Summary
Invariant natural killer T (iNKT) cells comprise a rare lymphocyte sublineage with phenotypic and functional properties similar to T and NK cells. Akin to conventional αβ T cells, their development occurs primarily in the thymus, where they originate from CD4+CD8+ double positive (DP) progenitors. However, the selection of iNKT cells is unique in that it is mediated by homotypic interactions of DP cells and recognition of glycolipid antigen-CD1d complexes. Additionally, iNKT cells acquire an activated innate-like phenotype during development that allows them to release cytokines rapidly following antigen exposure. Given their hybrid features, it is not surprising that the developmental program of iNKT cells partially overlaps with that of T and NK cells. Several recent reports have provided new and exciting insights into the developmental mechanisms that direct NKT cell lineage commitment and maturation. In this review, we provide a discussion of the NKT cell developmental program with an emphasis on the signaling mechanisms and transcription factors that influence the ontogeny of this lineage. Continued investigations of the complex interplay of these transcription factors and their relationship with other extracellular and intracellular signaling molecules will undoubtedly provide important clues into the biology of this unusual T-cell lineage.
Keywords: natural killer T cells, cell fate, development, transcription factors
Introduction
Natural Killer T (NKT) cells are a heterogeneous subset of T lymphocytes that are developmentally and functionally distinct from conventional CD4+ and CD8+ T cells. While T and NKT cells both originate in the thymus, the requirements for their selection are divergent. Specifically, conventional T cells are selected by peptide antigens in complex with major histocompatibility complex (MHC) class I or II molecules present on the surface of thymic epithelial cells, whereas NKT cells develop following selection by self glycolipid antigens in complex with the MHC class I–like molecule CD1d, when presented by CD4+CD8+ double positive (DP) thymocytes. Additionally, NKT cells are characterized by several distinguishing traits, including the expression of NK cell-specific surface markers, a restricted T-cell receptor (TCR) repertoire, and an innate memory-like phenotype. As a consequence of their activated phenotype, NKT cells rapidly secrete copious quantities of Th1 and Th2 cytokines after activation through the TCR and are now recognized to play an important role at the interface between the innate and adaptive immune systems. To acquire their functional properties, NKT cell progenitors must respond to cues provided to them in the thymus by regulating the expression of transcription factors that further promote their development. We review the steps involved in NKT cell ontogeny and discuss recent advances made in the field that identify transcription factors critical to the development of this unique cell lineage.
Phenotypic and functional properties of NKT cells
Several NKT cell subsets exist in humans and mice that differ in their pattern of expression of surface markers as well as their functional properties. Some of these subsets are discussed below and their salient features summarized in Tables 1 and 2.
Table 1.
Differences and similarities between conventional T cells, NKT cells and NKT-like cells
| Features | NKT cells | NKT-like (innate) cells | |||||
|---|---|---|---|---|---|---|---|
| αβ T | Type I | Type II | MAIT | T-CD4+ | CD8+$ | γδT @ | |
| TCR repertoire | Diverse | Vα14Jα18 Vβ8.2, Vβ7, Vβ2 |
Diverse, some have Vα3.2Jα9, Vα8Jα9, Vβ8 |
Vα19Jα33 Vβ8, Vβ6 |
Diverse | Diverse | Vγ1.1Vδ6.3 |
| MHC-restriction | MHC I or MHC II |
CD1d | CD1d | MR-1 | MHC II | MHC I | ND |
| Selecting cells | TEC | DP | DP | B cells or DCs? |
DP | DP | ND |
| TCR ligands | Peptide antigens |
iGb3, αGalCer |
Sulfatide, Lyso- sulfatide |
Hydrophilic antigens or lipids |
ND | ND | Non- peptidic antigens |
| Positive selection | Yes | Yes | Yes | Yes | Yes | Yes | No |
|
CD4/CD8
expression |
CD4+ or CD8+ | CD4+ or DN | CD4+ or DN | primarily DN | CD4+ | CD8+ | No |
|
Activated
Phenotype |
After antigen exposure |
Yes | Yes | Yes | Yes | Yes | Yes |
|
SLAM-SAP
dependent |
No | Yes | Yes | No | Yes | Yes | Yes |
| PLZF expression | No | Yes | Yes | Yes | Yes | Yes | Yes |
| α-GalCer reactivity | No | Yes | No | No | No | No | No |
NKT: natural killer T, TCR: T cell receptor, MHC: major histo-compatibility, MR-1: major histocompatibility molecule related-1, MAIT: mucosal associated invariant T, TEC: thymic epithelial cells, DP: double positive, DN: double negative, SLAM: signaling lymphocytic activation molecule, SAP: SLAM-associated molecule, iGb3: isoglobotrihexosylceramide, PLZF: promyelocytic zinc finger, α-GalCer: alpha-galactosylceramide, ND: not determined.
Innate-like CD8+ T cell subset found in Itk-deficient mice
Specific subset of γδ T cells defined by the expression of Vγ1.1+Vδ6.3+ TCR
Table 2.
Characteristics of mouse and human invariant NKT cells
| Feature | Mouse | Human |
|---|---|---|
| TCR α chain | Vα14Jα18 | Vα24Jα18 |
| TCR β chain | Vβ8.2, Vβ7, Vβ2 | Vβ11 |
| CD1d dependent | Yes | Yes |
| α-Gal-Cer-reactivity | Yes | Yes |
| Selecting Ligand | Controversial | Controversial |
| Phenotype | NK1.1+, NK1.1−, CD4+ or DN |
CD161+, CD4+, DN or CD8+ |
| Frequency in thymus | ~ 0.5% | <0.1% |
| Frequency in liver | 20-30% | ~1% |
| Frequency in spleen | ~1% | Unknown |
|
Cytokine production upon
TCR ligation |
IL-4, IFN-γ | IL-4, IFN-γ |
Type I NKT cells
In the mouse, type I NKT cells, also known as ‘classical’ or ‘invariant’ NKT cells (hereafter referred to as iNKT cells), express an invariant TCR that is composed of a common α-chain (Vα14-Jα18) in combination with a limited number of β-chains (Vβ8.2, Vβ7 or Vβ2) (1, 2). This invariant TCR confers reactivity to glycolipid antigens, as exemplified by the model iNKT cell agonist α-galactosylceramide (α-GalCer), which was originally isolated from the sea sponge Agelas mauritianus (3). The development and use of α-GalCer-loaded CD1d tetramers has greatly facilitated the study of iNKT cells and it is now clear that murine α-GalCer-CD1d tetramer reactive cells are made up of at least two populations based on their expression of CD4, including CD4+ and CD4−CD8− double negative (DN) subsets, as well as NK1.1 (NK1.1+ and NK1.1−) (1, 2, 4). An NK1.1− population exists in the thymus, which likely represents a pool of immature cells (5, 6), as well as in the periphery. Extra-thymic NK1.1− cells are comprised in part of immature cells that have emigrated from the thymus (5-7), as well as mature iNKT cells that have down-regulated the NK1.1 receptor following antigenic encounter. Indeed, in vitro stimulation NK1.1+ iNKT cells leads to the down regulation of NK1.1 expression (8).
Human type I NKT cells express a TCR that is made up of the Vα24-Jα18 and Vβ11 α- and β-TCR chains, which like its murine counterpart, reacts with lipid antigens such as α-GalCer (3, 4, 9-11). Mature human NKT cells from the blood express CD161 (a marker equivalent to murine NK1.1), and are CD4+, DN or CD8+(12, 13). It is not known why human NKT cells express the CD8 marker, as there is no evidence that this molecule is required to bind to CD1d during development in mice (14). The functions of different subsets of human peripheral blood iNKT cells are not yet well understood; however, the CD4+, DN or CD8+ subsets appear to differ in their profiles of gene expression, cytokine production, and ability to activate bystander cells (15-18).
In the mouse, iNKT cells are found primarily in the liver, thymus, and spleen, where they constitute up to 30-40%, 1%, and 0.5% of the lymphocytes within these organs, respectively (19-21). They are less abundant in peripheral lymph nodes. The distribution of NKT cells is less well studied in humans; however, iNKT cells comprise 0.008-1.176% of peripheral blood T cells (22), 0.001-0.01% of total human thymocytes (12, 13), and 1% of liver lymphocytes (19-21).
Type II NKT cells
Type II NKT cells are a CD1d-restricted population of cells that in the mouse exhibit heterogeneous TCR αβ chain usage with some cells expressing a recurrent Vα3.2-Jα9/Vβ8 or Vα8/Vβ8 chain combination (1, 4). Although type II NKT cells were first described over a decade ago, less progress has been made in their characterization compared to type I NKT cells. This lack of progress has been largely due to the absence of specific surface markers and agonistic antigens for this population. Recently, it was reported that sulfatide, a self-glycolipid derived from myelin, specifically stimulates a fraction of type II NKT cells and that these cells can be detected with tetramers composed of sulfatide-CD1d complexes (23). Using these tetramers, it has been determined that type II NKT cells are significantly fewer in number than their type I counterparts in the mouse spleen (23). However, it is important to note that the sulfatide-CD1d tetramers do not recognize the Vα3.2-Jα9 and Vα8 subsets (23), implying that the sulfatide-CD1d reactive cells do not include the entire population of type II cells. Type II NKT cells have been detected in the livers of human hepatitis patients (24). Other antigens, such as lyso-sulfatide, (an analog of sulfatide) and phenyl 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonate have been reported to recognize certain type II NKT cells in mice (25) and humans (26), respectively. In the mouse, type II NKT cells appear to be comprised of CD4+, CD4−CD8−, NK1.1+, and NK1.1− subpopulations (27, 28).
Functions of Type I and Type II NKT cells
Stimulation of murine type I NKT cells with α-GalCer or other bacterial or mammalian glycolipids results in the rapid production of cytokines and upregulation of co-stimulatory receptors, which activates numerous other immune cell types including dendritic cells, and macrophages, as well as NK, B and T cells (29, 30). As a result, activation of iNKT cells is known to modulate an array of normal and pathogenic immune responses (29, 30). Type I NKT cells mediate protection against certain infections and cancers by virtue of their production of Th1 cytokines (IFN-γ and IL-12) (29) and can also promote specific diseases through secretion of Th2 cytokines (IL-4 and IL-13), as in the case of asthmatic allergen-induced airway hypersensitivity (31). However, they also down regulate inflammatory responses in murine graft versus host disease (GVHD) (32) and human multiple sclerosis (33). Type II NKT cells also mediate both inflammatory and suppressive roles in different disease models (29). While they promote inflammatory responses in hepatitis (24, 34) and ulcerative colitis (35), they act as regulatory cells in experimental autoimmune encephalitis (EAE) (23), type I diabetes (36), and GVHD (37). Furthermore, in a recent study, Ambrosino et al. (38) demonstrated that not only do type I and type II NKT cells have opposite roles in tumor immunity but that they can also counter-regulate one another’s activity.
