Abstract
We have investigated the antibacterial activity and cytotoxicity of a series of amino-terminated poly(amidoamine) (PAMAM) dendrimers modified with poly(ethylene glycol) (PEG) groups. The antibacterial activity of the PAMAM dendrimers and their derivatives against the common ocular pathogens, Pseudomonas aeruginosa and Staphylococcus aureus, was evaluated by their minimum inhibitory concentrations (MICs). For the unmodified third and fifth generation (G3 and G5) amino-terminated dendrimers, the MICs against both P. aeruginosa and S. aureus were in the range of 6.3–12.5 μg mL−1, comparable to that of the antimicrobial peptide LL-37 (1.3–12.5 μg mL−1) and within the wide range of 0.047–128 μg mL−1 for the fluoroquinolone antibiotics. PEGylation of the dendrimers decreased their antibacterial activities, especially for the Gram-positive bacteria (S. aureus). The reduction in potency is likely due to the decrease in the number of protonated amino groups and shielding of the positive charges by the PEG chains, thus decreasing the electrostatic interactions of the dendrimers with the negatively-charged bacterial surface. Interestingly, localization of a greater number of amino groups on G5 vs. G3 dendrimers did not improve the potency. Significantly, even a low degree of PEGylation, e.g. 6% with EG11 on G3 dendrimer, greatly reduced the cytotoxicity towards human corneal epithelial cells while maintaining a high potency against P. aeruginosa. The cytotoxicity of the PEGylated dendrimers to host cells is much lower than that reported for antimicrobial peptides. Furthermore, the MICs of these dendrimers against P. aeruginosa are more than two orders of magnitude lower than other antimicrobial polymers reported to date. These results motivate further exploration of the potential of cationic dendrimers as a new class of antimicrobial agents that may be less likely to induce bacterial resistance than standard antibiotics.
Introduction
The potential biomedical applications of dendrimers has become an active area of research.1–3 Dendrimers are a new class of hyperbranched macromolecules possessing distinctive properties such as well-defined globular architecture, narrow polydispersity and tunability of surface functionalities.4,5 Their tunable nanometric size and chemical functionality offer versatility for incorporating a wide variety of functional moieties either through encapsulation in the interior of the dendrimer or by tethering onto the periphery via covalent modification or physisorption for drug/gene delivery and imaging.1–3
Poly(amidoamine) dendrimers (PAMAM) are arguably the most extensively studied dendrimers for biomedical applications,1–3,6 especially as carriers of biologically active agents.7,8 For drug delivery, pharmacokinetics and cytotoxicity of the system are important concerns. The polycationic PAMAM dendrimers are known to be cytotoxic.9 However, covalent attachment of acetyl groups,10 lauroyl groups,11 or poly(ethylene glycol) (PEG) chains11–14 to the peripheral amino groups of PAMAM dendrimers decreases their cytotoxicity to host cells, probably due to the reduction of the number of protonated amino groups and shielding of the positive charges on the dendrimers. In particular, in vitro experiments using various human and animal cell lines showed that increasing the coverage of PEG chains on amino-terminated PAMAM decreased the cytotoxicity of the dendrimers.11–14 In addition, PEGylation of PAMAM dendrimers has also been reported to greatly enhance the circulation time and improve biodistribution and biocompatibility.12,14–19
PAMAM and other dendrimers with peripheral amino groups have been used as carriers or scaffolds for the covalent attachment of antimicrobial agents, such as antibiotics,20–22 quaternary ammonium,23 and viral inhibitors.24 However, the antimicrobial activities of the PAMAM dendrimers themselves have only recently been discovered.25 During our preliminary study25 of using PAMAM dendrimers as carriers of antimicrobial peptides (AMPs), such as LL-37,26–28 we found that generation 5 (G5), amino-terminated PAMAM dendrimer was an effective antibacterial agent against common ocular pathogens, such as Pseudomonas aeruginosa and Staphylococcus aureus. As expected, the unmodified G5 dendrimer showed cytotoxicity to human corneal epithelial cells (HCECs).25 Significantly, the ~43% PEGylated G5 PAMAM became non-cytotoxic to HCECs and maintained a high potency against P. aeruginosa although it became inactive against S. aureus.25 This result was exciting since P. aeruginosa is one of the most common pathogens associated with bacterial keratitis, a serious ocular infection that may lead to blindness,29 and the causative organism has developed significant resistance to current antibiotics.30 AMPs are natural antibiotics that have remained effective against bacterial pathogens for millions of years.31 Despite extensive research on AMPs,32–38 the development of AMP-based anti-infective drugs has been hampered by factors such as their relatively low potency against pathogens, high cytotoxicity and high cost of manufacturing.32,34,38 It has been proposed that low affinity targeting of bacterial membranes themselves rather than a specific bacterial receptor is one of the means through which AMPs circumvent the evolution of bacterial resistance.31,35 Compared to AMPs, the amino-terminated PAMAM dendrimers possess a much higher number of positive charges, and thus have a higher affinity towards the negatively-charged bacterial surface. In addition, they are much cheaper to manufacture than AMPs. These considerations motivated us to investigate the potential of PEGylated PAMAM as a new type of antibacterial agent. We hypothesized that PAMAM dendrimers with a high charge density and large size (higher generation) would be more effective in killing bacteria, but also could be more toxic to host cells. On the other hand, PEGylation of the PAMAM dendrimers should reduce their toxicity to the host cells, but perhaps at the expense of lowering their antibacterial activity. Accordingly, we prepared a series of PEGylated PAMAM dendrimers designated as G3-%EGm and G5-%EGm that differ in generation (G3 or G5), degree (%) of PEGylation of the peripheral amino groups, and the length (EG7 or EG11) of the monodispersed PEG chains (Scheme 1). We measured the minimum inhibitory concentration (MIC) of these dendrimers for P. aeruginosa and S. aureus and their cytotoxicity to HCECs. We show that the generation, degree of PEGylation and length of the PEG chains of the PEGylated PAMAM dendrimers, to a limited extent, can be optimized to achieve efficient bactericidal activity with low cytotoxicity to the host cells.
