Abstract
The effects of prolactin (PRL) during the pathogenesis of breast cancer are mediated in part though Stat5 activity enhanced by its interaction with its transcriptional inducer, the prolyl isomerase cyclophilin B (CypB). We have demonstrated that knockdown of CypB decreases cell growth, proliferation, and migration, and CypB expression is associated with malignant progression of breast cancer. In this study, we examined the effect of CypB knockdown on PRL signaling in breast cancer cells. CypB knockdown with two independent siRNAs was shown to impair PRL-induced reporter expression in breast cancer cell line. cDNA microarray analysis was performed on these cells to assess the effect of CypB reduction, and revealed a significant decrease in PRL-induced endogenous gene expression in two breast cancer cell lines. Parallel functional assays revealed corresponding alterations of both anchorage-independent cell growth and cell motility of breast cancer cells. Our results demonstrate that CypB expression levels significantly modulate PRL-induced function in breast cancer cells ultimately resulting in enhanced levels of PRL-responsive gene expression, cell growth, and migration. Given the increasingly appreciated role of PRL in the pathogenesis of breast cancer, the actions of CypB detailed here are of biological significance.
Introduction
An increasing body of literature supports a significant role for the hormone prolactin (PRL) in the pathogenesis of human breast cancer. Epidemiologic analysis has revealed that PRL concentration is associated with an increased risk for breast cancer (Hankinson et al. 1999, Eliassen et al. 2007, Tworoger et al. 2007), particularly in post-menopausal women. These clinical observations are further substantiated by the PRL transgenic mouse model, which develops both estrogen receptor positive (ER+) and negative (ER−) mammary tumors after 12–18 months (Wennbo et al. 1997, Wennbo & Tornell 2000, Rose–Hellekant et al. 2003). Several lines of evidence have also indicated that PRL acts as both an endocrine and autocrine/paracrine progression factor for mammary carcinoma in vitro and in vivo in rodents and humans (Clevenger et al. 1995, Das & Vonderhaar 1997).
The effects of PRL are mediated by the interaction with its receptor (PRLR; Clevenger et al. 2003). Binding of PRL activates the pre-dimerized PRLR (Gadd & Clevenger 2006) and results in the activation of PRLR-associated signaling cascades such as Jak2/Stat5, Fyn/Src, Shc/Grb2, Sos/Raf/MAPK, and Tec/Nek3/Vav/Rac (Clevenger et al. 2003), resulting in the transactivation of PRL-responsive genes that include cyclin D1, CISH, and β-casein (Guyettek et al. 1979, Pezet et al. 1999, Brockman & Schuler 2005, Utama et al. 2006, Fang et al. 2008). The summation of these signaling events results in the terminal maturation of normal mammary tissues (Maus et al. 1999, Miller et al. 2007), and contributes to the PRL-induced growth (18–20) and motility (Maus et al. 1999, Miller et al. 2007) of malignant breast cells and tissues.
Previous research in our laboratory had demonstrated that some of the actions of PRL are directly mediated by the localization and function of this ligand within the nucleus (Clevenger et al. 1991, Rycyzyn et al. 2000). Yeast two-hybrid screening was used to identify binding partners involved in this process (Rycyzyn et al. 2000, Rycyzyn & Clevenger 2002) and identified that the peptidyl prolyl cis–trans isomerase, cyclophilin B (CypB), was a binding partner required for the nuclear translocation of PRL (Rycyzyn et al. 2000). Within the nucleus, the PRL–CypB complex was found to function as an inducer for the latent transcription factor Stat5, through its induction of the release of the Stat5 inhibitor, PIAS3 (Rycyzyn & Clevenger 2002). CypB has also been demonstrated to be associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer (Fang et al. 2009a).
The cyclophilins are members of a larger class of PPIase proteins widely expressed throughout the body, known as the immunophilins that are targets for the immunosuppressive agents FK506, cyclosporine A, and rapamycin (Kofron et al. 1991, Fischer et al. 1998). Although classically thought to assist in protein folding, immunophilins also serve as signaling switches via prolyl isomerization (Hunter 1998), regulating the activity of cell surface receptors transforming growth factor β (Huse et al. 1999), tyrosine kinases (Brazin et al. 2002, Zheng et al. 2008), and transcription factors such as c-Myb (Leverson & Ness 1998) and IRF4 (Mamane et al. 2000). In addition to its actions within the nucleus as a transcriptional inducer, CypB as a secreted protein is also thought to serve as a ligand for the CD147 receptor, thereby regulating the motility of cells expressing this receptor (Yurchenko et al. 2001, Yang et al. 2006, Pakula et al. 2007, Melchior et al. 2008). Indeed, a recent study has revealed that CypB present in the conditioned medium of the breast carcinoma cell line MDA-MB231 promoted chemotaxis of bone marrow-derived mesenchymal stromal cells (Lin et al. 2008). However, despite these insights into CypB action, the function of CypB during PRL-induced gene expression and action in breast cancer cells has remained undocumented.
In this study, for the first time, the effects of altering CypB levels in breast cancer cells during PRL stimulation were examined at the level of gene expression profiling, anchorage-independent growth, and cell migration. These studies demonstrate that expression levels of CypB significantly augmented PRL-induced action in breast cancer cells ultimately resulting in enhanced levels of growth and migration of breast cancer cells.
Materials and methods
Cell lines, vectors, and reagents
The human breast cancer cell lines (T47D and MCF7) from American Type Culture Collection (ATCC, Manassas, VA, USA) were maintained in DMEM (Hyclone, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), 50 μg/ml penicillin, and 50 μg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. The si-CypB cells (T47D siRNA CypB knockdown stable cells) and si-Luc cells (siRNA control cells for non-genomic targets) were described before (Fang et al. 2009a). Vectors used in this study are firefly luciferase reporter pGL4-CISH (Fang et al. 2008), pGL4-LHRE (Fang et al. 2009b), renilla luciferase reporter pGL4.73 (Promega), pGL410-SV40 (Fang et al. 2009a), pcDNA3.1 (Invitrogen), and pcDNA3.1-CypB (Rycyzyn et al. 2000). Human recombinant PRL was a gift from Dr Tony Kossiakoff (University of Chicago). Antibodies used in this study are anti-CypB (Invitrogen, 37-0600) and α-tubulin (Zymed, 32-2500, Carlsbad, CA, USA).