Other NKT-like cell populations
Additional ‘NKT-like’ T-cell populations have been described in humans and mice, including MHC molecule related-1 (MR-1)-restricted mucosal associated invariant T (MAIT) cells (39), thymocyte-selected CD4+ T cells (T-CD4+) (40), unique populations of innate-like CD8+ T cells that emerge in mice with impairments in TCR signaling such as animals lacking expression of the Tec kinase ITK (41) the Inhibitor of DNA binding factor Id3 (42) or the transcription factor KLF2 (43), or expressing the SLP-76 Y145F mutation (44), and finally the γ1.1+δ6.3+ subset of γδ T cells (45). Interestingly, many of these lineages share properties with iNKT cells, such as co-production of IL-4 and IFNγ upon activation, expression of the promyelocytic zinc finger (PLZF) transcription factor and reliance upon the signaling lymphocytic activation molecule (SLAM)-associated protein (SAP) for their development (see below). Some of the characteristics of these other NKT-like lineages are summarized in Table 2.
Models for iNKT cell development
Similar to conventional T cells, development of iNKT cells occurs in the thymus. These cells are absent in nude mice (5, 46), fail to develop in thymectomized mice (47, 48) and are first detected in the thymus in the perinatal period (6, 7, 49, 50). Historically, there have been two schools of thought regarding the intrathymic development of iNKT cells. While some investigators believe that iNKT cells develop from a distinct precursor, others support the idea of a common T cell precursor pool. Accordingly, two models have been proposed to explain the lineage choice of iNKT cell ontogeny (51). The ‘committed precursor’ model suggests that iNKT cells develop very early during embryogenesis, even prior to the development of the thymus or appearance of the conventional T cells. In contrast, the ‘TCR instructive’ model proposes that it is only after the Vα14 to Jα18 rearrangement that iNKT cell lineage commitment and further differentiation occurs (52). The application of CD1d tetramers has made it possible to clarify some of these original controversies. Consistent with the instructional model, it is now clear that both conventional αβ T cells and iNKT cells proceed through the same early triple negative (CD3− CD4−CD8−) stage and have almost identical signaling requirements for the development of their respective precursors (53) (Fig. 1). Rather, it is during the double positive (CD4+CD8+) stage that Vα14 to Jα18 rearrangement and invariant TCR expression occurs, which dictates NKT lineage commitment (54-57). Analysis of individual T-cell hybridomas with Vα14-Jα18 rearrangements on one chromosome revealed the presence of other Vα to Jα rearrangements on the homologous chromosome, implying that selection of the invariant α-chain rather than a directed rearrangement may be responsible for the restricted TCR repertoire (52).
Fig.1. Schematic view of iNKT cell versus conventional αβ T-cell development.
NKT cell precursors diverge from conventional αβ T cells at the CD4+CD8+ double positive (DP) stage. The invariant TCR α chain (iTCR) on iNKT cell precursors recognizes glycolipid antigens presented by the CD1d molecule expressed at the surface of ‘selecting’ DP thymocytes. In contrast, CD4+ and CD8+ T cells express diverse TCRs that recognize peptide antigens presented by MHC class II or class I molecules, respectively, expressed by thymic cortical epithelial cells (TEC). Mature thymic emigrants of the conventional αβ T cell lineage have a naive phenotype, as opposed to iNKT cells, which acquire a memory/effector phenotype during thymic development. Immature NKT cells can acquire the NK1.1 marker after emigration to peripheral tissues or within the thymus. While a subset of NK1.1+ iNKT cells remain in the thymus as ‘long term residents’, some also migrate to the periphery.
Positive and negative selection of iNKT cells
Conventional T cells undergo positive or negative selection, which is primarily determined by the avidity of TCR-ligand interactions. When T cells have low-to-intermediate reactivity to self-peptide/MHC class I or class II, they are positively selected and differentiate into mature CD8+ or CD4+ single positive (SP) T cells. On the other hand, T cells with high reactivity for self-peptide-MHC complexes are eliminated via negative selection. The net result of these interactions allows for the generation of T cells with low recognition for self and elimination of potentially auto-reactive cells. Although less well understood than inαβ T cells, it is currently believed that NKT cells also undergo positive and negative selection events, which allow for the elimination of highly autoreactive cells while preserving a functional pool of cells.
Positive selection
NKT cells are absent in CD1d−/− mice and in bone marrow chimeric mice that lack CD1d expression on DP thymocytes (47, 58, 59). Conversely, they develop normally in pLck-hCD1d transgenic mice (on a CD1d−/− background) that express CD1d under the control of the proximal Lck promoter, where CD1d is exclusively expressed on cortical thymocytes (60). These experiments demonstrate the critical requirement for CD1d expression, particularly on DP cortical thymocytes, for the positive selection of NKT cells. Additionally, for efficient NKT cell selection, CD1d must recycle through specific intracellular pathways, where it can be loaded with endosome- or lysosome- derived antigens (30). The adapter protein AP-3 and prosaposin play important roles in the loading of glycolipids into the CD1d groove. AP-3 binds to the cytoplasmic tail of CD1d and directs the trafficking of CD1d to lysosomes, where endogenous glycolipids are processed and loaded onto CD1d (61). Prosaposin is a protein precursor to four saposins (A, B, C and D), small lysosomal glycoproteins that are required for the processing and hydrolysis of sphingolipids (62). Indeed, antigen-presenting cells lacking AP-3 or prosaposin fail to properly activate iNKT cells (63)s and AP-3- and prosaposin-deficient mice exhibit marked impairments in iNKT cell development (63).
There is growing consensus in the field that the ligand for positive selection of iNKT cells is most likely a self-antigen. Although α-GalCer is a strong NKT cell agonist, it is not expressed in mammalian cells and thus is not considered to be a potential selecting ligand. Other glycosphingolipids share structural similarities to α-GalCer, such as the endogenous lipid antigen isoglobotrihexosylceramide (iGb3). iGb3 is a relatively weak agonist compared to α-GalCer and was initially favored as a candidate selecting antigen (64-67). However, evidence regarding the role of iGb3 in NKT cell development has been conflicting. Data supporting its role in NKT cell selection comes from the study of β-hexosaminidase-B-deficient mice, which exhibit a dramatic reduction in thymic NKT cell numbers (62). β-hexosaminidase-B-deficient bone marrow–derived DCs can normally present several complex derivatives of α-GalCer that require lysosomal processing prior to NKT cell recognition, but they are not able to process and present iGb4 (the precursor to iGb3), which requires removal of the outer β-branched hexosamine, for NKT cell recognition (68). On the contrary, another study argued that it was not the lack of lysosomal iGb3 but rather a more general defect in glycosphingolipid processing that disrupted NKT cell development these animals (69). Furthermore, mice deficient in iGb3 synthase, an enzyme essential for iGb3 biosynthesis, exhibit normal NKT cell development (70). Thus is seems unlikely that iGb3 is responsible for NKT cell selection. Several other classes of molecules such as phospholipids, glycosphingolipids, gangliosides and galactosylceramides and their derivatives have been proposed as possible candidates for positive selection (30); however, due to lack of compelling evidence, their role in selection has remained questionable.
The TCR β chains of iNKT cells belong to a restricted set of Vβ families including Vβ8, Vβ7 and Vβ2 in mice and Vβ11 in humans (1). This biased Vβ usage could reflect the inability of the invariant TCR α chain to pair with other Vβs or instead it could be the consequence of iNKT selection (65). Expression of a rearranged Vα14-Jα18 transgene in CD1d-deficient mice leads to the emergence of T cells with a diverse Vβ repertoire (65). In these T cells, the invariant TCR α chain pairs indiscriminately with a broad Vβ repertoire, ruling out the possibility of pairing biases. Furthermore, only the cells expressing the Vβ7, Vβ8 and Vβ2 could be stimulated by a putative endogenous ligand (CD1d expressed on DCs) and iGb3, with similar Vβ7>Vβ8>Vβ2 responses as is seen in primary iNKT cells (65). As this affinity hierarchy of Vβ chains directly correlates with their respective degree of enrichment during thymic selection, it was proposed that primarily the process of positive selection tailors the restricted Vβ repertoire of iNKT cells. Interestingly, two recent studies provide evidence further corroborating the notion that the restricted TCR β repertoire is shaped by positive selection (71, 72).
Negative selection
For conventional T cells, most of the developing thymocytes expressing TCRs with high avidity for self-peptides undergo negative selection. This step, however, is not as clearly defined for iNKT cells. Addition of α-GalCer to fetal thymic organ cultures (FTOC) results in a dose-dependent reduction in NKT cell numbers (73, 74). Treatment of mice with α-GalCer via intraperitoneal injection from days 3 to 14 post-birth results in depletion of α-GalCer loaded CD1d tetramer + cells, with no significant effect on any other T lymphocyte populations (74). However, addition of this antigen to FTOC or in vivo administration at a later stage after the development of iNKT cells does not impact the absolute numbers of cells (74). Overexpression of CD1d on thymic stromal, dendritic cells or DP thymocytes in transgenic mice causes a variable decrease in the number of iNKT cells (60, 73). In these mice, there are fewer iNKT cells expressing the higher affinity Vβ8.2 TCR and more expressing Vβ2, which is known to have a lower affinity for α-GalCer in the context of CD1d (73). Collectively, these studies suggest that NKT cells are susceptible to negative selection during their development. However, it must be recognized that the above-mentioned studies each showed a failure of iNKT cell development, as opposed to the deletion of formed iNKT cells. Thus, further studies are required to better understand whether iNKT cells actually undergo negative selection, and if so, at what stage of development and via which molecular mechanisms (2).
Signals regulating the positive selection of iNKT cells
Consistent with the importance of proper expression, processing and antigen loading of CD1d for iNKT cell selection, the generation of the appropriate invariant CD1d-restricted TCR is critical for the development of iNKT cells at this stage. NKT cells are absent from mice that are deficient in the Rag-1 or Rag-2 genes and in mice that lack the Jα18 TCRα segment (75), and are profoundly diminished or absent in mice with deletion of molecules that function to transmit TCR-generated signals, such as the CD3ζ chain, Lck, ZAP-70, SLP-76, ITK, LAT and Vav, (44, 76-81). Formation of the invariant TCR requires the proper rearrangement of the α-chain gene segments to generate the characteristic Vα14-Jα18 combination. Although the process of V(D)J recombination is stochastic, the probability of selection of individual V and J segments is not equal and for more distal segments, such as Jα18, requires more time to allow for the generation of multiple DNA excision circles (82). Therefore, deletion of the genes that shorten the lifespan of DP thymocytes, such as BcL-xL, RORγt, HEB, and cMyb (55, 56, 83, 84) (see below), reduce the likelihood that progenitors will survive long enough to rearrange the invariant TCR α chain with the end result being a reduction in iNKT cell number.
iNKT cell development at the DP stage is highly dependent on signals generated by engagement of the SLAM family (SLAMf) of surface receptors, which are expressed on developing NKT cell precursors as well as cortical DP thymocytes (Fig. 2). The SLAM receptors include SLAM (CD150) itself, 2B4 (CD244), CD84, Ly9 (CD229), NK- T- and B-cell antigen (Ly108 in the mouse), and CD2-like receptor activating cytotoxic cells (CRACC, CD319). With the exception of 2B4, which interacts with CD48 as its ligand, these receptors all function in a homotypic manner and bind to like receptors with high affinity (85). SLAM receptor signaling is mediated SAP, which binds via its phosphotyrosine-binding pocket to one or more conserved tyrosine-based motifs present within the SLAM receptor cytoplasmic tails (86). Using a distinct region of its SH2 domain centered at arginine 78, SAP also binds to the Src family tyrosine kinase Fyn, leading to an increase in kinase activity and generation of a SLAM receptor-SAP-Fyn-mediated tyrosine phosphorylation signal (87, 88).