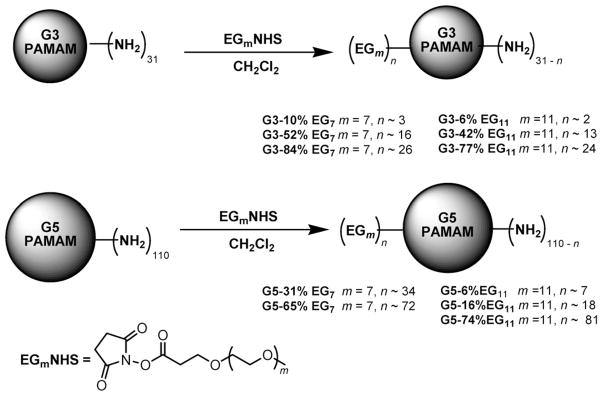
Scheme 1.
PEGylation of G3 and G5 PAMAM dendrimers with EGmNHS to provide PAMAM G3-%EGm and G5-%EGm. The amounts of EGmNHS are specified in Table 1. The degree of PEGylation and number of PEG chains (n) were calculated from the difference of Mws of the dendrimer before and after PEGylation, which were obtained from their MALDI-TOF mass spectra.
Experimental
Materials
Different generations (G3 and G5) of amino-terminated PAMAM dendrimers with ethylenediamine initiator core were purchased from Dendritech, Inc. EG7NHS and EG11NHS (NHS-m-dPEG™, Mw = 509 and 685, respectively) were from Quanta Biodesign. Prepacked Sephadex PD-10 columns used for gel filtration chromatography were obtained from GE Healthcare. trans-3-Indoleacrylic acid (≥99.0%) as matrix substance for MALDI-TOF mass spectrometry was purchased from Sigma-Aldrich.
The components of an approximately 1 L of SHEM-X media used in the culture assay include 500 mL of Minimum Essential Medium Eagle (Sigma-Aldrich M4655), 500 mL of F12 nutrient mixture (1×) (Gibco 31765-035), 5 mL of dimethyl sulfoxide (Sigma-Aldrich D2650), 10 mL of antibiotic and antimycotic (100×) (Gibco 15240-062), 5 μg of bovine insulin (Sigma-Aldrich I1882), 100 μL of cholera toxin (Sigma-Aldrich C8052), 100 μL of epidermal growth factor (EGF) (Sigma-Aldrich), 11.8 mL of HEPES buffer solution (1 M) (Gibco 15630-080) and 2 mL of Normocin™ (InvivoGen ant-nr-2).
Synthesis and characterization of PEG-PAMAM
The amounts of dry PAMAM and EGmNHS (m = 7 or 11) for the synthesis are listed in Table 1. The compounds were then dissolved in methanol and CH2Cl2, respectively. In a typical procedure, a 10% (w/v) solution of EGmNHS (Scheme 1) in CH2Cl2 was added to a stirred methanolic solution of the G3 (19% w/w) or G5 (21% w/w) PAMAM dendrimer in a vial. The mixture was stirred vigorously overnight, and then the solvent was removed under vacuum. The product was purified by gel filtration chromatography using a prepacked Sephadex PD-10 column with Millipore water as eluent, and then dried in vacuo.
Table 1.
Amounts of dry G3, G5 and EGmNHS used in the synthesis of the PEGylated PAMAM dendrimers
| PEGylated PAMAM dendrimer | PAMAM |
EGmNHS |
|||
|---|---|---|---|---|---|
| mg | μmol | mg | μmol | Eq. | |
| G3-10% EG7 | 10 | 1.5 | 10 | 20 | 13 |
| G3-52% EG7 | 11 | 1.6 | 18 | 35 | 22 |
| G3-84% EG7 | 10 | 1.5 | 43 | 84 | 56 |
| G3-6% EG11 | 6.0 | 0.87 | 5.5 | 8.0 | 9 |
| G3-42% EG11 | 4.9 | 0.72 | 9.2 | 13 | 18 |
| G3-77% EG11 | 5.3 | 0.78 | 31 | 45 | 58 |
| G5-31% EG7 | 10 | 0.37 | 11 | 22 | 59 |
| G5-65% EG7 | 5.7 | 0.21 | 20 | 39 | 186 |
| G5-6% EG11 | 5.9 | 0.22 | 4.6 | 6.7 | 30 |
| G5-16% EG11 | 5.3 | 0.20 | 8.4 | 12 | 60 |
| G5-74% EG11 | 5.9 | 0.22 | 31 | 45 | 205 |
The products were characterized by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry using a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems), operated in the positive ion linear mode with an accelerating voltage ranging from 20 kV to 25 kV with delayed extraction. The sample preparation protocol for MALDI-TOF MS of PAMAM is similar to that reported in the literature.39 Briefly, the matrix, trans-3-indoleacrylic acid, was dissolved in a 1: 1 (v/v) water–acetonitrile to prepare a 0.05 M stock solution. A series of dendrimer concentrations ranging from 100 to 1000 μM in methanol were prepared. The analytical sample was prepared by spotting the dendrimer solution followed by the matrix solution on the MALDI plate, then drying in ambient conditions. Each mass spectrum (see ESI†) was the average of the spectra obtained from 200 randomly located laser shots on the same sample. As an example, Fig. 1 displays the MALDI spectrum of an EG7-modified G3 PAMAM, showing a measured molecular weight (Mw) of 17 247 Da, corresponding to approximately 84% PEGylation. Hence the compound was designated as G3-84% EG7.
Fig. 1.
