Summary
GATA family transcription factors play multiple vital roles in hematopoiesis in many cell lineages, and in particular, T cells require GATA-3 for execution of several developmental steps. Transcriptional activation of the Gata3 gene is observed throughout T-cell development and differentiation in stage-specific fashion. GATA-3 has been described as a master regulator of T-helper 2 (Th2) cell differentiation in mature CD4+ T cells. During T-cell development in the thymus, its roles in the CD4 vs. CD8 lineage choice and at the β-selection checkpoint are the best characterized. In contrast, its importance prior to β-selection has been obscured both by the developmental heterogeneity of double negative (DN) 1 thymocytes and the paucity of early T-lineage progenitors (ETPs), a subpopulation of DN1 cells that contains the most immature thymic progenitors that retain potent T-lineage developmental potential. By examining multiple lines of in vivo evidence procured through the analysis of Gata3 mutant mice, we have recently demonstrated that GATA-3 is additionally required at the earliest stage of thymopoiesis for the development of the ETP population. Here, we review the characterized functions of GATA-3 at each stage of T-cell development and discuss hypothetical molecular pathways that mediate these functions.
Keywords: GATA-3, ETP, β-selection, CD4 SP, Th2
T-cell development in thymus
T-cell production in the thymus is supported by continuous, life-long migration of T-cell progenitors derived from hematopoietic stem cells (HSCs) in the bone marrow (1). Most if not all current models for lineage specification from HSCs derive from the concept that, as cells mature, they progressively lose more and more differentiation potential (Fig. 1). HSC and multi-potential progenitors (MPPs) reside in the phenotypically defined LSK [lineage− (Lin−)Sca1+cKithi] population (Table 1), and early developmental induction of cell surface fms-like tyrosine kinase 3 (Flt3) expression within this population is accompanied by a loss of self-renewal potential (2, 3). In comparison to MPPs, lymphoid primed MPPs (LMPPs) have markedly reduced erythroid potential (4–7). While most hematopoietic lineage cells (specifically erythroid, myeloid, and B lymphoid cells and all of their progenitors) develop in the fetal liver and/or bone marrow, T lymphoid cells develop in the thymus, which provides unique microenvironmental cues that are essential for T-cell development (8).
Fig. 1. A model for T lymphopoiesis.
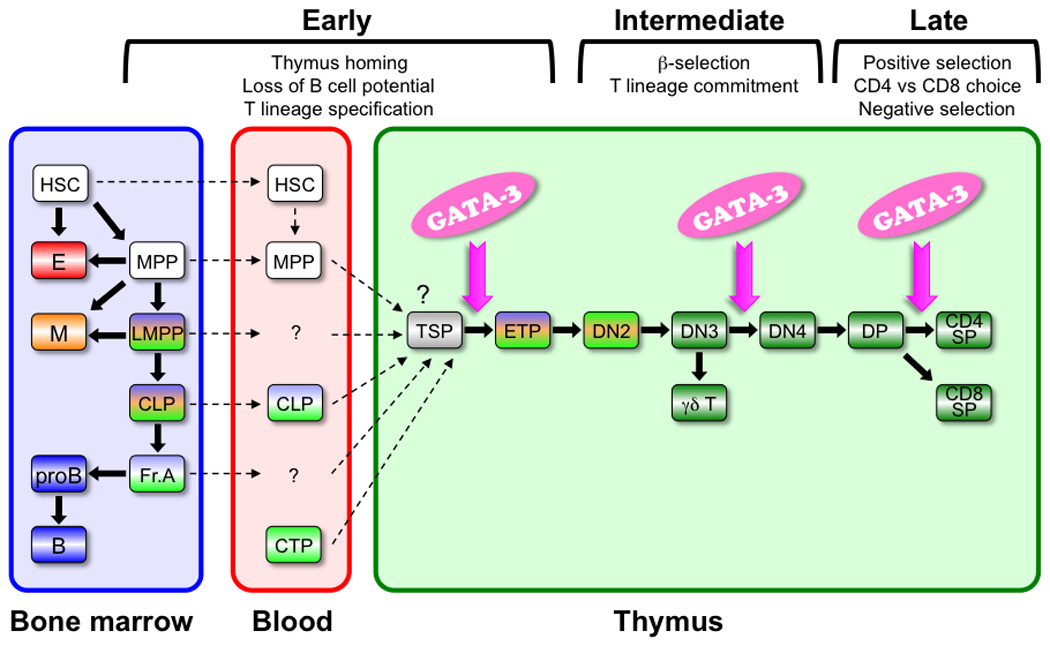
Popular models for lineage specification from HSCs derive from the concept that, as cells mature, they progressively lose differentiation potential. Myeloid, B, and T-lineage potential are shown in orange, blue, and green, respectively. Cells that are committed to the T lineage are shown in dark green. The identity of the thymus homing cells remains controversial. Possible homing cells are shown in dashed lines. GATA-3 is required for the development of ETP, DN4, and CD4 SP T cells in the thymus (magenta arrow). E, erythroid lineage cells; M, myeloid lineage cells; B, B lymphoid lineage cells
Table 1.
Hematopoietic populations and abbreviations (mouse)
| Cell population | Abbreviation | Cell surface phenotype |
|---|---|---|
| Lineage negative cells | Lin− | |
| LSK | Lin−Sca1+cKithi | |
| Hematopoietic stem cell | HSC | CD150+CD48−CD34−Flt3−LSK |
| Multipotent progenitors | MPP | Flt3loLSK |
| Lymphoid-primed multipotent progenitors | LMPP | Flt3hiLSK |
| Common lymphoid progenitors | CLP | Lin−IL7Rα+Sca1locKitlo |
| Early thymic progenitors | ETP | CD8−CD3−TCRβ−γδTCR−NK1.1−CD11c− Mac1−Gr1−B220−CD19−Ter119− cKithiCD25− |
| Double negative T cells, stage 1 | DN1 | CD4−CD8−CD44+CD25− |
| Double negative T cells, stage 2 | DN2 | Either Lin−cKithiCD25+ or Lin−CD44+CD25+ |
| Double negative T cells, stage 3 | DN3 | Either Lin−cKitlo/−CD25+ or Lin−CD44−CD25+ |
| Double negative T cells, stage 4 | DN4 | Either Lin−cKitlo/−CD25− or Lin−CD44−CD25− |
| Double positive | DP | CD4+CD8+ |
| Single positive | SP | Either CD4+CD8− or CD4−CD8+ |
T-lymphocyte progenitors migrate into the thymus from the fetal liver or bone marrow through the blood stream. Chemokines, cell adhesion molecules, and their receptors CCL25/CCR9, CCL19-CCL21/CCR7, and P-selectin/PSGL-1 regulate this migration (9–15). The identity of thymus-homing cells continues to be controversial (reviewed in 16). These cells have been proposed to reside in several different bone marrow compartments that are distinguishable by their expression of different cell surface markers: LSK (17–19), Lin−cKitloSca1loIL7Rα+ (CLP) (20, 21), and in the B220+CD19− population (22, 23). Regardless of their cellular origin, very rare (and at this point, hypothetical) thymus-seeding progenitors (TSPs) upon encountering the thymic epithelium are thought to amplify to generate ETPs, the most immature T-cell progenitors in the thymus that have been characterized (24–26).
Intrathymic ETPs constitute a cKithiLin− fraction that harbors all of the most potent T-cell developmental potential but comprises only a subset of about 2–4% of all double negative 1 (DN1) stage cells (27). Very shortly after thymic entry, these progenitors lose their B-cell potential and become specified to the T-cell lineage. The most immature subset (Flt3+) of adult ETPs retains modest B-cell developmental potential (27, 28). Thymic epithelial cells express the Notch ligand Delta-like 4 and activate Notch1-mediated signals, which in turn promote T-cell development while at the same time repressing further B and myeloid cell development (27–32). However, the existence of T-restricted progenitors has also been reported to emerge from multiple other physiological sites: in fetal and adult peripheral blood (33, 34), in the fetal liver (35, 36), in the spleen shortly after bone marrow transplantation (37, 38), and even in the normal adult mouse spleen (39). In addition, inactivation of the transcriptional repressor LRF in HSCs activates high levels of Notch signaling in these cells and leads to the emergence of T-cell development in the bone marrow as well as in other extrathymic tissues (40). These data, taken together, suggest that prethymic T-lineage commitment may indeed occur at reduced incidence or under certain unusual circumstances but that T-cell development in tissues other than the thymus is actively suppressed under non-pathological conditions.
ETPs progress through the DN2 stage to the DN3 stage of development. While ETPs and DN2 cells retain myeloid lineage potential, DN3 cells have lost that potential and become fully committed to the T lineage (41, 42). During these intermediate stages of T-cell development, the Tcrg, Tcrd, and Tcrb loci undergo rearrangement (43, 44). At the same time, preT-cell antigen receptor-α (Ptcra) and Cd3e transcripts increase (45, 46). CD3, TCRβ, and preTCRα make up the pre-TCR complex (reviewed in 47). Cells that have formed a functional preTCR complex can develop into double positive (DP) cells, while cells that have failed to initially produce a functional complex rearrange the other Tcrb allele in a second attempt to generate a functional preTCR complex; if rearrangements on both alleles fail to generate an active TCRβ protein, those cells are eliminated by apoptosis. This step is referred as the β-selection checkpoint and is essential for the development of αβ T cells. Complex transcriptional inputs, including from RBPJ, MYB, TCF1, LEF1, E2A, HEB, GFI1, IKAROS, RUNX/CBFβ, PU.1, and GATA-3 form a network under the direct and indirect influence of Notch signaling to support T-cell specification and commitment from multi-potential progenitors (reviewed in 48). Other essential players include cytokines and their receptors on hematopoietic cells, such as interleukin 7 (IL7)/IL7R, stem cell factor (SCF)/cKit, Flt3 ligand, and Flt3 (27, 49–54).