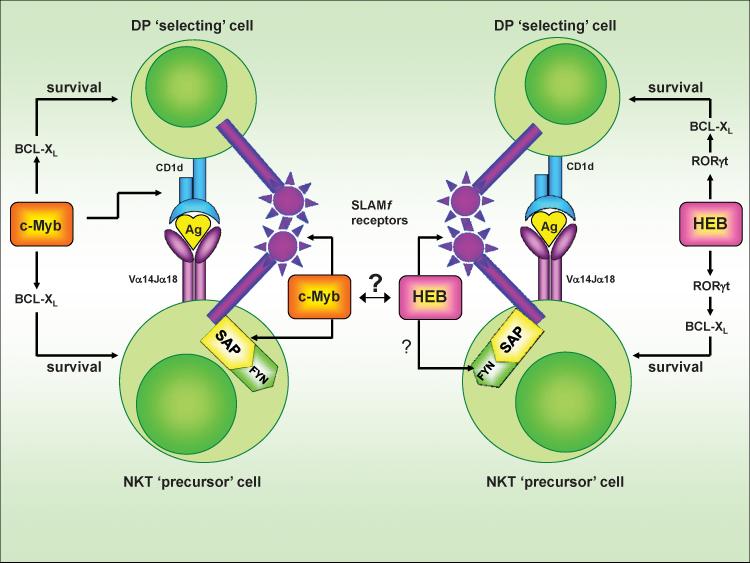
Fig. 2. The SLAM-SAP-Fyn signaling axis during iNKT cell development.
‘Selecting’ DP thymocytes and iNKT ‘precursor’ cells communicate via invariant TCR-CD1d and SLAM family (SLAM, Ly108) interactions. Engagement of the SLAM family receptors at the cell surface leads to recruitment of the adaptor molecule SAP and the Src family tyrosine kinase Fyn. The SLAMf receptor-SAP-Fyn signaling complex triggers activation of NFκB via protein kinase θ (PKCθ) and Bcl10. Additionally, SLAMf receptor-SAP-Fyn signaling also phosphorylates and recruits the SH2 domain–containing inositol phosphatase (SHIP), Dok1/2 adaptor proteins, and the Ras GTPase-activating protein (RasGAP). By binding to RasGAP, Dok1/2 inhibits Ras-MAPK activation. It is possible that SLAMf receptor-induced signals cooperate with those induced by the invariant TCR to modulate NFκB-Ras pathway activity in a manner required for iNKT cell development.
Evidence for the crucial role of SLAM-SAP signaling in iNKT cell development came when we and other groups showed that patients with the immunodeficiency X-linked lymphoproliferative disease (XLP), who harbor inactivating mutations in the gene encoding SAP, and Sap−/− mice, lack iNKT cells (89-91). Interestingly, similar results are seen in mice lacking Fyn or expressing a mutant version of SAP that cannot bind to Fyn (SapR78A) (77, 92). Recently, the block in iNKT cell development in Sap−/− mice was identified as occurring early in ontogeny, just at or after positive selection (93). Based on these findings, it was hypothesized that SAP might interact with one or more of the SLAM receptors in immature progenitors to promote NKT cell ontogeny at this stage. In support of this notion, combined interference with Ly108 and SLAM signaling profoundly limits NKT cell development (93). Together, these data suggest that SLAM and Ly108 function via SAP and Fyn to promote signals critical for commitment to the iNKT cell lineage. In line with this notion, the Slamf1 (encoding SLAM) and Slamf6 (encoding Ly108) genes were recently identified as candidate loci controlling iNKT cell numbers in NOD mice, which are known to exhibit quantitative and qualitative defects in this lineage (94). In this study, it was demonstrated that there is reduced expression of SLAM at the DP stage, which might interfere with iNKT cell production.
It is currently unclear as to how SLAMf receptor-SAP signals coordinate with those emanating from the TCR to foster proper iNKT cell development. In T cells, activation of the SLAMf receptor-SAP-Fyn pathway results in recruitment of the SH2 domain–containing inositol phosphatase (SHIP), the Dok1/2 adaptor proteins, and the Ras GTPase-activating protein (RasGAP) (95, 96). As Dok1/2 inhibits Ras-MAPK activation by binding to RasGAP (95, 96), it is possible that in the absence of SAP, there is a failure to negatively regulate Ras signaling, which might lead to impairment in iNKT cell development. Although it has been put forth that iNKT cell development is normal in transgenic mice expressing inhibitors of Ras signaling during thymocyte development (97), these studies were completed prior to the availability of α-GalCer-CD1d tetramers and it would be of great interest to re-examine these animals using this reagent. The SLAM-SAP–Fyn complex also links to NFκB via protein kinase θ (PKCθ) and the Bcl10 adaptor protein (98). SAP- or Fyn-deficient T cells fail to recruit PKCθ and Bcl10 to the immunological synapse, have impaired IκBα degradation and reduced NFκBp50 nuclear translocation in response to TCR stimulation, suggesting that this pathway may interact directly with TCR signaling cascades (98). Mice deficient in PKCθ or Bcl10 have severely reduced iNKT cell numbers in the thymus and spleen, respectively (99, 100). Additionally, mice expressing a dominant negative IκBα transgene and those lacking NFκBp50 contain severely diminished iNKT cell populations. Taken together, PKCθ, Bcl-10, and NFκBp50 appear to critically regulate the iNKT cell ontogeny, although the precise stage(s) at which these molecules affect iNKT cell development and/or homeostasis remains unclear (100, 101) (see below).
NKT cell maturation
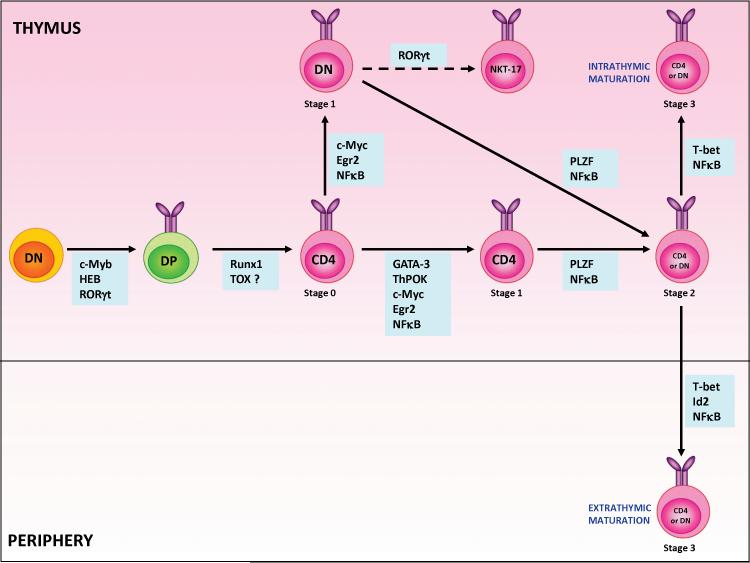
Following positive selection, NKT cells progress down a pathway of maturation that is marked by the sequential acquisition of specific patterns of surface marker expression. Currently, it is believed that maturation is comprised of four stages based on expression of CD24 (the heat stable antigen), CD44 and NK1.1 (Fig. 3). The most immature NKT cells are defined as CD24hiCD44loNK1.1− (Stage 0), followed by CD24loCD44loNK1.1− (Stage 1), CD24loCD44hiNK1.1− (Stage 2), and finally CD24loCD44hiNK1.1 + (Stage 3) (30). Stage 0 NKT cells are small in size and present in very low numbers (approximately 1 in every million thymocytes) (2), consistent with the observation that they do not undergo proliferation. On the contrary, cells in Stage 1 proliferate briskly, suggesting that the post-selection expansion of lineage committed iNKT cells occurs at this stage. Transition from Stage 0 to Stage 1 marks another unique phase of NKT cell maturation, where a proportion of cells down regulate CD4, giving rise to a subset of NKT cells that are DN. As cells progress to Stage 2, they upregulate CD44 and thus acquire a memory or activated phenotype. A majority of the CD24loCD44hiNK1.1− cells emigrate to peripheral tissues, where they stop proliferating and rapidly express NK1.1 and other NK lineage markers such as NKG2D, Ly49A, C/I, and G2 as well as markers of T cell activation such as CD69 and CD122 (102). Interestingly, a fraction of CD44hi and NK1.1− cells remain in the thymus as terminally mature long-lived resident cells (103, 104). Early after positive selection and during their maturation in the thymus, iNKT cells become functionally competent and can respond to TCR engagement, although the pattern of cytokines they produce varies depending upon the developmental stage. For example, Stage1 NKT cells typically produce IL-4 following TCR stimulation in vitro, where as Stage 2 NKT cells produce IL-4 and IFN-γ and mature NKT cells (Stage 3) produce more IFN-γ than IL-4 (7). Other molecules, such as cytolytic effectors (perforin, granzyme B and FAS ligand) and specific chemokines and their receptors are more prominent in the Stage 3 cells (102).
Fig. 3. Stages of NKT cell development.
NKT cell development occurs in the thymus from a common precursor pool of CD4+CD8+ DP thymocytes. Thymocytes that express the canonical invariant TCR are selected following interaction with CD1d+ SLAMf receptor-expressing DP thymocytes and commit to the NKT cell lineage. After positive selection, immature iNKT cells undergo lineage expansion and acquire a memory phenotype and markers of the NK cell lineage. The intermediate stages of the differentiation are characterized by differences in expression of CD24, CD44 and NK1.1. Although most immature NKT cells emigrate from the thymus at Stage 2 and acquire NK1.1 expression in the periphery, a subset of Stage 2 cells acquire this marker in the thymus and remains as long-term residents.
Transcriptional regulation of lineage choice
In order for iNKT cells to develop and function properly, signals emanating from TCR-CD1d and SLAM receptor interactions must culminate in the coordinated regulation of genes that allow for the specification and maturation of this lineage. Several studies, including some quite recent, have provided valuable insights into the transcriptional mechanisms regulating iNKT cell ontogeny. Some of these studies are discussed here with a focus on how specific transcription factors regulate aspects of iNKT cell development, such as selection, expansion, maturation and acquisition of effector function (Fig. 4).
Fig. 4. Transcriptional regulation of different stages of iNKT cell development.
The developmental program of iNKT cells is controlled by the activity of several transcriptional factors, some of which regulate multiple steps of this process. The definitive stages at which some of these factors are expressed during iNKT cell development remains unclear. Based on recent data, we have placed specific factors along the iNKT cell developmental program at the earliest stage where the activity of a given transcription factor appears critical for iNKT cell development. It is likely that these factors could also be regulating the later stages of ontogeny or differentiation. As individual members of the NFκB family of transcription factors impact different stages, they are placed at multiple points in the developmental pathway.