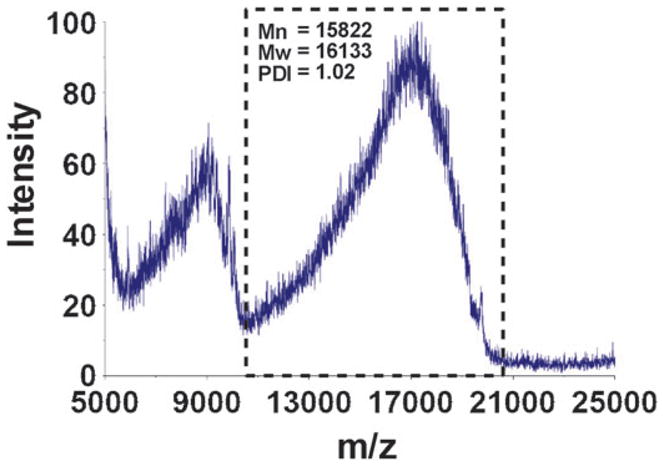
MALDI-TOF mass spectrum of G3 PAMAM modified with EG7 showing a measured molecular weight of 17 247 Da. The peak at ~8600 Da corresponds to the double charged molecular ion. The boxed regions were selected for the number and weight average molecular weight (Mn and Mw) and polydispersity index (PDI) calculations.
Note that PAMAM dendrimers are known to have defects.40,41 The Mw of a G3 PAMAM dendrimer is 6909 Da. However, the MALDI-TOF MS measurement showed that the commercial G3 PAMAM used in this study had a measured Mw of 6859 Da, which is probably due to the failure of attaching the last ethylenediamine to most dendrimer molecules during the synthesis. Hence, the number of peripheral amino groups was estimated to be 31 (not 32 for the ideal G3). Similarly, the measured Mw of the commercial G5 PAMAM dendrimer was 26 373 Da, lower than the theoretical Mw of 28 828 Da. According to the data provided by the manufacturer and literature,42 the number of peripheral amino groups in the commercial G5 PAMAM dendrimer used in this study was approximately 110 (not 128 for the ideal G5). The difference in the measured Mws of PEG-modified PAMAM and the unmodified dendrimers provides the approximate number of PEG chains attached on PEG-PAMAM, and thus the degree of PEGylation.
Preparation of bacterial solutions
P. aeruginosa (PA ATCC 19 66043) and S. aureus (SA ATCC 29 213) were the laboratory strains used in this study. We used P. aeruginosa and S. aureus in this study, since both are common ocular pathogens, which have been known to cause bacterial keratitis in human patients.29,44 Single colonies of P. aeruginosa and S. aureus were inoculated in 5 mL of Difco’s nutrient broth (NB) and then incubated overnight at 37 °C while shaking at 180 rpm. A small volume (~300 μL) of this overnight growth was resuspended to 1 mL of NB and the optical density was adjusted to about 0.20 at 620 nm, which corresponds to approximately 1.0 × 107 colony-forming units (CFU) per mL. This solution was then diluted in NB to prepare a bacterial stock solution having a concentration of 2.0 × 105 CFU per mL.
Preparation of dendrimer solutions
The unmodified and PEGylated PAMAM dendrimers were dissolved in water to prepare 8.0 mg mL−1 dendrimer stock solutions. Serial double dilution of the stock solution was performed. For the MIC assay, the stock solution was diluted with an equal volume of 0.02% acetic acid containing 0.40% bovine serum albumin (BSA) to obtain a concentration of 4.0 mg mL−1. Repeating the double dilution using 0.01% acetic acid with 0.20% BSA provided a series of concentrations ranging from 2000 down to 31 μg mL−1. The final test concentration range used for the incubation with the bacteria was 4000 down to 31 μg mL−1. For the MTT cytotoxicity assay, different dendrimer concentrations (1000, 500, 100, 50, 10, 5, 1, 0.5, 0.1 and 0.05 μg mL−1) were prepared by diluting the stock solution in serum-free SHEM-X media.
Antibacterial assay
The minimum inhibitory concentration (MIC) of the PAMAM derivatives, that is, the lowest concentration of the compounds that inhibits the visible growth of P. aeruginosa and S. aureus after 18–24 h of incubation, was determined using a broth microdilution technique.45,46 Briefly, 11 μL of each of the above prepared solutions of the PAMAM and PEG-PAMAM dendrimers was added to the wells containing 100 μl of 2.0 × 105 CFU per mL of the bacterial solution in a 96-well sterile polypropylene microtiter plate. Control wells were composed of 100 μL of the 2.0 × 105 CFU per mL bacterial solutions without dendrimers and 100 μL of NB medium without bacteria. To reduce evaporation during incubation which is critical to the assay, each of the empty outer wells of the microtiter plate was filled with sterile water to increase the water vapor pressure in the incubator.45 The microtiter plates were incubated at 37 °C while shaking at 150 rpm for 18–24 h. At the end of the incubation, the pellets formed were resuspended prior to scanning the optical densities using a bioassay reader (Perkin Elmer HTS 7000, Norwalk, CT) with a wavelength of 635 nm, which is proportional to the number of bacteria in suspension. The data were plotted using Origin 7 (OriginLab, Northampton, MA). The MIC value is defined as the concentration at which a sharp decline in optical density reaches the minimum (close to zero).47
Cytotoxicity assay
Cytotoxicity of the dendrimers to SV40-transformed human corneal epithelial cells (SV40-HCEC),48 a gift from Dr Kaoru Araki-Sasaki (Tane Memorial Eye Hospital, Osaka, Japan), was determined by following a standard MTT assay (MTT is 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole). Briefly, cells were maintained in SHEM-X media with 10% fetal bovine serum (FBS). HCECs (7000 cells per well) were grown for 36 h at 37 °C to confluence in 96-well sterile microtiter plates.49,50 The media with FBS was removed and the cells were washed with 1× phosphate buffer saline (PBS) and serum-starved for 4 h by incubating in SHEM-X media alone at 37 °C. The media in the plate were replaced with 100 μl of the prepared dendrimer solutions. Then, the plate was incubated at 37 °C for 24 h. Untreated HCECs in culture media alone were used as negative control while HCECs treated with 10 μL of 0.2% benzalkonium chloride (bac) served as a positive control. Cytotoxicity was assayed with an MTT kit (Chemicon International, Temecula, CA) following the manufacturer’s instruction. This assay is based on the enzymatic conversion of the yellow tetrazolium salt (MTT reagent) into the purple formazan precipitate by viable cells. The concentration of formazan formed is proportional to the number of viable cells. To measure this concentration, the formazan precipitate in the plate was dissolved by adding a solution of 0.04 N HCl in isopropanol, and the absorbance at 590 nm with a reference wavelength of 635 nm was immediately measured using a FLUOstar Omega multi-mode plate reader (BMG Labtech Incorporated, NC).
