified during the 1980s as Philippine Xoo race 6 (Khush and Angeles, 1999), and xa13 was first discovered as a resistant allele against PXO99 in Indian rice variety BJ1 (Ogawa et al., 1987). IRBB13 carrying xa13 is resistant to >50% of Xoo strains/isolates collected from major rice-growing areas of China and India (Shanti et al., 2001; Singh et al., 2003; Li et al., 2009). These results suggest that Xoo strains, which overcome rice by activating Xa13, may have become the dominant pathogens in at least China and India. Elucidation of the biochemical function of XA13, COPT1, and COPT5 and examining the molecular mechanisms of coevolution of rice and Xoo will help to formulate effective strategies for controlling this bacterial blight. For example, we may use a tissue-specific or Xoo-induced promoter to suppress the expression of Xa13, COPT1, or COPT5 to improve rice disease resistance.
METHODS
Protein–Protein Interaction in Yeast Cells
The yeast two-hybrid assays were conducted using Clontech Matchmaker GAL4 Two-Hybrid System 3 according to the manufacturer’s instructions. Different cDNA fragments encoding the MtN3.1 domain of XA13 were obtained by PCR amplification from rice variety Minghui 63 (Oryza sativa ssp indica) using different primers (see Supplemental Table 4 online).
The split-ubiquitin system was used to investigate the interaction of full-length and truncated membrane proteins. The yeast two-hybrid membrane protein system kit (MoBiTec) was used for this assay according to the manufacturer’s instructions. The full-length and 3′ end of Xa13 cDNA and COPT full-length cDNAs were amplified from rice variety Zhonghua 11 (O. sativa ssp japonica) using specific primers (see Supplemental Table 4 online). Please see Supplemental Methods online for additional details.
Protein Topology Analyses
The subcellular localization of COPT1 and COPT5 was analyzed by fusion of the target protein with the GFP using different primers (see Supplemental Table 5 online). Transient expression of the fusion genes in white onion (Allium cepa) epidermal cells was performed by Agrobacterium tumefaciens–mediated transformation (Ge et al., 2006). The locations of COPT1-FLAG and COPT5-FLAG in transgenic plants were also analyzed by detection of membrane-protein fraction using anti-FLAG antibody. The transmembrane regions and their orientation of XA13 protein were analyzed by the split-ubiquitin system.
The localization of Os COPT1, COPT5, and XA13 proteins in Saccharomyces cerevisiae was analyzed by fusion of these proteins with GFP. Plasmids harboring the fusion genes were transformed into yeast MPY17 cells (Pena et al., 2000). The fluorescence signal was visualized with a LEICA DM4000B fluorescent microscope. Please see Supplemental Methods online for additional details.
Rice Transformation
Agrobacterium-mediated transformation was performed using calli derived from mature embryos of japonica rice variety Zhonghua 11 or Mudanjiang 8 (O. sativa ssp japonica) or indica variety IRBB13 (O. sativa ssp indica) according to published protocols (Lin and Zhang, 2005; Ge et al., 2006). Please see Supplemental Methods online for additional details.
Protein–Protein Interaction in Plants
BiFC assays were applied to study the interaction of XA13 and COPT1 or COPT5 based on the procedure reported previously (Walter et al., 2004). The full-length cDNAs of these genes were amplified using specific primers (see Supplemental Table 4 online) and cloned into the pS1301nYFP or pS1301cYFP vectors. All the plasmids were transformed into tobacco (Nicotiana benthamiana) plants via A. tumefaciens strain GV3101-pM90. YFP signal in leaf epidermal cells was observed using a Leica TCS SP2 AOBS confocal microscope (Leica Microsystems). Coimmunoprecipitation assays were also conducted to study the interaction of XA13 and COPT1 or COPT5. The leaves of rice plants carrying COPT1-FLAG or COPT5-FLAG at maximum-tillering stage were analyzed using anti-XA13 antibody (Yuan et al., 2009) and anti-FLAG antibody (Sigma-Aldrich) by the procedure reported previously (Yuan et al., 2009). Please see Supplemental Methods online for additional details.
Pathogen Inoculation
To evaluate bacterial blight disease, plants were inoculated with Philippine Xoo strain PXO99 (race 6) at the booting (panicle development) stage by the leaf-clipping method (Sun et al., 2004). Disease was scored by measuring the percentage lesion area (lesion length/leaf length) at 10 or 14 d after inoculation.
For studying the effect of Cu on disease development, rice plants at booting stage were inoculated with PXO99 that was premixed with 0.1 to 100 μM CuSO4 for 30 min.
