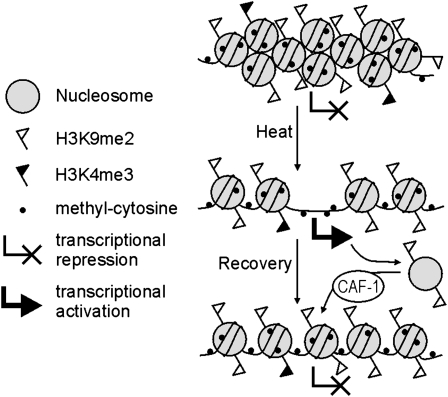
This work shows that prolonged heat stress leads to activation of specific endogenous repeats. Unexpectedly, this heat-induced transcriptional activation does not require DNA demethylation or change of histone modification, but rather involves a dramatic reduction of nucleosome association with the DNA.
Abstract
Epigenetic factors determine responses to internal and external stimuli in eukaryotic organisms. Whether and how environmental conditions feed back to the epigenetic landscape is more a matter of suggestion than of substantiation. Plants are suitable organisms with which to address this question due to their sessile lifestyle and diversification of epigenetic regulators. We show that several repetitive elements of Arabidopsis thaliana that are under epigenetic regulation by transcriptional gene silencing at ambient temperatures and upon short term heat exposure become activated by prolonged heat stress. Activation can occur without loss of DNA methylation and with only minor changes to histone modifications but is accompanied by loss of nucleosomes and by heterochromatin decondensation. Whereas decondensation persists, nucleosome loading and transcriptional silencing are restored upon recovery from heat stress but are delayed in mutants with impaired chromatin assembly functions. The results provide evidence that environmental conditions can override epigenetic regulation, at least transiently, which might open a window for more permanent epigenetic changes.
INTRODUCTION
Terrestrial plants are inevitably exposed to temperature changes, and their sessile lifestyle requires that they deal with daily and seasonal temperature fluctuations in situ. In addition to sophisticated adaptation mechanisms for these regular variations, they have developed additional signaling, repair, and response functions that are activated upon heat stress exerted by exceptionally high temperatures. Key components of this heat response, among several other pathways involved in protecting various cellular functions and induced upon extreme heat, are heat shock proteins and their corresponding heat shock transcription factors (Kotak et al., 2007). Interestingly, heat stress leads to increased genetic instability and higher rates of somatic homologous recombination (Lebel et al., 1993; Pecinka et al., 2009). Since somatic homologous recombination is, at least partially, controlled by the configuration of chromatin at the target loci (Takeda et al., 2004; Endo et al., 2006; Kirik et al., 2006), heat stress could potentially exert its effect on genetic stability through modification of chromatin configuration and the accessibility of DNA for repair and recombination. Recently, a specific variant of histone H2A has been identified as a thermosensor, regulating temperature-dependent gene expression (Kumar and Wigge, 2010). Furthermore, it has been claimed that heat-induced acclimation can be transmitted to subsequent generations via an epigenetic mechanism (Whittle et al., 2009), although heat-induced somatic recombination rates were not elevated beyond the exposed generation (Pecinka et al., 2009). Thus, a connection between heat stress, chromatin, and epigenetically regulated gene expression is widely thought to occur but as yet has been poorly studied.
We chose to address this topic in Arabidopsis thaliana, which is sensitive to elevated temperatures (Binelli and Mascarenhas, 1990) and has a wide range of well-characterized epigenetic regulators and target genes (for review, see Henderson and Jacobsen, 2007). The numerous repetitive transgenic markers and endogenous repeats in Arabidopsis are especially suitable for studying epigenetic regulatory mechanisms. In general, expression of repeats is suppressed by transcriptional gene silencing (TGS), concomitant with high levels of DNA methylation, inactive chromatin marks, and chromatin compaction (e.g., Soppe et al., 2002). However, repetitive elements can be activated upon developmental reprogramming during pollen and seed development (Mosher et al., 2009; Slotkin et al., 2009) or due to a lack of several trans-acting epigenetic regulators (e.g., Lippman et al., 2003). Thus, they represent suitable indicators to score interference with epigenetic regulation under stress conditions.
Here, we show that prolonged heat stress leads to a transient transcriptional activation of transgenic as well as specific endogenous repeats that are regulated by TGS. These changes are independent of senescence, DNA repair, and heat stress signaling. Unexpectedly, heat-induced transcriptional activation does not require DNA demethylation. Whereas histone modifications show only minor variation upon heat stress, there is evidence for a dramatic reduction in the number of nucleosomes associated with DNA. This reduction in nucleosome density is not restricted to heat stress–activated sequences but occurs throughout the genome. Efficient resilencing of some of the activated targets during a recovery phase seems to require the Chromatin Assembly Factor 1 (CAF-1) complex (Kaya et al., 2001), probably for its activity in reloading nucleosomes. Nevertheless, the higher order of heterochromatin is lost during prolonged heat stress, and this effect persists in exposed tissue beyond transcriptional resilencing.
RESULTS
Long Heat Stress Alleviates TGS
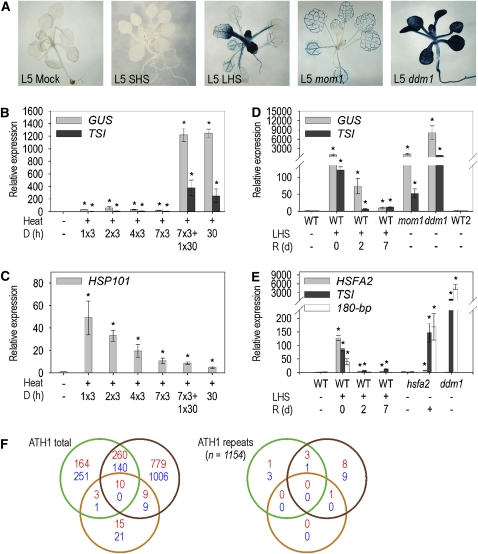
To investigate whether heat stress has an effect on epigenetically regulated transcription, we exposed 21-d-old in vitro grown plants of line L5, carrying a single insert of a multicopy P35S:GUS gene suppressed by TGS (Morel et al., 2000; Probst et al., 2004), to different regimes of elevated temperature and screened for transcriptional activation of β-glucuronidase (GUS) by histochemical staining. Whereas short heat stress (SHS) for 3 h at 37°C had no visible effect, very strong GUS expression was achieved with long heat stress (LHS) for 30 h at 37°C (Figure 1A). Quantitative RT-PCR revealed minor activation after SHS but more than 1000× induction after LHS (Figure 1B). The effect of LHS could not be recapitulated by multiple repetitions of SHS on subsequent days, and prior SHS did not significantly change the amount of GUS transcript upon subsequent LHS (Figure 1B). The same applies to TRANSCRIPTIONALLY SILENT INFORMATION (TSI), an endogenous family of repeats regulated by TGS (Steimer et al., 2000) (Figure 1B) and centromeric 180-bp repeats (see Supplemental Figure 1A online). By contrast, HEAT SHOCK PROTEIN101 (HSP101) was induced by a single SHS pulse and adaptively declined upon repeated SHS or LHS in all heat treatments (Figure 1C). To determine the kinetics of activation, we quantified GUS, TSI, and HSP101 transcripts at short time intervals from 1 to 48 h at 37°C (see Supplemental Figures 1C and 1D online). As expected, HSP101 was induced after 1 h of stress but strongly reduced at later time points despite ongoing heat treatment (see Supplemental Figure 1D online). GUS and TSI were notably activated only upon stress exposure longer than 12 or 18 h, respectively, and longer stress generally correlated with higher expression of these repeats (see Supplemental Figure 1C online). The extent and duration of activation of several marker genes were determined immediately after LHS as well as after 2 and 7 d of recovery and compared with levels in the TGS mutants decrease in dna methylation1 (ddm1) (Vongs et al., 1993) and morpheus’ molecule1 (mom1) (Amedeo et al., 2000). LHS-induced GUS, TSI, and 180-bp transcripts reached levels comparable to those in mom1 but not in ddm1 (Figure 1D; see Supplemental Figure 1B online). A recovery phase of only 2 d led to the disappearance of the majority of marker gene transcripts, revealing restoration of TGS. Therefore, TGS of several repetitive sequences can be transiently alleviated by an extended period of heat stress.