Results and discussion
Antibacterial activities of PAMAM and PEG-PAMAM
As shown in Scheme 1, the G3 and G5 PAMAM dendrimers, containing ~31 and ~110 peripheral amino groups, were PEGylated with the monodispersed EGmNHS of varied chain lengths (EG7 and EG11). The coverage of PEG chains on the resulting PEG-PAMAM was varied by the amount of EGmNHS. The degree of PEGylation on the PEG-PAMAM was calculated by obtaining the difference between the Mws of the PEGylated derivatives and the unmodified dendrimers measured by MALDI-TOF mass spectrometry (ESI†), and the data on % of PEGylation and number of PEG chains (n values) are summarized in Scheme 1. It should be noted, however, that it was difficult to obtain precise control of the PEG coverage. In particular, a high coverage of G5 PAMAM with EG11 is difficult to achieve as compared to the shorter EG7, likely due to steric hindrance.
Several measures of antibacterial activity, particularly the concentration needed to kill half of the bacteria (EC50) and the minimum inhibitory concentration (MIC) that inhibits the visible growth of the bacteria, have been widely used for evaluating the susceptibility of potential drugs against specific pathogens. In our previous work, we have determined the EC50 of G5 PAMAM with or without 43% PEGylation by counting the bacterial colonies grown on agar plates.25 In the present study, we used a standard, more efficient and reproducible assay to assess the activities of the dendrimer derivatives. Specifically, we measured the MIC values of these compounds using a modified broth microdilution assay in a 96-well plate format.51 After incubation of the bacteria with the dendrimer derivatives, the optical density at 635 nm, which is proportional to the number of bacteria, was measured with a plate reader. The MIC value of the dendrimer was then obtained from the plot of the optical densities vs. the concentrations of the dendrimer. We used P. aeruginosa and S. aureus in this study, since both are common ocular pathogens, which have been known to induce in vivo bacterial keratitis particularly among contact lens wearers.29,44
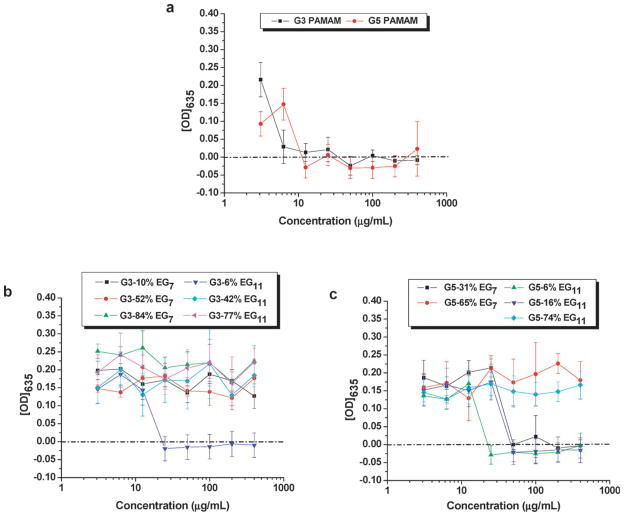
Fig. 2 shows the plots of the optical densities and MICs of P. aeruginosa after an overnight incubation with the unmodified G3 and G5 dendrimers (a), PEGylated G3 (b) and PEGylated G5 (c). Results showed that both G3 and G5 PAMAM were effective against P. aeruginosa in a concentration-dependent manner (Fig. 2a). The MIC values are 6.3 μg mL−1 (same result from four independent experiments) for G3, and 12.5 μg mL−1 (same result from six independent experiments) for G5. These observed MIC values are higher than those reported for the currently used drugs for the treatment of bacterial keratitis, namely ciprofloxacin (Ciloxan®), gatifloxacin (Zymar®), levofloxacin (Levaquin®) and moxifloxacin (Vigamox®) (1, 2, 2 and 2 μg mL−1, respectively).52 However, certain bacterial keratitis isolates have developed resistance to these fluoroquinolone antibiotics, including ciprofloxacin.53,54 The MIC values for dendrimers G3 and G5 were comparable to those reported MICs for the antimicrobial peptide LL-37 (ranging from 1.3 to 5.7 μg mL−1, depending upon the strain of P. aeruginosa used).55 Furthermore, we previously determined an EC50 value of 1.5 ± 0.1 μg mL−1 25 for G5 PAMAM, which is comparable with a reported EC50 value of 2.8 ± 1.3 μg mL−1 for LL-37 against P. aeruginosa.28
Fig. 2.
Plots of the optical densities at 635 nm vs. concentrations of unmodified PAMAM dendrimers (a), and PEGylated G3 (b) and G5 (c) PAMAM dendrimers in Difco’s nutrient broth after incubation with P. aeruginosa ATCC 19 660 for 18–24 hours. The data in each plot were obtained from a representative experiment where each data point corresponds to a mean of three replicates ± standard deviation.
The MIC values derived from Fig. 2 are summarized in Table 2, showing the effect of dendrimer generation, degree of PEGylation and PEG chain length. Also included in Table 2 are the minimum molar concentrations (MICPA/Mw) of the dendrimers that are needed to inhibit the growth of P. aeruginosa, and the corresponding molar concentrations of the amino groups on them. These concentrations are proportional to the minimum number of the molecules and their amino groups needed to inhibit growth of a bacterial cell.
Table 2.