Knockdown of CypB
For these studies, two separate knockdown approaches were utilized for cross-validation purposes to exclude off-target effects. The first approach used the lentiviral si-CypB sequence (this siRNA sequence is against the 3′-end region of the CypB coding sequence; Robida et al. 2007). The si-CypB cells (T47D siRNA CypB knockdown stable cells) and si-Luc cells (siRNA control cells for non-genomic targets; Fang et al. 2009a) were previously used. For independent validation of the effects of siRNA knockdown of CypB, a second approach used a different siRNA (si-CypB-T, against the central region of the CypB coding sequence, Cat no.: D-001136-01-05, Dharmacon, Lafayette, CO, USA) that was transiently transfected in T47D and MCF7 cells in parallel with si-control RNA (Cat no. D-001210-01 from Dharmacon) using RNAiMAX (Invitrogen). After 48 h, cells were harvested for RNA isolation.
Luciferase assay, western blot, microarray and data analysis, RT-PCR and real-time PCR, soft agar growth, and cell motility assay
Dual luciferase assay was conducted according to Fang et al. (2008), and details are described in the figure legends. Microarray was conducted on Illumina Human Ref-6 Version 2 Expression Chip (Illumina, San Diego, CA, USA). T47D si-CypB cells were cultured in the growth medium for 3 days followed by 24 h arrest prior to PRL treatment (100 ng/ml) for 2 h. RNA isolation for microarray analysis was conducted as described in Fang et al. (2009a) and in the supplementary data, see section on supplementary data given at the end of this article. Primers for real-time PCR are listed in Supplemental Table 1, see section on supplementary data given at the end of this article. Microarray data were deposited in Gene Expression Omnibus database with accession number GSE15505 (GEO, http://www.ncbi.nlm.nih.gov/geo/). Soft agar growth and cell motility assay were conducted according to Zheng et al. (2008). The details of methods are in Supplemental Figure 1, see section on supplementary data given at the end of this article.
Statistical analysis
All experiments described here were performed at least three times. Statistical analysis was performed on GraphPad Prism 4 (GraphPad Software, La Jolla, CA, USA), and specified in the figure legends. The results are shown as the means with error bars depicting ±s.e.m. P<0.05 is considered as statistically significant.
Results
The genes regulated by PRL contribute to essential cell functions
In addition to its well-recognized regulation of genes associated with milk production and lactation (Clevenger et al. 2008), PRL has been shown to induce expression of many genes involved in cell proliferation and survival including BCL2 (Beck et al. 2002), CEBPβ (Nanbu-Wakao et al. 2000), CISH (Pezet et al. 1999, Utama et al. 2006), c-Myc (Dominguez-Caceres et al. 2004), and cyclin D1 (Brockman et al. 2002). To systematically query the effects of PRL on global gene expression in human breast cancer cells, the ER+ T47D breast cancer cell line was used for microarray analysis with or without PRL treatment. Two hours of PRL stimulation (100 ng/ml) was selected as the optimal time course length, given the preliminary data from our laboratory that had shown maximal RNA expression from the PRL-responsive CISH, c-Myc, and cyclin D1 gene loci at that time (Fang et al. 2009b). RNA isolated from these cells was subject to cDNA synthesis, labeling, and hybridization to the Illumina human cDNA chip. In order to reduce false positives, probes with measurement value below the background level (detection P value <0.01) in all hybridizations were filtered out. For subsequent statistical analysis, 17 901 probes were kept. To establish a physiologically relevant cut point for analysis of PRL-induced gene expression, the RNA levels of the PRL-induced c-Myc gene (Dominguez-Caceres et al. 2004) were assessed. While microarray analysis demonstrated a 1.4-fold induction of c-Myc expression, real-time PCR revealed that c-Myc was induced 2.4-fold following PRL treatment. Similar phenomenon was also observed for BCL3, CEBPβ, CISH, and cyclin D1. In addition, as documented by the manufacturer, the Illumina cDNA array is sensitive to detect as little as 1.3-fold change in either direction (up or down). Given this, the criterion to filter the microarray data was set to 1.3-fold (up- or down-regulation) for subsequent analysis.
To simultaneously indicate the size of biological effects (log2 fold change as X-axis) and the statistical significance (−log10 P value as Y-axis) at global level, a volcano plot was used to compare the difference of gene expression between two groups (Fig. 1A). As seen in volcano map, the red dots represent the selected differentially expressed genes significantly regulated by PRL treatment (P<0.05, false discovery rate (FDR)<0.05, fold change ≥1.3 up or down). Two-dimensional hierarchical clustering was applied to these filtered probes to generate a global overview of gene expression map in the form of a heat map. Heat map analysis showed a remarkable difference in gene expression pattern between PRL treatment and non-PRL treatment groups. It also indicated, in a global view, highly consistent results among the triplicates in each group (Fig. 1B). Of 28 significantly differentially expressed genes, 19 were up-regulated and 9 were down-regulated. Many genes listed therein have not been reported upon before as PRL-responsive genes in breast cancer, including BCL3 and BCL6 (Fig. 1B). Subsequent real-time PCR analysis confirmed the PRL-regulated mRNA up-regulation of BCL3 expression and down-regulation of BCL6 expression (Fig. 1C and D). BCL3 plays an important role in cell proliferation (Na et al. 1999), and sequence analysis using TFsearch program revealed ten Stat-binding sites within the 10 kb region including promoter/enhancer upstream of transcriptional start point, as well as within the exons, introns, and the 3′-untranslated region. BCL6 is a zinc finger transcription factor and found to be expressed in breast cancer cells (Logarajah et al. 2003), and overexpression of Stat5 has been found to repress BCL6 expression (Scheeren et al. 2005, Walker et al. 2007). Interestingly, the regulation of menin expression via PRL regulation of BCL6 regulates pancreatic β-cell growth in pregnant mice; loss of this process may contribute to gestational diabetes mellitus (Karnik et al. 2007).
Figure 1.
