Abstract
BACKGROUND
Multiplex arrays are increasingly used for measuring protein biomarkers. Advantages of this approach include specimen conservation, limited sample handling, and decreased time and cost, but the challenges of optimizing assay format for each protein, selecting common dilution factors, and establishing robust quality control algorithms are substantial. Here, we use measurements of 15 protein biomarkers from a large study to illustrate processing, analytic, and quality control issues with multiplexed immunoassays.
METHODS
We contracted with ThermoScientific for duplicate measurements of 15 proteins in 2322 participants from a community-based cohort, a plasma control, and recombinant protein controls using 2 custom planar microarrays with 6 (panel A) or 9 (panel B) capture antibodies printed in each well. We selected constituent analytes in each panel based on endogenous concentrations and assay availability. Protocols were standardized for sample processing, storage, and freeze-thaw exposures. We analyzed data for effects of deviations from processing protocols, precision, and bias.
RESULTS
Measurements were within reportable ranges for each of the assays; however, concentrations for 7 of the 15 proteins were not centered on the dose–response curves. An additional freeze-thaw cycle and erroneous sample dilution for a subset of samples produced significantly different results. Measurements with large differences between duplicates were seen to cluster by analyte, plate, and participant. Conventional univariate quality control algorithms rejected many plates. Plate-specific medians of cohort and plasma control data significantly covaried, an observation important for development of alternative quality control algorithms.
CONCLUSIONS
Multiplex measurements present difficult challenges that require further analytical and statistical developments.
Protein immunoassays provide information about quantities and forms of endogenous proteins. Uniplex enzyme immunoassays have been the workhorse for protein measurement for decades, but they can be laborious and expensive and consume relatively large amounts of specimen. Multiplex analysis of protein biomarkers is an attractive strategy for obtaining large numbers of measurements rapidly. Multiplex approaches have already been successfully used in gene expression microarrays (1). For example, Mammaprint concurrently quantifies expression of 70 genes to evaluate breast cancer prognosis (2). Thus, there is considerable interest in protein multiplex arrays that enable simultaneous quantification of circulating proteins to improve disease diagnosis and prognosis (3).
Presently, antibody-based platforms are the core technology for protein multiplex arrays (4). Assay formats include suspension arrays and planar arrays that use traditional immunometric principles. Several planar multiplex platforms capable of simultaneously measuring up to 16 proteins are commercially available (5, 6). Multiplexed immunoassays overcome some of the limitations of uniplex assays. For example, measuring several proteins from a single sample conserves specimen, limits sample handling, decreases throughput time, and reduces labor costs. Concerns have been raised, however, about the reliability and consistency of multiplexed protein immunoassays. Analytic challenges include difficulty in optimizing a shared format that works well for each component protein (e.g., capture/detection system, incubation time, and washing steps) as well as addressing variability in reagent lots and the manufacturing processes. Selection of a single sample dilution factor that enables measurements in physiological ranges for each constituent assay on a panel is often not feasible. Lack of robust multivariate QC algorithms and guidelines for multiplexed protein array data rejection criteria are problematic.
Multiplexing presents additional QC challenges compared with uniplex analyses. The failure of 1 constituent assay to meet QC specifications results in rejection of results for all assays on the panel. Samples failing QC specifications should be retested using the same measurement system, because substitution of a uniplex assay may introduce bias due to differences in assay format. However, the probability of all assays simultaneously meeting QC specifications is much lower than the probability of a uniplex test passing QC. At present, reference guidelines for multiplex QC programs are under development by the Food and Drug Administration (FDA)4 (7).
This article explores assay performance and data integrity of multiplexed immunoassays in a large community-based study. We measured 15 circulating protein biomarkers in duplicate wells across 2 multiplex panels using a contracted laboratory service. Our study design included 3 levels of recombinant protein controls as well as internal replicates of a volunteer’s plasma sample on each plate. This resulted in a total of 95 040 measurements for standards, controls, and cohort samples across 132 microplates. We used multivariate data analyses to analyze analyte drift and uncontrolled sample selection bias caused by poor randomization. This case study provides technical insights into the capabilities and limitations of a commercial multiplexed immunoassay platform.