Minimum inhibitory weight (MICPA) and molar concentrations (MICPA/Mw) of the G3 and G5 PAMAM dendrimers against P. aeruginosa, and the corresponding concentrations of the peripheral amino groups (MICPA × NNH2/Mw)
| Compounda | Measured Mws from MALDIb/Da | PDIc | NNH2d | MICPA/μg mL−1 | MICPAMw−1/μM | MICPA × NNH2/Mw−1/μM |
|---|---|---|---|---|---|---|
| G3-84% EG7 | 17247 | 1.02 | 5 | >400 | — | — |
| G3-77% EG11 | 20464 | 1.02 | 7 | >400 | — | — |
| G3-52% EG7 | 13106 | 1.02 | 15 | >400 | — | — |
| G3-42% EG11 | 14424 | 1.02 | 18 | >400 | — | — |
| G3-10% EG7 | 8139 | 1.02 | 28 | >50 | — | — |
| G3-6% EG11 | 8282 | 1.02 | 29 | 25 | 3.0 | 87 |
| G3 | 6859 | 1.02 | 31 | 6.3 | 0.92 | 29 |
| G5-74% EG11 | 72841 | 1.01 | 29 | >400 | — | — |
| G5-65% EG7 | 55006 | 1.02 | 38 | >400 | — | — |
| G5-31% EG7 | 39713 | 1.03 | 76 | 50 | 1.3 | 99 |
| G5-16% EG11 | 36499 | 1.03 | 92 | 50 | 1.4 | 129 |
| G5-6% EG11 | 30599 | 1.03 | 103 | 25 | 0.82 | 84 |
| G5 | 26374 | 1.02 | 110 | 12.5 | 0.47 | 52 |
Percent PEGylations were calculated using the measured Mws from the MALDI spectra of the dendrimers.
Mws of the highest frequency fragment in the major peak from the MALDI spectra were obtained using the Voyager Data Explorer software.
PDIs were calculated using the Mn and Mw derived from the boxed regions selected in the MALDI-TOF mass spectra of the dendrimers.
NNH2 is the approximate number of the free amino groups.
We hypothesize that, similar to AMPs, the cationic nature of the dendrimers is essential for their initial electrostatic interaction with the negatively-charged components of the bacterial membrane, such as lipopolysaccharide (LPS) or lipoteichoic acid.33 Indeed, a relatively narrow range of the minimum molar concentrations of the amino groups (MICPA × NNH2/Mw = 29–129 μM) on the dendrimers correlated with the effective inhibition of bacterial growth. Hence, a higher molar concentration (MICPA/Mw) of the G3 vs. G5 dendrimers is needed to inhibit bacterial growth. To achieve an antimicrobial effect, the dendrimers may need to do more than the initial binding to the bacterial membrane; penetration and diffusion toward the cytoplasm may be required. To facilitate this step, a balance between the net positive charge, hydrophobicity and size may be necessary.32,34,56 G5 dendrimer has more peripheral NH2 groups than G3 (~110 vs. ~31), resulting in a higher surface charge density on the molecule.57 However, its larger size compared to G3 possibly reduced its capability to penetrate toward the bacterial membrane. The ease of penetration through the cytoplasmic membrane might be the reason that, against our initial hypothesis, G3 was more potent than G5 (MICPA = 6.3 μg mL−1 vs. 12.5 μg mL−1, MICPA × NNH2/Mw = 29 vs. 52 μM) although G5 localizes a higher number of NH2 groups (positive charge).
PEGylation resulted in a reduction of the antibacterial activity against P. aeruginosa (Table 2). Although the threshold values were not determined, a high PEG coverage deactivated the dendrimers against P. aeruginosa. One possible explanation for this observation is the decrease in the number of protonated NH2 groups, i.e. the charge density, and the partial shielding of the charges by the PEG chains, thus decreasing the electrostatic interactions with the negatively-charged bacterial surface. To inhibit the growth of the bacterium, a higher number of amino groups (MICPA × NNH2/Mw in Table 2) would then be necessary to compensate the shielding by the PEG chains. Interestingly, longer PEG chains slightly enhance the antibacterial activity. For instance, G3-10% EG7 did not show activity while G3-6%EG11 displayed an activity with a MIC/Mw of 3.0 μM. It is possible that in combination with the cationic character of the dendrimer, the dual hydrophobic/hydrophilic character of PEG chains to some extent enhances the permeation through the cell membrane.
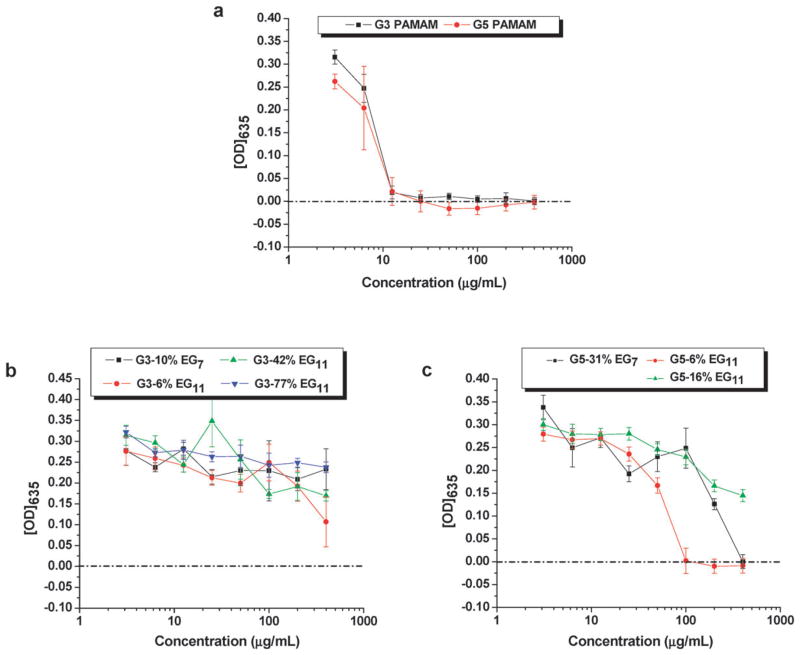
The results for the Gram-positive S. aureus are shown in Fig. 3. The MIC value derived from Fig. 3a for both G3 and G5 was 12.5 μg mL−1 (same result from three independent experiments). This MIC value was comparable to the reported MICs for LL-37 (2.9–12.5 μg mL−1,55 depending upon the strain of S. aureus used), but substantially higher than those of common fluoroquinolone antibiotics, including gatifloxacin (0.22 μg mL−1), levofloxacin (0.38 μg mL−1), moxifloxacin (0.047 μg mL−1), and ciprofloxacin (0.5 μg mL−1).