Figure 1.
Long Heat Stress Transiently Abolishes TGS.
(A) GUS-stained L5 plantlets after mock, short (SHS) and long (LHS) heat stress, and nontreated after crossing to mom1 and ddm1 mutants.
(B) to (E) qRT-PCR for RNA of TGS targets (GUS and TSI) and heat stress marker genes (HSP101 and HSFA2) in the wild type (WT = Col-0; WT2 = Col-0/Zh) and mutants (mom1, ddm1, and hsfa2; see text for description) after heat stress (D = frequency of application × duration in hours and R = recovery time in days), LHS = 30 h. Error bars indicate sd of triplicate measurement. Statistically significant differences between mock-treated wild types and stressed (or mutant) samples are indicated by asterisks (t test, P < 0.05).
(F) Differential gene expression (log fold changes of ≥2 [red] and ≤−2 [blue]) between mock and SHS (SHS R0, green circle) and mock versus LHS without (LHS R0, brown circle) or with (LHS R2, orange circle) recovery. ATH1 total, all probe sets; ATH1 repeats, probe sets representing repetitive elements (Slotkin et al., 2009).
LHS was effective but permitted survival (see Supplemental Figure 1F online) and seed set. To exclude that the transcriptional activation of normally silent genes was a side effect of DNA damage and/or senescence, we assayed transcript levels of the corresponding marker genes RAD51 (Doutriaux et al., 1998) and OXIDOREDUCTASE At4g10500 (Schmid et al., 2005), respectively. RAD51 was unaffected by LHS, and the oxidoreductase was induced only after recovery when GUS/TSI/180-bp transcripts had already disappeared (see Supplemental Figure 1E online). Moreover, the observed activation does not depend on heat stress signaling since mutants lacking HEAT SHOCK FACTOR A2 (HSFA2) (Nishizawa et al., 2006) express TSI and 180-bp after LHS as efficiently as the wild type (Figure 1E). Thus, the activation of repeats is independent of DNA repair, senescence, and heat signaling.
LHS Affects a Subset of Transcriptionally Silenced Endogenous Targets
To test the genome-wide effect on TGS targets, we performed transcriptome profiling on ATH1 Affymetrix arrays from mock- and LHS-treated plants directly (LHS R0) or after 2 d of recovery (LHS R2) and compared the results with published data from treatments for 3 h at 38°C (Kilian et al., 2007), here referred to as short heat stress (SHS R0). After LHS R0, 1058 and 1155 probe sets defining transcription units were significantly up- or downregulated (log2 fold change ≥2 or ≤−2, respectively) compared with the control. Among these, only 270 and 140 probe sets were up- or downregulated also after SHS (Figure 1F), indicating that many responses are specific for LHS. However, LHS-induced changes were transient, since only 19 (1.8%) and 9 (0.8%) genes remained up- or downregulated, respectively, after recovery. To focus on sequences that are known to be under epigenetic regulation, we extracted the data for the 1154 probe sets corresponding to repeats (Slotkin et al., 2009). These were barely affected by SHS (four each up- or downregulated) and only moderately by LHS (12 and 10 up- or downregulated) (Figure 1F, Table 1; see Supplemental Table 1 online). However, the majority (nine up- and nine downregulated) responded specifically to LHS. Reexamination of transcription of COPIA78, MULE2, ATHILA6A, CYP40, and ATLANTYS2A by quantitative RT-PCR (qRT-PCR) indeed verified significantly higher expression after LHS. With the exception of COPIA78, all returned to their previous levels during early recovery (Table 1; see Supplemental Figures 2A to 2C online). COPIA78, an long terminal repeat retrotransposon family, represents an interesting exception: it is not regulated by DDM1 and MOM1, showed a strong response to LHS, and had delayed resilencing during recovery (Table 1; see Supplemental Table 1 and Supplemental Figure 2A online). GP2NLTR, TA11, COPIA41, and IS112A were downregulated by LHS and regained or even surpassed their original level of expression during recovery (Table 1; see Supplemental Figures 2E and 2F online). We further analyzed expression of IG/LINE and soloLTR, two targets of RNA-directed DNA methylation (RdDM) (Huettel et al., 2006) that are strongly activated upon mutation of RNA-DEPENDENT RNA POLYMERASE2 (RDR2). After LHS, they were transcribed even more than in rdr2 and silenced after recovery (Table 1; see Supplemental Figure 2D online). In short, LHS activated several repeats that are not transcribed after SHS. The patterns of response suggest a transient, complex, and divergent disturbance of epigenetic silencing pathways.
Table 1.
The Effects of LHS, mom1, and ddm1 on the Transcriptional Activity of Repeats after 0, 2, and 7 d of Recovery
| Transcriptional Fold Changesa |
||||||||
| Wild-Type LHSb |
mom1 |
ddm1 |
||||||
| Target | ORF | R0 | R2 | R7 | Mock | LHS R0 | Mock | LHS R0 |
| COPIA78 | Multiple | +++ | +++ | +++ | 0 | +++ | 0 | +++ |
| GUS (L5) | – | +++ | ++ | + | +++ | +++ | +++ | +++ |
| TSI | Multiple | +++ | + | + | +++ | +++ | +++ | +++ |
| IG/LINE | At5g27845 | +++ | ++ | 0 | ++ | +++ | 0 | +++ |
| soloLTR | – | +++ | + | 0 | +++ | +++ | 0 | +++ |
| MULE2 | At2g15800 | +++ | 0 | 0 | +++ | +++ | +++ | +++ |
| ATHILA | Multiple | ++ | + | 0 | ++ | ++ | +++ | +++ |
| HPT (A-line) | – | ++ | 0 | 0 | + | n.d. | +++ | n.d. |
| 180-bp | Multiple | ++ | 0 | 0 | + | ++ | +++ | +++ |
| CYP40 | At2g15790 | + | 0 | 0 | + | + | + | + |
| ATLANTYS2A | At3g60930 | + | 0 | 0 | 0 | + | + | ++ |
| IS112A | At5g35490 | − | + | ++ | + | − | + | − |
| COPIA4I | At4g16870 | −c | 0 | - | - | − | + | ++ |
| TA11 | At1g72920 | − | 0 | + | 0 | - | 0 | 0 |
| GP2NLTR | At2g15040 | − | 0 | + | 0 | - | 0 | 0 |
R, recovery; ORF, open reading frame; n.d., not determined.
qRT-PCR data: +++, >400; ++, 40 to 400; +, 4 to 40; 0, −2 to 4; -, −4 to −2; −, −8 to −4.