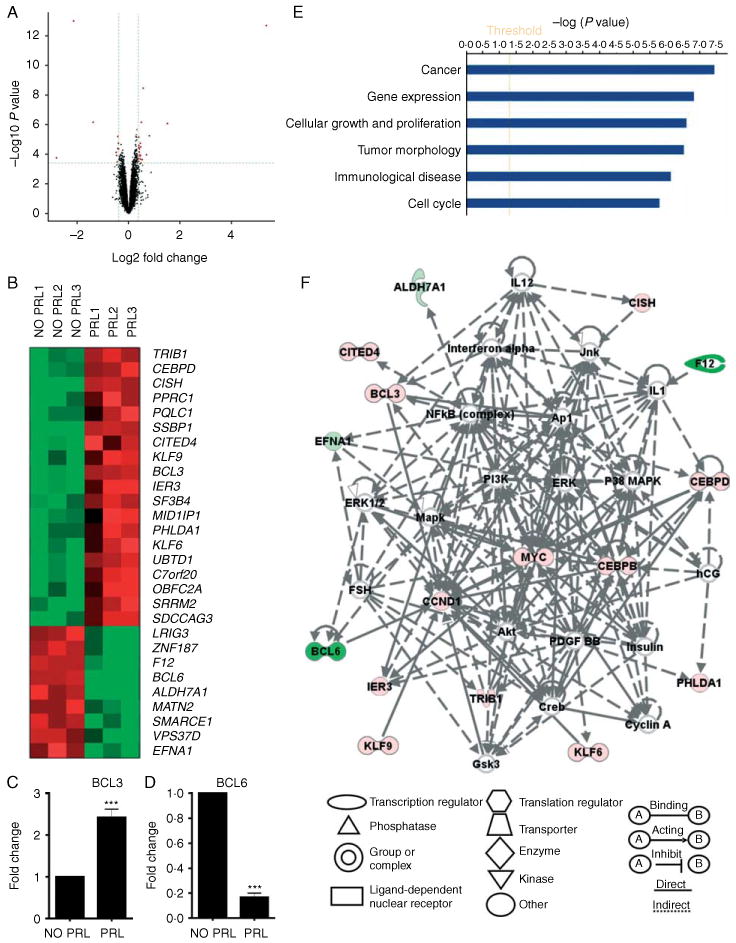
Microarray analysis demonstrates the effects of PRL on global gene expression. (A) Volcano map demonstrates the relationship between the observed fold change in gene expression and the P value significance of such changes in PRL-treated cells. The dotted lines represent the P value and fold change cutoffs. The red dots represent the selected genes filtered by criteria (fold change ≥1.3 up or down, P<0.05, FDR<0.05). (B) Heat map analysis reveals a global view of genes up- and down-regulated in PRL-treated cells. (C and D) Real-time PCR validates microarray results for BCL3 (C) and BCL6 (D). Statistical analysis was performed using Student's t-test. (E) The biological functional categories were obtained from the molecular and cellular function in the IPA database. (F) The top interaction networks generated using IPA analysis included genes associated with ‘cancer, hematological disease, and cell cycle’. The color indicates up-regulation (red), down-regulation (green), and complexes (gray) of genes. ***P<0.001.
Ingenuity pathway analysis (IPA) has been widely used for analysis of gene expression, proteomics, and metabolic data to elucidate tumor progression, bio-marker discovery, and drug discovery (Ganter & Giroux 2008). To assess the global effects of PRL on gene expression, IPA was used to systematically visualize the relationships of genes regulated by PRL. The transcripts from microarray results were filtered, and 28 transcripts were inputted into the IPA database (see Materials and methods). IPA was used to overlay the PRL-regulated genes onto the global networks developed from the information contained in the Ingenuity Pathways Knowledge Base. The genes regulated by PRL were categorized into different biological functions, with the ‘cancer’ biological function shown as the most significant category (Fig. 1E). When the 28 transcripts were mapped into the IPA Knowledge Base, the most significant modulated interaction network was associated with those genes implicated in the pathogenesis of ‘cancer, hematological disease, and cell cycle’ (Fig. 1F). This interaction network revealed that many of these PRL-regulated genes interact with each other, and in doing so may enhance PRL-induced tumorigenesis. Network analysis of these cancer-associated genes reveals a central role for many gene products involved in PRL signaling, such as CISH, MAPK, CEBPβ, ERK, and c-Myc.
Ectopic expression of CypB enhanced Stat5-responsive reporter expression
PRL action is mediated in the cells through distinct signaling pathways including the Jak2/Stat5 signaling pathway. In this pathway, the intranuclear PRL–CypB complex acts as a Stat5 transcriptional inducer to regulate gene expression (Rycyzyn et al. 2000, Rycyzyn & Clevenger 2002). To characterize the role of CypB during PRL-induced expression of Stat5-responsive genes in breast cancer cells, reporter assay was used to test the effect of CypB ectopic expression on such promoters in T47D cells. This cell line was selected for our initial screening studies, given its regulatory robust expression of both the PRLR and CypB (data not shown). For these studies, the pGL4-CISH luciferase reporter was used for PRL-induced reporter assay (Fang et al. 2008); this expression construct contained the 1 kb CISH promoter region-fused upstream of firefly luciferase reporter gene. These results showed that ectopic expression of CypB itself had no effect on luciferase expression of the pGL4-CISH reporter in the absence of PRL (Fig. 2A). In the presence of PRL, CypB enhanced PRL-induced luciferase expression of pGL4-CISH reporter (Fig. 2A). As a parallel control experiment, we also co-transfected pGL4-CISH with the pcDNA3.1-CypB-PPIase construct (a CypB mutant construct lacking activity of peptidyl prolyl cis–trans isomerase (PPIase or PPI)) in T47D cells. Results showed that compared to non-PRL treatment, PRL-induced luciferase fold change is 22.5±3.3 and 22.0±3.0 respectively in pcDNA3.1 and pcDNA3.1-CypB-PPIase transfectants, suggesting that the CypB-mediated enhancement of PRL-induced luciferase expression was dependent upon PPI activity. These results confirmed the effects of CypB on PRL-induced gene expression (Rycyzyn & Clevenger 2002) in T47D breast cancer cells.
Figure 2.