Fig. 3.
Plots of the optical densities at 635 nm vs. concentrations of unmodified PAMAM dendrimers (a), and PEGylated G3 (b) and G5 (c) PAMAM dendrimers in Difco’s nutrient broth after incubation with S. aureus ATCC 29 213 for 18–24 hours. The data in each plot were obtained from a representative experiment where each data point corresponds to a mean of three replicates ± standard deviation.
Unfortunately, even a low degree of PEGylation nearly completely deactivated the dendrimer against S. aureus especially for the smaller dendrimer G3 (Fig. 3b and c). The result is in agreement with our previous observation that G5 with 43% coverage of EG7 was inactive against S. aureus.25
The reason for the lower efficiency of the PAMAM dendrimers in killing Gram-positive compared with Gram-negative bacteria is not clear. Similar phenomena have been observed for certain antimicrobial peptides.58 The antibacterial action of the dendrimers probably involves disruption of the cytoplasmic membrane of the bacteria. For the Gram-negative bacteria, the top of the plasma membrane is bound by a thin periplasmic layer covered by an outer membrane containing negatively-charged LPS. Gram-positive bacterial cell walls are quite different. In Gram-positive bacteria, the outermost layer is thick and rigid, consisting of cross-linked peptidoglycan. Accumulation of polycationic antimicrobial peptides on the outer membrane of the Gram-negative bacteria may facilitate membrane permeation and hence killing of the bacteria.32–34 However, the ability of certain antimicrobial peptides to bind to lipoteichoic acid in the peptidoglycan layer of the Gram-positive bacteria does not correspond to their potency against Gram-positive bacteria.58 Perhaps the thick and cross-linked peptidoglycan layer on the Gram-positive bacteria greatly hinders the subsequent actions of some AMPs and the dendrimers to disrupt the cytoplasmic membrane, thus protecting the cytoplasmic membrane from disruption.
Toxicity of PEG-PAMAM to HCECs
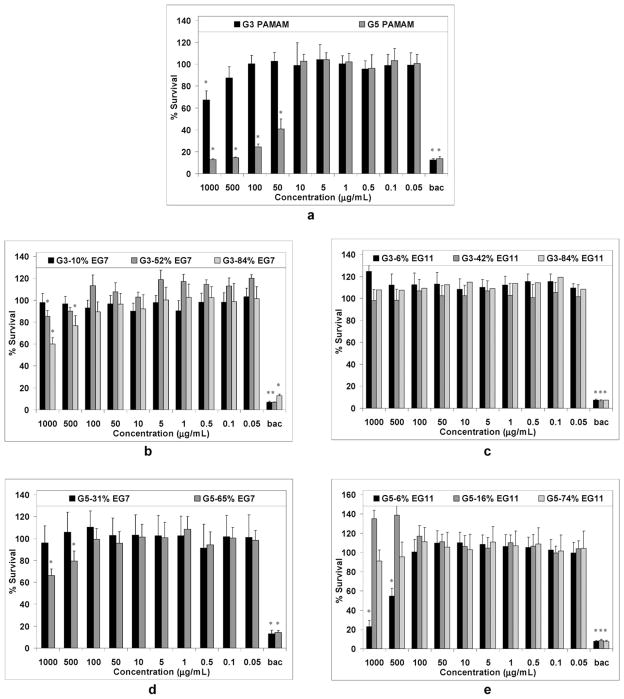
The cell viability (expressed as percent survival of the treated HCECs in comparison to untreated HCECs) vs. the concentration of the dendrimers is plotted in Fig. 4. Interestingly, the unmodified G3 dendrimer was only slightly toxic to HCECs even at the highest concentration of 1000 μg mL−1 (Fig. 4a). On the other hand, a decrease in cell viability was observed upon treatment with the unmodified G5 dendrimer at concentrations higher than 10 μg mL−1. For instance, at 50 μg mL−1, only 41% ± 9% of the cells relative to the negative control (untreated cells) were found to be viable.
Fig. 4.
Percent survival of HCECs after incubating with various concentrations of unmodified dendrimers G3 and G5 (a), G3 EG7-modified dendrimers (b), G3 EG11-modified dendrimers (c), G5 EG7-modified dendrimers (d), and G5 EG11-modified dendrimers for 24 hours. The benzalkonium chloride (bac)-treated HCECs were used as the positive control. Each plot was obtained from a representative experiment, where the data point is the mean of four replicates ± standard deviation. The values were compared to the untreated control at *P < 0.05 using the Tukey test.
At concentrations greater than 100 μg mL−1, G5 had approximately the same cytotoxicity as the positive control (cell viability < 15%). This result is consistent with the report by Hong et al.,59 that the formation of nanoholes on eukaryotic cells is promoted by high PAMAM concentrations. Phase contrast images (ESI†) of HCECs showed noticeable cell contraction and possible rupture upon treatment with 10 μg mL−1 of G5. At higher concentrations (≥50 μg mL−1), a significant detachment of the cells from the glass surface was observed, resulting in fewer cells viewed under the microscope. A preliminary study using a fluorescence-based assay (in situ cell death detection kit) to detect fragmented DNA suggested that cell death did not include an apoptotic component (data not shown).