The cells that survive β-selection develop into DN4 cells and then into immature single positive (CD8+CD4−) and DP stages of development. DP cells undergo rearrangement of the Tcra locus, and this rearrangement results in the formation of the mature TCRαβ complex. Next, the DP cells develop into either CD4−CD8+ (CD8 SP) or CD4+CD8− (CD4 SP) cells, or alternatively are eliminated by apoptosis. This CD8 versus CD4 lineage choice determines mature T-cell fate and is controlled by an intricate interplay between an increasingly well understood transcription factor network, including the GATA-3, TOX, Th-POK, and RUNX, all under the influence of TCR signaling (55–66). Those cells bearing TCR complexes that are able to bind to MHC survive (positive selection), while other cells are eliminated by apoptosis (death by neglect). A second round of negative selection at the DP and SP stages eliminates by apoptosis the developing T cells that respond to self-antigens.
Immature T-cell migration through the thymus ensures an orderly progression of T-cell development by providing non-cell-autonomous cues in discrete subregions of the thymus (reviewed in 67). Mature CD4 SP or CD8 SP T cells exit the thymus and home through the bloodstream to secondary lymphoid organs, where mature naive lymphocytes are maintained and where adaptive immune responses are initiated. Cytokines, their signaling pathways, and tissue-restricted transcription factors form an elaborately orchestrated network that maintains the proper continuous production of T cells. GATA-3 is one of the essential factors for T-cell development and differentiation, and its importance has been demonstrated from the beginning (in thymic ETPs) to the end of T-cell life (in peripheral Th2 CD4+ T cells).
Transcription factor GATA-3 is vital for T-cell development
At the time we originally cloned GATA-3, we found that it was the sole member of the GATA zinc-finger-type transcription factor family expressed in T lymphocyte cells (68). Six GATA factors have been identified in mammals (68–71), and all members appear to bind to a WGATAR recognition sequence found in the promoters and/or enhancers of literally thousands of tissue-restricted genes (72, 73). Hematopoietic cells (as do some additional tissues) express the ‘hematopoietic’ factors: GATA-1, GATA-2, and GATA-3. Each tissue and cell lineage expresses only very specific GATA factors, and only at very specific stages of maturation or development (Table 2). GATA-1 is essential for the development of erythroid cells, for the proliferation and/or maturation of megakaryocytes, for the production of circulating blood platelets, for the differentiation of eosinophils, and for mast cell differentiation (74–78). GATA-2 is required for proliferation/survival of early hematopoietic progenitors as well as for mast cell formation (79–82). GATA-3 is vital for the development of T cells in thymus (56, 57, 83–85), Th2 differentiation of peripheral CD4+ T cells (86–99), maturation of bone marrow natural killer (NK) cells (100), and development of thymic NK cells (101).
Table 2.
Expression of GATA factors in various hematopoietic lineages
| Hematopoietic progenitors | GATA-1, GATA-2, GATA-3 |
| Erythroid cells | GATA-1 |
| Megakaryocytes | GATA-1, GATA-2 |
| Mast cells | GATA-1, GATA-2 |
| Eosinophils | GATA-1 |
| T cells | GATA-3 |
| NK cells | GATA-3 |
GATA-2 and GATA-3 are also expressed and play key developmental roles in a wide variety of non-hematopoietic tissues. GATA-3 is essential for noradrenalin biosynthesis in sympathetic ganglia and the adrenal gland, and consequently Gata3 null mutant fetuses die at around e11 of a secondary cardiac insufficiency (102–104). This was definitively shown to be the case by rescuing Gata3 null mutant embryos to birth by feeding of pregnant intercrossed dams with catecholamine intermediates (103, 105). GATA-3 has also been shown to participate in development of the Wolffian duct and metanephric kidney (103, 104, 106), in hair follicle development and skin cell lineage differentiation (107, 108), in development of the luminal epithelium of the mammary gland (109, 110), as well as in the differentiation of multiple tissues derived from the pharyngeal arches, such as the jaw, parathyroid gland, and thymus (103, 111).
Expression of GATA-3 during T-cell development
During hematopoiesis, the Gata3 gene is first transcriptionally activated in HSCs, defined variously as either IL7Rα−Thy1.1loLSK (112) or CD38+CD34−LSK (113) or Thy1.1loLSK (114) or CD34−Flt3−LSK (7). The VCAM1−Flt3hi sub-fraction of MPPs expresses Gata3 mRNA (6). CLPs (112, 115) and pre-pro-B cells (Hardy fraction A) (115), which retain both T-cell and B-cell developmental potential, also express Gata3 mRNA in the bone marrow. In the thymus, Gata3 mRNA is expressed in the earliest ETPs that have been identified (27). Gata3 mRNA levels gradually increase between the ETP and DN3 stages and slightly diminish again in DN4 cells (46, 116). At the DP stage, Gata3 mRNA and protein are both repressed and then Gata3 transcription is strongly induced in late DP stage and remains high at the CD4 SP stage, while diminishing in CD8 SP cells (56, 84, 117).
GATA-3 is required at very early stages of T-cell development
The involvement of GATA-3 during early stages of thymopoiesis was first suggested in 1996 following the analysis of hematopoietic cells that had diminished GATA-3 expression as a result of anti-sense experiments (118). These investigators introduced antisense oligonucleotides into e13 Lin−cKit+ fetal liver cells or thymocytes and examined their development into Thy1+ cells in fetal thymus organ culture. The introduction of antisense oligonucleotide into fetal liver progenitors resulted in diminished Thy1+ cell development to less than 20% of control (nonsense oligonucleotide-transfected) cells. At the same time, the introduction of antisense oligonucleotides into fetal cKit+ thymocytes resulted in a ca. 50% reduction in Thy1+ cell development. Although alternative explanations were possible, the greater T-cell attenuation observed among the transfected fetal liver progenitors suggested that GATA-3 might be important for T lymphopoiesis, even before thymic entry.
Additional in vivo evidence for the involvement of GATA-3 in early T-cell development was provided in experiments that generated blastocyst chimeras starting with Gata3-null ES cells (either Gata3−/z null or control Gata3+/z heterozygous ES cells, both carrying a LacZ knock-in allele) (84). The contribution of the mutant cells was detected using infused FDG, a substrate that fluoresces upon cleavage by β-galactosidase. These investigators observed 2.2% FDG+ cells in DN1 thymocytes of Gata3+/z heterozygous ES chimera, but only 0.4% in Gata3−/z (null mutant) ES chimeras. The data indicated that the contribution of Gata3+/z heterozygous ES cells to the T-cell lineage was about 10% in blastocyst chimeras, as compared to 50–70% in the rest of the body (heart, brain, kidney, and lungs). Furthermore, the contribution of Gata3−/z null ES cells to the DN1 population was even lower, suggesting a dosage-sensitive requirement for GATA-3 in DN1 thymocytes. However, since relatively abundant NK1.1+ cells are also contained within the DN1 fraction (27, 85, 119) in which ETPs are sparse (2–4% of DN1 cells) (27), and given the fact that GATA-3 activity is known to be required for thymic NK cell development (101), the data were sufficiently equivocal that it was impossible to conclude decisively that GATA-3 was required for the development of early T-cell progenitors.
To gain more precise insight into the earliest stage at which GATA-3 becomes required for T-cell development prior to the β-selection checkpoint, we examined the development of immature T cells in multiple Gata3 mutant backgrounds: in Gata3 hypomorphic mutant embryos, in irradiated adult mice reconstituted with Gata3 null or hypomorphic mutant hematopoietic progenitor cells, and in adult mice in which the Gata3 gene was conditionally ablated (85). In reconstituted mice, Gata3z/z (null) mutant hematopoietic cells showed normal contributions to the bone marrow HSC, LMPP, CLP and Fr. A early hematopoietic compartments, all of which retain T-cell developmental potential. In contrast, the absolute number of Gata3z/z ETPs in HSC-repopulated recipient mice was less than 10% of heterozygous controls. In those reconstituted animals, development of DN2 and later stage T cells was even more significantly impaired, to less than 1% of controls. Consistent with these adoptive transfer experiments, conditional compound mutant (Gata3flox/flox:TgMx1cre) mice bearing activated Cre generated fewer than 3% of normal ETP numbers when compared to control (Gata3flox/+:TgMx1cre) Cre-activated animals.
To examine the possible direct involvement of GATA-3 in the development of fetal ETPs, we took advantage of Gata3 hypomorphic mutant (Gata3g/g) mice. Earlier experiments showed that GATA-3 is expressed during embryogenesis in the third pharyngeal pouch, from which the thymic epithelium develops (our unpublished observations). While drug-rescued Gata3 null mutant embryos develop no detectable thymus (103), homozygous Gata3 hypomorphic mutants indeed develop small thymi, and within those hypomorphic Gata3g/g thymi, embryos generated 16% or 13% of normal ETP numbers when compared to wildtype at e15.5 or e18.5, respectively.