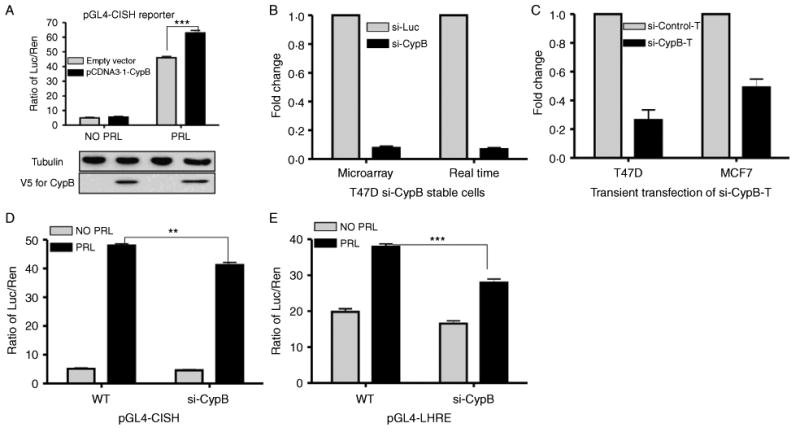
The effects of CypB overexpression and CypB knockdown on a Stat5-responsive reporter. (A) Luciferase assay using pGL4-CISH reporter. Cells were transfected with pGL4-CISH reporter, the renilla luciferase control (pGL4.73), and pcDNA3.1-CypB expression vector. Transfectants were cultured in the minimal defined medium for 24 h, followed by 24 h of PRL stimulation prior to luminescence assay. (B and C) CypB knockdown in T47D cells confirmed by real-time PCR and microarray (B), and transient transfection (C). (D and E) Luciferase assay using pGL4-CISH (D) and pGL4-LHRE (E). T47D parental cells (wt) or si-CypB cells were co-transfected with 100 ng pGL4-CISH (D) or pGL4-LHRE (E), along with 2 ng renilla luciferase control (pGL4.73) and 400 ng pcDNA3.1-CypB expression vector, and maintained in the FBS-containing growth medium overnight. Transfectants were then starved in the FBS-free minimal defined medium for 24 h, followed by 24 h of PRL (10 ng/ml for pGL4-CISH and 100 ng/ml for pGL4-LHRE) stimulation prior to luminescence assay. Statistical analysis was performed using two-way ANOVA. **P<0.01; ***P<0.001.
Knockdown of CypB impaired the expression of Stat5-responsive reporters
To complement the overexpression-based studies, the effects of CypB knockdown on the PRL/Stat5 signaling pathway were assessed in T47D transfected stably with siRNA against a control (luciferase; termed si-Luc) or CypB (termed si-CypB; Fang et al. 2009a). Results from real-time PCR and microarray confirmed a significant knockdown of CypB (Fig. 2B). A second different sequence directed against CypB (termed as si-CypB-T, and control as si-control-T) was transiently transfected into T47D and MCF7 cells to validate that the effects noted in si-CypB stable transfectants were not due to off-target action (Fang et al. 2009a). The effect of transient transfection of this siRNA also resulted in significantly reduced CypB levels (Fig. 2C). This second siRNA was also used later in gene profile validation studies (see Fig. 3). Two PRL-induced, Stat5-responsive promoter reporter constructs, termed pGL4-LHRE (a synthetic construct containing six Stat5-responsive elements) and pGL4-CISH, were then introduced into the stable T47D si-CypB cells to evaluate the effects of CypB knockdown on PRL signaling. Results (Fig. 2D) showed that, compared to wild-type cells, the luciferase expression of pGL4-CISH in si-CypB cells had little change in the absence of PRL. In the presence of PRL, the luciferase expression of pGL4-CISH in si-CypB cells decreased significantly compared to wild-type cells. Similar results were observed for pGL4-LHRE (Fig. 2E). These findings revealed that CypB knockdown decreased PRL-induced expression luciferase in si-CypB cells, further demonstrating the potentiating effects of CypB on PRL-mediated gene expression.
Figure 3.
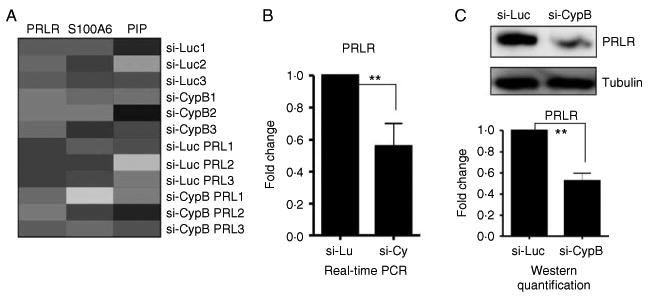
The characterization of PRL-related genes affected by CypB knockdown. (A) Heat map showed down-regulation of PRLR, S100A6, and PIP in si-CypB cells compared to si-Luc control cells. (B and C) Real-time PCR (B) and western blot (C) confirmed the PRLR down-regulation in si-CypB cells. Statistical analysis was performed using Student's t-test. **P<0.01.
Knockdown of CypB impaired the PRLR and PRL-inducible endogenous gene expression
To determine the effects of CypB knockdown on PRL-regulated gene expression, microarray analysis was conducted using si-CypB cells treated with or without PRL. To characterize the effect of CypB on PRL signaling in this analysis, ‘PRL’ was used as the key word in the ‘Gene Ontology (GO)’ program to search for PRL-related genes significantly regulated by CypB knockdown. Results revealed that the expression of the PRLR, S100A6, and the PRL-inducible protein (PIP) was significantly altered in si-CypB cells compared to si-Luc cells (Fig. 3A), a finding further confirmed by both real-time PCR and western blot analysis for PRLR gene (Fig. 3B and C).
Microarray analysis and subsequent validation studies suggested that the panel of the PRL-induced genes was regulated differently by PRL in si-Luc and si-CypB cells. Real-time PCR confirmed that CEBPβ, c-Myc, and cyclin D1 mRNA expression was significantly decreased in si-CypB stable cells compared to that of si-Luc stable cells, while little effect on BCL3 and CISH was observed (Fig. 4A–E). Real-time PCR results also showed that the PRL-repressed BCL6 mRNA expression was significantly decreased in si-CypB cells compared to that of si-Luc cells (Fig. 4F). The effect of transient transfection of a second differing siRNA (used in Fig. 2E, termed as si-CypB-T, and control as si-control-T) into both T47D and MCF7 cells was also tested in the context of PRL to further rule out off-target effects. The PRL-induced CEBPβ and cyclin D1 mRNA expression was significantly decreased in cells with CypB knockdown (Fig. 4G–L). While si-CypB-T cells resulted in a decrease in c-Myc and BCL6 levels, these results did not achieve statistical significance compared to results from the si-CypB stable T47D cells. This may be due to cell line variance (high PRLR level in T47D and modest PRLR in MCF7) and difference in knockdown efficiency (stable versus transient). Taken together, these findings indicate that reductions in CypB levels significantly impacted on PRL-induced gene expression at both global and locus-specific levels.
Figure 4.