It is well known that a high degree of PEGylation with long PEG chains, e.g. 22–28% PEG550, PEG750 and PEG2000, reduces the toxicity of amino-terminated PAMAM dendrimers to mammalian cells.11–15,17–19 Significantly, we found that even a low degree of PEGylation rendered the PAMAM dendrimers, particularly G3, nontoxic to HCECs (Fig. 4b–e). For instance, the viability of HCECs after 24 hours incubation with 1 mg mL−1 of G3-6% EG11 was 125% _ 5%. In comparison, the percent survival of HCECs exposed to 1.25 mg mL−1 of moxifloxacin and levofloxacin for 24 hours was 76% ± 7% and 106% ± 0%, respectively.60 The G5 derivative G5-6% EG11, being more cytotoxic than the above antibiotics, was still less cytotoxic compared to LL-37 (cytotoxic at concentrations > 100 μg mL−1 vs. > 10 μg mL−1).49
Conclusions
The localization of a greater number of amino groups on the higher generation (G5 vs. G3) dendrimers did not improve the potency against the bacteria. The MIC values against P. aeruginosa for the unmodified dendrimers and those with a low PEG coverage were in the range of 6.3–25 μg mL−1, comparable to that of the antimicrobial peptide LL-37. PEGylation of the dendrimers decreased the antibacterial activity especially for the Gram-positive bacteria (S. aureus). However, PEGylation was necessary to reduce the cytotoxicity of the dendrimers, particularly the G5 dendrimers, towards the host cells (HCECs) to the level comparable to the antibiotics. Fortunately, even a low degree of PEGylation, e.g. 6% of EG11 on G3, can achieve this task while maintaining a high potency against P. aeruginosa.
This work has contributed to the development of antimicrobial polymers in general.61 The MIC values of several PAMAM and PEG-PAMAM dendrimers reported here are significantly lower, mostly by more than two orders of magnitude, than that of other antimicrobial polymers reported to date.61 In particular, we identified that G3 dendrimers modified with a low coverage (<10%) of relatively long PEG chains (EG11) are promising antimicrobial agents that may be less likely to promote the development of bacterial resistance than the standard antibiotics. Although these dendrimers are less potent than many currently used antibiotics, the MICs reported for antibiotics are based upon levels achievable in the bloodstream. The concentrations of antimicrobials that can be achieved through direct topical application in the eye are thousands of times greater.62 Furthermore, they may potentially be used as antimicrobial coatings for medical implants, such as contact lenses, and for treatment of eye or skin infections.
Supplementary Material
Acknowledgments
Financial support from the Welch Foundation, the National Science Foundation (DMR-0706627), a training fellowship to AIL from the Keck Center Nanobiology Training Program of the Gulf Coast Consortia (NIH 5R90DK071054-03), the Texas Center for Superconductivity at the University of Houston, National Institutes of Health (HD058985), and the National Eye Institute (EY13175 to AM) are gratefully acknowledged.
Footnotes
Electronic supplementary information (ESI) available: The MALDI-MS spectra of the PAMAM dendrimers and their PEGylated derivatives, phase contrast images of HCECs treated with different concentrations of G5 PAMAM. See DOI: 10.1039/b904746h
References
- 1.Svenson S, Tomalia DA. Adv Drug Delivery Rev. 2005;57:2106–2129. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Stiriba SE, Frey H, Haag R. Angew Chem, Int Ed. 2002;41:1329–1334. doi: 10.1002/1521-3773(20020415)41:8<1329::aid-anie1329>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Lee CC, MacKay JA, Frechet JMJ, Szoka FC. Nat Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 4.Tomalia DA. Prog Polym Sci. 2005;30:294–324. [Google Scholar]
- 5.Archut A, Vogtle F. Chem Soc Rev. 1998;27:233–240. [Google Scholar]
- 6.Esfand R, Tomalia DA. Drug Discovery Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 7.Dufes C, Uchegbu IF, Schaetzlein AG. Adv Drug Delivery Rev. 2005;57:2177–2202. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Boas U, Heegaard PMH. Chem Soc Rev. 2004;33:43–63. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- 9.Duncan R, Izzo L. Adv Drug Delivery Rev. 2005;57:2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Kolhatkar RB, Kitchens KM, Swaan PW, Ghandehari H. Bioconjugate Chem. 2007;18:2054–2060. doi: 10.1021/bc0603889. [DOI] [PubMed] [Google Scholar]
- 11.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D’Emanuele A. Int J Pharm. 2003;252:263–266. doi: 10.1016/s0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y, Klutz AM, Jacobson KA. Bioconjugate Chem. 2008;19:1660–1672. doi: 10.1021/bc700483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Lopina ST, DiPersio LP, Schmidt SP. J Mater Sci: Mater Med. 2008;19:1991–1997. doi: 10.1007/s10856-007-3278-0. [DOI] [PubMed] [Google Scholar]
- 14.Luo D, Haverstick K, Belcheva N, Han E, Saltzman WM. Macromolecules. 2002;35:3456–3462. [Google Scholar]
- 15.Malik N, Wiwattanapatapee R, Klopsch R, Lorenz K, Frey H, Weener JW, Meijer EW, Paulus W, Duncan R. J Controlled Release. 2000;65:133–148. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 16.Harris JM, Chess RB. Nat Rev Drug Discovery. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 17.Okuda T, Kawakami S, Maeie T, Niidome T, Yamashita F, Hashida M. J Controlled Release. 2006;114:69–77. doi: 10.1016/j.jconrel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Gajbhiye V, Kumar PV, Tekade RK, Jain NK. Curr Pharm Des. 2007;13:415–429. [Google Scholar]
- 19.Kim TI, Seo HJ, Choi JS, Jang HS, Baek J, Kim K, Park JS. Biomacromolecules. 2004;5:2487–2492. doi: 10.1021/bm049563j. [DOI] [PubMed] [Google Scholar]