Materials and Methods
SAMPLE COLLECTION
Research participants included 2335 subjects (1166 African-Americans and 1169 non-Hispanic whites) in the Genetic Network of Arteriopathy study (8), as well as a healthy volunteer used as an internal assay control. The study was approved by the Institutional Review Boards of the Mayo Clinic (Rochester, MN) and the University of Mississippi Medical Center (Jackson, MS). Blood was collected by venipuncture after an overnight fast, centrifuged, aliquoted within 2 h, and stored at −80 °C. For multiplex analyte measurements, 300 μL of stored samples were thawed on ice and aliquoted into corresponding bar-coded Eppendorf tubes. Plasma control aliquots were interspersed after every 26th sample to create internal, blinded controls on each plate. We randomized samples by ethnicity such that a block of 40 African-American samples was followed by a block of 40 non-Hispanic white samples. Aliquots were refrozen at −80 °C, and shipped to SearchLight™ (ThermoScientific, Inc).
EXPERIMENTAL MEASUREMENTS
We selected 15 proteins for which SearchLight offered validated assays and grouped them into 2 multiplex panels based on endogenous analyte abundance and cross-reactivity. Panel A included 6 low-abundance proteins: interleukin-6 (IL-6), IL-18, tumor necrosis factor receptor-I (TNF-RI), TNF-related apoptosis-inducing ligand (TRAIL), P-selectin, and receptor of advanced glycation end products (RAGE); panel B comprised 9 relatively high-abundance proteins: monocyte chemoattractant protein-1 (MCP-1), TNF-RII, matrix metalloproteinase-9 (MMP-9), MMP-2, tissue inhibitor of metalloproteinases-1 (TIMP-1), TIMP-2, RANTES (regulated upon activation, normal T cell expressed, and secreted protein), E-selectin, and intracellular adhesion molecule-1 (ICAM). SearchLight printed plates for each panel of our study using noncontact Piezo-electric robotic arrayers to discretely immobilize 6 (panel A) or 9 (panel B) different capture antibodies within each well of 96-well microplates. All plates were printed within 2 days to minimize potential systematic error attributed to manufacturing.
ThermoScientific agreed to measure the 15 proteins in 2425 EDTA plasma samples using standardized operating procedures in their Clinical Laboratory Improvement Amendment–accredited service core (ID 22D1053083) after 1:2 dilution for panel A and 1:100 dilution for panel B using tissue culture medium supplemented with 10% fetal calf serum. On panel B, however, samples on plates 9–20 were erroneously diluted 1:200, and samples on plates 1–8, 24, 65, and 66 were exposed to an additional freeze-thaw cycle. Three levels of recombinant protein QC materials were assayed without dilution on each plate across the study. Standards, QC materials, and samples were plated in duplicate wells and measured on a total of 66 plates for each panel. Our assays were performed by 2 SearchLight technicians within a 2-week period (5, 6).
ANALYSES OF SEARCHLIGHT MICROARRAY DATA
We assessed whether dynamic range and sample dilution factor were adequate for each assay by selecting representative standard curves and evaluating the positions of cohort data (5th–95th percentiles) and median study values of the recombinant protein and plasma controls. For panel B, we calculated plate-specific least square means of cohort data (after adjusting for age, sex, and ethnicity) and plate-specific means of the plasma control measurements. We stratified these data by dilution factor and freeze-thaw cycle. To determine whether differences in sample handling affected test values, we calculated the ratio of concentrations obtained from plates diluted 1:200 to plates diluted 1:100 and the ratio of concentrations from plates exposed to 2 freeze-thaw cycles to plates exposed to a single freeze-thaw cycle. We tested the significance of differences in means using the nonparametric Wilcoxon rank sum statistic.