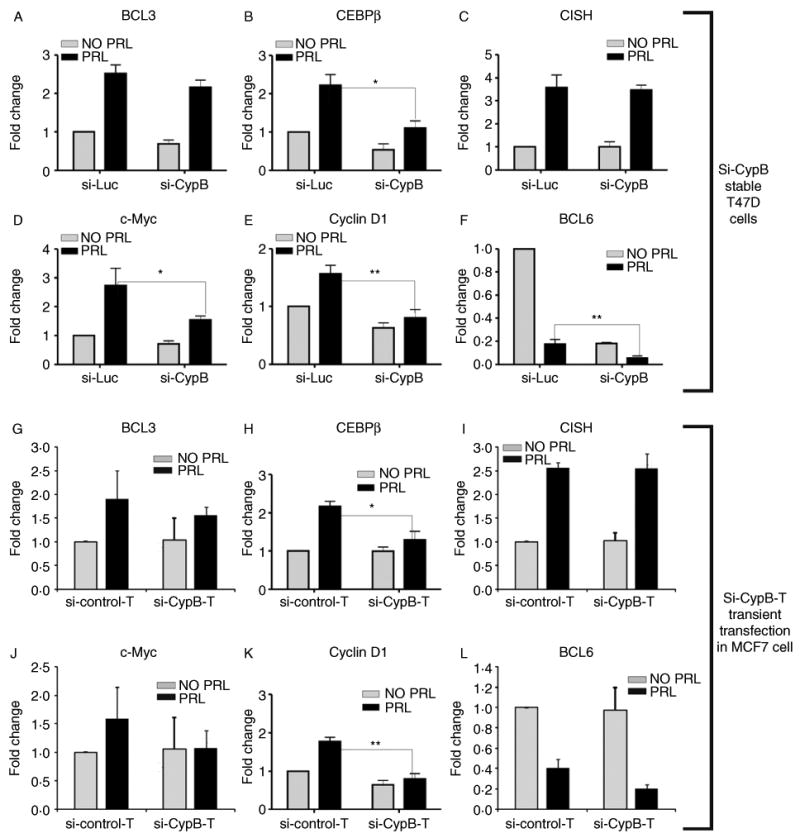
Real-time PCR validated the impairment of PRL-induced gene expression by CypB knockdown in T47D si-CypB stable cells (A–F) and in MCF7 cells (G–H) with transient transfect of si-CypB-T. The y-axis label ‘fold change’ is defined in the Materials and methods section. Statistical analysis was performed using Student's t-test. *P<0.05; **P<0.01.
CypB knockdown impaired the PRL-induced colony growth on the soft agar
Given that CypB knockdown significantly modulated the expression of the PRL-regulated CEBPβ, c-Myc, cyclin D1, and BCL6 genes in T47D cells, the effect of CypB knockdown on PRL-induced anchorage-dependent cell growth was tested using soft agar assay. These analyses revealed that colony number (counted by single colonies) and size (determined by the colony area) of si-Luc cells were increased in the presence of PRL. Colony number and size of si-CypB cells were significantly attenuated, showing that CypB is involved in the anchorage-independent cell growth (Fig. 5A–F).
Figure 5.
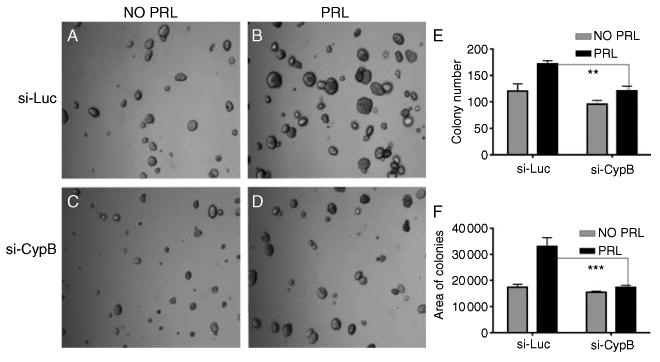
CypB knockdown results in the decreased PRL-induced soft agar growth and cell motility. Cells were grown on soft agar for 2 weeks, and the pictures were taken under phase contrast microscopy. (A) si-Luc without PRL treatment, (B) si-Luc with PRL treatment (200 ng/ml), (C) si-CypB without PRL treatment, (D) si-CypB with PRL treatment (200 ng/ml), (E) colony number on the soft agar, and (F) the total colony area on the soft agar. **P<0.01; ***P<0.001.
CypB knockdown impaired the PRL-induced cell motility
PRL has also been shown to stimulate T47D cell motility (Maus et al. 1999, Miller et al. 2007). To investigate whether CypB knockdown affected PRL-induced cell motility, Boyden chamber cell migration assay was conducted using PRL as a chemoattractant. These data revealed that PRL stimulated si-Luc cell motility, as previously described (Miller et al. 2007). The PRL-induced motility of the si-CypB cells was significantly decreased, revealing a contribution of CypB to PRL-mediated cell motility (Fig. 6). In our previous publication (Fang et al. 2009a), we have shown that knockdown of CypB impaired the FBS-inducible and estradiol-inducible cell migration, suggesting that the impairment of cell motility in si-CypB cells is due to global effects on gene expression involved in motility regulation. It is also noted that CypB knockdown decreased migration and colony formation even without PRL. This input may be due to the inhibition of the actions of autocrine PRL produced by these cells. In the presence of PRL, CypB knockdown impaired PRL-induced migration and colony formation (Figs 5 and 6).
Figure 6.
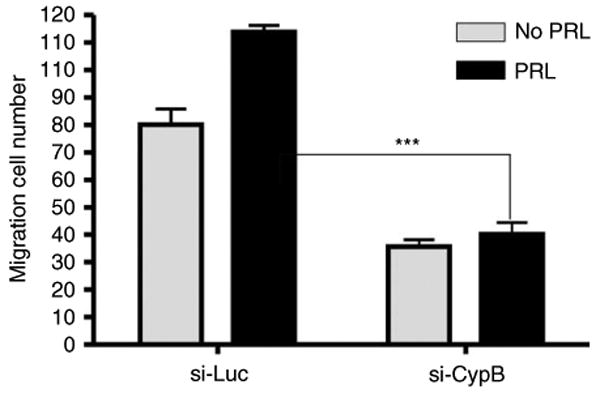
CypB knockdown results in the decreased PRL-induced cell motility. Cell motility was assayed using Boyden chamber migration assay. The inserts were coated with collagen I overnight. T47D cells were arrested in the FBS-free medium and placed in the inserts. Cells were cultured for 20 h, and the migrated cells were counted under a microscope. Statistical analysis was performed using two-way ANOVA. ***P<0.001.
Discussion
Given the increasingly appreciated role of PRL in the pathogenesis of breast cancer, surprisingly little is known in regards to the regulation of gene expression in such cells. In this context, this study sought to examine the effects of both PRL stimulation and CypB knockdown on PRL-induced gene expression by validated gene profiling, and relate these observations to PRL-induced growth and motility. CypB has been shown previously to play an important role in cell survival (Kim et al. 2008), cancer progression, cell proliferation, and cell growth (Fang et al. 2009a). Our results indicate that the absence of CypB significantly impairs the patterns of gene expression induced by PRL in breast cancer cells.