Two different wild types (WT and WT2) were included to match the different mutants as closely as possible. Unless stated otherwise, expression of the target did not differ significantly, and they are shown together.
−2 to 4 for WT2.
Another striking result from the microarray analysis was the detection of a gene cluster located close to the centromere of chromosome 2 in which 29 out of 32 genes represented on the ATH1 array were upregulated upon LHS. This cluster represents mitochondrial DNA inserted in the nuclear genome, where it has acquired some polymorphisms that allow nuclear and organelle copies to be distinguished (Stupar et al., 2001). Several mitochondrial transcripts were shown to accumulate transiently upon SHS (Adamo et al., 2008). After LHS, nuclear transcripts were also found for two of three tested genes (see Supplemental Table 2 online). The nuclear copies seem to have maintained the ancestral potential to respond to heat, but the heterochromatic neighborhood of the cluster may prevent transcription upon SHS.
Transcriptional Activation Occurs Independently of DNA Demethylation
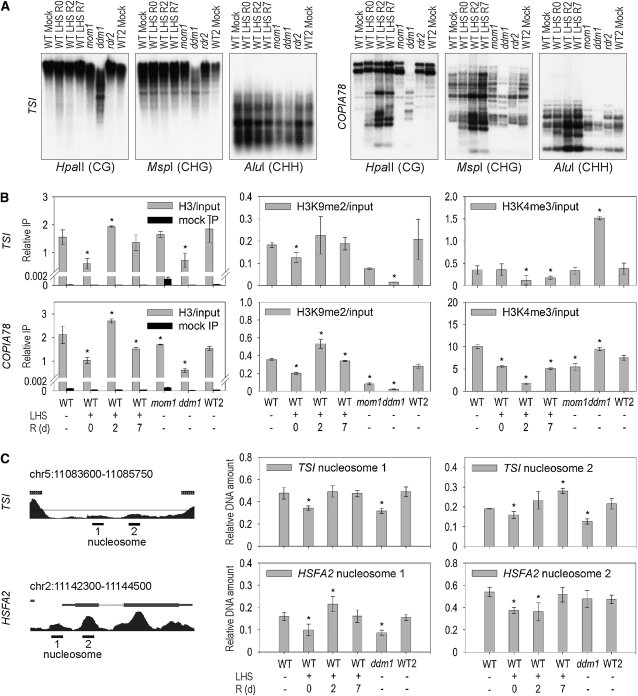
Release of TGS is often, but not obligatorily, correlated with loss of inactivating chromatin marks, such as DNA methylation and/or histone modifications. We therefore assayed both parameters after LHS. The total amount of 5-methyl deoxycytidine, reduced to one-third in ddm1, was at wild-type levels (6.4%) with or without LHS (see Supplemental Figure 3A online). DNA gel blots with methylation-sensitive restriction enzymes did not reveal demethylation at TSI, GUS, or 180-bp repeats (all highly methylated in the wild type) after LHS treatment or during recovery (Figure 2A; see Supplemental Figure 3B online). Even the CG-containing transcription factor binding site in the cauliflower mosaic virus 35S promoter of the GUS gene, demethylated in ddm1, remains methylated despite LHS-induced transcription (see Supplemental Figure 3C online). By contrast, smaller bands indicating nonmethylated CG, CHG, and CHH sites in COPIA78 appeared upon LHS (Figure 2A). Strikingly, maximum demethylation was reached only after 2 d of recovery when RNA levels were already declining, implying that it follows rather than precedes activation. Thus, LHS-induced activation of several TGS targets occurs despite DNA methylation, although this modification can be removed temporarily from a specific subset of targets.
Figure 2.
Chromatin Analysis after LHS.
(A) Methylation analysis of TSI and COPIA78 by DNA gel blotting of LHS samples without (LHS R0) or with recovery for 2 or 7 d (LHS R2 and LHS R7).
(B) Histone H3 occupancy and modifications (H3K9me2 and H3K4me3), relative to input, were assessed by ChIP and qPCR.
(C) Nucleosome occupancy analysis by MNase I sensitivity assay at a representative TSI locus and at HSFA2. The positions of the PCR-amplified regions with respect to nucleosomes are indicated (left).
(B) and (C) Error bars indicate sd of triplicate measurement. R, recovery time in days. Statistically significant differences between mock-treated wild types and stressed (or mutant) samples are indicated by asterisks (t test, P < 0.05).
Transcriptional Activation Does Not Persist into the Next Generation
We tested whether activation of the TGS markers in heat-exposed plants would also affect their progeny. However, no transcriptional activation was detected for TSI, GUS, or COPIA78 in the first poststress generation (S1) of mock- and LHS-treated plants (see Supplemental Figure 4A online). Congruently, all repeats were fully methylated in S1, including the originally demethylated COPIA78 (see Supplemental Figure 4B online). This suggests that heat stress–induced transcriptional activation is not heritable, even for the exceptional sequences that had partially lost DNA demethylation upon stress treatment.