Duplicate measurements of each sample allowed us to identify outliers as well as intraassay imprecision. We calculated the percentage of samples with imprecision between duplicate measurements using 3 cutpoints (CV >10%, >20%, and >30%). For each panel, we calculated the percentage of measurements with CV >30% and tabulated the number of proteins with CV >30% for each participant to evaluate the proportion of samples affected by disparate values. We performed these analyses without the TRAIL data for panel A because this analyte had high imprecision that affected nearly half of the measurements. We also calculated plate-specific percentages of cohort samples with CV >30% between duplicate measurements and analyzed ratios as described above.
We identified microplates that contained a large proportion of samples with high intraassay imprecision by calculating the predicted binomial variance of disparate values for each analyte and comparing this value to the observed number of samples with CV >30% on each plate. We visually inspected plate images on which all constituent proteins had greater intraassay imprecision than predicted (plates 9, 17, 22, 23, 24, 25, 37, 42, 45, and 65 on panel A; plates 9, 10, 11, 19, 25, 26, and 43 on panel B) as well as the images of plates with expected binomial variance.
We evaluated interassay variability and trending of the data across the study. We calculated plate-specific medians of the cohort values for each analyte and then calculated CVs for these plate-specific medians as well as for plasma control and recombinant QC material measurements as indicators of interplate imprecision. We explored whether fluctuations in cohort data were attributable to the biasing effects of variable demographic mix across plates by fitting general linear models that included age, sex, and ethnicity for each analyte (log-transformed) to the cohort test results. We then determined whether these demographic variables were randomly distributed across microplates for each panel using ANOVA for age and by calculating global χ2 values for sex and ethnicity. Using the plasma control measurements, we performed a 2-way ANOVA for each panel that included plate number and constituent analyte concentrations to determine whether there were systematic differences between plates across the study. We also investigated plate-to-plate variations in analyte concentrations by plotting plate-specific least square means of cohort data and the plasma control measurements for each protein. We determined whether the plasma control measurements covaried with cohort data across plates using the SAS “autoreg” procedure.
We divided QC materials into 3 sets: (1) the study volunteer’s plasma control, (2) recombinant QC1 and QC2 that were developed by SearchLight for our study, and (3) QC3, a recombinant protein control used routinely by SearchLight to monitor assay performance. Target values for QC analyses were set as the median study value for each analyte for the plasma control and the QC1 and QC2 set because prospective targets were not available. SearchLight provided target values for QC3 that had been established from previous studies. We set the QC target ranges at median ±20% for each analyte based on the maximum error generally acceptable in clinical laboratories. The QC1 and QC2 set was analyzed using a multirule approach such that QC specifications were exceeded if either QC measurement was 3 CVs above or below the target value or if both measurements were 2 CVs beyond the target value in the same direction. The plasma control and QC3 data sets were considered acceptable if the control value was within 3 CVs of the target value. Using the 3 sets of QC materials, we analyzed each protein as an independent assay and then each panel as a unit, such that all constituent assays were considered out of control if QC specifications were exceeded for at least 1 analyte. Statistical analyses were performed using SAS v.9.2.
Results
ANALYTIC MEASUREMENT RANGES
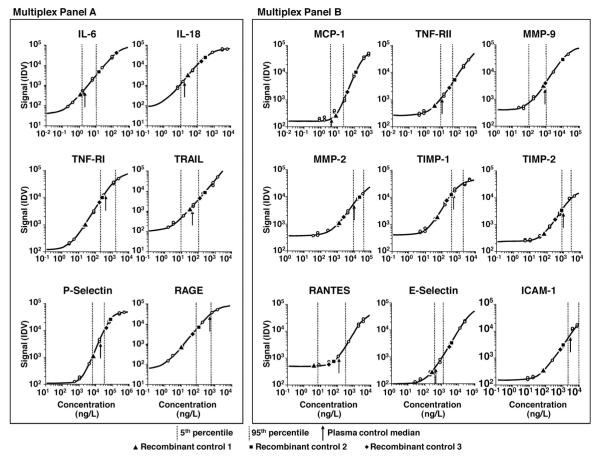
The 5th–95th percentiles of the cohort data and the recombinant protein and plasma control medians are plotted on representative standard curves for each assay (Fig. 1). Overall, study measurements were within reportable assay limits, confirming that targeted assay ranges and sample dilution factors were appropriate for our study. Cohort ranges were in the higher ends of the curves, however, for some assays, such as MMP-2, TIMP-1, TIMP-2, and ICAM, and in the lower ends of the curves for other assays, such as MCP-1, MMP-9, and RANTES. Our plasma control median was within the distribution of cohort values for each analyte except MMP-9. The recombinant protein QC materials were not consistently within the cohort data range, however; 10, 9, and 12 of the 15 proteins for QC1, QC2, and QC3, respectively, were outside the cohort value ranges.