Gene profiling analysis performed here and elsewhere (Fang et al. 2009a) showed that CypB knockdown regulated PRL-related genes that included the PRLR. It is important to note that given widely varying effects of CypB knockdown on PRL-induced genes (see Fig. 3), we believe that the effects of CypB extend beyond that of a mere reduction in PRLR levels; the PIP mRNA (PIP or GCPD15) protein was noted to be increased twofold in si-CypB cells. PIP was first cloned from T47D cells and was induced by 5 days of PRL and/or GH treatment (Murphy et al. 1987). Higher mRNA levels of PIP have a high correlation with the expression of ERα, progesterone receptor, and low-grade tumors (Clark et al. 1999); mRNA for the PRL regulatory element binding protein (PREB, also known as SEC12 or MGC3467) mRNA was up-regulated by PRL only in si-CypB cells (not in si-Luc cells). PREB acts as a transcriptional activator on PRL promoter region to regulate PRL expression (Fliss et al. 1999). S100A6 mRNA is a S100 calcium-binding protein, also known as PRLR-associated protein, and is down-regulated in si-CypB cells (Fang et al. 2009a). S100A6 has been observed in PRLR immunoprecipitates (Murphy et al. 1988). S100A6 is up-regulated in breast cancer cells and tissues (Maelandsmo et al. 1997), and knockdown of this gene appears to decrease both cell proliferation and motility (Breen & Tang 2003). The c-Myb proto-oncogene mRNA was down-regulated in PRL-stimulated si-CypB cells (P<0.05). c-Myb is associated with cell differentiation and proliferation (Weston 1999). c-Myb protein level is increased in in situ and invasive breast cancers (McHale et al. 2008). The c-Myb mRNA levels were regulated by cyclophilin isomerase activity (Leverson & Ness 1998). c-Myb is a Stat5a co-activator during PRL/Stat5a-driven gene expression (Fang et al. 2009b). The CEBPβ transcription factor mRNA was also down-regulated in si-CypB cells. CEBPβ is bZIP transcription factor acting as a Stat5 co-activator and is also a Stat5-regulated gene (Nanbu-Wakao et al. 2000). The PRLR promoter has CEBPβ-binding sites, and overexpression of CEBPβ up-regulates PRLR expression (Hu et al. 1998). This suggests that CypB has a profound impact on PRL-related gene expression.
Our previous work using a matched, progressive-based breast tissue microarray (Fang et al. 2009a) showed that CypB levels were increased in malignant breast epithelium, suggesting that the up-regulation of CypB in breast cancer could significantly modulate the biology of this disease. siRNA-mediated knockdown of CypB was found by gene profiling to significantly regulate genes related to cell proliferation, motility, and receptors (Fang et al. 2009a). Since CypB mediates PRL-responsive gene expression with profound effects, we hypothesized that CypB is multifaceted, serving as an activator of receptor expression (i.e. PRLR), a chaperone for ligand (i.e. PRL), and an inducer for the transcriptional factor (i.e. Stat5; Rycyzyn & Clevenger 2002), all of which may collectively contribute to the regulation of the PRL-responsive genes. Given the significant effect of CypB knockdown on the PRL-induced anchorage-independent cell growth and cell motility, these results would suggest that the molecular actions of CypB within the cell during PRL-induced signaling impact significantly on the biology of breast cancer cells.
The precise role of Stat5 in the pathogenesis of breast cancer remains to be fully clarified. While data from mouse models of mammary cancer clearly indicate that loss of Stat5 function results in a significant delay in the pathogenesis of malignancy at this site (Ren et al. 2002), data from human tissues have shown that phosphorylated/nuclear Stat5 is associated with a favorable histopathology (Sultan et al. 2005). Although CypB could influence Stat5 function both through its indirect actions at the cell surface and by its direct interaction with Stat5, it is interesting to note that many of the gene transcripts influenced by reduction in CypB expression in this manuscript demonstrate multiple Stat5-binding sites within their proximal promoter regions. As such, these findings would suggest that inhibitors that target both PRL-induced signals and CypB may have a synergistic potential at the level of Stat5 function in the treatment of breast cancer.
Supplementary Material
Acknowledgments
We thank Dr Hengli Tang at Florida State University for providing si-Luc and si-CypB pHIV-7/Puro vectors. We thank Drs Simon Lin and Pan Du in the Biostatistics Core for microarray data analysis.
Funding: This work was supported by National Institute of Health (RO1CA-102682), the Avon and Lynn Sage Foundations, and the Zell Scholar's Fund.
Footnotes
Supplementary data: This is linked to the online version of the paper at http://dx.doi.org/10.1677/JME-09-0140.