Enforced expression of GATA-3 by retroviral transduction has been shown to induce cKit expression (45, 120), and so to ask whether the observed reduction in mutant ETP and DN2 cells might be a simple consequence of reduced cKit cell surface expression, we analyzed the number of Lin−DN1 cells in Gata3 mutant animals. Indeed, careful gating on Lin− thymocytes has been shown to enrich for bona fide ETPs within the DN1 population (27). The absolute number of those Lin−DN1 cells was reduced in both the Gata3g/g hypomorphic mutant embryos and in Cre-induced (Gata3flox/flox:TgMx1cre) Gata3 conditionally mutant animals (85). The summary data consistently demonstrated that GATA-3 is required for the development of ETPs, while at the same time it appears to be dispensable for the development of pre-thymic T-cell progenitors.
How does GATA-3 control ETP development? Since we observed no developmental impairment in pre-thymic progenitors but a severe reduction in the earliest Gata3 null T-cell population, we considered four possibilities: that GATA-3 controls (i) homing of pre-thymic progenitors into the thymus; (ii) the differentiation of TSPs to ETPs; (iii) ETP proliferation, and/or (iv) ETP viability. Since Gata3 mutant fetal livers generated fewer Lin−CD27+cKithiCD25− cells (corresponding to pre-thymic lymphoid progenitors plus thymic ETPs) (17, 121) in vitro in OP9-DL1 co-cultures, we tentatively concluded that a homing deficiency was unlikely to be a primary cause of the reduced ETP numbers observed in the Gata3 mutants (85). However, we have not directly examined the thymus homing potential of Gata3 mutant progenitor cells, so a role for GATA-3 in thymus homing remains possible.
Blood-borne thymic seeding progenitors (TSPs) are thought to be extremely rare and their exact identity is unknown. The most immature subset of adult ETPs expresses the Flt3 cytokine receptor (27). To determine whether the reduced number of ETPs in the Gata3 mutants was caused by a differentiation arrest from early to late ETP subsets, we hoped to analyze the development of early ETP (Flt3+ETP) and late ETP (Flt3−ETP) in adoptive transfer recipients. Unfortunately the very low frequency of ETP recovery from the mutants (less than 10% of the normal adult ETP population) precluded recovery of consistent, reliable data. Analysis of Gata3g/g hypomorphic mutant cells in adoptive transfer recipients should be a more useful approach in this regard.
To address the third possibility, we examined the cell cycle status of Gata3 mutant cells. The percentage of e18.5 Gata3g/g ETP in S phase was very slightly higher than in Gata3+/+ ETP (our unpublished data). To examine the cell-cycle status in the complete absence of GATA-3, we again employed OP9-DL1 co-cultures. When compared to wild type Gata3, we again observed a slightly higher percentage of cells in S phase among Gata3z/z Lin−CD27+cKithiCD25− cells. Thus, decreased cycling also cannot explain the dearth of ETPs in Gata3-deficient mice.
We also examined the viability of Gata3 mutant T-cell progenitors. Since rapid clearance of dead cells by phagocytes in the thymus (122) might conceal small differences in cell longevity, we analyzed the viability of mutant cells in OP9-DL1 co-cultures by Annexin V staining. We observed no increase in the percentage of Annexin V+ cells in comparison to wildtype progenitors. Taken together, these data deductively suggest that the major GATA-3 cell-autonomous role in thymopoiesis is to control the differentiation of TSPs to ETPs and other downstream progenitors rather than participating in cell survival or proliferation, although further studies will be required to confirm or refute this hypothesis.
Since GATA-3 has been implicated in the control of differentiation at early stages of T-cell development, one might wonder if enforced expression of GATA-3 in multipotent pre-thymic progenitors might trigger precocious or unusually robust T-lineage development. However, the opposite was found to be true. Retroviral overexpression of GATA-3 in mouse bone marrow Thy1.1loLSK cells reportedly induced megakaryocyte and erythroid differentiation (123). Forcibly overexpressed GATA-3 by retroviral infection of mouse fetal liver progenitors caused impaired development of DN2, DN3, and DP T cells in fetal thymus organ culture (45). In a third study, retrovirally forced expression of GATA-3 in mouse fetal liver LSK or dedicated lymphoid progenitors was reported to block the development of DN2 and DN3 T cells in OP9-DL1 co-cultures (120). These results collectively suggest that GATA-3 activity must be kept low in pre-thymic progenitors in order to induce the initiation of T-cell fate and that the activation of Notch signaling, which is able to promote T-cell development (29, 124), cannot overcome the repressive effects of non-physiologically elevated levels of GATA-3.
Gata3 null mutant ES cells contribute to both B and myeloid cells in ES chimeric mouse bone marrow (83), and Gata3 null mutant e11.5 fetal liver cells generated similar numbers of B220+ cells as control progenitors in OP9 co-cultures (125). Gata3 null and Gata3 hypomorphic mutant hematopoietic progenitor cells showed neither increased nor decreased development of B or myeloid cells in the bone marrow and peripheral blood in adoptive transfer experiments (85). Furthermore, Gata3 hypomorphic embryos displayed normal development of B, myeloid, and erythroid cells in progenitors recovered from e18.5 fetal liver or spleen (85). These data demonstrate that GATA-3 is dispensable for the development of all B-lineage and myeloid cells.
GATA-3 is required at intermediate stages (DN2 to DN4) of T-cell development
The contribution of GATA-3 to intermediate stages of T-cell development has been demonstrated by conditional Gata3 ablation in DN2–DN3 thymocytes using the Lck promoter to direct the expression of Cre recombinase (Gata3flox/flox:TgLckcre) (57). This mutant mouse accumulated abnormally abundant DN3 cells and a reduced number of DN4 and later stage T cells. Gata3flox/flox:TgLckcre DN3 thymocytes rearranged the Tcrb gene, but DN4 cells had less intracellular TCRβ protein than wildtype and an increased percentage of Annexin V+ apoptotic cells (57). These data suggest that Gata3 deficient cells fail to form sufficient pre-TCR complex and are eliminated by apoptosis. A transgenic rescue attempt with DO11.10, which carries an major histocompatibility complex (MHC) class II-restricted rearranged TCR, failed to rescue reduced DP and SP T cells in the mutants (57). This failure suggested that GATA-3 must play an unidentified role in addition to the pre-TCR complex formation for the progression from the DN3 to DN4 stage. In contrast to thymocytes at the adult stage, the introduction of ROG (purported to be a natural inhibitor of GATA-3 in Th2 cells) (126), into mouse e14.5 fetal thymocytes (composed mostly of DN1–DN3 cells that have not yet been subject to β-selection) had little effect on TCRβ+ cell numbers in fetal thymus organ cultures (56). In our studies, Gata3g/g hypomorphic embryos had a normal DN4/DN3 profile compared to controls, but Gata3g/g cells indeed showed developmental defects at the adult DN3 to DN4 transition in adoptive transfer recipients (85). These data suggest that β-selection is less sensitive to reduced GATA-3 protein abundance in fetal than in adult thymi. Successful rearrangement of Tcrd and Tcrg genes at the DN2–DN3 stage is also necessary for the generation γδT cells in the thymus. GATA-3 is expressed in γδT cells (85), but its requirement or role in thymic γδT cells is not known.
Forced expression of GATA-3 in human CD34+CD3−CD4−CD8− immature thymocytes (prior to Tcrb gene rearrangement) (127) caused enhanced initial development toward the DP stage in fetal thymus organ cultures, while inducing a severe reduction in thymic cellularity after day 10 (128). In contrast, retroviral forced expression of GATA-3 in e14.5 to e15.5 DN1 and DN2 thymocytes inhibited differentiation into DP cells in fetal thymus organ cultures and in OP9-DL1 co-cultures (45, 120). In the absence of Notch signaling, forced expression of GATA-3 in mouse DN1 and DN2 fetal thymocytes promoted mast cell differentiation, a latent intrinsic potential of normal DN1 and DN2 thymocytes (120). Under normal physiological conditions, both GATA-2 and GATA-1 are expressed in mast cells and play essential roles in their differentiation (78, 80). Since GATA family proteins can apparently all bind to the GATA core DNA element (72, 73) and have been shown to be at least partially compensatory (129, 130), it is possible that more abundant GATA-3 generated in the forced expression models drives ectopic expression of genes that are under the control of either GATA-1 or GATA-2. We conclude that finely tuned levels of GATA-3 activity are required to survive the β-selection step in adult thymi. In support of this argument, we have shown previously that transcriptional regulation can be exquisitely sensitive to the absolute abundance of a transcription factor in vivo (131).