- 20.Yang H, Lopina ST. J Biomater Sci, Polym Ed. 2003;14:1043–1056. doi: 10.1163/156856203769231556. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Qu H, Ma M, Xu Z, Xu P, Fang Y, Xu T. Eur J Med Chem. 2007;42:1032–1038. doi: 10.1016/j.ejmech.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Ma M, Cheng Y, Xu Z, Xu P, Qu H, Fang Y, Xu T, Wen L. Eur J Med Chem. 2007;42:93–98. doi: 10.1016/j.ejmech.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 23.Chen CZ, Beck-Tan NC, Dhurjati P, van Dyk TK, LaRossa RA, Cooper SL. Biomacromolecules. 2000;1:473–480. doi: 10.1021/bm0055495. [DOI] [PubMed] [Google Scholar]
- 24.Bourne N, Stanberry LR, Kern ER, Holan G, Matthews B, Bernstein DI. Antimicrob Agents Chemother. 2000;44:2471–2474. doi: 10.1128/aac.44.9.2471-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calabretta MK, Kumar A, McDermott AM, Cai C. Biomacromolecules. 2007;8:1807–1811. doi: 10.1021/bm0701088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang LC, Jean D, Proske RJ, Reins RY, McDermott AM. Curr Eye Res. 2007;32:595–609. doi: 10.1080/02713680701446653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDermott AM. Ocul Surf. 2004;2:229–247. doi: 10.1016/s1542-0124(12)70111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon YJ, Huang LC, Romanowski EG, Yates KA, Proske RJ, McDermott AM. Curr Eye Res. 2005;30:385–394. doi: 10.1080/02713680590934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willcox MDP, Holden BA. Biosci Rep. 2001;21:445–461. doi: 10.1023/a:1017991709846. [DOI] [PubMed] [Google Scholar]
- 30.McGhee Charles NJ, Niederer R. Clin Exp Ophthalmol. 2006;34:3–5. doi: 10.1111/j.1442-9071.2006.01164.x. [DOI] [PubMed] [Google Scholar]
- 31.Peschel A, Sahl HG. Nat Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 32.Hancock REW, Sahl HG. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 33.Zaslo M. Nature. 2002;415:389–395. [Google Scholar]
- 34.Giuliani A, Pirri G, Nicoletto SF. Cent Eur J Biol. 2007;2:1–33. [Google Scholar]
- 35.Boman HG. J Intern Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 36.Toke O. Biopolymers. 2005;80:717–735. doi: 10.1002/bip.20286. [DOI] [PubMed] [Google Scholar]
- 37.Theuretzbacher U, Toney JH. Curr Opin Invest Drugs. 2006;7:158–166. [PubMed] [Google Scholar]
- 38.Zhang L, Falla TJ. Expert Opin Pharmacother. 2006;7:653–663. doi: 10.1517/14656566.7.6.653. [DOI] [PubMed] [Google Scholar]
- 39.Zaluzec EJ, Gage DA, Watson JT. Protein Expression Purif. 1995;6:109–123. doi: 10.1006/prep.1995.1014. [DOI] [PubMed] [Google Scholar]
- 40.Shi X, Majoros IJ, Patri AK, Bi X, Islam MT, Desai A, Ganser TR, Baker JR., Jr Analyst. 2006;131:374–381. doi: 10.1039/b515624f. [DOI] [PubMed] [Google Scholar]
- 41.Peterson J, Allikmaa V, Subbi J, Pehk T, Lopp M. Eur Polym J. 2003;39:33–42. [Google Scholar]
- 42.Shi X, Banyai I, Rodriguez K, Islam MT, Lesniak W, Balogh P, Balogh LP, Baker JR., Jr Electrophoresis. 2006;27:1758–1767. doi: 10.1002/elps.200500818. [DOI] [PubMed] [Google Scholar]
- 43.Pillar CM, Hobden JA. Invest Ophthalmol Visual Sci. 2002;43:1437–1444. [PubMed] [Google Scholar]
- 44.Bourcier T, Thomas F, Borderie V, Chaumeil C, Laroche L. Br J Ophthalmol. 2003;87:834–838. doi: 10.1136/bjo.87.7.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegand I, Hilpert K, Hancock REW. Nat Protocols. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 46.Andrews JM. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 47.Devienne KF, Raddi MSG. Braz J Microbiol. 2002;33:166–168. [Google Scholar]
- 48.Araki-Sasaki K, Ohashi Y, Sasabe T, Hayashi K, Watanabe H, Tano Y, Handa H. Invest Ophthalmol Visual Sci. 1995;36:614–621. [PubMed] [Google Scholar]
- 49.Huang LC, Petkova TD, Reins RY, Proske RJ, McDermott AM. Invest Ophthalmol Visual Sci. 2006;47:2369–2380. doi: 10.1167/iovs.05-1649. [DOI] [PubMed] [Google Scholar]
- 50.Nagaoka I, Hirota S, Yomogida S, Ohwada A, Hirata M. Inflammation Res. 2000;49:73–79. doi: 10.1007/s000110050561. [DOI] [PubMed] [Google Scholar]
- 51.Dominguez MC, De la Rosa M, Borobio MV. J Antimicrob Chemother. 2001;47:391–398. doi: 10.1093/jac/47.4.391. [DOI] [PubMed] [Google Scholar]
- 52.Schlech Barry A, Alfonso E. Surv Ophthalmol. 2005;50(Suppl 1):S7–S15. doi: 10.1016/j.survophthal.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein MH, Kowalski RP, Gordon YJ. Ophthalmology. 1999;106:1313–1318. [PubMed] [Google Scholar]
- 54.Kowalski RP, Dhaliwal DK, Karenchak LM, Romanowski EG, Mah FS, Ritterband DC, Gordon YJ. Am J Ophthalmol. 2003;136:500–505. doi: 10.1016/s0002-9394(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 55.Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shai Y. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 57.Islam MT, Shi X, Balogh L, Baker JR., Jr Anal Chem. 2005;77:2063–2070. doi: 10.1021/ac048383x. [DOI] [PubMed] [Google Scholar]
- 58.Scott MG, Gold MR, Hancock REW. Infect Immun. 1999;67:6445–6453. doi: 10.1128/iai.67.12.6445-6453.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X, Balogh L, Orr BG, Baker JR, Jr, Banaszak Holl MM. Bioconjugate Chem. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 60.Kim S-Y, Lim Jung A, Choi J-S, Choi E-C, Joo C-K. Cornea. 2007;26:720–725. doi: 10.1097/ICO.0b013e3180515251. [DOI] [PubMed] [Google Scholar]
- 61.Kenawy ER, Worley SD, Broughton R. Biomacromolecules. 2007;8:1359–1384. doi: 10.1021/bm061150q. [DOI] [PubMed] [Google Scholar]
- 62.Barnes SD, Pavan-Langston D, Azar DT. Principles and Practice of Infectious Diseases. Elsevier, Inc; Philadelphia, PA: 2005. pp. 1387–1406. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.