Heat Stress Reduces Nucleosome Occupancy
We analyzed the chromatin of LHS-treated plants by chromatin immunoprecipitation (ChIP) for hallmarks of inactive repeats, the presence of lysine 9 dimethylation (H3K9me2), and lack of lysine 4 trimethylation (H3K4me3) at histone H3 subunits (Fuchs et al., 2006). As described (Gendrel et al., 2002), histones at repeats in ddm1 lose H3K9me2 and gain H3K4me3; this includes the promoters of the nonactivated COPIA78 and HSP101 (Figure 2B; see Supplemental Figure 5 online). A significant reduction of H3K9me2, but no gain of H3K4me3, was observed directly after LHS (Figure 2B; see Supplemental Figure 5 online). Remarkably, ChIP with antibodies recognizing H3 irrespective of modifications revealed reduced nucleosome loading in ddm1, but also after LHS. All the Arabidopsis sequences analyzed had partially lost H3 association, regardless of whether they were transcribed, remained silent, or were intergenic (Figure 2B; see Supplemental Figure 5B online). An independent experiment using an antibody recognizing histone H4 (see Supplemental Figure 6 online) gave a similar result, indicating that the loss was not specific for H3 but rather was due to reduced overall nucleosome occupancy. The loss of nucleosomes was transient; all analyzed target sequences regained H3 and H4 association fully or to a large extent during recovery. In some cases, values during recovery were even higher than after mock treatments (Figure 2B). DNA fragments obtained by ChIP cover sequences in the range of 200 to 1000 bp. To obtain higher resolution, we analyzed sensitivity of defined regions by partial digestion of chromatin with Micrococcal Nuclease I (MNase I), followed by qPCR with primers located at defined nucleosome binding sites. These regions were chosen according to the genome-wide nucleosome positioning map of Arabidopsis (Chodavarapu et al., 2010) or on the basis of bioinformatic prediction (Segal et al., 2006). The assay confirmed reduced nucleosome occupancy at TSI repeats and COPIA78 as well as at the 5′ prime regions of three genes strongly upregulated after LHS (HSFA2, eEF1Balpha1, and UBIQUINOL-CYTOCHROME C REDUCTASE; Figure 2C; see Supplemental Figure 7 online). In all cases, higher sensitivity was detected immediately after LHS at the nucleosome overlapping the transcription start site and also (except for eEF1Balpha1) for the next nucleosome downstream (Figure 2C; see Supplemental Figures 7D and 7E online). In agreement with the ChIP data, nucleosomes tended to be reloaded, and sometimes even hyperaccumulated, during recovery (Figure 2C; see Supplemental Figures 7D and 7E online). Only an intergenic region that had reduced nucleosome occupancy, as evident from ChIP, did not show increased MNase I sensitivity. Thus, LHS causes an immediate and prevalent reduction in nucleosome occupancy, followed by reloading upon return to ambient temperatures.
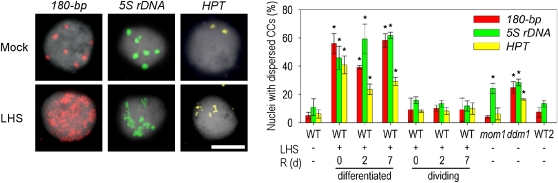
Heat Stress Causes Loss of Chromocenter Organization
The significant loss of nucleosomes after LHS prompted us to investigate global chromatin organization by fluorescence in situ hybridization (FISH). 180-bp and 5S rDNA repeats as well as a HYGROMYCIN PHOSPHOTRANSFERASE (HPT) multicopy transgene (all transcriptionally upregulated after LHS) form compact heterochromatic chromocenters (CCs) in >90% of interphase nuclei (Fransz et al., 2002; Probst et al., 2003). These were significantly dispersed in ~50% of nuclei from LHS-treated leaves (Figure 3). This rate is even higher than in ddm1, indicating substantial heterochromatin decondensation. The LHS-induced CC dissociation was persistent throughout recovery for up to 1 week (Figure 3) when leaves started to become senescent. Interestingly, decondensation was not observed in nuclei from meristematic tissue or in leaves grown after the LHS treatment (Figure 3).
Figure 3.
LHS Leads to Loss of Heterochromatin Compaction.
Heterochromatin condensation was analyzed by FISH with 180-bp (red, left), 5S rDNA (green, middle), and HPT (yellow, right) probes in nuclei (n = 240/experimental point) of mock- and LHS-treated plants and mutant controls. Bar = 5 μm. Error bars indicate sd of triplicate measurement. R, recovery time in days. Statistically significant differences between mock-treated wild types and stressed (or mutant) samples are indicated by asterisks (t test, P < 0.05).
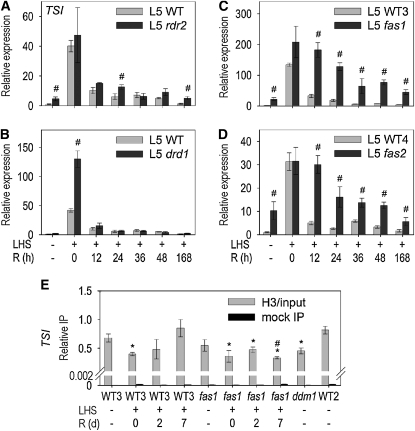
CAF-1 Is Required for Efficient Resilencing
To identify how epigenetic regulation is reestablished after persistent heat stress, we compared the LHS response in mutants lacking well-defined epigenetic regulators. The extent of TSI induction by LHS and the kinetics of resilencing were similar between the wild type, rdr2 (Figure 4A), and other RdDM mutants. Only drd1, which lacks a plant-specific putative chromatin remodeling factor of the SNF2 family (Kanno et al., 2004), showed enhanced LHS-induced transcription (Figure 4B). Nevertheless, the time course of resilencing in drd1 was comparable to that in the wild type (Figure 4B), rendering involvement of RdDM unlikely. By contrast, fas1 and fas2 expressed LHS-induced TSI sequences long after these have been silenced in the wild type (Figures 4C and 4D). These mutants lack different subunits of CAF-1, which loads nucleosomes onto freshly replicated DNA (Kaya et al., 2001). Using ChIP, we tested the kinetics of nucleosome occupancy on TSI repeats in heat-stressed wild-type and fas1 plants (Figure 4E). Wild-type plants lost nucleosomes immediately after stress, with the original level being restored during recovery. By contrast, fas1 plants had already mildly reduced nucleosome occupancy in mock-treated samples. This was further reduced after LHS, and there was no recovery even after 7 d. This is in agreement with leaky TSI silencing in fas1 mutants (Figures 4C and 4D) and may explain the delayed TSI resilencing in CAF-1 mutants, suggesting that CAF-1 is important for efficient restoration of silencing after LHS (Figure 5).
Figure 4.
Involvement of CAF-1 in Resilencing.
(A) to (D) Kinetics of TSI expression after LHS quantified by qRT-PCR during recovery (R = recovery time in hours) in the wild type, RdDM, and CAF-1 mutants (see text for description). WT = Col-0, WT3 = Enk/Col-0, and WT4 = Ler/Col-0.
(E) Histone H3 occupancy (relative to input) was assessed by ChIP and qPCR. R = recovery time in days.
(A) to (E) Error bars indicate sd of triplicate measurement.
(A) to (D) Statistically significant differences between wild-type and mutant samples at the same time points are indicated by # (t test, P < 0.05).
(E) Statistically significant differences between mock-treated and heat-stressed plants (wild type or mutants, respectively) are indicated by asterisks (t test, P < 0.05).
Figure 5.
Model of Heat Stress–Induced Epigenetic Changes.
Transcriptionally inactive repeats reside in compact, heavily DNA-methylated heterochromatin with substantial H3K9 dimethylation and low levels of H3K4 trimethylation (top); after LHS, nucleosomes are partially removed rather than their modifications being altered, while heterochromatin becomes decondensed and transcriptionally active (middle). During recovery, nucleosomes are reloaded (partially via CAF-1 activity) and dimethylated at H3K9, but without reconstituting compact heterochromatin (bottom).