Declaration of interest: The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Beck MT, Peirce SK, Chen WY. Regulation of bcl-2 gene expression in human breast cancer cells by prolactin and its antagonist, hPRL-G129R. Oncogene. 2002;21:5047–5055. doi: 10.1038/sj.onc.1205637. [DOI] [PubMed] [Google Scholar]
- Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. PNAS. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen EC, Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. Journal of Cellular Biochemistry. 2003;88:848–854. doi: 10.1002/jcb.10398. [DOI] [PubMed] [Google Scholar]
- Brockman JL, Schuler LA. Prolactin signals via Stat5 and Oct-1 to the proximal cyclin D1 promoter. Molecular and Cellular Endocrinology. 2005;239:45–53. doi: 10.1016/j.mce.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Molecular Endocrinology. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- Clark JW, Snell L, Shiu RP, Orr FW, Maitre N, Vary CP, Cole DJ, Watson PH. The potential role for prolactin-inducible protein (PIP) as a marker of human breast cancer micrometastasis. British Journal of Cancer. 1999;81:1002–1008. doi: 10.1038/sj.bjc.6690799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV, Altmann SW, Prystowsky MB. Requirement of nuclear prolactin for interleukin-2 – stimulated proliferation of T lymphocytes. Science. 1991;253:77–79. doi: 10.1126/science.2063207. [DOI] [PubMed] [Google Scholar]
- Clevenger CV, Chang WP, Ngo W, Pasha TL, Montone KT, Tomaszewski JE. Expression of prolactin and prolactin receptor in human breast carcinoma. Evidence for an autocrine/paracrine loop. American Journal of Pathology. 1995;146:695–705. [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV, Furth PA, Hankinson SE, Schuler LA. The role of prolactin in mammary carcinoma. Endocrine Reviews. 2003;24:1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV, Zheng J, Jablonski EM, Galbaugh TL, Fang F. From bench to bedside: future potential for the translation of prolactin inhibitors as breast cancer therapeutics. Journal of Mammary Gland Biology and Neoplasia. 2008;13:147–156. doi: 10.1007/s10911-008-9074-8. [DOI] [PubMed] [Google Scholar]
- Das R, Vonderhaar BK. Prolactin as a mitogen in mammary cells. Journal of Mammary Gland Biology and Neoplasia. 1997;2:29–39. doi: 10.1023/a:1026369412612. [DOI] [PubMed] [Google Scholar]
- Dominguez-Caceres MA, Garcia-Martinez JM, Calcabrini A, Gonzalez L, Porque PG, Leon J, Martin-Perez J. Prolactin induces c-Myc expression and cell survival through activation of Src/Akt pathway in lymphoid cells. Oncogene. 2004;23:7378–7390. doi: 10.1038/sj.onc.1208002. [DOI] [PubMed] [Google Scholar]
- Eliassen AH, Tworoger SS, Hankinson SE. Reproductive factors and family history of breast cancer in relation to plasma prolactin levels in premenopausal and postmenopausal women. International Journal of Cancer. 2007;120:1536–1541. doi: 10.1002/ijc.22482. [DOI] [PubMed] [Google Scholar]
- Fang F, Antico G, Zheng J, Clevenger CV. Quantification of PRL/Stat5 signaling with a novel pGL4-CISH reporter. BMC Biotechnology. 2008;8:11. doi: 10.1186/1472-6750-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Flegler AJ, Du P, Lin S, Clevenger CV. Expression of cyclophilin B is associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer. American Journal of Pathology. 2009a;174:297–308. doi: 10.2353/ajpath.2009.080753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Rycyzyn MA, Clevenger CV. Role of c-Myb during PRL-induced Stat5a signaling in breast cancer cells. Endocrinology. 2009b;150:1597–1606. doi: 10.1210/en.2008-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Tradler T, Zarnt T. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Letters. 1998;426:17–20. doi: 10.1016/s0014-5793(98)00242-7. [DOI] [PubMed] [Google Scholar]
- Fliss MS, Hinkle PM, Bancroft C. Expression cloning and characterization of PREB (prolactin regulatory element binding), a novel WD motif DNA-binding protein with a capacity to regulate prolactin promoter activity. Molecular Endocrinology. 1999;13:644–657. doi: 10.1210/mend.13.4.0260. [DOI] [PubMed] [Google Scholar]
- Gadd SL, Clevenger CV. Ligand-independent dimerization of the human prolactin receptor isoforms: functional implications. Molecular Endocrinology. 2006;20:2734–2746. doi: 10.1210/me.2006-0114. [DOI] [PubMed] [Google Scholar]
- Ganter B, Giroux CN. Emerging applications of network and pathway analysis in drug discovery and development. Current Opinion in Drug Discovery and Development. 2008;11:86–94. [PubMed] [Google Scholar]
- Guyette WA, Matusik RJ, Rosen JM. Prolactin-mediated transcriptional and post-transcriptional control of casein gene expression. Cell. 1979;17:1013–1023. doi: 10.1016/0092-8674(79)90340-4. [DOI] [PubMed] [Google Scholar]
- Hankinson SE, Willett WC, Michaud DS, Manson JE, Colditz GA, Longcope C, Rosner B, Speizer FE. Plasma prolactin levels and subsequent risk of breast cancer in postmenopausal women. Journal of the National Cancer Institute. 1999;91:629–634. doi: 10.1093/jnci/91.7.629. [DOI] [PubMed] [Google Scholar]
- Hu ZZ, Zhuang L, Meng J, Dufau ML. Transcriptional regulation of the generic promoter III of the rat prolactin receptor gene by C/EBPbeta and Sp1. Journal of Biological Chemistry. 1998;273:26225–26235. doi: 10.1074/jbc.273.40.26225. [DOI] [PubMed] [Google Scholar]
- Hunter T. Prolyl isomerases and nuclear function. Cell. 1998;92:141–143. doi: 10.1016/s0092-8674(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Huse M, Chen YG, Massague J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- Karnik SK, Chen H, McLean GW, Heit JJ, Gu X, Zhang AY, Fontaine M, Yen MH, Kim SK. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science. 2007;318:806–809. doi: 10.1126/science.1146812. [DOI] [PubMed] [Google Scholar]
- Kim J, Choi TG, Ding Y, Kim Y, Ha KS, Lee KH, Kang I, Ha J, Kaufman RJ, Lee J, et al. Overexpressed cyclophilin B suppresses apoptosis associated with ROS and Ca2+ homeostasis after ER stress. Journal of Cell Science. 2008;121:3636–3648. doi: 10.1242/jcs.028654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis–trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- Leverson JD, Ness SA. Point mutations in v-Myb disrupt a cyclophilin-catalyzed negative regulatory mechanism. Molecular Cell. 1998;1:203–211. doi: 10.1016/s1097-2765(00)80021-0. [DOI] [PubMed] [Google Scholar]