To analyze the response of Xoo to Cu, PXO99 and other Philippine Xoo strains, PXO61 (race 1) and PXO86 (race 2), were first grown in PSA liquid medium (10 g/L tryptone, 10 g/L sucrose, and 1 g/L l-glutamic acid, pH 7.0) at early logarithmic phase and were then grown for 12 h in PSA liquid medium containing 0 to 100 μM CuSO4. Bacterial cell densities were spectrometrically measured at 600 nm.
Gene and Protein Expression Analysis
RNA gel blot analysis was performed as described previously (Zhou et al., 2002). In brief, 20 μg of total RNA was fractionated by electrophoresis and transferred to a nylon filter. The filter was hybridized in hybridization buffer containing [α-32P]dCTP–labeled probe. The hybridization signal on the filter was detected using a fluorescent image analyzing system (Fujifilm FLA-5100). Quantitative RT-PCR (qRT-PCR) was conducted as described by Qiu et al. (2007). In brief, total RNA was treated with DNase I (Invitrogen) to remove contaminating DNA. The cDNA was synthesized from the treated total RNA by M-MLV reverse transcriptase using oligo(dT)15 primer (Promega). Quantitative real-time PCR was performed using the SYBR Premix Ex Taq kit (TaKaRa Biotechnology) on the ABI 7500 Real-Time PCR system (Applied Biosystems) following the manufacturer’s instructions. PCR primers for qRT-PCR are listed in Supplemental Table 6 online. The expression level of rice actin gene was used to standardize the RNA sample for each qRT-PCR. Each qRT-PCR assay was repeated at least twice with similar results, with each repetition having three replicates. Protein expression analysis was performed as described previously (Yuan et al., 2009).
Plant Growth and Treatment
The seeds of rice variety Zhonghua 11 were germinated for 3 d at room temperature on paper soaked with distilled water. After germination, the seedlings were transferred to a net floating on distilled water in a growth chamber in greenhouse with 14 h light and 10 h night for hydroponic culture. After 3 d, seedlings were transferred to a plastic container containing a nutrient solution consisting of 0.7 mM K2SO4, 0.1 mM KCl, 0.1 mM KH2PO4, 2.0 mM Ca(NO3)2, 0.5 mM MgSO4, 10 μM H3BO3, 0.5 μM MnSO4, 0.2 μM CuSO4, 0.5 μM ZnSO4, 0.05 μM Na2MoO4, and 0.1 mM Fe-EDTA for hydroponic culture. The nutrient solution was adjusted daily to pH 5.5 with 1 M HCl and renewed weekly (Ishimaru et al., 2006). To induce a deficiency of Cu, 3-week-old plants were grown for a further 2 weeks in the solution without Cu.
Xylem Sap Collection
The xylem sap was collected as described previously (Ueno et al., 2009; Uraguchi et al., 2009). In brief, rice plants at booting stage were decapitated and xylem exudate was collected using a micropipette for 10 min in the morning. To avoid contamination by symplastic Cu, the initial exudate was discarded.
Determination of Metal Concentration
Rice tissues were washed with distilled water, dried for 1 week at 80°C, and then wet ashed with 11 n HNO3. Concentrations of Cu, Fe, Mn, and Zn were measured using atomic absorption spectroscopy (SPECTR AA220) at wavelengths of 324.8, 248.3, 279.5, and 213.8 nm, respectively.
Heterologous Functional Complementation
Plasmid DNA harboring target gene was transformed into the yeast (S. cerevisiae) ctr1Δctr3Δ double mutant strain MPY17 (MATα,ctr1::ura3::kanR, ctr3::TRP1, lys2-801,his3) using the lithium acetate procedure (Pena et al., 1998). This mutant strain cannot grow on ethanol/glycerol media (YPEG) because it possesses a defective mitochondrial respiratory chain and because of the inability of cytochrome c oxidase to obtain its Cu factor (Pena et al., 1998). The mutant can be complemented by expression of Ctr-type Cu transporters from various organisms. Please see Supplemental Methods online for additional details.
Bioinformatics
Web-based topology prediction programs SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), HMMTOP (http://www.enzim.hu/hmmtop/), Phobius (http://phobius.sbc.su.se/), TMpred (http://www.ch.embnet.org/software/TMPRED), and TopPred (http://www.sbc.su.se/∼erikw/toppred2/) were used to predict the orientation of the transmembrane regions of XA13 protein.