Fig. 1. Representative dose–response curves.
Vertical dotted lines demark the 5th and 95th percentiles of cohort data. Arrows, squares, triangles, and diamonds depict the median values of the plasma control, recombinant protein quality control 1 (QC1), recombinant protein QC2, and recombinant protein QC3, respectively.
ASSAY IMPRECISION
All measurements were in duplicate. The proportions of cohort samples with high imprecision between the duplicate measurements are shown in Table 1. Average percentages of samples with high measurement imprecision (ranges across analytes) were 35.2% (18.7%–78.6%) at a 10% limit; 18.2% (5.3%–60.9%) at a 20% limit; and 11.6% (2.5%–46.3%) at a 30% limit. The poorest agreement between duplicates was observed in the TRAIL assay, in which 46.3% of the measurements had imprecision >30%. On both panels, the majority of the imprecision between duplicate measurements was observed in a limited number of samples. On panel A, after exclusion of TRAIL, 4.8% (1019 of 11 650 duplicate measurements) of the data differed by >30%. Although 23.9% of the 2335 samples had disparate measurements for at least 1 protein, >70% of these imprecise measurements were observed in only 10.6% of the samples. On panel B, 9.3% (1944 of 20 912 duplicate measurements) of the data differed by >30%. High imprecision was observed for at least 1 protein in 34.9% of the samples; on the other hand, more than 75% of the disparate measurements were observed in only 15.9% of the samples. We also observed that actual variance was an average 7.1-fold (range 2.6- to 14.7-fold) higher than the binomial variance across analytes. A large number of samples with high intraassay imprecision were observed for all of the constituent analytes on 10 plates for panel A and on 7 plates for panel B. Notably, 42.8% and 47.2% of the total disparate measurements for panel A and panel B, respectively, were located on these plates. Visual inspection of the plate images revealed differences in chemiluminescent signal intensity between duplicate wells on both panels (Supplemental Fig. 1, which accompanies the online version of this article at www.clinchem.org/content/vol55/issue6). On panel A, capture antibodies against different proteins were spotted at 6 discreet locations in each well; however, chemiluminescent signal was observable at 7 discreet locations.
Table 1.
Percentage of duplicate cohort measurements with imprecision (CV) >10%, >20%, or >30%.
