Abstract
In virtually every cell, neutral lipids are stored in cytoplasmic structures called lipid droplets (LDs) and also referred to as lipid bodies or lipid particles. We developed a rapid high-throughput assay based on the recovery of quenched BODIPY-fluorescence that allows to quantify lipid droplets. The method was validated by monitoring lipid droplet turnover during growth of a yeast culture and by screening a group of strains deleted in genes known to be involved in lipid metabolism. In both tests, the fluorimetric assay showed high sensitivity and good agreement with previously reported data using microscopy. We used this method for high-throughput identification of protein phosphatases involved in lipid droplet metabolism. From 65 yeast knockout strains encoding protein phosphatases and its regulatory subunits, 13 strains revealed to have abnormal levels of lipid droplets, 10 of them having high lipid droplet content. Strains deleted for type I protein phosphatases and related regulators (ppz2, gac1, bni4), type 2A phosphatase and its related regulator (pph21 and sap185), type 2C protein phosphatases (ptc1, ptc4, ptc7) and dual phosphatases (pps1, msg5) were catalogued as high-lipid droplet content strains. Only reg1, a targeting subunit of the type 1 phosphatase Glc7p, and members of the nutrient-sensitive TOR pathway (sit4 and the regulatory subunit sap190) were catalogued as low-lipid droplet content strains, which were studied further. We show that Snf1, the homologue of the mammalian AMP-activated kinase, is constitutively phosphorylated (hyperactive) in sit4 and sap190 strains leading to a reduction of acetyl-CoA carboxylase activity. In conclusion, our fast and highly sensitive method permitted us to catalogue protein phosphatases involved in the regulation of LD metabolism and present evidence indicating that the TOR pathway and the SNF1/AMPK pathway are connected through the Sit4p-Sap190p pair in the control of lipid droplet biogenesis.
Introduction
In virtually every cell, neutral lipids are stored in cytoplasmic structures called lipid droplets (LDs) (rev. [1]). In the budding yeast S. cerevisiae, these structures contain a neutral lipid core composed of triacylglycerols (TAGs) and sterol esters (SEs) in a 1∶1 ratio [2]. This core is delimited by a phospholipid monolayer associated with a diversity of proteins, which have structural functions and also show enzymatic activity [3], [4]. LDs are thought to originate in the endoplasmic reticulum [5], [6]. It is thought that as TAG and SE are synthesized, they accumulate between the internal and external endoplasmic reticulum membrane leaflets until LDs bud from the endoplasmic reticulum [7]. This hypothesis is reinforced by the fact that the enzymes that contribute to the esterification of fatty acids (FAs) and sterols (STEs) e.g., diacylglycerol acyltransferases Dga1p [8] and Lro1p [9], and acyl-CoA:sterol acyltransferases, Are1p and Are2p are localized to the endoplasmic reticulum [10]. Among these enzymes, only Dga1p is found in LDs [11]. Regarding the utilization of stored lipids, five enzymes responsible for the mobilization of stored lipids are localized to the LD. Three of these enzymes are TAG lipases – Tgl3p, Tgl4p and Tgl5p [12], [13] – and two are SE hydrolases – Yeh1p and Tgl1p [14]. Remarkably, the major enzyme contributing to SE hydrolase activity (Yeh2p) is localized at the plasma membrane [15]. This raises the question of whether the LD is more than a mere neutral lipid deposit. In fact, equilibrium between mobilization and LD synthesis seems to be important in various aspects of metabolism, such as cell cycle progression, that requires lipolysis [16], membrane trafficking and “lipid buffering” to regulate levels of free FA [17]. Once synthesized, free FAs and free STEs would represent a lipotoxic threat [18] if not converted by esterification into more biologically inert lipids, such as TAG and SEs, and then stored in LDs. In yeast, deletion of the acyltransferases that esterify fatty acids into TAGs and SEs lead to high sensitivity to unsaturated fatty acids [19]. In mammals, LDs control the synthesis and secretion of inflammatory mediators [20] and are implicated in virus propagation [21]. Different stimuli and signaling pathways are involved in the biogenesis of LDs; for example, in macrophages, leptin-induced LD formation is mediated by phosphatidylinositol 3-kinase (PI3-K) and the mammalian target of rapamycin (mTOR) pathways [22].
In yeast, LDs are quite dynamic organelles. It was observed that they are consumed as a response to a proliferative stimulus, such as the addition of rich medium to stationary cells. During the lag phase of growth [23], when nutrients are abundant, lipid mobilization provides construction blocks for the synthesis of more complex lipids such as phospholipids. When cells enter the exponential phase of growth, LDs are re-synthesized [23]. Nevertheless, little is known about the regulators and pathways driving these dynamics. It is known that Snf1, a homolog to mammalian AMP-activated kinase (AMPK), is a negative regulator of fatty acid synthesis and, once phosphorylated, mediates acetyl-CoA carboxylase phosphorylation and subsequent inhibition [24], [25]. In order to determine the role of protein phosphorylation in the regulation of LD dynamics, we screened yeast mutant strains in which protein phosphatases (including catalytic and regulatory subunits) had been knocked out (no phosphatase has been reported to regulate lipid droplet metabolism). Since the techniques used in the study of lipids are laborious and require time-consuming microscopic analysis, our group developed a new fluorimetric method suitable for high-throughput experiments. Nile red is an excellent fluorescent probe for detection of intracellular lipid droplets by fluorescence microscopy and flow cytofluorometry [26]–[28]. BODIPY® (4,4-difluoro-3a,4a,-diaza-s-indacene) has also been used in LD microscopic studies, and has previously been used for a screening of a yeast deletion mutant collection for LD abnormalities [29]. Although both molecules preferentially stain LDs, the latter seems to be more specific [26]. Based on BODIPY fluorescence, we were able to rapidly evaluate LD content without lipid extraction or microscopy analysis. Our results showed that this method is suitable for following LD dynamics during yeast culture growth and can be employed to screen a large number of mutants.