GATA-3 is required at late stages (DP to SP) of T-cell development in the thymus
GATA-3 expression is re-induced between early DP and late DP stages and remains high at the CD4 SP stage, while diminishing in CD8 SP T cells (56, 84, 117). It was found that forced transgenic expression of GATA-3 throughout T-cell development, achieved by placing it under the control of the human CD2 promoter/LCR (132, 133), did not impede thymocyte development but induced apoptosis and inhibited the final maturation of CD8 SP T cells in the thymus (134). Forced expression of GATA-3 in e16.5 thymocytes that were enriched for DP cells resulted in reduced generation of abundant TCR cell surface expression and increased CD4/CD8 ratios in fetal thymus organ cultures (56). Mice that were conditionally ablated for Gata3 at the DP stage and beyond using a Cd4-directed Cre transgene (Gata3flox/flox:TgCd4cre) generated reduced numbers of CD4 SP cells but normal numbers of CD8 SP cells (57). Inhibition of GATA-3 by retroviral introduction of ROG or anti-GATA-3 shRNAs into e14.5 to e16.5 thymocytes decreased the percentage of CD4 SP cells among the transduced TCRβhi cells that emerged from fetal thymus organ cultures (56). These results demonstrate that GATA-3 is required for CD4 SP cell development after positive selection.
GATA-3 promotes and is required for Th2 differentiation of peripheral CD4+ T cells
Peripheral naive helper CD4+ T cells differentiate into several subsets that produce distinct patterns of cytokines (reviewed in 135, 136). GATA-3 is highly expressed in Th2 cells, but not in Th1 cells (86, 87). Enforced expression of GATA-3 by either retroviral infection or transgenic means promotes Th2 differentiation and inhibits Th1 differentiation (88, 92, 94), while inhibition of Gata3 using either small interfering RNA (siRNA) or conditional Gata3 deletion significantly limits Th2 differentiation (95, 96, 98). Furthermore, GATA-3 plays roles in the chromatin modification of Th2 cytokine loci, including the Il4, Il5, and Il13 genes (90, 91, 93, 97, 99). These data indicate that GATA-3 is a master regulator of Th2 differentiation.
Target genes of GATA-3 in T cells
GATA-3 is presumed, or in a few cases known, to control T-cell development by trans-activation of various stage-specific target genes. What are the functional targets of GATA-3 during development from pre-thymic progenitors to ETPs? Although this is of course a critical question since this is the first currently documented stage to require GATA-3 activity in early hematopoiesis, no conclusive data are available so far from GATA-3 loss- or gain-of-function experiments. The very low number of ETPs and developmental arrest produced at or before ETP stages in Gata3 mutants make it difficult to discriminate loss of direct control by GATA-3 from loss of certain cell populations in the absence of GATA-3. The gain-of-function experiments (45, 120, 123) do not appear to have identified target genes that would explain the crucial importance of GATA-3 in these early stages.
During intermediate stages of T-cell development, one important cluster of target genes that are regulated by GATA-3 is the Tcr genes. We and others have shown that GATA-3 binds to enhancer and promoter elements of the Tcrb, Tcrg, and Tcrd genes (137–139). However, adult mice that were conditionally ablated for Gata3 at the DN2–DN3 transition (in Gata3flox/flox:TgLckcre mice) showed normal levels of Tcrb mRNA but reduced intracellular TCRβ protein in DN3 and DN4 cells (57). These data suggest that GATA-3 may somehow regulate TCRβ expression post-transcriptionally. GATA-3 binds to the Rag2 promoter in gel mobility shift assays (140), but conditionally mutant Gata3 mice show normal rearrangement of the Tcrb gene (57), indicating that GATA-3 is dispensable for Tcrb gene rearrangement. A transgenic rescue attempt with DO11.10, which carries an MHC class II-restricted rearranged TCR, failed to rescue reduced DP and SP T cells in the mutants (57). This failure suggested that GATA-3 must control a currently unidentified gene(s) that is critical for the development from the DN3 to DN4 stage.
To identify transcriptional target genes of GATA-3 prior to β-selection, GATA-3 was forcibly expressed by recombinant retroviral infection of e14.5 fetal thymocytes (which are principally composed of DN cells that have not yet undergone β-selection), and changes in gene expression were analyzed 24 hours post-infection (45, 120). Abundant ectopic expression of GATA-3 in these progenitor T cells activated Hes1, a direct target of the Notch signaling pathway that is known to be important for DN cell development (141) and at the same time represses the Sfpi1 gene encoding PU.1. PU.1 gradually diminishes in abundance from the DN1 to DN3 stages and has been shown to block T-cell development when forcibly expressed in T-lymphocyte progenitors (142). Based on the short-term response to Notch signaling, PU.1 and GATA-3, Georgescu et al. (143) proposed a basic gene regulatory network during the DN stages. It will be important to provide new inputs and insights to this proposed framework and to experimentally assess the relationships amongst these factors. In contrast, forced expression of GATA-3 in T-cell progenitors exerted little effect on many other genes that are important for T-cell development, including Il7r, Ptcra, Rag1, Rag2, CD3e, Myb, Tcf7, Lef1, E2a, Gfi1, Ikzf1, Runx1, and Notch1 (45, 120). Remarkably, forced GATA-3 expression also activated mast cell genes such as cKit, Gata1, Gata2, Mitf, Cpa3, and Tal1 (120). Although Hes1 was induced, an overall increase in Notch signaling was not observed after forced expression of GATA-3, because Dtx1 transcripts (another downstream target of Notch signaling pathway) (144) were reduced in this experiment (45, 120). However, forced expression of GATA-3 by recombinant retroviral infection of bone marrow cells isolated from 5-FU treated mice induced Dtx1 (145). Since Dtx1 and Dtx2 compound knock-out mice have no defect in T-cell development (146), any putative role for GATA-3 in Dtx1 expression needs to be analyzed more carefully at each T-cell developmental step (assuming that the Dtx1 mutant animals are indeed true nulls) (147). In addition to gain-of-function experiments, loss-of-function experiments will be essential to identify bona fide GATA-3 targets. In this regard, the Gata3 hypomorphic mutant mice (85) should prove to be invaluable, since ETP development is reduced but not completely missing in these mice and the development of the hypomorphic DN4 cells from DN3 cells is compromised in adoptive transfer animals.
During the CD4 vs. CD8 lineage choice, GATA-3 directly regulates the Zbtb7b gene, which encodes the zinc finger transcription factor Th-POK (66). Transgenic forced expression of Th-POK in DP and SP T cells under the control of the human CD2 promoter/LCR inhibited CD8 SP cell development and caused re-direction of MHC class I-restricted CD8 SP cells to differentiate into CD4 SP cells. Furthermore, the loss of DNA binding activity of Th-POK caused by point mutation of the zinc finger coding sequence in the Zbtb7b gene redirected MHC class II-restricted CD4 SP cells to differentiate into CD8 SP cells (60, 61). Since transgenic expression of Th-POK failed to rescue the defect in CD4 SP cell development that is observed in the absence of Gata3 (66), a gene(s) in addition to Th-POK that regulates CD4 SP cell development downstream of GATA-3 is likely to exist. Although GATA-3 binds to upstream regulatory sequences of the Cd8a gene (148), neither cell surface protein analysis nor reverse transcriptase polymerase chain reaction revealed any change in expression of CD8 or CD4 in Gata3 mutant DP cells (56, 57).
During Th2 activation, GATA-3 induces the expression of Th2 cytokine genes (Il4, Il5, and Il13) coincident with multiple changes in chromatin modification (86, 87, 90, 91, 93, 97, 99). GATA-3 induces the Bhlhe41 gene that encodes the basic helix-loop-helix transcription factor DEC2, which is specifically expressed in Th2 cells, is essential for Th2 differentiation, and promotes the expression of Th2 cytokines (149). GATA-3 also restricts Th1 differentiation by inhibiting Stat4 transcription (88, 150) and directly represses Ifng (99). ChIP experiments showed that GATA-3 binds to conserved non-coding sequences of the Th1-specific Il18r1 gene in Th2 cells (151). Forced GATA-3 expression in naive CD4+ T cells from wildtype or Il4-deficient mice represses Foxp3 mRNA, the key transcription factor gene in regulatory T cells (152). Similar repression is observed in human naive CD4+CD45RA+ T cells, and ChIP experiments revealed the binding of GATA-3 to a palindromic GATA site of the FOXP3 gene in Th2 cells but not in induced Treg cells (153). These data initially indicate that Foxp3 might be directly repressed by GATA-3.
In attempting to identify direct targets of GATA-3 and TBET in human Th1 and Th2 cells, GATA-3 was immunoprecipitated in ChIP assays followed by hybridization of the recovered DNA to microarrays containing probes for 8 kbp surrounding the transcription start sites of 18,450 protein-coding genes (ChIP-Chip)(154). This experiment identified 344 GATA-3-bound genes, including both Th2 and Th1. Thus although GATA-3 target genes are beginning to be characterized in peripheral Th2 cells, the functionally important targets of GATA-3 remain enigmatic during the early, intermediate, and late stage of T-cell development in the thymus, with only a few exceptions (Fig. 2). Systematic identification of these downstream target genes will be required to understand these regulomes.
Fig. 2. A hypothetical transcriptional hierarchy mediated through and by GATA-3.