| CV >10% | CV >20% | CV >30% | |
|---|---|---|---|
| Panel A | |||
| IL-6 | 33.1 | 12.8 | 6.0 |
| IL-18 | 50.7 | 26.1 | 14.9 |
| TNF-RI | 40.6 | 17.3 | 7.9 |
| TRAIL | 78.6 | 60.9 | 46.3 |
| P-selectin | 23.1 | 8.1 | 3.8 |
| RAGE | 38.4 | 18.3 | 11.1 |
| Panel B | |||
| MCP-1 | 38.8 | 21.1 | 13.1 |
| TNF-RII | 29.1 | 14.6 | 9.8 |
| MMP-9 | 45.8 | 28.0 | 20.9 |
| MMP-2 | 31.0 | 14.7 | 10.2 |
| TIMP-1 | 18.7 | 5.3 | 2.5 |
| TIMP-2 | 20.7 | 9.4 | 6.5 |
| RANTES | 29.9 | 14.8 | 8.4 |
| E-selectin | 19.1 | 6.9 | 3.8 |
| ICAM | 28.0 | 13.3 | 8.0 |
| Average | 35.2 | 18.2 | 11.6 |
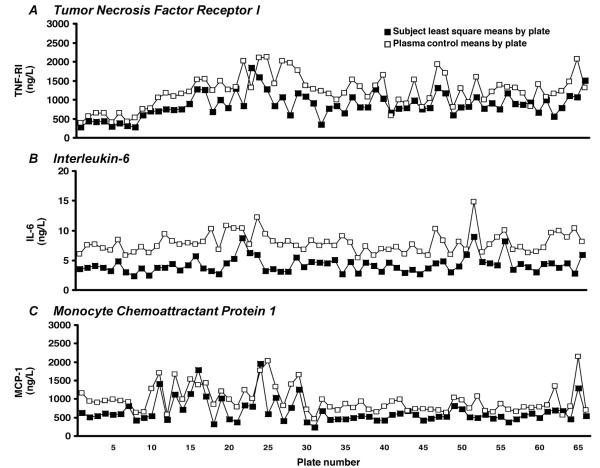
We evaluated interplate imprecision of the study materials using CV as a measure of variability. CVs were >20% in most assays for cohort data, plasma control, and recombinant protein QC materials. The exception was the P-selectin assay, for which the CV was 19.8% in the cohort data. We hypothesized that interplate fluctuations in cohort data could be attributed to biasing effects of demographic mix due to poor randomization. Using multivariable regression analyses, we identified that protein concentrations were significantly influenced by participant age (12 of 14 proteins), sex (6 of 14 proteins), and ethnicity (10 of 14 proteins) in an analyte-specific manner. We observed significant variation in all 3 variables: F = 33.4 for age, χ2 = 154.2 for sex, and χ2 = 1583.1 for ethnicity (P < 0.0001 for all characteristics). Using 2-way ANOVA for each panel, we determined that systematic fluctuations in protein concentrations were significant across plates in the plasma control data (F = 3.66 for panel A; F = 5.42 for panel B; P < 0.0001 for both panels). Plate-specific means of cohort data calculated after adjustment for age, ethnicity, and sex covaried with plasma control materials (Fig. 2). Covariance of cohort samples and plasma control levels was significant for all analytes across the study (P < 0.001).
Fig. 2. (A), (B), and (C) illustrate the covariance of plate-specific population-normalized cohort means and plasma control means across 66 microplates for 3 representative proteins.
PREANALYTICAL AND ANALYTICAL FACTORS IMPACT PROTEIN CONCENTRATIONS AND MEASUREMENT PRECISION
We observed that nonstandard sample dilutions or additional freeze-thaw cycles resulted in analyte-specific changes in protein concentrations in the cohort and plasma control data (Table 2). Measurements of MCP-1, TNF-RII, TIMP-1, E-selectin, and ICAM were significantly higher in cohort samples diluted 1:200 compared with samples diluted 1:100 (P < 0.05). Similarly, TIMP-1 and E-selectin concentrations were significantly higher in plasma control samples diluted 1:200 (P < 0.05). MMP-9, TIMP-1, and RANTES concentrations were significantly lower in both cohort and plasma control samples that were subjected to additional freeze-thaw cycles (P < 0.05). We also explored whether differences in dilution factor and freeze-thaw cycle were associated with poor imprecision. The 1:200 dilution factor was associated with a significant increase in the percentage of samples per plate with imprecision >30% for most analytes (P < 0.05), excluding MCP-1, TIMP-1, and E-selectin. Notably, 44.7% of the disparate measurements were obtained from samples diluted 1:200 on panel B. The additional freezethaw cycle did not significantly affect imprecision, with the exception of MCP-1.
Table 2.