PLZF, a regulator of iNKT development and function
Studies from our laboratory and others have identified that the broad complex tramtrack bric-a-brac-zinc finger (BTB-ZF) transcription factor PLZF is a key regulator of iNKT cell development and function (105, 106). This protein is member of a family of transcription factors that regulate a variety of immunological processes, such as the germinal center reaction (Bcl6), CD4 versus CD8 lineage commitment and T versus B-cell fate decisions (107-110). In the thymus, PLZF expression is largely restricted to iNKT cells, where it is highest in the immature stage 0/1 population (105, 106). Plzf−/− mice or animals harboring a natural mutation in Zbtb16, the gene encoding PLZF, exhibit a ~90% reduction in iNKT cell number in the thymus (105, 106), but show no defects in the development of conventional αβ T cells. In the thymus, PLZF-deficient cells do not acquire the characteristic iNKT cell phenotype. In the periphery, PLZF-deficient iNKT cells fail to acquire the ability to co-secrete Th-1 and Th-2 cytokines upon primary activation. However, upon secondary activation PLZF-deficient iNKT cells can produce IL-4 and IFN-γ (105, 106).
It is currently not understood how the loss of PLZF impairs iNKT cell development or effector functions. PLZF-deficient iNKT progenitors express the invariant TCR and do not appear to exhibit an increase in apoptosis or a proliferative defect, at least as demonstrated by incorporation of BRDU by specific precursor populations (105). Nonetheless, PLZF is expressed in the most immature CD24hiCD44− iNKT cells, indicating that it might function in ontogeny early after lineage commitment (105). Given the unique requirement for PLZF during iNKT cell development, we considered that it might play a role downstream of the SLAMf receptor-SAP-Fyn signaling axis to direct the development of iNKT cells. However, the remaining rare α-GalCer-CD1d tetramer positive cells in the thymus of SAP- or Fyn-deficient mice express normal levels of PLZF (105, 106). Transgenic expression of PLZF in T cells results in the acquisition of memory like qualities in peripheral CD4+ and CD8+ T cells, such as increased CD44 and diminished CD62L expression, but no acquisition of NK or NKT cell – associated markers [NKG2D, NK1.1, DX5, 2B4 (106, 111, 112)]. Transgenic T cells exhibit altered patterns of cytokine production and enhanced proliferation compared to non-transgenic cells (111) that are independent of SAP- and Fyn-mediated signals (111). Taken together, these studies demonstrate that PLZF is selectively required for the development of iNKT but not conventional αβ T cells, where it also promotes the acquisition of an effector/memory phenotype and functions.
We and others (45, 113) also recently demonstrated that PLZF is required for the functional maturation of an iNKT-like population of γδ T cells. γδ T cells comprise a heterogeneous lineage of T cells that emerge in the thymus directly from DN precursors. These cells develop in waves during fetal and neonatal life, and upon exiting from the thymus, migrate to specific tissues, such as the skin, musocal surfaces and secondary lymphoid organs. Following activation, γδ T cells primarily produce IFNγ; however, they also serve as a source of IL-4 and IL-17 (114-119). Like iNKT cells, a unique subset of γδ T cells expressing the Vγ1.1+Vδ6.3+ TCR can produce both IL-4 and IFNγ following antigenic stimulation (119, 120). Based on their iNKT-like properties, we investigated whether PLZF might be expressed in and/or required for the development or function of these Vγ1.1+Vδ6.3+ T cells. We observed that some Vγ1.1+Vδ6.3+ T cells expressed high levels of PLZF (45), particularly those cells capable of producing IL-4 or both IL-4 and IFNγ (45). Loss of PLZF expression did not significantly reduce the percentage of Vγ1.1+Vδ6.3+ T cells (45, 113); however, it impaired the ability of these cells to produce multiple cytokines and chemokines, including IL-4 and IFNγ (45, 113). Curiously, SAP- but not Fyn-deficient mice, exhibit a reduction in Vγ1.1+Vδ6.3+ T cells, and in those cells that are present, there is reduced PLZF expression (45). Thus, while initially thought to be a unique regulator of iNKT cell function, PLZF is now believed to be pivotal for the acquisition of the effector/memory phenotype characteristic of both iNKT and iNKT-like Vγ1.1+Vδ6.3+ T cells (45, 105, 106, 113, 121). Furthermore, the selective requirement for SAP, but not Fyn, for the development of this PLZF-expressing γδ T cell population suggests that SAP uses distinct signaling mechanisms to promote development of Vγ1.1+Vδ6.3+ T cells versus iNKT cells.
Transcription factors that exclusively regulate iNKT cell lineage development
While mutations in many of the transcription factors that regulate conventional T cell development also affect the iNKT cell lineage; there are some transcription factors that appear to exclusively regulate iNKT cell ontogeny. Thus, alterations that interfere with the expression of these genes, such as naturally occurring mutations, targeted germline gene mutations or mutations that allow for temporal or conditional deletion within the T cell lineage selectively impair iNKT cell ontogeny with little or no effect on the intrathymic development of αβ T cells.
The NFκB family of transcription factors
The NFκB family of transcription factors consists of NFκB1 (p50), NFκB2 (p52), RelA (p65), RelB, and c-Rel (122). In quiescent cells, NFκB proteins are sequestered in the cytoplasm by inhibitory IκB⟨ molecules. In response to numerous extracellular stimuli, activation of the IκB kinase (IKK) results in phosphorylation and ubiquitin-dependent degradation of IκB⟨ and release of NFκB proteins. NFκB homo- and heterodimeric complexes subsequently translocate into the nucleus, where they regulate expression of specific target genes that are important for cellular development and function (122-124). An alternative pathway involves NFκB inducing kinase (NIK)-mediated phosphorylation of the inhibitory p100 precursor, its processing to p52 and the nuclear translocation of p52-RelB heterodimers (125). Manipulations such as targeted deletion of individual NFκB members or interference with classical NFκB pathway activity only modestly impair conventional T cell development (126-129). On the contrary, distinct members of the NFκB family are required in both hematopoietic and non-hematopoietic cells to promote the ontogeny of iNKT cells at multiple stages (101, 130-132) (Fig. 4). Mice expressing a T-cell-specific dominant-negative IκBα transgene (101, 131) or lacking expression of p50/ NFκB1 or p52/ NFκB2 (101) exhibit a dramatic reduction in iNKT cells. Furthermore, activation of NFκB via the classical pathway is required in a cell-intrinsic manner to allow transition of NK1.1− precursors to mature NK1.1+ NKT cells (101, 132) and to promote the production of cytokines in response to α-GalCer (100). Although expression of the pro-survival molecule Bcl-XL rescues NKT cell number in animals expressing the dominant-negative IκB transgene, it fails to restore the defect in activation-induced cytokine production (131). Therefore, NFκB signaling to downstream targets other than Bcl-XL is important for NKT cell function (131). Defects in the alternative NFκB pathway also inhibit NKT cell development, as mice with a deficiency in RelB expression in thymic stroma have severely reduced NK1.1− and NK1.1+ NKT cell numbers (130). Additionally, RelA but not c-Rel, regulates lineage expansion of immature NKT cells and their terminal maturation by inducing expression of IL-15 and IL-7 (132). Finally, NFκB regulates homeostasis of mature NKT cells by regulating TCR-induced lymphotoxin α and β expression in a cell-intrinsic manner (132).
In a recent study, it was demonstrated that mice lacking expression of CYLD, a deubiquitinating enzyme that acts as a negative regulator of NFκB, exhibit massive apoptosis of iNKT cells, which leads to a severe reduction in the total number of NKT cells in the thymus and periphery (133). iNKT cells lacking CYLD exhibit reduced IL-7Rα expression. As the result, cells do not properly phosphorylate STAT5 or upregulate ICOS in response to IL-7 treatment. The diminished IL-7 signaling and ICOS expression likely contribute to the reduced iNKT cell numbers in these mice by impairing survival and/or other homeostatic mechanisms (134). Remarkably, inhibition of NFκB signaling via expression of a dominant-negative IκB⟨ restores expression of IL-7Rα and ICOS (133). Collectively, these studies suggest that derangements that reduce or enhance NFκB activity impair the development of iNKT cells. Thus, the function of this transcription factor family must be tightly regulated to allow for proper iNKT cell ontogeny.
Early growth response 2 (Egr2)
Engagement of the TCR induces numerous signaling events, including release of calcium from intracellular stores and activation of the phosphatase calcineurin, which results in the dephosphorylation and subsequent nuclear translocation of NFAT family member and the subsequent regulation of NFAT target genes (135). Although many studies have documented the importance of the calcineurin-NFAT pathway in conventional T cell development, the first evidence for the role of this pathway in iNKT cell development comes from a recent study in which targeted deletion of Cnb1, the gene encoding the regulatory component of calcineurin, resulted in a dramatic reduction in thymic and peripheral iNKT cell numbers (136). To understand the mechanisms by which calcineurin-NFAT signaling might influence iNKT cell development, the authors examined the involvement of the Egr family of transcription factors (specifically Egr1, Egr2 and Egr3), known targets of NFAT, using mice deficient for the expression of these proteins in all tissues (Egr1−/−, Egr3−/−) or selectively within hematopoietic cells (Egr2−/− fetal liver chimeras). Surprisingly, absence of Egr2, but not Egr1 or Egr3, leads to significant reductions in iNKT cells, but does not affect development of CD4+ or CD8+ T cells. Egr2 does not appear to play a role in lineage commitment or positive selection of iNKT cells, as normal percentages and numbers of CD24+ cells (Stage 0) were present in Egr2-deficient fetal liver chimeras. However, fewer cells downregulated CD24 and more retained a CD44−NK1.1− phenotype, implicating a partial block at the transition from Stage 0 to Stage 1 of ontogeny (Fig. 4). A higher proportion of Egr2-deficient iNKT cell progenitors appear to be proliferating, yet at the same time, they also exhibit an in increase in apoptosis. As Egr2-deficient thymocytes express normal levels of pro-survival cytokines, including IL-2, IL-15 and IL-7, and anti-apoptotic proteins, such as Bcl-2 and Bcl-XL (136), it is unclear why the loss of Egr2 leads to the increased death of immature iNKT cells just after positive selection.
Transcription factors that regulate iNKT cell ontogeny at the DP stage
Since conventional αβ T cells and iNKT cells originate from a pool of common precursors, it is not unexpected that transition through the DN to DP stage of thymocyte development is controlled by transcription factors that regulate events common to both lineages, including rearrangement of the TCR α- and β-chains and proliferation. However, targeted deletion of specific transcription factors at the DP stage also profoundly impairs iNKT cell lineage commitment. Indeed, seminal studies in this area demonstrated a critical role for the RORγt transcription factor, which functions to prolong the survival of DP thymocytes by upregulating expression of Bcl-XL, thereby allowing sufficient time for rearrangement of the more distal Vα-Jα gene segments typical of the invariant TCR (55, 56). The Runt related transcription factor 1 (Runx1) is also a crucial regulator of iNKT cell development at DP stage and in its absence iNKT cell development is blocked at the earliest detectable iNKT committed subset (56). In contrast to RORγt, Vα14-Jα18 rearrangements can be detected in DP cells lacking Runx1 (56), indicating that its activity is required for different aspects of iNKT development, perhaps during positive selection or for expansion of post-selection precursors. Here, we highlight several recent reports that provide new insights into the roles of transcription factors that play critical, yet selective, roles in iNKT development at the DP stage.