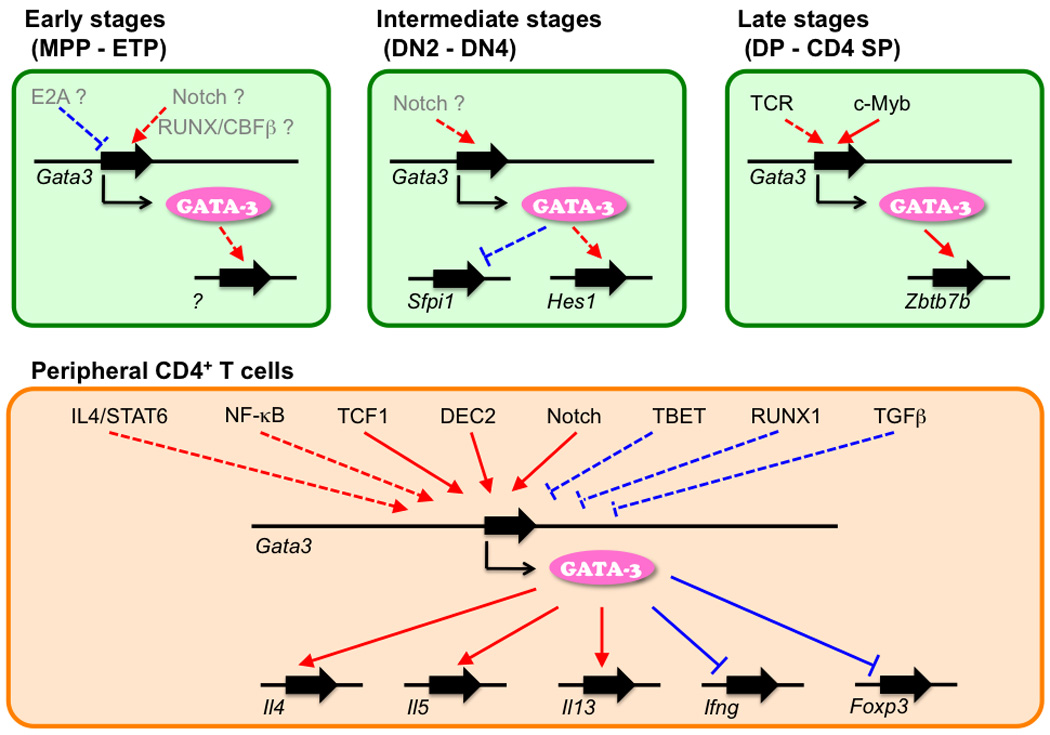
Factors that regulate Gata3 gene at each stage are shown. Positive regulators are shown in red arrow and negative regulators are shown in blue arrow. Factors that have been shown to directly bind Gata3 gene are shown by a solid line, while other factors are shown with a dashed line. Factors that are regulated by GATA-3 protein positively (red arrow) or negatively (blue arrow) are shown. Genes that have been show to be directly bound by GATA-3 are shown with a solid line, while genes for which evidence for direct regulation is lacking or questionable are shown with dashed lines. Factors shown in gray are suggested from correlative data.
Upstream regulators of Gata3 expression in T cells
An equally important unanswered question is the nature of the factor(s) that act upstream of Gata3 and control its expression throughout T-cell development. Is Gata3 gene activation spurred by a single predominant factor using a single enhancer throughout T-cell life? Or, do multiple factors drive Gata3 gene expression at each stage through individually discrete stage-specific enhancers? Although information is not yet available to answer this question, the currently scarce emerging data suggests the involvement of several factors that act directly or indirectly to regulate Gata3 at each developmental stage.
Activation of the Notch signaling pathway in hematopoietic progenitors induces T-cell development and multiple gene expression changes that are important for early and intermediate stages of T-cell development (29, 124). In the absence of Notch signaling, ETPs are not generated, while LSK progenitors circulating in the peripheral blood develop normally (27). Taghon et al. (121) performed a detailed kinetic study of fetal liver progenitors after exposure to Notch ligands in the OP9-DL1 co-culture system. Under these conditions, DN2 and DN3 T cells first emerge at days 3 and 4, respectively. Induction of Gata3 mRNA was first observed on day 3 in parallel with elevated transcription from the Tcf7, Ptcra, and Cd3e genes, while upregulated expression of the Notch target genes Hes1 and Hes5 was apparent even on day 1. Since Gata3 mRNA increases gradually between the DN1 and DN3 stages (46, 116), these data suggest that increasing Gata3 mRNA correlates with T-cell development but not in direct response to Notch signaling. Downregulation of the RUNX/CBFβ complex activity impairs the generation/expansion of ETPs, as well as their differentiation into DN2 and DN3 cells (155). Exposure of fetal liver progenitors to Notch ligand for 3–5 days in OP9-DL1 co-cultures induced Gata3 mRNA in DN1 cells, and this induction was attenuated in Cbfbrss/− hypomorphic mutant hematopoietic cells (156). One interpretation of these data is that Notch signaling may drive Gata3 gene expression through the RUNX/CBFβ complex during the earliest stages of T-cell development; a second is that development of Gata3-expressing cells may be impaired in Cbfbrss/− hypomorphic mutant cells. We note that a direct effect of the RUNX complex on Gata3 transcription has not been demonstrated to date.
The E2a gene encodes two alternatively spliced isoforms, E12 and E47. E47 cooperates with Notch to promote T-cell development from bone marrow progenitors (157) and E2a-deficient mice develop T-cell lymphomas that have a DN3-DP phenotype (158, 159). GFI1b, which is positively regulated by E2A, represses Gata3 in an E2a−/− lymphoma, and inhibition of Gata3 by siRNA or by forced Gfi1b expression prevents the expansion and survival of E2a−/− lymphoma cells (159). Repression of E protein activity in transgenic mice expressing Id1 under the control of the Lck gene causes induction of Gata3 mRNA in the CD4−CD8−CD44+CD25lo cells that aberrantly accumulate in these mice (145). These observations suggest that E2A proteins might negatively regulate Gata3 gene through Gfi1b. Since a role for endogenous Gfi1b, which is transiently induced at the DN3 stage, has not been reported and since the observation that forced transgenic expression of Gfi1b under the control of the Lck promoter had little effect on DP T-cell development (160), the physiological significance of Gata3 repression by Gfi1b will need to be examined much more extensively.
In DP thymocytes, GATA-3 is upregulated in response to TCR stimulation, and its expression level correlates with the strength of TCR signaling (56). The transcription factor MYB is required for T-cell development from the DN3 to DN4 stage, for survival and/or proliferation of DP cells, and for development of CD4 SP cells in the thymus (161, 162). Mice in which Myb was conditionally ablated at the DP and later stages (Mybflox/flox:TgCd4cre) showed reduced expression of Gata3 from DP through CD4 SP cells (163). Transgenic mice that express v-Myb, the oncogenic form of c-Myb, in T cells under the control of the CD2 promoter/LCR displayed about twofold higher levels of Gata3 mRNA in DP cells after positive selection (163). These data all implicate MYB in the regulation of Gata3 during the DP to CD4 SP cell transition.
IL4/signal transducer and activator of transcription 6 (STAT6) and IL2/STAT5 pathways are required for Th2 CD4+ T-cell differentiation (reviewed in 135). Forced ectopic expression of STAT6 in Th1 cells by retroviral introduction of a STAT6-ER fusion protein followed by tamoxifen treatment induced Gata3 and the Th2 cytokine genes Il4 and Il5 (164). Induction of Gata3 under Th2-favoring conditions was abrogated in the absence of Stat6 (165). Induction of the Il4 gene by GATA-3 (86) enhances Gata3 activation by a positive-feedback loop. In contrast, retrovirally enforced expression of a constitutively active form of STAT5a did not induce Gata3 expression but did trigger Th2 differentiation in the absence of IL4/STAT6 (166). Since this Th2 induction was abrogated in Gata3-deficient cells, endogenous GATA-3 appears to be required for STAT5-driven Th2 differentiation (98). GATA family proteins induce Gata3 mRNA in Stat6-deficient peripheral T cells (167, 168). These data indicate that autoactivation supports Gata3 activation in Th2 cells. Mice deficient in Nfkb1 (encoding the p50 subunit of NF-κB) show impaired production of Th2 cytokines after challenge with ovalbumin and reduced induction of GATA-3 (169). SAP and Viperin contribute to Th2 differentiation through NF-κB (170, 171). These data indicate that NF-κB-dependent Gata3 induction is required for Th2 differentiation.
Inactivation of Notch signaling by germline deletion of Rbpj or transgenic expression of a dominant negative form of its cofactor, Mastermind-like1, or by inactivation of Notch1 and Notch2 compromises Th2 responses (172–176). Retroviral transduction of the activated form of Notch in naive Stat6-deficient CD4+ T cells induced Gata3 mRNA and GATA-3-dependent Th2 responses (175, 176). The activated form of Notch induces transcription of Gata3 from the Ia promoter (177) but not from the Ib promoter, and ChIP experiments showed binding of RBPJ to the Gata3 Ia promoter (175, 176). These data indicate that the Notch signaling pathway induces Th2 responses at least in part by directly activating the Gata3 gene. Although Notch signaling activates the Ia promoter, it has also been shown that Ib transcripts are 100-fold more abundant than Ia transcripts during Th2 activation (178). Activation of Gata3 by the Notch signaling pathway is also observed at earlier stages of T-cell development (156), and loss of either factor resulted in impaired ETP development (27, 85). Furthermore, both Notch signaling and GATA-3 play important roles that are independent of pre-TCR complex formation required to pass the β-selection checkpoint (57, 179). Although direct regulation of the Gata3 gene by Notch effectors has not been shown except during peripheral stages, these data suggest that collaborative roles for Notch and GATA-3 are required repeatedly for T-cell development from the beginning to the end of T-cell development.