Effects of changes in dilution factor and freeze-thaw (F/T) cycle on protein concentrations.
| Ratio of values from plates with 1:200 dilution to values from plates with 1:100 dilution |
Ratio of values from plates with 2 F/T cycles to plates with 1 F/T cycle |
|||
|---|---|---|---|---|
| Panel B | Least square cohort mean |
Plasma control mean |
Least square cohort mean |
Plasma control mean |
| MCP-1 | 1.10a | 1.14 | 1.24 | 1.34 |
| TNF-RII | 1.16a | 1.11 | 1.07 | 1.09 |
| MMP-9 | 2.00 | 1.87 | 0.67b | 0.64b |
| MMP-2 | 1.03 | 0.99 | 0.90 | 0.90 |
| TIMP-1 | 1.26a | 1.36b | 1.25a | 1.19a |
| TIMP-2 | 0.96 | 0.86 | 1.12 | 1.12 |
| RANTES | 0.90 | 0.78 | 1.24a | 1.33b |
| E-selectin | 1.30b | 1.26a | 0.97 | 1.01 |
| ICAM | 1.16a | 0.91 | 1.0 | 1.05 |
P < 0.05.
P < 0.01.
QUALITY CONTROL ANALYSES
We retrospectively analyzed the QC data separately for plasma control, QC1 and QC2, and QC3 measurements (Table 3). The percentage of plates that exceeded QC specifications was >10% for 13, 12, and 8 of the 15 analytes using plasma control, recombinant QC1 and QC2, and QC3 measurements, respectively. Analytes for which QC results exceeded target ranges for <10% of the plates included IL-16 using the plasma control, RAGE, and E-selectin using the QC1 and QC2 set, and P-selectin, RAGE, MMP-2, TIMP-2, RANTES, and ICAM using QC3. When panel A was analyzed as a unit, 43.9%, 57.8%, and 43.8% of the plates exceeded QC target ranges using the plasma control, the QC1 and QC2 set, and QC3, respectively. When panel B was analyzed as a unit, 56.1%, 54.6%, and 66.7% of the plates exceeded QC target ranges for using the plasma control, the QC1 and QC2 set, and QC3, respectively. On panel A, 47 of 330 measurements exceeded QC target ranges, and nearly 50% of these measurements were located on 8 plates (23, 24, 25, 39, 52, 56, 57, and 66). On panel B, 111 of 594 measurements exceeded QC target ranges, with over half of these measurements located on 9 plates (9, 13, 23, 26, 41, 43, 44, 47, and 50). Thus, a low percentage of microplates contained a high proportion of analytes that exceeded QC specifications. Also, 44% and 58% of the cohort samples with intraassay imprecision >30% were located on these plates for panel A and panel B, respectively.
Table 3.
Number of microtiter plates that exceeded QC target ranges across 66 plates.
| Plasma control |
Recombinant QC1 and QC2 |
Recombinant QC3 |
|
|---|---|---|---|
| Panel A | |||
| IL-6 | 6 | 9 | 20 |
| IL-18 | 8 | 20 | 11 |
| TNF-RI | 16 | 12 | 10 |
| P-selectin | 9 | 21 | 6 |
| RAGE | 8 | 5 | 5 |
| TRAIL | 33 | 43 | 33 |
| Any of first 5 | 29 | 38 | 29 |
| Any of the 6 | 47 | 56 | 48 |
| Panel B | |||
| MCP-1 | 16 | 24 | 25 |
| TNF-RII | 11 | 9 | 17 |
| MMP-9 | 16 | 24 | 31 |
| MMP-2 | 9 | 12 | 4 |
| TIMP-1 | 14 | 13 | 7 |
| TIMP-2 | 10 | 12 | 6 |
| RANTES | 16 | 17 | 5 |
| E-selectin | 10 | 3 | 10 |
| ICAM | 10 | 7 | 2 |
| Any of the 9 | 37 | 36 | 44 |
Discussion
Major advantages of multiplexed protein immunoassays include specimen conservation, reduced sample handling, and decreased throughput time. In our study, 15 proteins were measured using <300 μL of EDTA plasma in 2335 samples in less than 2 weeks. However, bias introduced by inconsistent handling of our samples illustrates that this is an important threat to data validity in biomarker research (9). In microarrays, errors in sample handling are perpetuated across measurements of multiple analytes: erroneous dilution of 376 samples affected 3384 measurements, and additional freeze-thawing of 427 samples affected 3843 measurements. Changes in protein concentrations due to these variations in sample handling were analyte specific; these effects should be well characterized for constituent proteins.