Statistical Analysis
The significant differences between control and treatment of the samples were analyzed by the pairwise t test installed in the Microsoft Office Excel program.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: COPT1 (GQ387494), COPT5 (GQ387495), Xa13 (DQ421395), HMA9 (NP_001058305), IRT1 (AB070226), ZIP4 (AB126089), and actin (X15865). Additional accession numbers for putative COPT family members in rice and other species can be found in Supplemental Tables 2 and 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of the Transmembrane Regions and Their Orientations of XA13 Protein by Split-Ubiquitin System.
Supplemental Figure 2. A Fragment (87 to 161 Amino Acids) of COPT1 Interacted with the MtN3.1 Domain but Not the MtN3.2 Domain of XA13 Protein in the Yeast Two-Hybrid Assay.
Supplemental Figure 3. Analysis of the Interaction between Full-Length XA13 and COPT by Split-Ubiquitin Assays.
Supplemental Figure 4. Plasma Membrane Localization of COPT1 and COPT5.
Supplemental Figure 5. Transgenic Plants Carrying COPT1-FLAG or COPT5-FLAG and Location of COPT1 and COPT5 Proteins.
Supplemental Figure 6. Analyses of Homomeric and Heteromeric Interactions of OsCOPT1, OsCOPT5, and XA13 Proteins by Split-Ubiquitin System.
Supplemental Figure 7. Expression of Xa13, COPT1, and COPT5 in Rice Root Was Influenced by Copper (Cu) Level.
Supplemental Figure 8. Manipulation of COPT1 and COPT5 Expression.
Supplemental Figure 9. Modulating COPT1, COPT5, and Xa13 Expression Influenced Copper (Cu) Accumulation in Rice Roots at Booting Stage.
Supplemental Figure 10. Modulating COPT1, COPT5, and Xa13 Expression Did Not Influence Iron (Fe), Manganese (Mn), and Zinc (Zn) Contents in Rice Shoots at Booting Stage.
Supplemental Figure 11. The Expression Patterns of HMA9, COPT1, COPT5, Xa13, IRT1, and ZIP4 in the Shoots of Transgenic Plants Analyzed by Quantitative RT-PCR.
Supplemental Figure 12. Suppressing COPT1 or COPT5 Increased Accumulation of XA13 Protein in T2 Transgenic Lines Analyzed Using Anti-XA13 Antibody.
Supplemental Figure 13. Enhanced Resistance Was Associated with Reduced Expression of COPT1 or COPT5 in Susceptible Zhonghua 11 and Resistant IRBB13 Backgrounds.
Supplemental Figure 14. Xylem Sap from Resistance IRBB13 Infected with Xoo Strain PXO99 Suppressed PXO99 Growth.
Supplemental Table 1. Prediction of Transmembrane Helices and Their Orientations of XA13 Protein by Different Web-Based Software.
Supplemental Table 2. Putative COPT Family Members in Rice Genome.
Supplemental Table 3. Sequence Identity and Similarity of COPT1 and COPT5 with the Ctr and COPT Proteins from Other Species
Supplemental Table 4. PCR Primers Used for Protein–Protein Interaction Assays.
Supplemental Table 5. PCR Primers Used for Protein Topology Analyses.
Supplemental Table 6. PCR Primers Used for Quantitative RT-PCR Assays.
Supplemental Table 7. PCR Primers Used for Yeast Complementation Experiments.
Acknowledgments
We thank D.J. Thiele for providing yeast mutant and plasmids as well as L.G. Ma for the helpful suggestions and protocol for coimmunoprecipitation assay. This work was supported by grants from the National Program on the Development of Basic Research in China (2006CB101904), the National Natural Science Foundation of China (30930063 and 30921091), and the National Program of High Technology Development of China (2006AA10A103).