CD4+CD25− T cells isolated from mice ablated for the Tcf7 gene (encoding transcription factor TCF1) or from transgenic mice expressing ICAT, a specific inhibitor of the interaction of TCF1 and its cofactor β-catenin, showed impaired Th2 differentiation and reduced expression of Gata3 mRNA transcribed from the Gata3 Ib promoter compared to control cells, while transgenic mice expressing β-catenin showed the opposite response (178). ChIP experiments revealed binding of TCF1 and β-catenin to the Gata3 Ib promoter in stimulated CD4+ T cells (178). These data indicate that TCF1-dependent Gata3 activation is required for Th2 differentiation.
Mice ablated for Bhlhe41 (encoding transcription factor DEC2) showed impaired Th2 differentiation, and the CD4+ T cells isolated from those mutant mice had reduced GATA-3 mRNA in Th0 or Th2 cultures (149). Retrovirally directed expression of DEC2 greatly induced Gata3 transcription under Th2- or Th17-polarizing conditions, but it was only slightly induced under Th1-polarizing conditions (149). ChIP experiments using cells isolated from transgenic mice expressing FLAG-tagged DEC2 under the control of the CD2 promoter/LCR showed DEC2 binding to Gata3 (149). These data indicate that DEC2-dependent Gata3 induction is required for Th2 differentiation.
The transcription factor TBET, a master regulator of Th1 cell differentiation, represses the Gata3 gene (180). In addition to exerting transcriptional control by Gata3 repression, TBET may bind directly to GATA-3 protein and thereby sequester it from binding to target DNA sites (181). RUNX1, which promotes Th1 differentiation, represses the Gata3 gene (182). Additionally, RUNX3, which cooperates with TBET to promote Th1 differentiation, interacts with GATA-3 and suppresses its transcriptional activity (183). In contrast to its repressive effect at the Th1 stage, the RUNX/CBFβ complex may actually function as an activator of Gata3 at early stages (156). Since both GATA-3 and RUNX complexes play important roles at multiple stages (early, late, and peripheral) of T lymphopoiesis, it will be intriguing to elucidate the similarities and differences of their possible interplay at each stage. TGFβ supports Th17 differentiation by repressing Gata3 and Stat4 (184). The aryl hydrocarbon receptor (AhR) and its agonist M50354 also seem to skew the Th1/Th2 balance toward Th1 dominance by repressing Gata3 (185).
These upstream factors have all been implicated to exert direct control over Gata3 transcription through the promoter or putative enhancers of the Gata3 gene (Table 3), or indirectly by controlling other direct regulators or by modifying chromatin status within the locus (Fig. 2). We have found that Gata3 is expressed in up to two dozen discrete tissue types, and each expression pattern appears to be driven by a discrete, individual tissue-specific enhancer element (103, 106, 186–188, our unpublished data). To initially localize many of these regulatory elements, mice harboring reporter transgenes containing as little as 300 bp to mice bearing as much as 662 kbp of Gata3 genomic DNA have been examined in transfection and transgenic reporter assays. These studies identified regulatory elements for Gata3 that specify its expression in the 1st and 2nd branchial arches, the genital tubercle/cloaca, in mammary epithelial cells, hair follicles, vestibulocochlear ganglia, the CNS (three separate enhancers), the eye, the ectoplacental cone/placenta, the thyroid gland, the mesonephric and metanephric tubules of the embryonic kidney, the semicircular canals of the inner ear and primordia specifying the endocardial cushions. Perhaps surprisingly, these studies also indicated that the regulatory elements that control Gata3 expression in the sympathetic nervous system probably lie beyond 451 kbp 5′ or 211 kbp 3′ (as defined by one transgenic YAC; the genome sequence-revised endpoints) with respect to the GATA-3 translational start site. However, since all these studies were performed between days 10.5 and 14.5 of embryogenesis, the element specifying Gata3 expression in mature T cells was not examined (since the first DP cells develop at around e16.5) (189). We have recently found that this same 662 kbp Gata3 YAC does not harbor sufficient sequence information to direct reporter gene expression in adult thymocytes, but intriguingly we have now identified multiple overlapping BACs that can activate a Gata3-directed reporter gene in T cells in vivo (our unpublished observations). The developmental and temporal specificity of this regulatory sequence(s) may reveal profound new insights into the regulation of Gata3, and therefore indirectly into the molecular mechanisms controlling T-cell development.
Table 3.
Binding sites of proposed upstream transcription factors
| Transcription factor |
Gata3 regulation | Recovered region by ChIP | ChIP | Reference |
|---|---|---|---|---|
| MYB | CD4 SP | 382–618 bps upstream of Ib | total thymocytes | (163) |
| RBPJ | Th2 | 291–398 bps upstream of Ia | CD4+ T | (175) |
| RBPJ | Th2 | 154–310 bps upstream of Ia | stimulated CD4+ T | (176) |
| TCF1 | Th2 | 550–650 bps upstream of Ib | stimulated CD4+ T | (178) |
| DEC2 | Th2 | 438–1072 bps downstream of Ib (1st intron and 2nd exon) |
naïve CD4+ T | (149) |
Conclusions
We have reviewed here the requirements for and known functions of GATA-3 in T-lineage cells from the earliest T-cell progenitors in the thymus all the way to peripheral CD4+ Th2 cells. Growing evidence has begun to reveal the hierarchical positioning of GATA-3 and other molecules in these important developmental processes, especially in peripheral Th2 cell activation. In contrast, only a handful of factors have been demonstrated to interact with GATA-3 during T-cell development in the thymus. The development of next-generation sequencing technology has made it possible to comprehensively identify direct targets of a transcription factor through whole genome ChIP sequencing and whole genome mRNA sequencing. Subsequent molecular biological confirmation of the functional importance of these genomic indications will be essential to understand the transcriptional hierarchy mediated by and through GATA-3. To these ends, the Gata3 hypomorphic mutant as well as the Gata3 null and conditional alleles should prove to be indispensable tools. As described above, a skeletal network description for immature T-cell development has been published and should serve as an outstanding framework for probing aspects of transcriptional regulation during immature T-cell development (145). Although much more data will be required to elaborate the influences and nuances in this network that will produce a precisely refined picture of the process as a whole, overwhelming evidence supports the hypothesis that GATA-3 must play a central role at multiple stages of T-cell development, and therefore, it must be a fundamental contributor to many steps in the overall framework from the birth to the death of T cells. It will be fascinating both to test mechanistically how the network is maintained as well as to learn what (known and currently unknown) molecules participate in the establishment of T-cell development.
Acknowledgements
T. Hosoya was supported in part by a fellowship from the Astellas Foundation for Research on Metabolic Disorders. I. Maillard was supported by the Damon Runyon Cancer Research Foundation and the American Society of Hematology. This research was initially supported by National Institutes of Health research grant R01 GM28896 to J.D. Engel.