Blood is an ideal source of biomarkers because it contains proteins shed by diseased tissues and is readily accessible. The wide range of protein concentrations in blood poses a challenge to assay configurations, however. An integral step in assay development includes adjusting calibration curves to represent physiological ranges for each assay. For multiplexed assays, technologists must also identify a single sample dilution factor such that constituent protein concentrations fall within dynamic assay ranges. If assays cannot be configured to meet the constraints of a common dilution factor, splitting the panel based on circulating protein abundance is necessary.
Other investigators have evaluated multiplex assays to measure inflammatory markers in research studies (10). In smaller studies that used solid-phase multiplexed immunoassays, intraassay imprecision ranged from 4.2% to 28.1% and interassay imprecision ranged from 9.4% to 28.1% (11–15). However, reliability and consistency of multiplexed protein arrays in large-scale studies is debatable, and allowable levels of imprecision for multiplex immunoassays have not yet been defined. In regard to imprecision between duplicate wells, we evaluated the proportion of cohort data with disparate measurements at 3 cutoff thresholds. We observed that when we allowed 30% imprecision between duplicate measurements, fewer than 15% of the cohort duplicates had disparate values for most analytes. Exceptions were MMP-9 and TRAIL assays, for which 46.3% and 20.9% of the cohort duplicates, respectively, had measurements with imprecision >30%. Additional development efforts would benefit these assays. In our study, a small proportion of samples accounted for a large percentage of disparate values on both panel A and panel B. On panel B, almost half of the measurements with high imprecision were erroneously diluted 1:200. This further illustrates the impact of preanalytical factors on study data. Samples with high intraplate imprecision across analytes were located on a small proportion of microplates on both panels. In regard to interassay imprecision, we observed high variability (CV >20% across 66 microtiter plates for each panel) for most analytes in the cohort data, the plasma control, and all 3 levels of recombinant protein QC materials. We identified that age, sex, and ethnicity, all significant determinants of protein concentrations, were not distributed randomly across the study, and that these differences affected the plate-specific median test results. Even after we adjusted for demographic bias, however, we observed signal fluctuations across microplates that significantly covaried with fluctuations in plasma control values, indicating systematic error across the study.
Standardization of data reporting, common analysis tools, and useful controls are under development for genetic testing by the Microarray Quality Control Project to provide confidence in the consistency and reliability of the gene expression platforms (16–18). In addition, initiatives for developing sustainable reference standards needed for performance validation efforts and quality control are in progress (16, 18). Similar initiatives would benefit multiplexed protein platforms.
The FDA has classified multiplex arrays as in vitro diagnostic multivariate index assays (IVDMIAs) (7). The draft FDA guidance document suggests that preclinical reviews of multiplex assay performance should include reproducible measurement of the input variables, including analyte concentrations (7), yet there are no formal validation guidelines available for multiplex assays. Findlay et al. (19) developed a standard operating procedure for pharmacokinetic immunoassays based on FDA guidelines in which acceptable intra- and interassay precision (based on QC material data) should have <30% variation; Urbanowska et al. (15) proposed expanding these precision criteria to include multiplex panels. Application of these criteria to our study would result in rejection of a substantial portion of measurements.
Our study illustrates multiple concerns about long-term robustness, contributions of nonbiological variation to results, and the need for better QC algorithms. In our opinion, algorithms that correct multiplexed protein immunoassay data for analytic variability need to be developed because retesting will often not be feasible for many large studies.