AP-1 transcription factors
The contribution of AP-1 transcriptional complexes to T-cell development and activation has been an area of intense investigation for many years. AP-1 complexes function as dimers composed of Jun (c-Jun, JunD, Jun B) and Fos proteins (c-Fos, FosB, Fra-1 and Fra-2) as well as members of the ATF family of basic leucine zipper proteins. AP-1 activity is integral to many cellular processes, including proliferation and apoptosis, and within mature T cells, AP-1 activity lies downstream of the TCR to regulate the expression of many genes, including those encoding IL-2 and IL-4. In developing thymocytes, AP-1 demonstrates DNA binding and transcriptional activity within DN and SP T cells (137-139). Interestingly, AP-1 activity is not detected in DP thymocytes, suggesting that tight regulation of its function and the subsequent expression of AP-1-specific target genes, is critical at this stage of thymocyte development (137). In earlier studies, a transgene encompassing the Lck proximal promoter was used to drive expression of B-cell activating transcription factor (BATF), a negative regulator of AP-1 activity, early during thymocyte development. BATF-transgenic mice exhibit normal thymic cellularity and subset distribution; however, they demonstrate a reduced number of both CD4+ and DN iNKT cells within the thymus and peripheral organs (140, 141). Analysis of developmental markers reveal that fewer transgenic iNKT cells upregulate NK1.1, indicating a delay in entering the latest stage of iNKT cell maturation (141). The authors also demonstrate that transcripts encoding proteins belonging to the Fos and Jun families are present in iNKT cells and display a characteristic sequence-specific DNA binding associated with AP-1 transcriptional activity within hours of stimulation of the invariant TCR (141).
A recent report examined mice in which the Fos family protein Fra-2 was deleted at the DP stage. Contrary to forced expression of BATF, loss of Fra-2 leads to a selective expansion of thymic and splenic iNKT cells (142). Furthermore, each of the developmental subsets (as defined by CD44 and NK1.1 expression) was over-represented in Fra-2-deficient iNKT cells. Based on this observation, the authors argue that Fra-2 regulates iNKT cell development during or immediately after lineage selection. NKT cells that develop in the absence of Fra-2 have increased use of the lower affinity Vβ8.2 receptor, suggesting that Fra-2 deficient NKT cells are more likely to undergo positive selection. In the absence of Fra-2, iNKT cells produce unusually high amounts of IL-2 and IL-4, and exhibit increased proliferation. These data indicate that Fra-2 expression, and hence AP-1 directed gene expression, is also necessary for proper regulation of iNKT cell functions (142). This notion is consistent with the observation that loss of Fra-2 led to a 10-fold up-regulation of Jun (an activating member of the AP-1 family), and downregulation of BATF (142). Based on these studies, one can conclude that the coordinated regulation of AP-1 transcriptional activity is important for iNKT cell development and function, with inhibition of this pathway perturbing iNKT cell development, and loss of Fra-2 having the opposite effect. It is interesting to note that conventional T cells that lack Fra2 expression also produce large amounts of cytokines immediately following activation (142). This latter observation suggests that Fra-2 may also be important for the development or function of memory or other innate-like T cell populations. Indeed, previous studies have shown a role for Jun in αβ/γδ T-cell development (143).
Thymocyte selection-associated HMG box (TOX) protein
The thymocyte selection-associated HMG box protein (TOX) is induced in DP thymocytes as the result of TCR-calcineurin-dependent signaling as these cells transition to SP cells (144, 145). Appreciation for the requirement for TOX in iNKT cell development was obtained following the examination of Tox−/− mice, which exhibit a striking reduction in thymic iNKT cells (146). Although the specific stage of NKT cell development affected by TOX-deficiency is not described, the near to complete absence of CD1d-tetramer+ cells suggests a developmental arrest at an early stage (Fig. 4). The dependence on TOX is not unique to the iNKT cell lineage as CD4+ T cells and Foxp3+ regulatory T cells are also absent; however, emergence of CD8+ T cells is largely unaffected (146). The zinc finger transcription factor GATA-3 is also required for the development of conventional CD4+ T cells (147); however, TOX deficiency does not impair GATA-3 expression in DP cells (146). Interestingly, expression of the transcription factor ThPOK (c-krox), which acts downstream of GATA-3 (148) during commitment to the CD4+ lineage (147, 149), is completely absent. Thus, it remains possible that TOX might exert its effects in CD4+ T cell lineage commitment via effects on ThPOK expression. Since absence of ThPOK leads to less severe defects in iNKT cell development than loss of TOX (see below), TOX likely regulates the expression of other target genes critical for iNKT cell development.
Myelocytomatosis oncogene (c-Myc)
The basic-helix-loop-helix zip (bHLHZip) transcription factor c-Myc plays important roles in regulating cell proliferation, survival and apoptosis (150). Within the T-cell lineage, c-Myc is expressed at high levels in immature DN thymocytes and its expression decreases as cells differentiate into DP cells (151). Conditional deletion of c-Myc early during T-cell development leads to a severe block in T cell development as the result of profound defects in pre-TCR-induced proliferation (152). Recently, two groups used the system of CD4-Cre-mediated gene deletion to investigate the requirement for this transcription factor during iNKT cell development (153, 154). In both of these reports, c-Myc deficiency in DP thymocytes selectively impairs NKT cell ontogeny without influencing the development of conventional or natural regulatory T cells. In the absence of c-Myc, iNKT cells appear to be arrested at the CD24loCD44− stage and this defect was cell-intrinsic (Figure 4). Although the requirement for c-Myc in iNKT cell development is consistent in both studies, the reports differed in some aspects. According to Mycko et al. (154), in the absence of c-Myc, immature iNKT cells (CD24loCD44−) fail undergo lineage expansion, where as Dose et al. (153) demonstrated that c-Myc is critical for a later stage of intrathymic proliferation that precedes memory acquisition. Nonetheless, both studies show that the iNKT cell developmental block is not due to impaired survival, as transgenic expression of Bcl-2 does not rescue the defect. Although the reason for the discrepancy between these reports is not clear, they both demonstrate that c-Myc is indispensible for acquisition of a mature CD44+NK1.1+ phenotype (154). c-Myc deficient iNKT cells have normal levels of PLZF (153, 154), indicating that the defect in maturation is not due to a failure to properly express this factor.
The HEB transcription factor
The E protein family of basic helix-loop-helix transcription factors is comprised of the members HEB, E12, E47 (E2A) and E2-2, which bind as homo- or heterodimers to DNA at E-box sites and control the expression of genes essential for lineage development. Within developing T cells, the activity of both E2A and HEB promote TCR gene rearrangement and control thymocyte survival, proliferation and positive selection (155-158). Until a recent report by D’Cruz et al. (83), it was not known whether specific E proteins also played a role in iNKT cell development. In this report, deletion of HEB in DP thymocytes results in a major block in iNKT cell ontogeny at the CD24hi CD44−NK1.1− stage but has minimal effects on the development of CD4+ or CD8+ T cells. In contrast, deletion of E2A has no discernable effect on iNKT cell development. Interestingly, loss of expression of both HEB and E2A impairs T cell development, but does not further exacerbate the iNKT cell defect beyond that occurring in the absence of HEB. Thus, there is a unique requirement for HEB in the development of iNKT cells. Loss of HEB results in diminished levels of RORγt and Bcl-XL mRNA, poor survival of DP thymocytes and impairments in generation of distal Jα gene rearrangements including Vα14 Jα18. Directed expression of Bcl-XL or a rearranged V⟨14 TCR results in the emergence of phenotypically and functionally mature iNKT cells, providing further evidence that HEB influences iNKT cell ontogeny by driving the expression genes regulate thymocyte survival, thereby allowing for distal TCRα rearrangements (83)(Figs 4, 5).
Fig. 5. Relationship between the SLAMf receptor-SAP-Fyn signaling axis and the transcriptional activity of c-Myb and HEB.
c-Myb and HEB regulate the earliest steps in NKT cell development, just prior to and perhaps following the point of positive selection. c-Myb and HEB both facilitate the prolonged survival of DP thymocytes to allow for distal Va14-Ja18 TCR rearrangements. RORγt is a direct downstream target of HEB, but not of c-Myb, and CD1d appears to be a target of c-Myb, but not HEB. c-Myb and HEB function at a similar point in iNKT cell ontogeny as the SLAMf receptor-SAP-Fyn signaling complex. Remarkably, c-Myb regulates the expression of SLAM, Ly108 and SAP while HEB might modulate the expression of Fyn and SLAM. Thus, these transcription factors likely promote iNKT cell development by inducing the expression of proteins involved in the SLAMf receptor-SAP-Fyn signaling complex. It is unknown if c-Myb and HEB are expressed simultaneously or if they regulate one another’s activity at this point in iNKT cell development.
The proto-oncogene c-Myb
The Myb proto-oncogene encodes a nuclear DNA binding protein that is expressed primarily in hematopoietic tissues and acts as an activator and repressor of transcription. Although deletion of the Myb gene in the germline is embryonic lethal, examination of chimeric mice generated from Rag blastocyts reveals that cells lacking c-Myb have a profound developmental defect at the DN stage (159). Abrogation of c-Myb expression at distinct stages of T-cell development reveals additional roles for c-Myb activity in DP survival and differentiation into CD4+ SP cells (160). While the role of c-Myb in conventional T cells has been well established, it only recently became clear that this molecule is also critical for iNKT cell ontogeny. Similar to HEB, deletion of c-Myb at the DP stage leads to a marked cell-intrinsic reduction in the generation of iNKT cells that is associated with impaired TCRα gene rearrangements (84) (Fig. 4). Although Bcl-XL expression is reduced in c-Myb-deficient cells and retrovirus-mediated expression of this molecule allows for restoration of normal secondary TCRα gene rearrangements, iNKT cell development remains impaired. Consistent with the notion that other factors might be contributing to the iNKT cell developmental defect, c-Myb-deficient DP cells also exhibit defects in the expression of SLAM, Ly108 and SAP and fail to ‘provide help’ to CD1d-deficient cells during iNKT cell development in mixed bone marrow chimeric mice (84). Taken together, the results of this study suggest that c-Myb functions to regulate a distinct set of downstream targets that function at in critical and non-overlapping manners to regulate iNKT cell selection at the DP stage of development.
Transcription factors that regulate development of the CD4+ iNKT cell lineage
Depending upon their surface expression of the CD4 co-receptor, iNKT cells can be sub-divided into CD4+ and DN subsets, which exhibit unique functional characteristics. The exact mechanisms by which a subset of the CD4+ iNKT cells downregulate their CD4 expression is elusive. However, recent studies have indentified at least two transcription factors important during development of this iNKT cell subset.
GATA-3
GATA-3 is a C2C2 type zinc finger transcription factor whose expression within the immune system is restricted to NK, T and iNKT cells (161). Within the T-cell lineage, GATA-3 functions at multiple levels, including differentiation of common lymphoid progenitors into early DN cells, progression through β-selection and during the development of CD4+ T cells (162, 163). GATA-3 is also essential for the differentiation of CD4+ T cells into Th2 cytokine producing cells (162, 163). Loss of GATA-3 at the DP stage leads to a cell-intrinsic impairment in the generation of CD4+ iNKT cells (161). This defect is apparent in mice as young as 2 weeks of age and involves both NK1.1− and NK1.1+ precursors. Curiously, in the absence of GATA-3, extra-thymic NKT cells also undergo increased apoptosis, resulting in reduced numbers of splenic and more notably liver iNKT cells (161). In the periphery, iNKT cells fail to express CD69 and do not produce Th1 or Th2-type cytokines in response to a-GalCer (161). CD69 upregulation and IFN-γ secretion are restored when proximal TCR signals are bypassed using PMA and ionomycin; however, this treatment fails to restore IL-4 and IL-13 production. The data from this paper highlight several important properties regarding GATA-3 in iNKT cells. First, GATA-3 is critical for generation of the CD4+ iNKT cell lineage and production of Th2-type cytokines. This latter observation is consistent with the role of GATA-3 as a direct transactivator of Th2 cytokine genes. Second, GATA-3 is important for iNKT cell survival, particularly in the liver. It remains to be determined whether this defect is the result of altered regulation of genes involved in controlling heptatic iNKT cell apoptosis, such as CXCR6 (164). Third, given the observation that PMA and ionomycin restore TCR-induced upregulation of CD69 and IFN-γ secretion, once can also conclude that GATA-3 likely functions downstream of the TCR to regulate iNKT cell activation.