References
- Ayres P.G. (2004). Alexis Millardet: France’s forgotten mycologist. Mycologist 18: 23–26 [Google Scholar]
- Barnhill J.C., Stokes A.J., Koblan-Huberson M., Shimoda L.M.N., Muraguchi A., Adra C.N., Turner H. (2004). RGA protein associates with a TRPV ion channel during biosynthesis and trafficking. J. Cell. Biochem. 91: 808–820 [DOI] [PubMed] [Google Scholar]
- Beaudoin J., Laliberté J., Labbé S. (2006). Functional dissection of Ctr4 and Ctr5 amino-terminal regions reveals motifs with redundant roles in copper transport. Microbiology 152: 209–222 [DOI] [PubMed] [Google Scholar]
- Bender C.L., Malvick D.K., Conway K.E., George S., Pratt P. (1990). Characterization of pXV10A, a copper resistance plasmid in Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 56: 170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkow G., Gabbay J. (2004). Putting copper into action: Copper-impregnated products with potent biocidal activities. FASEB J. 18: 1728–1730 [DOI] [PubMed] [Google Scholar]
- Bughio N., Yamaguchi H., Nishizawa N.K., Nakanishi H., Mori S. (2002). Cloning an iron-regulated metal transporter from rice. J. Exp. Bot. 53: 1677–1682 [DOI] [PubMed] [Google Scholar]
- Burkhead J.L., Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. (2009). Copper homeostasis. New Phytol. 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Chu Z., Fu B., Yang H., Xu C., Li Z., Sanchez A., Park Y.J., Bennetzen J.L., Zhang Q., Wang S. (2006a). Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 112: 455–461 [DOI] [PubMed] [Google Scholar]
- Chu Z., Yuan M., Yao J., Ge X., Yuan B., Xu C., Li X., Fu B., Li Z., Bennetzen J.L., Zhang Q., Wang S. (2006b). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20: 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D.A. (1990). Genetics of bactericide resistance in plant pathogenic bacteria. Annu. Rev. Phytopathol. 28: 201–219 [Google Scholar]
- Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., Wang S. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.C., Henriques F.S. (1991). Biochemical, physiological, and structural effects of excess copper in plants. Bot. Rev. 57: 246–273 [Google Scholar]
- Ge X., Chu Z., Lin Y., Wang S. (2006). A tissue culture system for different germplasms of indica rice. Plant Cell Rep. 25: 392–402 [DOI] [PubMed] [Google Scholar]
- Gu K., Yang B., Tian D., Wu L., Wang D., Sreekala C., Yang F., Chu Z., Wang G.L., White F.F., Yin Z. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435: 1122–1125 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Suzuki M., Kobayashi T., Takahashi M., Nakanishi H., Mori S., Nishizawa N.K. (2005). OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 56: 3207–3214 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45: 335–346 [DOI] [PubMed] [Google Scholar]
- Kampfenkel K., Kushnir S., Babiychuk E., Inzé D., Van Montagu M. (1995). Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J. Biol. Chem. 270: 28479–28486 [DOI] [PubMed] [Google Scholar]
- Kay S., Bonas U. (2009). How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12: 37–43 [DOI] [PubMed] [Google Scholar]
- Kay S., Hahn S., Marois E., Hause G., Bonas U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651 [DOI] [PubMed] [Google Scholar]
- Khush G.S., Angeles E.R. (1999). A new gene for resistance to race 6 of bacterial blight in rice, Oryza sativa L. Rice Genet. Newsl. 16: 92–93 [Google Scholar]
- Kim B.E., Nevitt T., Thiele D.J. (2008). Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4: 176–185 [DOI] [PubMed] [Google Scholar]
- Lee J., Peña M.M., Nose Y., Thiele D.J. (2002). Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 277: 4380–4387 [DOI] [PubMed] [Google Scholar]
- Lee J., Prohaska J.R., Dagenais S.L., Glover T.W., Thiele D.J. (2000). Isolation of a murine copper transporter gene, tissue specific expression and functional complementation of a yeast copper transport mutant. Gene 254: 87–96 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim Y.Y., Lee Y., An G. (2007). Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 145: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Song C.F., Pang X.M., Yang Y., Wang J.S. (2009). Analysis of pathotypic and genotypic diversity of Xanthomonas oryzae pv. oryzae in China. J. Phytopathol. 157: 208–218 [Google Scholar]
- Lin Y.J., Zhang Q. (2005). Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 23: 540–547 [DOI] [PubMed] [Google Scholar]
- Ma W., Guttman D.S. (2008). Evolution of prokaryotic and eukaryotic virulence effectors. Curr. Opin. Plant Biol. 11: 412–419 [DOI] [PubMed] [Google Scholar]
- Niño-Liu D.O., Ronald P.C., Bogdanove A.J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7: 303–324 [DOI] [PubMed] [Google Scholar]
- Nose Y., Rees E.M., Thiele D.J. (2006). Structure of the Ctr1 copper trans’PORE’ter reveals novel architecture. Trends Biochem. Sci. 31: 604–607 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Lin L., Tabien R.E., Khush G.S. (1987). A new recessive gene for resistance to bacterial blight of rice. Rice Genet. Newsl. 4: 98–100 [Google Scholar]