DISCUSSION
We have shown that several classes of repetitive elements in the Arabidopsis genome that are silenced by epigenetic regulation at ambient temperature were transcriptionally activated upon exposure of plants to prolonged periods of heat stress. These conditions also caused differential expression of a subset of protein-coding genes. Although there was some overlap with the response to SHS pulses, the pattern and kinetics of altered expression were surprisingly different. This was not due to the detrimental effects of the prolonged application of stress, since plants could recover completely from the stress, and the transcriptional response was transient and independent of DNA damage signaling and senescence. The fact that transcriptional activation was limited to heat stress of >24 h suggests that it is a rather specific response, distinct from that of the regular diurnal changes in environmental conditions.
The consequences of long-lasting heat treatment were also distinct from genetic interference with transcriptional silencing. While several targets showed responses under heat stress similar to those of epigenetic mutants, others reacted differently. COPIA4I and IS112A elements were downregulated by long exposure to heat but were weakly affected by mom1 and upregulated by ddm1 (Table 1). In addition, heat stress activated the RdDM targets IG/Line and soloLTR to an extent beyond that seen in the rdr2 mutant. There were also unexpected differences in mechanistic aspects. In contrast with several other stress effects (reviewed in Madlung and Comai, 2004; Chinnusamy and Zhu, 2009), or upon loss of the epigenetic regulators DDM1, MET1, HOG1, CMT3, and VIM1 (Chan et al., 2005; Woo et al., 2007), LHS-induced transcriptional activation of repeats occurred without loss of DNA methylation, thereby resembling the effect of mutations in MOM1, FAS1, FAS2, BRU1, and RPA2 (Amedeo et al., 2000; Takeda et al., 2004; Elmayan et al., 2005). The only specifically LHS-induced element (COPIA78), although repetitive and with heterochromatic marks, was not expressed in the ddm1 mutant, and demethylation here followed rather than preceded transcription. This is similar to the Tam3 transposon of Antirrhinum majus, which is activated and demethylated at low temperature (15°C) and in which DNA demethylation coupled to replication is a consequence of transcriptional activation (Hashida et al., 2003, 2006). Methylation within the body of a tobacco (Nicotiana tabacum) gene has been shown to be reduced by heavy metal and oxidative stress (Choi and Sano, 2007). However, it is not clear whether this is required for activation since the promoter was also unmethylated prior to stress application (Choi and Sano, 2007). Transcriptional activation without demethylation can occur also upon other stress treatments (Lang-Mladek et al., 2010). In general, neither demethylation nor removal of histone modifications appears to be essential for the activation of several repeats by heat stress. Together with the relatively unaffected (according to microarray data) expression levels of known TGS genes in LHS-treated plants, this indicates that heat stress causes a complex transcriptional response not limited to a specific pathway or factor in the regulation of repeat silencing.
Looking for common features of genes differentially expressed after LHS, it was striking that six out of 10 downregulated repeats (COPIA4I, COPIA4LTR, IS112A, TA11, TAT1, and GP2NLTR) belong to loci known to determine disease resistance, and some of these genes also had reduced transcript levels. For example, the RECOGNITION OF PERONOSPORA PARASITICA4 (RPP4) locus, associated with COPIA4 repeats, contains three assigned open reading frames: At4g16860 (RPP4 + COPIA4LTR), At4g16870 (COPIA4I), and At4g16880, all of which are downregulated after heat stress. This resembles the finding that genes within resistance clusters, including neighboring repeats, are often coregulated (Yi and Richards, 2007). Therefore, sequences in such a genomic neighborhood may be affected by LHS only indirectly and could reflect heat stress effects on the resistance genes, followed by spreading of transcriptional silencing to the close vicinity. Indeed, even moderately increased temperatures can reduce resistance to biotic stress by pathogens (Wang et al., 2009), although the expression levels of these genes were not analyzed in this study. The upregulated and coregulated cluster of what were originally mitochondrial genes integrated into the nuclear genome may have maintained its heat response, with additional epigenetic regulation imposed by its heterochromatic environment.
Changes in histone modifications and/or expression levels of the enzymes exerting these changes have been described for stress responses in several experimental systems (reviewed in Chinnusamy and Zhu, 2009). The reduction in the inactivating chromatin mark H3K9me2 relative to the input in our experiments could be interpreted as confirmation of such a correlation, as could the small increase of H3K9me2 at some targets after 2 d of recovery, which is in agreement with an increased expression level of histone methyltransferase KYP1 immediately after heat stress (according to microarray data). However, the quantification of H3 and H4 association and cleavage efficiency by MNase I document that prolonged heat stress resulted in a partial dissociation of histones from DNA. This would explain the apparent loss of both H3K9me2 and H3K4me3 (e.g., Figure 2B) compared with input values. Considering the reduction in nucleosome occupancy by normalizing the values to H3, it is clear that the levels of modifications on the remaining histones remained relatively unchanged. qPCR after ChIP experiments reveals nucleosome association ± 1000 bp around the primer binding sites due to the size of DNA fragments used, whereas PCR after MNase I assays reveals chromatin organization with less coverage but higher resolution. These independent assays both indicate substantial nucleosome loss at most regions analyzed. Differences between neighboring nucleosomes or remaining nucleosomes at individual targets nevertheless indicate a potential specificity of the response. Reduced nucleosome density may facilitate access of the transcriptional machinery to the promoters of repetitive elements, thus allowing their expression, similar to nucleosome depletion at HSP70 promoters in Drosophila melanogaster upon heat stress (Petesch and Lis, 2008). Even more support for the role of histone-mediated transcriptional regulation in the temperature response comes from the recent discovery of the important role of the histone H2A.Z variant in Arabidopsis (Kumar and Wigge, 2010). At moderately high temperatures, tight wrapping of H2A.Z and the amount of H2A.Z are reduced at the promoter of heat-responsive genes, such as HSP70, which is associated with their increased transcriptional activity and with decreased expression of certain other targets. However, this cannot explain heat stress activation of TGS targets, since heavy DNA methylation at their promoters is mutually antagonistic with the H2A.Z modification in Arabidopsis (Zilberman et al., 2008). Therefore, the more extreme and lasting heat stress in our experiments seems to destabilize and/or remove entire nucleosomes, including those containing canonical histones. Our data are in agreement with the suggestion by Kumar and Wigge (2010) that the removal of nucleosomes is independent of transcription since individual nucleosomes are not removed in spite of transcription, while other, nontranscribed, parts of the genome also showed a reduction in H3 association. Whether this removal of nucleosomes is an active process or a passive response to the elevated temperature remains to be elucidated. A requirement for active reloading, in parallel to regaining epigenetic regulation of the repeats and restoring the original nucleosome loading upon recovery, is suggested by the delayed resilencing of some repeats in mutants with reduced CAF-1 functionality, which generally have lower nucleosome density (Kirik et al., 2006).