ThPOK
Recently it was reported that ThPOK, a member of the BTB-POZ family of zinc finger transcription factors, is important for the generation of CD4+ iNKT cells. Specifically, helper deficient mice homozygous for a spontaneous mutation in Zbtb7b, the gene encoding ThPOK (108, 165) have normal numbers and percentages of thymic and peripheral iNKT cells, yet mature iNKT cells completely lack CD4 expression (14). Interestingly, a proportion of ThPOK-mutant iNKT cells express CD8, revealing that loss of this transcription factor either impairs repression of CD8+ after selection or redirects iNKT cells to re-acquire CD8+ expression at a later point in development. ThPOK-deficient iNKT cells exhibit defects in TCR-induced cytokine mRNA expression and secretion, including production of IL-4 and to a lesser extent IFNγ. It has been suggested that GATA-3 may function upstream of ThPOK in CD4+ T cell lineage commitment (148). Therefore, it is possible that GATA-3 and ThPOK might function in a similar linear pathway in iNKT cells to promote development of the CD4+ subset as well as acquisition of specific effector functions.
RORγt: a transcription factor associated with IL-17 producing NK1.1− iNKT cells
Although it is well established that RORγt facilitates iNKT cell development at the DP stage (49, 55), recent studies highlight a novel role for this transcription factor in the differentiation of a unique subpopulation of iNKT cells capable of producing large quantities of the pro-inflammatory cytokine IL-17 in response to TCR stimulation (166). These cells, known as iNKT-17 cells, exhibit a NK1.1− CD4− phenotype and are present in the thymus as well as peripheral organs, including the liver, spleen, lymph node and lungs (166-168). Thymic and peripheral iNKT-17 cells express RORγt, similar to what is observed for conventional CD4+ Th-17 cells (167). However, unlike CD4+ Th-17 cells, iNKT cells do not require IL-6 and TGF-β or IL-23 for the induction or maintenance of IL-17 production (168). It is currently not clear whether IL-17 producing NK1.1− iNKT cells represent a terminally differentiated population or if they retain the capacity to further differentiate. In one report, tracking studies reveal that iNKT cell emigrants are negligible in the lymph node, a site of robust iNKT cell-associated IL-17 production (167). While these data suggest that iNKT-17 cells may not necessarily represent the immediate progeny of cells directly out of the thymus, this same report demonstrates that CD4− NK1.1− cells can give rise to NK1.1+CD4− cells in FTOC (167). Based on these findings, the authors suggest that CD4− NK1.1− cells branch off of the iNKT cell developmental pathway prior to NK1.1 upregulation, perhaps at a time when CD44 expression goes from low to high (Figure 4). These results differ from a second report, where it was observed that neither CD4+ nor CD4− NK1.1− iNKT cells could generate NK1.1+ cells in FTOC (166). In this latter study, cultured cells include specifically the CD44+ NK1.1− subset (as opposed to total NK1.1− cells), which might represent a more terminally differentiated population incapable of NK1.1 upregulation. Nonetheless, both groups demonstrate that presence of RORγt is tightly linked to the expression of IL-17 by the DN iNKT cell population.
Transcription factors that regulate terminal iNKT cell maturation or homeostasis
The terminal maturation of iNKT cells involves the upregulation of specific markers of the NK cell lineage, such as NK1.1, NKG2D and members of the Ly-49 family (2, 4), as well as acquisition of innate-like effector functions. iNKT cells must receive and respond to cues that favor their survival and accumulation at specific sites. The mechanisms regulating iNKT cell homeostasis remain poorly understood. There is some evidence that homeostasis is highly dependent on the cytokine IL-15 (169, 170), and to a lesser extent on IL-7 (169, 171). Other factors that regulate iNKT cell homeostasis include the chemokine receptor CXCR6 and the integrin LFA-1; however, these latter factors exclusively regulate homeostasis of hepatic but not splenic or thymic iNKT cells (164, 172-174).
The T-box transcription factor T-bet
T-bet is a transcription factor originally shown to mediate Th1 lineage specification that also plays an indispensable role in the final maturation stages of iNKT cells (102, 175). In the absence of T-bet, iNKT cell numbers are reduced and development is blocked at CD44hi NK1.1− (Stage 2) (Fig. 4). Thymic T-bet−/− iNKT cells do not express CD122, a component of the IL-15 receptor. As a consequence, iNKT cells lacking T-bet fail to proliferate in response to IL-15 (175). In addition, T-bet-deficient cells do not acquire characteristic iNKT cell effector functions, such as the ability to produce IFN-γ in response to TCR stimulation or to exhibit cytolytic activity (102, 175, 176). Consistent with these developmental and functional data, T-bet expression increases from Stage 1 to 3 of iNKT cell maturation and T-bet directly regulates the activation of genes associated with mature iNKT cell functions, such a perforin, FasL and IFNγ, among others (102). T-bet deficiency results in a similar developmental block in NK cells (175) indicating that T-bet operates to control the maturation of both these immune cell lineages.
Inhibitor of DNA-binding proteins (Id)
The inhibitor of DNA binding (Id) factors dimerize with E proteins and interfere with their DNA binding and transcriptional activity (177). Id2, a member of the Id protein family, is crucial for development of the immune system, as is evident by the analysis of Id2-deficient mice, which lack peripheral lymph nodes, Peyer’s patches and mature NK cells (178, 179). Examination of these mice has also revealed that Id2 is important for the development of CD8α+ dendritic cells, intraepithelial T lymphocytes and Langerhans cells and for promoting optimal effector and memory CD8+ T-cell responses (178, 180). Recent studies demonstrate that iNKT cell survival and homeostasis are also under the regulation of Id2 (178, 180-182). Specifically, in the absence of Id2, iNKT cell numbers are drastically and selectively reduced in the liver, yet normal in the thymus and spleen. While there is a perturbation in the percentages of Stage 2 and Stage 3 cells in the liver (with increased Stage 2 and decreased Stage 3), the absolute number of both of these iNKT cell subsets is significantly reduced. Examinations into the mechanisms underlying the reduced number of hepatic cells reveal no defects in homing to the liver; rather, Id2-deficient cells demonstrate greater apoptosis. Additionally, Id2 deficient cells have a lower expression of CXCR6 (164, 172), which as noted above, is implicated in the survival of liver iNKT cells (181). As the iNKT cell phenotype is more severe in cells lacking Id2 compared to CXCR6, the authors sought additional mechanisms to explain the increase in cell death. Indeed, hepatic Id2-deficient iNKT cells also demonstrate reduced expression of Bcl-2 and Bcl-XL and genetic ablation of the pro-apoptotic molecule Bim in Id2-deficient cells rescues iNKT cell numbers. As these molecules are known targets of E proteins, it is likely that Id2 modulates iNKT cell homeostasis in the liver by regulating E protein activity to maintain a favorable balance between pro- and anti-apoptotic signals. Furthermore, since the HEB-deficient phenotype manifests at a much earlier stage of iNKT cell development than loss of Id2 (Fig. 4), it does not seem probable that these 2 proteins antagonize each other’s activity in iNKT cells, at least in the earlier stages of development.
Conclusions
Although it has been more than 20 years since the initial descriptions of an unusual population of T cells that also express the NK1.1 marker, it has not been until recently that we have come to appreciate the complexity and importance of what are now known as iNKT cells for the generation of normal as well as pathological immune responses. Indeed, the past 5 years have seen an explosion in our understanding of the molecular mechanisms that regulate the development of iNKT cells from a pool of precursors common to conventional αβ T cells. Thanks to the development and use of α-GalCer-loaded CD1d tetramers, one can now identify iNKT cells early in ontogeny and track progression through the subsequent stages of development. As a result, it is possible to examine how manipulations that affect the expression of genes, such as germline or conditional interruption of specific transcription factors, influence iNKT cell lineage commitment and differentiation. Collectively, these studies have identified transcription factors selectively required for the development of iNKT cells, but not conventional T or NK cells, as well as factors that share developmental requirements with other lineages. It is also now apparent that the activity of some transcription factors is critical at a unique stage in the developmental program of iNKT cells, while others function at multiple points in development. Since iNKT cells acquire a memory like phenotype early in development, it is well appreciated that transcription factors associated with the activation of other lineages are essential to the differentiation of iNKT cells.
Despite the progress made, many questions remain regarding the molecular mechanisms controlling iNKT cell development and maturation. For example, how do signals generated by the TCR, SLAM family and possibly other surface receptors, cooperate to activate or repress specific transcription factors that promote commitment to the iNKT cell lineage? Although the SLAM-SAP signaling axis is linked to the NFκB pathway in conventional T cells, we find that forced expression of the NFκB target gene, Bcl-XL, or the inhibitory NFκB kinase (IKKβ), a catalytic subunit of the IκB kinase complex essential for NFκB activation, fails to restore NKT cell development in Sap−/− mice (183). Thus, it remains possible that the SLAM-SAP complex functions via other mechanisms, such as the Ras-MAPK pathway, to promote the induction of specific transcription factors crucial for iNKT cell development. In DP cells, does SLAM-SAP signaling induce HEB or c-Myb activity, or do these transcription factors preferentially function upstream of the SLAM-SAP signaling axis, as is suggested by the reduced expression of Slam, Ly108, SAP and Fyn in cells lacking c-Myb (84) or possibly HEB (83) (Fig. 5). What is the functional relationship between these crucial early transcription factors and what dictates the activity of these and other transcriptional regulators that function later in iNKT cell development or in the homeostasis of iNKT cells? What are the critical target genes regulated by these transcription factors? Given the pace of the last 5 years, it is likely that future studies will provide answers to these questions and lead to a greater understanding of the transcriptional networks regulating iNKT cell development. These studies are also likely to define whether these networks are distinct from, or overlap with, the networks controlling the ontogeny of NK, iNKT-like or conventional αβ T cells.
An expanding number of iNKT cell subsets are being identified in humans; however, little is known regarding their precise developmental requirements and mechanisms of regulation. This information is of significance due to the involvement of, and counter-regulatory roles for, unique iNKT cell subsets in normal immunity and the etiology of certain diseases. So far, most of our knowledge about these subsets stems from the study of mice. Will the results of these studies translate to humans? Are there specific genetic or non-genetic (i.e. environmental) factors that contribute to the marked variability in iNKT cell numbers between individuals? Which of these factors also influence iNKT cell tissue distribution or function? The limited available information regarding these issues presents a serious challenge in the field of human iNKT cell biology. Ultimately, it is anticipated that identification of the regulatory mechanisms guiding human iNKT cell development will provide insights into how iNKT cell numbers and/or functions can be manipulated to enhance normal immunity and to treat clinical conditions such as immunodeficiency, allergy, autoimmunity and cancer.