- Lin SY, Yang J, Everett AD, Clevenger CV, Koneru M, Mishra PJ, Kamen B, Banerjee D, Glod J. The isolation of novel mesenchymal stromal cell chemotactic factors from the conditioned medium of tumor cells. Experimental Cell Research. 2008;314:3107–3117. doi: 10.1016/j.yexcr.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L, Venkitaraman A, Wagner S. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 2003;22:5572–5578. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- Maelandsmo GM, Florenes VA, Mellingsaeter T, Hovig E, Kerbel RS, Fodstad O. Differential expression patterns of S100A2, S100A4 and S100A6 during progression of human malignant melanoma. International Journal of Cancer. 1997;74:464–469. doi: 10.1002/(sici)1097-0215(19970822)74:4<464::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Sharma S, Petropoulos L, Lin R, Hiscott J. Posttranslational regulation of IRF-4 activity by the immunophilin FKBP52. Immunity. 2000;12:129–140. doi: 10.1016/s1074-7613(00)80166-1. [DOI] [PubMed] [Google Scholar]
- Maus MV, Reilly SC, Clevenger CV. Prolactin as a chemoattractant for human breast carcinoma. Endocrinology. 1999;140:5447–5450. doi: 10.1210/endo.140.11.7245. [DOI] [PubMed] [Google Scholar]
- McHale K, Tomaszewski JE, Puthiyaveettil R, Livolsi VA, Clevenger CV. Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Modern Pathology. 2008;21:565–571. doi: 10.1038/modpathol.2008.7. [DOI] [PubMed] [Google Scholar]
- Melchior A, Denys A, Deligny A, Mazurier J, Allain F. Cyclophilin B induces integrin-mediated cell adhesion by a mechanism involving CD98-dependent activation of protein kinase C-delta and p44/42 mitogen-activated protein kinases. Experimental Cell Research. 2008;314:616–628. doi: 10.1016/j.yexcr.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Miller SL, Antico G, Raghunath PN, Tomaszewski JE, Clevenger CV. Nek3 kinase regulates prolactin-mediated cytoskeletal reorganization and motility of breast cancer cells. Oncogene. 2007;26:4668–4678. doi: 10.1038/sj.onc.1210264. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Tsuyuki D, Myal Y, Shiu RP. Isolation and sequencing of a cDNA clone for a prolactin-inducible protein (PIP). Regulation of PIP gene expression in the human breast cancer cell line, T-47D. Journal of Biological Chemistry. 1987;262:15236–15241. [PubMed] [Google Scholar]
- Murphy LC, Murphy LJ, Tsuyuki D, Duckworth ML, Shiu RP. Cloning and characterization of a cDNA encoding a highly conserved, putative calcium binding protein, identified by an anti-prolactin receptor antiserum. Journal of Biological Chemistry. 1988;263:2397–2401. [PubMed] [Google Scholar]
- Na SY, Choi JE, Kim HJ, Jhun BH, Lee YC, Lee JW. Bcl3, an IkappaB protein, stimulates activating protein-1 transactivation and cellular proliferation. Journal of Biological Chemistry. 1999;274:28491–28496. doi: 10.1074/jbc.274.40.28491. [DOI] [PubMed] [Google Scholar]
- Nanbu-Wakao R, Fujitani Y, Masuho Y, Muramatu M, Wakao H. Prolactin enhances CCAAT enhancer-binding protein-beta (C/EBP beta) and peroxisome proliferator-activated receptor gamma (PPAR gamma) messenger RNA expression and stimulates adipogenic conversion of NIH-3T3 cells. Molecular Endocrinology. 2000;14:307–316. doi: 10.1210/mend.14.2.0420. [DOI] [PubMed] [Google Scholar]
- Pakula R, Melchior A, Denys A, Vanpouille C, Mazurier J, Allain F. Syndecan-1/CD147 association is essential for cyclophilin B-induced activation of p44/42 mitogen-activated protein kinases and promotion of cell adhesion and chemotaxis. Glycobiology. 2007;17:492–503. doi: 10.1093/glycob/cwm009. [DOI] [PubMed] [Google Scholar]
- Pezet A, Favre H, Kelly PA, Edery M. Inhibition and restoration of prolactin signal transduction by suppressors of cytokine signaling. Journal of Biological Chemistry. 1999;274:24497–24502. doi: 10.1074/jbc.274.35.24497. [DOI] [PubMed] [Google Scholar]
- Ren S, Cai HR, Li M, Furth PA. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- Robida JM, Nelson HB, Liu Z, Tang H. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. Journal of Virology. 2007;81:5829–5840. doi: 10.1128/JVI.02524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-Hellekant TA, Arendt LM, Schroeder MD, Gilchrist K, Sandgren EP, Schuler LA. Prolactin induces ERalpha-positive and ERalpha-negative mammary cancer in transgenic mice. Oncogene. 2003;22:4664–4674. doi: 10.1038/sj.onc.1206619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV. The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. PNAS. 2002;99:6790–6795. doi: 10.1073/pnas.092160699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycyzyn MA, Reilly SC, O'Malley K, Clevenger CV. Role of cyclophilin B in prolactin signal transduction and nuclear retrotranslocation. Molecular Endocrinology. 2000;14:1175–1186. doi: 10.1210/mend.14.8.0508. [DOI] [PubMed] [Google Scholar]
- Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, Wijnands E, Gimeno R, Vyth-Dreese FA, Blom B, Spits H. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nature Immunology. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–760. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- Tworoger SS, Eliassen AH, Sluss P, Hankinson SE. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. Journal of Clinical Oncology. 2007;25:1482–1488. doi: 10.1200/JCO.2006.07.6356. [DOI] [PubMed] [Google Scholar]
- Utama FE, LeBaron MJ, Neilson LM, Sultan AS, Parlow AF, Wagner KU, Rui H. Human prolactin receptors are insensitive to mouse prolactin: implications for xenotransplant modeling of human breast cancer in mice. Journal of Endocrinology. 2006;188:589–601. doi: 10.1677/joe.1.06560. [DOI] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- Wennbo H, Tornell J. The role of prolactin and growth hormone in breast cancer. Oncogene. 2000;19:1072–1076. doi: 10.1038/sj.onc.1203349. [DOI] [PubMed] [Google Scholar]
- Wennbo H, Kindblom J, Isaksson OG, Tornell J. Transgenic mice overexpressing the prolactin gene develop dramatic enlargement of the prostate gland. Endocrinology. 1997;138:4410–4415. doi: 10.1210/endo.138.10.5461. [DOI] [PubMed] [Google Scholar]
- Weston K. Reassessing the role of C-MYB in tumorigenesis. Oncogene. 1999;18:3034–3038. doi: 10.1038/sj.onc.1202728. [DOI] [PubMed] [Google Scholar]
- Yang JM, O'Neill P, Jin W, Foty R, Medina DJ, Xu Z, Lomas M, Arndt GM, Tang Y, Nakada M, et al. Extracellular matrix metalloproteinase inducer (CD147) confers resistance of breast cancer cells to Anoikis through inhibition of Bim. Journal of Biological Chemistry. 2006;281:9719–9727. doi: 10.1074/jbc.M508421200. [DOI] [PubMed] [Google Scholar]
- Yurchenko V, O'Connor M, Dai WW, Guo H, Toole B, Sherry B, Bukrinsky M. CD147 is a signaling receptor for cyclophilin B. Biochemical and Biophysical Research Communications. 2001;288:786–788. doi: 10.1006/bbrc.2001.5847. [DOI] [PubMed] [Google Scholar]
- Zheng J, Koblinski JE, Dutson LV, Feeney YB, Clevenger CV. Prolyl isomerase cyclophilin A regulation of Janus-activated kinase 2 and the progression of human breast cancer. Cancer Research. 2008;68:7769–7778. doi: 10.1158/0008-5472.CAN-08-0639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