Despite unchanged (DNA and histone methylation) or only transiently modified (transcription and histone loading) attributes, one parameter of chromatin organization was not restored to prestress conditions. The massive dissociation of heterochromatin, which exceeded even that in ddm1 mutants (Mittelsten Scheid et al., 2002; Soppe et al., 2002), remained in nuclei of differentiated tissue that had been exposed to LHS, beyond the recovery phase when silencing and nucleosomes had been reinstalled and until exposed leaves started to show signs of senescence. Together with the general loss of nucleosomes, LHS-induced decondensation of chromocenters could increase the accessibility of DNA to transcription complexes. This seems likely in Drosophila, where heat stress induces puffing of chromosomes at HSP70 loci. The process requires poly(ADP)ribose polymerase and is essential for high levels of HSP70 and thermotolerance being reached (Tulin and Spradling, 2003). Decondensed heterochromatin in Arabidopsis was found in 2-d-old seedlings, in response to dedifferentiation in cell culture or floral transition in development (Mathieu et al., 2003; Tessadori et al., 2007a, 2007b), but regular chromocenters were formed in a stepwise process after a longer period in culture. Heterochromatin decondensation per se was not sufficient for repeat activation (Tessadori et al., 2007b). More permanent and even repeat-specific heterochromatin decondensation has been described for plants grown at low light intensity (Tessadori et al., 2009). Since this was specific for ecotypes that originate from low geographical latitudes with naturally high light intensity, this can also be seen as a stress response. While life-long culture of these plants at higher light intensity could eliminate the phenotype of CC decondensation (Tessadori et al., 2009), the study does not address whether already decondensed chromatin could revert to the regular configuration by a switch in light conditions, which is a question of interest in the context of our data. Nevertheless, decondensation of heterochromatin is not a general response to stress, since we did not observe this phenotype after freezing (−4°C for 24 h) or UV-C irradiation (3000 J/m2). It also does not affect all tissues equally, since meristematic nuclei were excluded from LHS-induced decondensation. This may indicate an additional safeguarding mechanism to minimize epigenetic and possibly genetic damage in the germ line. It is possible that decondensation is a controlled process that allows increased transcriptional activity of heterochromatin-embedded targets that are important for heat stress tolerance in differentiated cells, while preventing repeat activation in dividing cells and upon the formation of subsequent generations. However, the open chromatin after prolonged heat exposure could allow occasional expression switching and may contribute to a potential influence of environmental factors on the epigenetic landscape.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana line L5 (Morel et al., 2000; Elmayan et al., 2005) is in the Columbia-0 (Col-0) background and was crossed with mom1-1 in Zh (Amedeo et al., 2000), ddm1-5 in Zh (Mittelsten Scheid et al., 1998), fas1-1 in Enk and fas2-1 in Landsberg erecta (Ler) (Kaya et al., 2001), rdr2-1 in Col-0 (Xie et al., 2004), and drd1-6 in Col-0 (Kanno et al., 2004). Furthermore, we used hsfa2-1 (Charng et al., 2007) in Col-0 and Line A (Mittelsten Scheid et al., 1998) either as wild type or crossed with mom1-1 or ddm1-5 (all in Zh). WT refers to Col-0. WT2/3/4 F3 hybrids between Col-0 and Zh, Enk, or Ler, respectively, were used to match the outcrossed lines as closely as possible.
Plants were grown for 21 d after sowing on GM medium in vitro at 21°C under long-day conditions (16 h light/8 h dark) prior to stress. For heat stress, plants were transferred to 37°C for 3 h (SHS) or 30 h (LHS) starting in the light period and allowed to recover under prestress growth conditions.
GUS Staining
GUS histochemical staining was performed as described (Pecinka et al., 2009).
Primers
The primers used in this study are listed and their use is specified in Supplemental Table 3 online.
DNA Methylation Analysis
Genomic DNA was isolated using a DNeasy Plant Maxi Kit (Qiagen). For DNA gel blot assays, 5 μg of DNA were digested with 20 units of HpaII, MspI, or AluI (MBI Fermentas), separated on 1.2% agarose gels, depurinated in 250 mM HCl for 10 min, denatured in 0.5 M NaOH and 1.5 M NaCl for 30 min, and neutralized in 0.5 M Tris, 1.5 M NaCl, and 1 mM EDTA at pH 7.2 for 2× 15 min. DNA was blotted onto Hybond N+ membranes (Amersham) with 20× SSC, washed, and UV cross-linked with a Stratalinker (Stratagene). Hybridization was performed as described (Church and Gilbert, 1984). Sequence-specific probes (for details, see Supplemental Table 3 online) radioactively labeled with 50 μCi of dCT-α-32P (Amersham) were synthesized by the Rediprime II Random Prime Labeling System (GE Healthcare) and purified via G50 Probequant (Amersham) columns. Signals were detected using phosphor imager screens (Amersham) and scanned by a Molecular Imager FX (Bio-Rad). For the specific methylation assay at the ASF-1 transcription factor binding site, 200 ng of genomic DNA were digested with 5 units of TaiI (MBI Fermentas) and used as a template for PCR with primers qP35-TaiI-F/qP35-TaiI-R (amplicon 1), qP35-TaiI-2F/qP35-TaiI-R (amplicon 2) and qPCR-GUS-F/pPCR-GUS-R (control). Global DNA methylation quantification was performed in technical triplicate by cation-exchange HPLC as described (Rozhon et al., 2008).
RNA Extraction and cDNA Synthesis
Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen). cDNA was synthesized with random hexamer primers and the RevertAid M-MuLV Reverse Transcriptase kit (MBI Fermentas).
qPCR Analysis
qRT-PCR analysis was performed in technical triplicate and with a minimum of two biological replicates using the SensiMix Plus SYBR kit (PEQLAB Biotechnologie) and iQ5 equipment (Bio-Rad). The expression values were calculated according to Pfaffl (2001) and normalized to the expression of the UBC28 gene, which is not changed under heat stress conditions. For ChIP data, relative signal ratios of immunoprecipitated samples were normalized to those of corresponding input or histone H3 samples, as indicated. For MNase I sensitivity assays, the means of individual MNase I–treated samples were multiplied by a correction factor to compensate for different amounts of DNA and compared.
ChIP
ChIP was performed as described (http://www.epigenome-noe.net/researchtools/protocol.php?protid=13) with the antibodies rabbit polyclonal to histone H3 (Abcam; ab1791), rabbit polyclonal to histone H4 (Abcam; ab10158), mouse monoclonal to histone H3 dimethyl K9 (Abcam; ab1220), rabbit antiserum to histone H3 trimethyl K4 (Upstate; 07-473) and quantified by qPCR. Relative values were calculated with input DNA, for H4 set aside prior to immunoprecipitation (60 μL) and for H3 after mock treatment without antibody (500 μL).