Acknowledgements
This work was supported by a grant from the XLP Research Trust and the National Institute of Health (KEN).
References
- 1.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 3.Kawano T, et al. CD1d-restricted and TCR-mediated activation of Valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 4.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name. Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 5.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1− CD4+ CD1d-dependent precursor stage. J Exp Med. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 7.Gadue P, Stein PL. NK T cell precursors exhibit differential cytokine regulation and require Itk for efficient maturation. J Immunol. 2002;169:2397–2406. doi: 10.4049/jimmunol.169.5.2397. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Huang H, Paul WE. NK1.1+ CD4+ T cells lose NK1.1 expression upon in vitro activation. J Immunol. 1997;158:5112–5119. [PubMed] [Google Scholar]
- 9.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4−8− T cells. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8-−alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baev DV, et al. Distinct homeostatic requirements of CD4+ and CD4− subsets of Valpha24-invariant natural killer T cells in humans. Blood. 2004;104:4150–4156. doi: 10.1182/blood-2004-04-1629. [DOI] [PubMed] [Google Scholar]
- 13.Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol. 2005;35:1399–1407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 14.Engel I, et al. Co-receptor choice by V alpha14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010;207:1015–1029. doi: 10.1084/jem.20090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuling H, et al. EBV-induced human CD8+ NKT cells suppress tumorigenesis by EBV-associated malignancies. Cancer Res. 2009;69:7935–7344. doi: 10.1158/0008-5472.CAN-09-0828. [DOI] [PubMed] [Google Scholar]
- 16.Lin H, Nieda M, Nicol AJ. Differential proliferative response of NKT cell subpopulations to in vitro stimulation in presence of different cytokines. Eur J Immunol. 2004;34:2664–2671. doi: 10.1002/eji.200324834. [DOI] [PubMed] [Google Scholar]
- 17.Lin H, Nieda M, Rozenkov V, Nicol AJ. Analysis of the effect of different NKT cell subpopulations on the activation of CD4 and CD8 T cells, NK cells, and B cells. Exp Hematol. 2006;34:289–295. doi: 10.1016/j.exphem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Lin H, Nieda M, Hutton JF, Rozenkov V, Nicol AJ. Comparative gene expression analysis of NKT cell subpopulations. J Leukoc Biol. 2006;80:164–173. doi: 10.1189/jlb.0705421. [DOI] [PubMed] [Google Scholar]
- 19.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond KJ, et al. CD1d-restricted NKT cells: an interstrain comparison. J Immunol. 2001;167:1164–1173. doi: 10.4049/jimmunol.167.3.1164. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh RA, et al. Patients with X-linked lymphoproliferative disease due to BIRC4 mutation have normal invariant natural killer T-cell populations. Clin Immunol. 2009;132:116–123. doi: 10.1016/j.clim.2009.03.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Exley MA, et al. Cutting edge: Compartmentalization of Th1-like noninvariant CD1d-reactive T cells in hepatitis C virus-infected liver. J Immunol. 2002;168:1519–1523. doi: 10.4049/jimmunol.168.4.1519. [DOI] [PubMed] [Google Scholar]
- 25.Roy KC, Maricic I, Khurana A, Smith TR, Halder RC, Kumar V. Involvement of secretory and endosomal compartments in presentation of an exogenous self-glycolipid to type II NKT cells. J Immunol. 2008;180:2942–2950. doi: 10.4049/jimmunol.180.5.2942. [DOI] [PubMed] [Google Scholar]
- 26.Van Rhijn I, et al. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci U S A. 2004;101:13578–13583. doi: 10.1073/pnas.0402838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berzofsky JA, Terabe M. NKT cells in tumor immunity: opposing subsets define a new immunoregulatory axis. J Immunol. 2008;180:3627–3635. doi: 10.4049/jimmunol.180.6.3627. [DOI] [PubMed] [Google Scholar]
- 28.Park SH, Weiss A, Benlagha K, Kyin T, Teyton L, Bendelac A. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terabe M, Berzofsky JA. The role of NKT cells in tumor immunity. Adv Cancer Res. 2008;101:277–348. doi: 10.1016/S0065-230X(08)00408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 31.Akbari O, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9:582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 32.Pillai AB, George TI, Dutt S, Teo P, Strober S. Host NKT cells can prevent graft-versus-host disease and permit graft antitumor activity after bone marrow transplantation. J Immunol. 2007;178:6242–6251. doi: 10.4049/jimmunol.178.10.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Araki M, Kondo T, Gumperz JE, Brenner MB, Miyake S, Yamamura T. Th2 bias of CD4+ NKT cells derived from multiple sclerosis in remission. Int Immunol. 2003;15:279–288. doi: 10.1093/intimm/dxg029. [DOI] [PubMed] [Google Scholar]
- 34.Vilarinho S, Ogasawara K, Nishimura S, Lanier LL, Baron JL. Blockade of NKG2D on NKT cells prevents hepatitis and the acute immune response to hepatitis B virus. 2007. pp. 18187–18192. [DOI] [PMC free article] [PubMed]
- 35.Fuss IJ, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113:1490–1497. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duarte N, et al. Prevention of diabetes in nonobese diabetic mice mediated by CD1d-restricted nonclassical NKT cells. J Immunol. 2004;173:3112–3118. doi: 10.4049/jimmunol.173.5.3112. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Choi EY, Chung DH. Donor bone marrow type II (non-Valpha14Jalpha18 CD1d-restricted) NKT cells suppress graft-versus-host disease by producing IFN-gamma and IL-4. J Immunol. 2007;179:6579–6587. doi: 10.4049/jimmunol.179.10.6579. [DOI] [PubMed] [Google Scholar]
- 38.Ambrosino E, et al. Cross-regulation between type I and type II NKT cells in regulating tumor immunity: a new immunoregulatory axis. J Immunol. 2007;179:5126–5136. doi: 10.4049/jimmunol.179.8.5126. [DOI] [PubMed] [Google Scholar]
- 39.Treiner E, Lantz O. CD1d- and MR1-restricted invariant T cells: of mice and men. Curr Opin Immunol. 2006;18:519–526. doi: 10.1016/j.coi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horai R, et al. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP Protein-Dependent Natural Killer T-like Cells Regulate the Development of CD8+ T Cells with Innate Lymphocyte Characteristics. Immunity. 2010;33:1–13. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jordan MS, et al. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alonzo ES, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2009;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levitsky HI, Golumbek PT, Pardoll DM. The fate of CD4-8- T cell receptor-alpha beta+ thymocytes. J Immunol. 1991;146:1113–1117. [PubMed] [Google Scholar]
- 47.Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol. 2000;164:2412–2418. doi: 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- 48.Hammond K, Cain W, van Driel I, Godfrey D. Three day neonatal thymectomy selectively depletes NK1.1+ T cells. Int Immunol. 1998;10:1491–1499. doi: 10.1093/intimm/10.10.1491. [DOI] [PubMed] [Google Scholar]
- 49.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makino Y, Kanno R, Koseki H, Taniguchi M. Development of Valpha4+ NK T cells in the early stages of embryogenesis. Proc Natl Acad Sci U S A. 1996;93:6516–6520. doi: 10.1073/pnas.93.13.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacDonald HR. Development and selection of NKT cells. Curr Opin Immunol. 2002;14:250–254. doi: 10.1016/s0952-7915(02)00329-1. [DOI] [PubMed] [Google Scholar]
- 52.Shimamura M, Ohteki T, Beutner U, MacDonald HR. Lack of directed V alpha 14-J alpha 281 rearrangements in NK1+ T cells. Eur J Immunol. 1997;27:1576–1579. doi: 10.1002/eji.1830270638. [DOI] [PubMed] [Google Scholar]
- 53.Pear WS, Tu L, Stein PL. Lineage choices in the developing thymus: choosing the T and NKT pathways. Curr Opin Immunol. 2004;16:167–173. doi: 10.1016/j.coi.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Dao T, et al. Development of CD1d-restricted NKT cells in the mouse thymus. Eur J Immunol. 2004;34:3542–3552. doi: 10.1002/eji.200425546. [DOI] [PubMed] [Google Scholar]
- 55.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci U S A. 2005;102:5114–5119. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 57.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 58.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu H, Chun T, Colmone A, Nguyen H, Wang CR. Expression of CD1d under the control of a MHC class Ia promoter skews the development of NKT cells, but not CD8+ T cells. J Immunol. 2003;171:4105–4112. doi: 10.4049/jimmunol.171.8.4105. [DOI] [PubMed] [Google Scholar]
- 60.Schumann J, Pittoni P, Tonti E, Macdonald HR, Dellabona P, Casorati G. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol. 2005;175:7303–7310. doi: 10.4049/jimmunol.175.11.7303. [DOI] [PubMed] [Google Scholar]
- 61.Elewaut D, et al. The adaptor protein AP-3 is required for CD1d-mediated antigen presentation of glycosphingolipids and development of Valpha14i NKT cells. J Exp Med. 2003;198:1133–1146. doi: 10.1084/jem.20030143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 63.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 64.Schumann J, Mycko MP, Dellabona P, Casorati G, MacDonald HR. Cutting edge: influence of the TCR Vbeta domain on the selection of semi-invariant NKT cells by endogenous ligands. J Immunol. 2006;176:2064–2068. doi: 10.4049/jimmunol.176.4.2064. [DOI] [PubMed] [Google Scholar]
- 65.Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia C, et al. Synthesis and biological evaluation of alpha-galactosylceramide (KRN7000) and isoglobotrihexosylceramide (iGb3) Bioorg Med Chem Lett. 2006;16:2195–2199. doi: 10.1016/j.bmcl.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 67.Cheng L, et al. Efficient activation of Valpha14 invariant NKT cells by foreign lipid antigen is associated with concurrent dendritic cell-specific self recognition. J Immunol. 2007;178:2755–2762. doi: 10.4049/jimmunol.178.5.2755. [DOI] [PubMed] [Google Scholar]
- 68.Chiu YH, et al. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 69.Gadola SD, et al. Impaired selection of invariant natural killer T cells in diverse mouse models of glycosphingolipid lysosomal storage diseases. J Exp Med. 2006;203:2293–2303. doi: 10.1084/jem.20060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porubsky S, Speak AO, Luckow B, Cerundolo V, Platt FM, Grone HJ. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci U S A. 2007;104:5977–5982. doi: 10.1073/pnas.0611139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mallevaey T, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chun T, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med. 2003;197:907–918. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pellicci DG, et al. Intrathymic NKT cell development is blocked by the presence of alpha-galactosylceramide. Eur J Immunol. 2003;33:1816–1823. doi: 10.1002/eji.200323894. [DOI] [PubMed] [Google Scholar]
- 75.Marrella V, et al. A hypomorphic R229Q Rag2 mouse mutant recapitulates human Omenn syndrome. J Clin Invest. 2007;117:1260–1269. doi: 10.1172/JCI30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arase H, et al. Developmental arrest of NK1.1+ T cell antigen receptor (TCR)-alpha/beta+ T cells and expansion of NK1.1+ TCR-gamma/delta+ T cell development in CD3 zeta-deficient mice. J Exp Med. 1995;182:891–895. doi: 10.1084/jem.182.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, et al. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 79.Chan G, Hanke T, Fischer KD. Vav-1 regulates NK T cell development and NK cell cytotoxicity. Eur J Immunol. 2001;31:2403–2410. doi: 10.1002/1521-4141(200108)31:8<2403::aid-immu2403>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 80.Iwabuchi K, et al. Defective development of NK1.1+ T-cell antigen receptor alphabeta+ cells in zeta-associated protein 70 null mice with an accumulation of NK1.1+ CD3− NK-like cells in the thymus. Blood. 2001;97:1765–1775. doi: 10.1182/blood.v97.6.1765. [DOI] [PubMed] [Google Scholar]