Supplementary Material
Acknowledgments
Research Funding: NIH grant UO1-HL-81331 from the National Heart, Lung, and Blood Institute.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
- FDA
- U.S. Food and Drug Administration
- IL
- interleukin
- TNF-R
- tumor necrosis factor receptor
- TRAIL
- TNF-related apoptosis-inducing ligand
- RAGE
- receptor of advanced glycation end products
- MCP
- monocyte chemoattractant protein
- MMP
- matrix metalloproteinase
- TIMP
- tissue inhibitor of metalloproteinases
- RANTES
- regulated upon activation, normal T cell expressed, and secreted protein
- ICAM
- intracellular adhesion molecule
- IVDMIA
- in vitro diagnostic multivariate index assay
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.MAQC Consortium The MicroArray Quality Control (MAQC) project shows interand intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–61. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van’t Veer LJDH, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature (Lond) 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 4.Schweitzer B, Kingsmore SF. Measuring proteins on microarrays. Curr Opin Biotechnol. 2002;13:14–9. doi: 10.1016/s0958-1669(02)00278-1. [DOI] [PubMed] [Google Scholar]
- 5. [Accessed November 2008];Endogen SearchLight. http://www.endogen.com/services/
- 6. [Accessed April 2009];Meso Scale Discovery. Instruments. http://www.mesoscale.com/CatalogSystemWeb/WebRoot/products/instruments.aspx.
- 7.US Food and Drug Administration [Accessed April 2009];Guidance for industry and FDA staff: pharmacogenetic tests and genetic tests for heritable markers. http://www.fda.gov/cdrh/oivd/guidance/1549.pdf.
- 8.Boerwinkle E. Multi-center genetic study of hypertension: the Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 9.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–9. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 10.Pollard HB, Srivastava M, Eidelman O, Jozwik C, Rothwell SW, Mueller GP, et al. Protein microarray platforms for clinical proteomics. Proteomics Clin Appl. 2007;1:934–52. doi: 10.1002/prca.200700154. [DOI] [PubMed] [Google Scholar]
- 11.Hsu HY, Silkewittemann, Schneider EM, Weiss M, Joos TO. Suspension microarrays for the identification of the response patterns in hyperinflammatory diseases. Med Eng Phys. 2008;30:976–83. doi: 10.1016/j.medengphy.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Montagnana M, Volpe A, Lippi G, Caramaschi P, Salvagno GL, Biasi D, et al. Relationship between matrix metalloproteinases/tissue inhibitors of matrix metalloproteinases systems and autoantibody patterns in systemic sclerosis. Clin Biochem. 2007;40:837–42. doi: 10.1016/j.clinbiochem.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Toedter G, Hayden K, Wagner C, Brodmerkel C. Simultaneous detection of eight analytes in human serum by two commercially available platforms for multiplex cytokine analysis. Clin Vaccine Immunol. 2008;15:42–8. doi: 10.1128/CVI.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liew M, Groll MC, Thompson JE, Call SL, Moser JE, Hoopes JD, et al. Validating a custom multiplex ELISA against individual commercial immunoassays using clinical samples. Biotechniques. 2007;42:327–8. 30–3. doi: 10.2144/000112332. [DOI] [PubMed] [Google Scholar]
- 15.Urbanowska T, Mangialaio S, Zickler C, Cheevapruk S, Hasler P, Regenass S, Legay F. Protein microarray platform for the multiplex analysis of biomarkers in human sera. J Immunol Methods. 2006;316:1–7. doi: 10.1016/j.jim.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Baker SCBS, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, et al. External RNA Controls Consortium. The External RNA Controls Consortium: a progress report. Nat Methods. 2005;2:731–4. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 17.Ball CASG, Parkinson H, Rocca-Sera P, Brooksbank C, Causton HC, Cavalieri D, et al. Microarray Gene Expression Data (MGED) Society. Standards for microarray data. Science (Wash DC) 2002;298:539. doi: 10.1126/science.298.5593.539b. [DOI] [PubMed] [Google Scholar]
- 18.Shi L. [Accessed March 2009];Executive summary: the MicroArray Quality Control (MAQC) project: toward consensus on “Best Practices” for the generation, analysis, and application of microarray data in the discovery, development, and review of FDA-regulated products. http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/docs/MAQC_Executive Summary_Nov212006.doc.
- 19.Findlay JW, Smith WC, Lee JW, Nordblom GD, Das I, DeSilva BS, et al. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective. J Pharm Biomed Anal. 2000;21:1249–73. doi: 10.1016/s0731-7085(99)00244-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.