MNase I Sensitivity Assay
MNase I sensitivity assay was performed as published (Ricardi et al., 2010) with the following modifications. For chromatin isolation, 1 g of frozen tissue was homogenized to a fine powder, resuspended in 10 mL of extraction buffer 1 (0.44 M sucrose, 10 mM Tris-HCl, pH 8.0, 5 mM β-mercaptoethanol, and 1× protease inhibitor cocktail = 1 mM PMSF and 1 Complete, Mini, EDTA Free protease inhibitor tablet [Roche]/20 mL buffer), filtered through Miracloth, and centrifuged at 2880g for 20 min. The pellet was resuspended in 10 mL of extraction buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and 1× protease inhibitor cocktail), incubated on ice for 10 min, and centrifuged at 2100g for 20 min. The pellet was dissolved in 4 mL of extraction buffer 2 without Triton X-100 and centrifuged at 2100g for 20 min. The pellet was then dissolved in 4 mL of Percoll extraction buffer (95% v/v Percoll, 0.25 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, and 1× protease inhibitor cocktail) and spun down for 10 min at 12,000g. The upper phase was transferred into a new tube, diluted at least five times with nuclei resuspension buffer (10% glycerol, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 10 mM β-mercaptoethanol, and 1× protease inhibitor cocktail), and centrifuged at 12,000g for 10 min. The pellet was dissolved in 4 mL of nuclei resuspension buffer and centrifuged at 12,000g for 10 min (repeated twice).
For MNase I digestion, the pellet was dissolved in 500 μL Micrococcal nuclease buffer (50 mM Tris-HCl, pH 8.5, 5 mM Mg acetate, 25% glycerol, and 1 mM CaCl2), and 100-μL aliquots were digested with 0, 3, 6, and 12 units of MNase I (Takara) at 37°C for 20 min. The reaction was terminated by adding 10 μL of 0.5 M EDTA, 20 μL of 1 M Tris-HCl, pH 6.8, and 1.5 μL of 14 mg/mL proteinase K and incubation at 45°C for 1 h.
For DNA recovery, DNA was recovered using standard phenol:chloroform extraction and precipitated with addition of yeast tRNA as a carrier. The pelleted nucleic acids were dissolved in 50 μL of water containing 10 μg/mL RNase A at 4°C overnight. Samples were analyzed by gel electrophoresis, and 10× diluted samples were used for qPCR. Quantitative analysis was performed on mock-treated samples (no nuclease, control for normalization) and samples treated with 12 units of MNase I (best preparation of mononucleosomes according to gel electrophoresis). Nucleosome-occupied regions were identified using the Methylome browser (http://epigenomics.mcdb.ucla.edu/Nuc-Seq/; Segal et al., 2006; Chodavarapu et al., 2010), and the primers were positioned within single sequencing reads.
FISH and Microscopy
Nuclei were extracted either from whole plants or specific tissues (meristems or leaves that developed after stress treatment) as described (Pecinka et al., 2004) and transferred to slides using a Cytospin (MPW Medical Instruments). Hybridization, posthybridization washes, and FISH detection were performed as described (Pecinka et al., 2004). 180-bp and 5S rDNA probes were amplified and labeled with Biotin-dUTP or Digoxigenin-dUTP via PCR using primers 180bpF/180bpR and 5SrRNAqF/5SrRNAqR, respectively. Plasmid pGL2 (Bilang et al., 1991) containing the HPT gene was labeled by nick translation. Microscopy was done with an AxioImager Z.1 (Zeiss), and the images were assembled in Photoshop (Adobe Systems).
Genome-Wide Expression Profiling
Biological duplicates of total RNA samples were submitted to the microarray service of the Nottingham Arabidopsis Stock Centre (http://affymetrix.Arabidopsis.info/). The data files from hybridization to Affymetrix ATH1 microarrays were analyzed using the Bioconductor solution (www.bioconductor.org) under the R platform (www.r-project.org). The expression values were normalized by the GeneChip Robust Multiarray Averaging method (gcRMA; Wu et al., 2004). Differential gene expression analysis was performed with an empirical Bayes moderated t test using linear modeling (LIMMA; Smyth, 2004). The differentially expressed genes were identified by false discovery rate–corrected P values (≤0.05) and a log2 fold change cutoff (≥2, downregulated; ≤−2, upregulated). The transcriptional profiles of SHS R0 originate from previously published experiments (Kilian et al., 2007).
Detection of Transcripts from Mitochondrial Insertion on Chromosome 2
Regions corresponding to ATH1 IDs 263504_s_at (AT2G07677+ATMG00940), 265227_s_at (AT2G07695+ATMG01280), and 257338_s_at (AT2G07711+ATMG00513) were amplified from cDNA with primers recognizing both nuclear and mitochondrial copies (see Supplemental Table 3 online). The PCR products were cloned and sequenced. Transcripts were assigned to nuclear or mitochondrial origin based on single nucleotide polymorphisms (see Supplemental Table 2 online).
Accession Numbers
Accession numbers of sequences relevant for this article are as follows: At1g64230 (UBC28), At1g65470 (FAS1), At1g72920 (TA11), At1g74310 (HSP101), At2g07677/Atmg00940 (263504_s_at), At2g07695/Atmg01280 (265227_s_at), At2g07711/Atmg00513 (257338_s_at), At2g15040 (GP2NLTR), At2g15790 (CYP40), At2g15800 (MULE2), At2g16390 (DRD1), At2g26150 (HSFA2), At3g60930 (ATLANTYS2), At4g05640 (ATHILA6A), At4g10500 (OXIDOREDUCTASE), At4g11130 (RDR2), At4g16870 (COPIA4I), At5g12110 (eEF1Balpha1), At5g20850 (RAD51), At5g25450 (UBIQUINOL-CYTOCHROME C REDUCTASE), At5g27845 (IG/LINE), At5g35490 (IS112A), and At5g64630 (FAS2). The microarray data are available under Gene Expression Omnibus accession number GSE18666.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. LHS Transiently Abolishes TGS.
Supplemental Figure 2. Expression of Endogenous TGS Targets under LHS.
Supplemental Figure 3. DNA Methylation Analysis after LHS.
Supplemental Figure 4.. LHS Activated TGS Targets Are Transcriptionally Silenced and DNA Is Methylated in the Next Generation.
Supplemental Figure 5. Analysis of Histone H3 Modification and Occupancy after LHS.
Supplemental Figure 6. Analysis of Histone H4 Occupancy after LHS.
Supplemental Figure 7. Analysis of Nucleosome Occupancy after LHS.
Supplemental Table 1. Repeats with Significantly Altered Expression after SHS and LHS without (R0) and after 2 d (R2) of Recovery.
Supplemental Table 2. Activation of Genes in a Nuclear Cluster of Mitochondrial Origin under SHS and LHS without (R0) and after 2 d (R2) of Recovery.
Supplemental Table 3. Primers Used in This Study.
NOTE ADDED IN PROOF
Parts of this work are consistent with data described in a parallel publication.