References
- 1.Donskoy E, Goldschneider I. Thymocytopoiesis is maintained by blood-borne precursors throughout postnatal life. A study in parabiotic mice. J Immunol. 1992;148:1604–1612. [PubMed] [Google Scholar]
- 2.Adolfsson J, et al. Upregulation of flt3 expression within the bone marrow lin(−)sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 3.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adolfsson J, et al. Identification of flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegué E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 6.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Månsson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Pantelouris EM. Absence of thymus in a mouse mutant. Nature. 1968;217:370–371. doi: 10.1038/217370a0. [DOI] [PubMed] [Google Scholar]
- 9.Liu C, et al. The role of CCL21 in recruitment of t-precursor cells to fetal thymi. Blood. 2005;105:31–39. doi: 10.1182/blood-2004-04-1369. [DOI] [PubMed] [Google Scholar]
- 10.Rossi FM, et al. Recruitment of adult thymic progenitors is regulated by p-selectin and its ligand PSGL-1. Nat Immunol. 2005;6:626–634. doi: 10.1038/ni1203. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, et al. Coordination between CCR7- and ccr9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 13.Gossens K, Naus S, Corbel SY, Lin S, Rossi FM, Kast J, Ziltener HJ. Thymic progenitor homing and lymphocyte homeostasis are linked via s1p-controlled expression of thymic p-selectin/CCL25. J Exp Med. 2009;206:761–778. doi: 10.1084/jem.20082502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Förster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2010;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 15.Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhandoola A, von Boehmer H, Petrie HT, Zúñiga-Pflücker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: Plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 18.Perry SS, Wang H, Pierce LJ, Yang AM, Tsai S, Spangrude GJ. L-Selectin defines a bone marrow analog to the thymic early t-lineage progenitor. Blood. 2004;103:2990–2996. doi: 10.1182/blood-2003-09-3030. [DOI] [PubMed] [Google Scholar]
- 19.Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc Natl Acad Sci U S A. 2007;104:6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 21.Inlay MA, et al. Ly6D marks the earliest stage of b-cell specification and identifies the branchpoint between b-cell and t-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin CH, Aifantis I, Scimone ML, von Andrian UH, Reizis B, von Boehmer H, Gounari F. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat Immunol. 2003;4:866–873. doi: 10.1038/ni965. [DOI] [PubMed] [Google Scholar]
- 23.Scimone ML, Aifantis I, Apostolou I, von Boehmer H, von Andrian UH. A multistep adhesion cascade for lymphoid progenitor cell homing to the thymus. Proc Natl Acad Sci U S A. 2006;103:7006–7011. doi: 10.1073/pnas.0602024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 25.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zúñiga-Pflücker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-t cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Benz C, Martins VC, Radtke F, Bleul CC. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J Exp Med. 2008;205:1187–1199. doi: 10.1084/jem.20072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambandam A, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 28.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 29.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 20.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 21.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch U, et al. Delta-Like 4 is the essential, nonredundant ligand for notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodewald HR, Kretzschmar K, Takeda S, Hohl C, Dessing M. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 1994;13:4229–4240. doi: 10.1002/j.1460-2075.1994.tb06743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–116. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamoto H, Ohmura K, Katsura Y. Direct evidence for the commitment of hematopoietic stem cells to T, B and myeloid lineages in murine fetal liver. Int Immunol. 1997;9:1011–1019. doi: 10.1093/intimm/9.7.1011. [DOI] [PubMed] [Google Scholar]
- 36.Masuda K, et al. Prethymic t-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancrin C, Schneider E, Lambolez F, Arcangeli ML, Garcia-Cordier C, Rocha B, Ezine S. Major T cell progenitor activity in bone marrow-derived spleen colonies. J Exp Med. 2002;195:919–929. doi: 10.1084/jem.20011475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maillard I, et al. Notch-Dependent t-lineage commitment occurs at extrathymic sites following bone marrow transplantation. Blood. 2006;107:3511–3519. doi: 10.1182/blood-2005-08-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arcangeli ML, Lancrin C, Lambolez F, Cordier C, Schneider E, Rocha B, Ezine S. Extrathymic hemopoietic progenitors committed to T cell differentiation in the adult mouse. J Immunol. 2005;174:1980–1988. doi: 10.4049/jimmunol.174.4.1980. [DOI] [PubMed] [Google Scholar]
- 40.Maeda T, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 42.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult t-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 43.Godfrey DI, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. Onset of tcr-beta gene rearrangement and role of tcr-beta expression during CD3-CD4-CD8- thymocyte differentiation. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 44.Capone M, Hockett RD, Zlotnik A. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) pro-t thymocytes. Proc Natl Acad Sci U S A. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias AM, Chen D, Rothenberg EV. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev Biol. 2002;246:103–121. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- 46.David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, Rothenberg EV. Progression of regulatory gene expression states in fetal and adult pro-t-cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Boehmer H. Unique features of the pre-t-cell receptor alpha-chain: Not just a surrogate. Nat Rev Immunol. 2005;5:571–577. doi: 10.1038/nri1636. [DOI] [PubMed] [Google Scholar]
- 48.Rothenberg EV, Moore JE, Yui MA. Launching the t-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peschon JJ, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodewald HR, Kretzschmar K, Swat W, Takeda S. Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity. 1995;3:313–319. doi: 10.1016/1074-7613(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 51.Rodewald HR, Ogawa M, Haller C, Waskow C, DiSanto JP. Pro-Thymocyte expansion by c-kit and the common cytokine receptor gamma chain is essential for repertoire formation. Immunity. 1997;6:265–272. doi: 10.1016/s1074-7613(00)80329-5. [DOI] [PubMed] [Google Scholar]
- 52.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sitnicka E, et al. Critical role of FLT3 ligand in IL-7 receptor independent T lymphopoiesis and regulation of lymphoid-primed multipotent progenitors. Blood. 2007;110:2955–2964. doi: 10.1182/blood-2006-10-054726. [DOI] [PubMed] [Google Scholar]
- 54.Jensen CT, et al. Permissive roles of hematopoietin and cytokine tyrosine kinase receptors in early t-cell development. Blood. 2008;111:2083–2090. doi: 10.1182/blood-2007-08-108563. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J. TOX: An HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 56.Hernández-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-lla J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 57.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 58.Woolf E, et al. Runx3 and runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aliahmad p, et al. TOX provides a link between calcineurin activation and CD8 lineage commitment. J Exp Med. 2004;199:1089–1099. doi: 10.1084/jem.20040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun G, et al. The zinc finger protein ckrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 61.He X, et al. The zinc finger transcription factor th-pok regulates CD4 versus CD8 t-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 62.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Egawa T, Littman DR. Thpok acts late in specification of the helper T cell lineage and suppresses runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muroi S, et al. Cascading suppression of transcriptional silencers by thpok seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 65.Setoguchi R, et al. Repression of the transcription factor th-pok by runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, et al. Distinct functions for the transcription factors GATA-3 and thpok during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ladi E, Yin X, Chtanova T, Robey EA. Thymic microenvironments for T cell differentiation and selection. Nat Immunol. 2006;7:338–343. doi: 10.1038/ni1323. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 69.Evans T, Felsenfeld G. The erythroid-specific transcription factor eryf1: A new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 70.Tsai SF, Martin DI, Zon LI, D'Andrea AD, Wong GG, Orkin SH. Cloning of cdna for the major dna-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 71.Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JB, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 72.Ko LJ, Engel JD. Dna-Binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merika M, Orkin SH. Dna-Binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pevny L, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 75.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity gata-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Migliaccio AR, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the gata-1low mouse mutant. J Exp Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 80.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 81.Ling KW, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodrigues NP, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 83.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the t-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 84.Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 85.Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 87.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine th1 and th2 cells and controls th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 88.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of th1 development mediated by GATA-3 through an il-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 89.Zhang DH, Yang L, Cohn L, Parkyn L, Homer R, Ray P, Ray A. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11:473–482. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 90.Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, Arai N. GATA-3 induces T helper cell type 2 (th2) cytokine expression and chromatin remodeling in committed th1 cells. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takemoto N, et al. Cutting edge: Chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for th2-specific cytokine gene cluster. J Immunol. 2000;165:6687–6691. doi: 10.4049/jimmunol.165.12.6687. [DOI] [PubMed] [Google Scholar]
- 92.Farrar JD, Ouyang W, Löhning M, Assenmacher M, Radbruch A, Kanagawa O, Murphy KM. An instructive component in T helper cell type 2 (th2) development mediated by GATA-3. J Exp Med. 2001;193:643–650. doi: 10.1084/jem.193.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 94.Nawijn MC, et al. Enforced expression of GATA-3 in transgenic mice inhibits th1 differentiation and induces the formation of a T1/st2-expressing th2-committed T cell compartment in vivo. J Immunol. 2001;167:724–732. doi: 10.4049/jimmunol.167.2.724. [DOI] [PubMed] [Google Scholar]
- 95.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Skapenko A, et al. GATA-3 in human T cell helper type 2 development. J Exp Med. 2004;199:423–428. doi: 10.1084/jem.20031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamashita M, et al. Essential role of GATA3 for the maintenance of type 2 helper T (th2) cytokine production and chromatin remodeling at the th2 cytokine gene loci. J Biol Chem. 2004;279:26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- 98.Zhu J, et al. Conditional deletion of gata3 shows its essential function in T(H)1–T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 99.Chang S, Aune TM. Dynamic changes in histone-methylation 'marks' across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 100.Samson SI, et al. GATA-3 promotes maturation, ifn-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 101.Vosshenrich CA, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 102.Pandolfi PP, et al. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 103.Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- 104.Moriguchi T, et al. Gata3 participates in a complex transcriptional feedback network to regulate sympathoadrenal differentiation. Development. 2006;133:3871–3881. doi: 10.1242/dev.02553. [DOI] [PubMed] [Google Scholar]