- Page M.D., Kropat J., Hamel P.P., Merchant S.S. (2009). Two Chlamydomonas CTR copper transporters with a novel Cys-Met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena M.M., Puig S., Thiele D.J. (2000). Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275: 33244–33251 [DOI] [PubMed] [Google Scholar]
- Peña M.M.O., Koch K.A., Thiele D.J. (1998). Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 2514–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Andrés-Colás N., García-Molina A., Peñarrubia L. (2007). Copper and iron homeostasis in Arabidopsis: Responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ. 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Qiu D., Xiao J., Ding X., Xiong M., Cai M., Cao Y., Li X., Xu C., Wang S. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Reinders A., Schulze W., Thaminy S., Stagljar I., Frommer W.B., Ward J.M. (2002). Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 10: 763–772 [DOI] [PubMed] [Google Scholar]
- Riggio M., Lee J., Scudiero R., Parisi E., Thiele D.J., Filosa S. (2002). High affinity copper transport protein in the lizard Podarcis sicula: Molecular cloning, functional characterization and expression in somatic tissues, follicular oocytes and eggs. Biochim. Biophys. Acta 1576: 127–135 [DOI] [PubMed] [Google Scholar]
- Römer P., Hahn S., Jordan T., Strauss T., Bonas U., Lahaye T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318: 645–648 [DOI] [PubMed] [Google Scholar]
- Römer P., Recht S., Lahaye T. (2009a). A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. USA 106: 20526–20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P., Recht S., Strauss T., Elsaesser J., Schornack S., Boch J., Wang S., Lahaye T. (2010). Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 187: 1048–1057 [DOI] [PubMed] [Google Scholar]
- Römer P., Strauss T., Hahn S., Scholze H., Morbitzer R., Grau J., Bonas U., Lahaye T. (2009b). Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150: 1697–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancenón V., Puig S., Mateu-Andrés I., Dorcey E., Thiele D.J., Peñarrubia L. (2004). The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Biol. Chem. 279: 15348–15355 [DOI] [PubMed] [Google Scholar]
- Sancenón V., Puig S., Mira H., Thiele D.J., Peñarrubia L. (2003). Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Shanti M.L., George M.L.C., Vera Cruz C.M., Bernardo M.A., Nelson R.J., Leung H., Reddy J.N., Sridhar R. (2001). Identification of resistance genes effective against rice bacterial blight pathogen in eastern India. Plant Dis. 85: 506–512 [DOI] [PubMed] [Google Scholar]
- Singh S., Sodhi M., Vikal Y., George M.L.C., Bala G.S., Mangat G.S., Garg M., Sidhu J.S., Dhaliwal H.S. (2003). DNA fingerprinting and virulence analysis of Xanthomonas oryzae pv. oryzae isolates from Punjab, northern India. Euphytica 130: 107–115 [Google Scholar]
- Stavrinides J., McCann H.C., Guttman D.S. (2008). Host-pathogen interplay and the evolution of bacterial effectors. Cell. Microbiol. 10: 285–292 [DOI] [PubMed] [Google Scholar]
- Stokes A.J., Wakano C., Del Carmen K.A., Koblan-Huberson M., Turner H. (2005). Formation of a physiological complex between TRPV2 and RGA protein promotes cell surface expression of TRPV2. J. Cell. Biochem. 94: 669–683 [DOI] [PubMed] [Google Scholar]
- Sun X., Cao Y., Yang Z., Xu C., Li X., Wang S., Zhang Q. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37: 517–527 [DOI] [PubMed] [Google Scholar]
- Ueno D., Koyama E., Kono I., Ando T., Yano M., Ma J.F. (2009). Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in rice. Plant Cell Physiol. 50: 2223–2233 [DOI] [PubMed] [Google Scholar]
- Uraguchi S., Mori S., Kuramata M., Kawasaki A., Arao T., Ishikawa S. (2009). Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 60: 2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Yang B., Sugio A., White F.F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 103: 10503–10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Chu Z., Li X., Xu C., Wang S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 50: 947–955 [DOI] [PubMed] [Google Scholar]
- Zhou B., Gitschier J. (1997). hCTR1: A human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. USA 94: 7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Peng K., Zhaohui C., Wang S., Zhang Q. (2002). The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci. China C Life Sci. 45: 449–467 [DOI] [PubMed] [Google Scholar]
- Zhou H., Cadigan K.M., Thiele D.J. (2003). A copper-regulated transporter required for copper acquisition, pigmentation, and specific stages of development in Drosophila melanogaster. J. Biol. Chem. 278: 48210–48218 [DOI] [PubMed] [Google Scholar]
- Zhou H., Thiele D.J. (2001). Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276: 20529–20535 [DOI] [PubMed] [Google Scholar]