Tittel-Elmer M., Bucher E., Broger L., Mathieu O., Paszkowski J., Vaillant I. (2010). Stress-induced activation of heterochromatic transcription. PLoS Genetics, in press
Acknowledgments
We thank H. Vaucheret for line L5, H. Hirt for the hsfa2 mutant, B. Dekrout for HPLC analysis, B. Wohlrab for technical assistance, as well as A. Förster-Sümecz for reading and M. Siomos and H. Rothnie for editing the manuscript. This work was supported by grants GEN-AU GZ 200.140-VI/1/2006 and FWF P18986-B17 from the Austrian Science Fund, by the European Union Network of Excellence “Epigenome” to O.M.S., and by Wiener Wissenschafts-, Forschungs- und Technologie Fonds to A. von Haeseler, cosupervisor of H.Q.D. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that there is no competing interest.
References
- Adamo A., Pinney J.W., Kunova A., Westhead D.R., Meyer P. (2008). Heat stress enhances the accumulation of polyadenylated mitochondrial transcripts in Arabidopsis thaliana. PLoS ONE 3: e2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedeo P., Habu Y., Afsar K., Mittelsten Scheid O., Paszkowski J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405: 203–206 [DOI] [PubMed] [Google Scholar]
- Bilang R., Iida S., Peterhans A., Potrykus I., Paszkowski J. (1991). The 3′-terminal region of the hygromycin-B-resistance gene is important for its activity in Escherichia coli and Nicotiana tabacum. Gene 100: 247–250 [DOI] [PubMed] [Google Scholar]
- Binelli G., Mascarenhas J. (1990). Arabidopsis: Sensitivity of growth to high temperature. Dev. Genet. 11: 294–298 [Google Scholar]
- Chan S.W., Henderson I.R., Jacobsen S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Charng Y.Y., Liu H.C., Liu N.Y., Chi W.T., Wang C.N., Chang S.H., Wang T.T. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu R.K., et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.S., Sano H. (2007). Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genomics 277: 589–600 [DOI] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux M.P., Couteau F., Bergounioux C., White C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Elmayan T., Proux F., Vaucheret H. (2005). Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 15: 1919–1925 [DOI] [PubMed] [Google Scholar]
- Endo M., Ishikawa Y., Osakabe K., Nakayama S., Kaya H., Araki T., Shibahara K.I., Abe K., Ichikawa H., Valentine L., Hohn B., Toki S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., de Jong J.H., Lysak M., Castiglione M.R., Schubert I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J., Demidov D., Houben A., Schubert I. (2006). Chromosomal histone modification patterns - From conservation to diversity. Trends Plant Sci. 11: 199–208 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Hashida S., Kitamura K., Mikami T., Kishima Y. (2003). Temperature shift coordinately changes the activity and the methylation state of transposon Tam3 in Antirrhinum majus. Plant Physiol. 132: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida S.N., Uchiyama T., Martin C., Kishima Y., Sano Y., Mikami T. (2006). The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 18: 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. (2007). Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Huettel B., Kanno T., Daxinger L., Aufsatz W., Matzke A.J.M., Matzke M. (2006). Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Mette M.F., Kreil D.P., Aufsatz W., Matzke M., Matzke A.J.M. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14: 801–805 [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara K., Taoka K., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kirik A., Pecinka A., Wendeler E., Reiss B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Larkindale J., Lee U., von Koskull-Doring P., Vierling E., Scharf K.D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lang-Mladek C., Popova O., Kiok K., Berlinger M., Rakic B., Aufsatz W., Jonak C., Hauser M.T., Luschnig C. (2010). Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 3: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E.G., Masson J., Bogucki A., Paszkowski J. (1993). Stress-induced intrachromosomal recombination in plant somatic cells. Proc. Natl. Acad. Sci. USA 90: 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1: 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Comai L. (2004). The effect of stress on genome regulation and structure. Ann. Bot. (Lond.) 94: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Jasencakova Z., Vaillant I., Gendrel A.V., Colot V., Schubert I., Tourmente S. (2003). Changes in 5S rDNA chromatin organization and transcription during heterochromatin establishment in Arabidopsis. Plant Cell 15: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Afsar K., Paszkowski J. (1998). Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Probst A.V., Afsar K., Paszkowski J. (2002). Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 13659–13662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J.B., Mourrain P., Beclin C., Vaucheret H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Pecinka A., Rosa M., Schikora A., Berlinger M., Hirt H., Luschnig C., Mittelsten Scheid O. (2009). Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Schubert V., Meister A., Kreth G., Klatte M., Lysak M.A., Fuchs J., Schubert I. (2004). Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113: 258–269 [DOI] [PubMed] [Google Scholar]
- Petesch S.J., Lis J.T. (2008). Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fransz P.F., Paszkowski J., Mittelsten Scheid O. (2003). Two means of transcriptional reactivation within heterochromatin. Plant J. 33: 743–749 [DOI] [PubMed] [Google Scholar]
- Ricardi M.M., Gonzalez R.M., Iusem N.D. (2010). Protocol: Fine-tuning of a Chromatin Immunoprecipitation (ChIP) protocol in tomato. Plant Methods 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon W., Baubec T., Mayerhofer J., Mittelsten Scheid O., Jonak C. (2008). Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 375: 354–360 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L.Y., Thastrom A., Field Y., Moore I.K., Wang J.P.Z., Widom J. (2006). A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzic M., Becker J.D., Feijo J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3 [DOI] [PubMed] [Google Scholar]
- Soppe W.J.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer A., Amedeo P., Afsar K., Fransz P., Mittelsten Scheid O., Paszkowski J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R.M., Lilly J.W., Town C.D., Cheng Z., Kaul S., Buell C.R., Jiang J.M. (2001). Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. USA 98: 5099–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Tadele Z., Hofmann I., Probst A.V., Angelis K.J., Kaya H., Araki T., Mengiste T., Mittelsten Scheid O., Shibahara K., Scheel D., Paszkowski J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F., Chupeau M.C., Chupeau Y., Knip M., Germann S., van Driel R., Fransz P., Gaudin V. (2007b). Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 120: 1200–1208 [DOI] [PubMed] [Google Scholar]
- Tessadori F., Schulkes R.K., van Driel R., Fransz P. (2007a). Light-regulated large-scale reorganization of chromatin during the floral transition in Arabidopsis. Plant J. 50: 848–857 [DOI] [PubMed] [Google Scholar]
- Tessadori F., et al. (2009). PHYTOCHROME B and HISTONE DEACETYLASE 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 5: e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A., Spradling A. (2003). Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299: 560–562 [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wang Y., Bao Z.L., Zhu Y., Hua J. (2009). Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Whittle C.A., Otto S.P., Johnston M.O., Krochko J.E. (2009). Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 87: 650–657 [Google Scholar]
- Woo H.R., Pontes O., Pikaard C.S., Richards E.J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.J., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917 [Google Scholar]
- Xie Z.X., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. (2007). A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. (2008). Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]