- 105.Zhou QY, Quaife CJ, Palmiter RD. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature. 1995;374:640–643. doi: 10.1038/374640a0. [DOI] [PubMed] [Google Scholar]
- 106.Hasegawa SL, Moriguchi T, Rao A, Kuroha T, Engel JD, Lim KC. Dosage-Dependent rescue of definitive nephrogenesis by a distant gata3 enhancer. Dev Biol. 2007;301:568–577. doi: 10.1016/j.ydbio.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaufman CK, et al. GATA-3: An unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurek D, Garinis GA, van Doorninck JH, van der Wees J, Grosveld FG. Transcriptome and phenotypic analysis reveals gata3-dependent signalling pathways in murine hair follicles. Development. 2007;134:261–272. doi: 10.1242/dev.02721. [DOI] [PubMed] [Google Scholar]
- 109.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 111.Grigorieva IV, et al. Gata3-Deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor gcm2. J Clin Invest. 2010;120:2144–2155. doi: 10.1172/JCI42021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 113.Zhong JF, et al. Gene expression profile of murine long-term reconstituting vs. Short-Term reconstituting hematopoietic stem cells. Proc Natl Acad Sci U S A. 2005;102:2448–2453. doi: 10.1073/pnas.0409459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 115.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 117.George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 118.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no b-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 120.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Taghon TN, David ES, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible t-lineage specification induced by notch/delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Surh CD, Sprent J. T-Cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. [PubMed] [Google Scholar]
- 123.Chen D, Zhang G. Enforced expression of the GATA-3 transcription factor affects cell fate decisions in hematopoiesis. Exp Hematol. 2001;29:971–980. doi: 10.1016/s0301-472x(01)00670-1. [DOI] [PubMed] [Google Scholar]
- 124.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 125.Hozumi K, et al. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur J Immunol. 2008;38:977–985. doi: 10.1002/eji.200737688. [DOI] [PubMed] [Google Scholar]
- 126.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 127.Ktorza S, Sarun S, Rieux-Laucat F, de Villartay JP, Debré P, Schmitt C. Cd34-Positive early human thymocytes: T cell receptor and cytokine receptor gene expression. Eur J Immunol. 1995;25:2471–2478. doi: 10.1002/eji.1830250910. [DOI] [PubMed] [Google Scholar]
- 128.Taghon T, De Smedt M, Stolz F, Cnockaert M, Plum J, Leclercq G. Enforced expression of GATA-3 severely reduces human thymic cellularity. J Immunol. 2001;167:4468–4475. doi: 10.4049/jimmunol.167.8.4468. [DOI] [PubMed] [Google Scholar]
- 129.Tsai FY, Browne CP, Orkin SH. Knock-In mutation of transcription factor GATA-3 into the GATA-1 locus: Partial rescue of GATA-1 loss of function in erythroid cells. Dev Biol. 1998;196:218–227. doi: 10.1006/dbio.1997.8842. [DOI] [PubMed] [Google Scholar]
- 130.Takahashi S, et al. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood. 2000;96:910–916. [PubMed] [Google Scholar]
- 131.Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative mare-dependent transcriptional regulation is determined by the abundance of small maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 132.Greaves DR, Wilson FD, Lang G, Kioussis D. Human CD2 3'-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 133.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 134.Nawijn MC, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol. 2001;167:715–723. doi: 10.4049/jimmunol.167.2.715. [DOI] [PubMed] [Google Scholar]
- 135.Zhu J, Paul WE. CD4 T cells: Fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marine J, Winoto A. The human enhancer-binding protein gata3 binds to several t-cell receptor regulatory elements. Proc Natl Acad Sci U S A. 1991;88:7284–7288. doi: 10.1073/pnas.88.16.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Joulin V, Bories D, Eléouet JF, Labastie MC, Chrétien S, Mattéi MG, Roméo PH. A t-cell specific TCR delta DNA binding protein is a member of the human GATA family. EMBO J. 1991;10:1809–1816. doi: 10.1002/j.1460-2075.1991.tb07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ko LJ, Yamamoto M, Leonard MW, George KM, Ting P, Engel JD. Murine and human t-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human t-cell receptor delta gene enhancer. Mol Cell Biol. 1991;11:2778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kishi H, Wei XC, Jin ZX, Fujishiro Y, Nagata T, Matsuda T, Muraguchi A. Lineage-Specific regulation of the murine RAG-2 promoter: GATA-3 in T cells and pax-5 in B cells. Blood. 2000;95:3845–3852. [PubMed] [Google Scholar]
- 141.Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bhlh gene hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-t cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 143.Georgescu C, et al. A gene regulatory network armature for T lymphocyte specification. Proc Natl Acad Sci U S A. 2008;105::20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yamamoto N, et al. Role of deltex-1 as a transcriptional regulator downstream of the notch receptor. J Biol Chem. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- 145.Wang HC, Perry SS, Sun XH. Id1 attenuates notch signaling and impairs T cell commitment by elevating deltex1 expression. Mol Cell Biol. 2009;29:4640–4652. doi: 10.1128/MCB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lehar SM, Bevan MJ. T cells develop normally in the absence of both deltex1 and deltex2. Mol Cell Biol. 2006;26:7358–7371. doi: 10.1128/MCB.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hsiao HW, Liu WH, Wang CJ, Lo YH, Wu YH, Jiang ST, Lai MZ. Deltex1 is a target of the transcription factor NFAT that promotes T cell anergy. Immunity. 2009;31:72–83. doi: 10.1016/j.immuni.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 148.Landry DB, Engel JD, Sen R. Functional GATA-3 binding sites within murine CD8 alpha upstream regulatory sequences. J Exp Med. 1993;178:941–949. doi: 10.1084/jem.178.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang XO, et al. Requirement for the basic helix-loop-helix transcription factor dec2 in initial TH2 lineage commitment. Nat Immunol. 2009;10:1260–1266. doi: 10.1038/ni.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses th1 development by downregulation of stat4 and not through effects on il-12rbeta2 chain or t-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 151.Yu Q, Chang HC, Ahyi AN, Kaplan MH. Transcription factor-dependent chromatin remodeling of il18r1 during th1 and th2 differentiation. J Immunol. 2008;181:3346–3352. doi: 10.4049/jimmunol.181.5.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wei J, Duramad O, Perng OA, Reiner SL, Liu YJ, Qin FX. Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2007;104:18169–18174. doi: 10.1073/pnas.0703642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mantel PY, et al. Gata3-Driven th2 responses inhibit tgf-beta1-induced FOXP3 expression and the formation of regulatory T cells. Plos Biol. 2007;5:e329. doi: 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jenner RG, et al. The transcription factors t-bet and GATA-3 control alternative pathways of t-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U S A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Talebian L, et al. T-Lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in cbfbeta dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development, and cooperate with notch signaling during T cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bain G, et al. E2A deficiency leads to abnormalities in alphabeta t-cell development and to rapid development of t-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu W, Kee BL. Growth factor independent 1B (gfi1b) is an E2A target gene that modulates gata3 in t-cell lymphomas. Blood. 2007;109:4406–4414. doi: 10.1182/blood-2006-08-043331. [DOI] [PubMed] [Google Scholar]
- 160.Doan LL, et al. Growth factor independence-1b expression leads to defects in T cell activation, IL-7 receptor alpha expression, and T cell lineage commitment. J Immunol. 2003;170:2356–2366. doi: 10.4049/jimmunol.170.5.2356. [DOI] [PubMed] [Google Scholar]
- 161.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 162.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci U S A. 2004;101:14853–14858. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maurice D, Hooper J, Lang G, Weston K. C-Myb regulates lineage choice in developing thymocytes via its target gene gata3. EMBO J. 2007;26:3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated stat6 induces the expression of th2-specific cytokines and transcription factors in developing th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 165.Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 166.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 167.Ouyang W, Löhning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-Independent GATA-3 autoactivation directs il-4-independent th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 168.Ranganath S, Murphy KM. Structure and specificity of GATA proteins in th2 development. Mol Cell Biol. 2001;21:2716–2725. doi: 10.1128/MCB.21.8.2716-2725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for nf-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 170.Cannons JL, et al. SAP regulates T(H)2 differentiation and pkc-theta-mediated activation of nf-kappab1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 171.Qiu LQ, Cresswell P, Chin KC. Viperin is required for optimal th2 responses and t-cell receptor-mediated activation of nf-kappab and AP-1. Blood. 2009;113:3520–3529. doi: 10.1182/blood-2008-07-171942. [DOI] [PubMed] [Google Scholar]
- 172.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 173.Tanigaki K, et al. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 174.Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Amsen D, et al. Direct regulation of gata3 expression determines the T helper differentiation potential of notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol. 2002;168:4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- 178.Yu Q, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Maillard I, et al. The requirement for notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-t cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O'Shea JJ, Strober W. T-Bet regulates th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of t-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 182.Komine O, et al. The runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the th2 lineage by repressing GATA3 expression. J Exp Med. 2003;198:51–61. doi: 10.1084/jem.20021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kohu K, et al. The runx3 transcription factor augments th1 and down-modulates th2 phenotypes by interacting with and attenuating GATA3. J Immunol. 2009;183:7817–7824. doi: 10.4049/jimmunol.0802527. [DOI] [PubMed] [Google Scholar]
- 184.Das J, et al. Transforming growth factor beta is dispensable for the molecular orchestration of th17 cell differentiation. J Exp Med. 2009;206:2407–2416. doi: 10.1084/jem.20082286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Negishi T, et al. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J Immunol. 2005;175:7348–7356. doi: 10.4049/jimmunol.175.11.7348. [DOI] [PubMed] [Google Scholar]
- 186.Lieuw KH, Li G, Zhou Y, Grosveld F, Engel JD. Temporal and spatial control of murine GATA-3 transcription by promoter-proximal regulatory elements. Dev Biol. 1997;188:1–16. doi: 10.1006/dbio.1997.8575. [DOI] [PubMed] [Google Scholar]
- 187.Lakshmanan G, Lieuw KH, Grosveld F, Engel JD. Partial rescue of GATA-3 by yeast artificial chromosome transgenes. Dev Biol. 1998;204:451–463. doi: 10.1006/dbio.1998.8991. [DOI] [PubMed] [Google Scholar]
- 188.Lakshmanan G, Lieuw KH, Lim KC, Gu Y, Grosveld F, Engel JD, Karis A. Localization of distant urogenital system-, central nervous system-, and endocardium-specific transcriptional regulatory elements in the GATA-3 locus. Mol Cell Biol. 1999;19:1558–1568. doi: 10.1128/mcb.19.2.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Penit C, Vasseur F. Cell proliferation and differentiation in the fetal and early postnatal mouse thymus. J Immunol. 1989;142:3369–3377. [PubMed] [Google Scholar]