- 81.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 82.Guo J, et al. Regulation of the TCRalpha repertoire by the survival window of CD4+CD8+ thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 83.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol. 2010;11:435–441. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma CS, Nichols KE, Tangye SG. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol. 2007;25:337–379. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- 86.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 87.Latour S, et al. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol. 2003;5:149–154. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 88.Chan B, et al. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 89.Nichols KE, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pasquier B, et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 92.Nunez-Cruz S, et al. Differential requirement for the SAP-Fyn interaction during NK T cell development and function. J Immunol. 2008;181:2311–2320. doi: 10.4049/jimmunol.181.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jordan MA, Fletcher JM, Pellicci D, Baxter AG. Slamf1, the NKT cell control gene Nkt1. J Immunol. 2007;178:1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- 95.Latour S, Gish G, Helgason CD, Humphries RK, Pawson T, Veillette A. Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product. Nat Immunol. 2001;2:681–690. doi: 10.1038/90615. [DOI] [PubMed] [Google Scholar]
- 96.Wang N, et al. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–1264. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alberola-Ila J, Hogquist KA, Swan KA, Bevan MJ, Perlmutter RM. Positive and negative selection invoke distinct signaling pathways. J Exp Med. 1996;184:9–18. doi: 10.1084/jem.184.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cannons JL, et al. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 99.Schmidt-Supprian M, et al. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci U S A. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stanic AK, Bezbradica JS, Park JJ, Van Kaer L, Boothby MR, Joyce S. Cutting edge: the ontogeny and function of Va14Ja18 natural T lymphocytes require signal processing by protein kinase C theta and NF-kappa B. J Immunol. 2004;172:4667–4671. doi: 10.4049/jimmunol.172.8.4667. [DOI] [PubMed] [Google Scholar]
- 101.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, Gapin L. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Berzins SP, McNab FW, Jones CM, Smyth MJ, Godfrey DI. Long-term retention of mature NK1.1+ NKT cells in the thymus. J Immunol. 2006;176:4059–4065. doi: 10.4049/jimmunol.176.7.4059. [DOI] [PubMed] [Google Scholar]
- 104.Drennan MB, et al. Cutting edge: the chemokine receptor CXCR3 retains invariant NK T cells in the thymus. J Immunol. 2009;183:2213–2216. doi: 10.4049/jimmunol.0901213. [DOI] [PubMed] [Google Scholar]
- 105.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 108.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 109.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 110.Maeda T, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant’Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 112.Raberger J, et al. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kreslavsky T, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Belles C, et al. Bias in the gamma delta T cell response to Listeria monocytogenes. V delta 6.3+ cells are a major component of the gamma delta T cell response to Listeria monocytogenes. J Immunol. 1996;156:4280–4289. [PubMed] [Google Scholar]
- 115.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 116.Yin Z, Chen C, Szabo SJ, Glimcher LH, Ray A, Craft J. T-Bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-gamma by gammadelta T cells. J Immunol. 2002;168:1566–1571. doi: 10.4049/jimmunol.168.4.1566. [DOI] [PubMed] [Google Scholar]
- 117.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006;177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 118.Jensen KD, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gerber DJ, Azuara V, Levraud JP, Huang SY, Lembezat MP, Pereira P. IL-4-producing gamma delta T cells that express a very restricted TCR repertoire are preferentially localized in liver and spleen. J Immunol. 1999;163:3076–3082. [PubMed] [Google Scholar]
- 120.Azuara V, Levraud JP, Lembezat MP, Pereira P. A novel subset of adult gamma delta thymocytes that secretes a distinct pattern of cytokines and expresses a very restricted T cell receptor repertoire. Eur J Immunol. 1997;27:544–553. doi: 10.1002/eji.1830270228. [DOI] [PubMed] [Google Scholar]
- 121.Lee YJ, et al. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 123.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 124.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 125.Pomerantz JL, Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 126.Kontgen F, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 127.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 128.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zheng Y, Vig M, Lyons J, Van Parijs L, Beg AA. Combined deficiency of p50 and cRel in CD4+ T cells reveals an essential requirement for nuclear factor kappaB in regulating mature T cell survival and in vivo function. J Exp Med. 2003;197:861–874. doi: 10.1084/jem.20021610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Elewaut D, et al. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J Exp Med. 2003;197:1623–1633. doi: 10.1084/jem.20030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stanic AK, et al. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol. 2004;172:2265–2273. doi: 10.4049/jimmunol.172.4.2265. [DOI] [PubMed] [Google Scholar]
- 132.Vallabhapurapu S, et al. Rel/NF-kappaB family member RelA regulates NK1.1− to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. Eur J Immunol. 2008;38:3508–3519. doi: 10.1002/eji.200737830. [DOI] [PubMed] [Google Scholar]
- 133.Lee AJ, et al. Regulation of natural killer T-cell development by deubiquitinase CYLD. EMBO J. 2010;29:1600–1612. doi: 10.1038/emboj.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Akbari O, et al. ICOS/ICOSL interaction is required for CD4+ invariant NKT cell function and homeostatic survival. J Immunol. 2008;180:5448–5456. doi: 10.4049/jimmunol.180.8.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 136.Lazarevic V, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen F, Chen D, Rothenberg EV. Specific regulation of fos family transcription factors in thymocytes at two developmental checkpoints. Int Immunol. 1999;11:677–688. doi: 10.1093/intimm/11.5.677. [DOI] [PubMed] [Google Scholar]
- 138.Rincon M, Flavell RA. Regulation of AP-1 and NFAT transcription factors during thymic selection of T cells. Mol Cell Biol. 1996;16:1074–1084. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen D, Rothenberg EV. Molecular basis for developmental changes in interleukin-2 gene inducibility. Mol Cell Biol. 1993;13:228–237. doi: 10.1128/mcb.13.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Williams KL, et al. BATF transgenic mice reveal a role for activator protein-1 in NKT cell development. J Immunol. 2003;170:2417–2426. doi: 10.4049/jimmunol.170.5.2417. [DOI] [PubMed] [Google Scholar]
- 141.Zullo AJ, Benlagha K, Bendelac A, Taparowsky EJ. Sensitivity of NK1.1 negative NKT cells to transgenic BATF defines a role for activator protein-1 in the expansion and maturation of immature NKT cells in the thymus. J Immunol. 2007;178:58–66. doi: 10.4049/jimmunol.178.1.58. [DOI] [PubMed] [Google Scholar]
- 142.Lawson VJ, Maurice D, Silk JD, Cerundolo V, Weston K. Aberrant selection and function of invariant NKT cells in the absence of AP-1 transcription factor Fra-2. J Immunol. 2009;183:2575–2584. doi: 10.4049/jimmunol.0803577. [DOI] [PubMed] [Google Scholar]
- 143.Riera-Sans L, Behrens A. Regulation of alphabeta/gammadelta T cell development by the activator protein 1 transcription factor c-Jun. J Immunol. 2007;178:5690–5700. doi: 10.4049/jimmunol.178.9.5690. [DOI] [PubMed] [Google Scholar]
- 144.Aliahmad P, et al. TOX provides a link between calcineurin activation and CD8 lineage commitment. J Exp Med. 2004;199:1089–1099. doi: 10.1084/jem.20040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 146.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4-versus CD8- lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 150.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 151.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 152.Dose M, et al. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood. 2006;108:2669–2677. doi: 10.1182/blood-2006-02-005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci U S A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mycko MP, et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J Immunol. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 155.Bain G, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Barndt R, Dai MF, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during alpha beta thymopoiesis. J Immunol. 1999;163:3331–3343. [PubMed] [Google Scholar]
- 157.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Allen RD, 3rd, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 161.Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, Ho IC. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 162.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 163.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 164.Germanov E, Veinotte L, Cullen R, Chamberlain E, Butcher EC, Johnston B. Critical role for the chemokine receptor CXCR6 in homeostasis and activation of CD1d-restricted NKT cells. J Immunol. 2008;181:81–91. doi: 10.4049/jimmunol.181.1.81. [DOI] [PubMed] [Google Scholar]
- 165.Dave VP, Allman D, Keefe R, Hardy RR, Kappes DJ. HD mice: a novel mouse mutant with a specific defect in the generation of CD4+ T cells. Proc Natl Acad Sci U S A. 1998;95:8187–8192. doi: 10.1073/pnas.95.14.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Michel ML, et al. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Coquet JM, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4−NK1.1− NKT cell population. Proc Natl Acad Sci U S A. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Michel ML, et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Matsuda JL, et al. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 170.Ranson T, et al. IL-15 availability conditions homeostasis of peripheral natural killer T cells. Proc Natl Acad Sci U S A. 2003;100:2663–2668. doi: 10.1073/pnas.0535482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sandberg JK, Stoddart CA, Brilot F, Jordan KA, Nixon DF. Development of innate CD4+ alpha-chain variable gene segment 24 (Valpha24) natural killer T cells in the early human fetal thymus is regulated by IL-7. Proc Natl Acad Sci U S A. 2004;101:7058–7063. doi: 10.1073/pnas.0305986101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Geissmann F, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ohteki T, Maki C, Koyasu S, Mak TW, Ohashi PS. Cutting edge: LFA-1 is required for liver NK1.1+ TCR alpha beta+ cell development: evidence that liver NK1.1+ TCR alpha beta+ cells originate from multiple pathways. J Immunol. 1999;162:3753–3756. [PubMed] [Google Scholar]
- 174.Emoto M, Mittrucker HW, Schmits R, Mak TW, Kaufmann SH. Critical role of leukocyte function-associated antigen-1 in liver accumulation of CD4+ NKT cells. J Immunol. 1999;162:5094–5098. [PubMed] [Google Scholar]
- 175.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 176.Matsuda JL, George TC, Hagman J, Gapin L. Temporal dissection of T-bet functions. J Immunol. 2007;178:3457–3465. doi: 10.4049/jimmunol.178.6.3457. [DOI] [PubMed] [Google Scholar]
- 177.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 178.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yokota Y, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 180.Cannarile MA, et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 181.Monticelli LA, et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc Natl Acad Sci U S A. 2009;106:19461–19466. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.D’Cruz LM, Yang CY, Goldrath AW. Transcriptional regulation of NKT cell development and homeostasis. Curr Opin Immunol. 2010;22:199–205. doi: 10.1016/j.coi.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cen O, et al. The adaptor molecule signaling lymphocytic activation molecule-associated protein (SAP) regulates IFN-gamma and IL-4 production in V alpha 14 transgenic NKT cells via effects on GATA-3 and T-bet expression. J Immunol. 2009;182:1370–1378. doi: 10.4049/jimmunol.182.3.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]