Using our new approach, we were able to identify 13 (out of 65) phosphatase knockout strains that presented abnormal LD levels. Among these, we further studied Sit4, a Ser/Thr protein phosphatase, as a positive regulator of LD metabolism. We propose that Sit4p, in association with its regulatory subunit Sap190p, regulates lipid metabolism by controlling the phosphorylation status of Snf1 kinase. In fact, the snf1-null strain showed increased LD levels, confirming unpublished data from Kohlwein and coworkers [30], while sit4- or sap190-null strains exhibited the opposite phenotype, which is consistent with the hyperphosphorylation of Snf1p observed in these strains.
Materials and Methods
Reagents
BODIPY® 493/503 (referred to here as BODIPY) was purchased from Invitrogen, prepared as a 10 mM stock solution in DMSO and kept at −70°C. Soraphen A was a gift from Rolf Müller (Helmoltz-Zentrum für Infektionsforschung) and kept as a 0.1 mg/ml stock solution in 10% methanol. All other reagents were from Sigma-Aldrich (St. Louis, MO).
Strains
Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4741- MATa gene deletion library (Open Biosystems). BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), dga1lro1 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 dga1::KanMX lro1::KanMX) and are1are2 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 are1::KanMX are2::KanMX) were kindly provided by Sepp D. Kohlwein (University of Graz, Austria).
Yeast growth and cell fixation
Saccharomyces cerevisiae strains were grown in YPD liquid-rich medium (1% yeast extract, 2% peptone and 2% glucose) at 30°C. For determination of LD content, aliquots were withdrawn during growth and cells were immediately fixed by adding 1/9 volume of formaldehyde (final concentration 3.7% v/v). After incubation for 15 min at 25°C, cells were collected by centrifugation (3000 rpm, 5 min) and washed once with distilled water. Cells were resuspended in water and adjusted to 5 O.D.600 nm/ml and kept at 4°C until use. For use in high-throughput screening selected strains from the BY4741- MATa gene deletion library (Open Biosystems) were cultured overnight in YPD medium in 96-well plates. Afterwards they were transferred to fresh YPD medium with the aid of a 96 -Pin Replicator and grown at 30°C with shaking. Growth was monitored during 48 hours to ensure that all deleted strains, even those that have a slow-growth phenotype, had attained the stationary phase. The OD600 varied between 2.8–6.3. The mean growth was OD600 = 4.2±0.6. Each sample was then fixed by the addition of 22 µl of formaldehyde, incubated for 15 min at 25°C and washed with distilled water. Fixed cells were harvested and resuspended in 200 µl of distilled water.
Liquid fluorescence recovery assay
LD content was determined by recovering the fluorescence of quenched BODIPY following the addition of fixed cells (Figure 1). For this purpose, 5 µl of a formaldehyde-fixed cell suspension was added to 200 µl of quenched reading buffer containing 5 µM BODIPY and 500 mM KI, all contained in a 96-well black-wall/clear-bottom plate (Costar). The settings for fluorescence were excitation 480 nm, emission 510 nm, cutoff filter at 495 nm, PMT automatic. Concomitant to each fluorescence reading, the quantity of cells was determined by measuring the absorbance at 600 nm. Each sample was read four times by adding subsequent 5-µl aliquots of a formaldehyde-fixed cell suspension to the same well. Reading blanks were acquired by incubating the reading buffer for 5 min at 30°C before the addition of cells. Data quality was analyzed by evaluating the linearity of measurements compared to cell number. From this curve, the relative fluorescence per number of cells was determined and denoted the lipid droplet index (LD index). If R-square <0.9, data were discarded (Figure S1 and Figure S2).
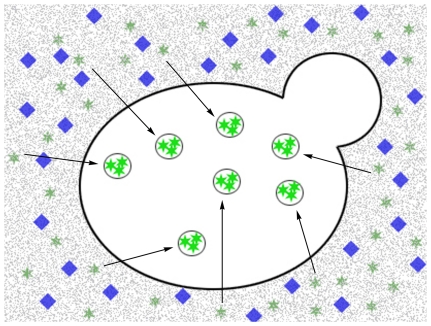
Figure 1. A schematic representation of the fluorescence recovery assay.
Based on the high specificity of the fluorescent probe BODIPY 493/503 for lipid droplets, we designed a rapid fluorimetric method for the evaluation of lipid droplets inside yeast cells. With this method, BODIPY's fluorescence is quenched in solution and, when cells are added, fluorescence is recovered due to passive BODIPY dislocation from the medium to lipid droplets, from which the quencher is excluded.
Statistical analysis of high-throughput experiments
In order to classify strains as high lipid droplet content (hlc) or low lipid droplet content (llc) with respect to wild-type, LD indexes were compared as follows. For each independent experiment, the LD index of each individual strain was normalized to the mean LD index of all samples contained in the same plate. Then, the mean LD index from four independent experiments was compared to the mean LD index of the wild-type (WT) strain (n = 14). The F-test was performed to determine whether the variability in the data collected for each deletion strain was similar to that observed for the wild-type strain; a homoscedastic or heteroscedastic t-test was employed accordingly. Strains with values statistically different (p<0.05) were catalogued as hlc or llc strains when the normalized LD index was at least 20% higher or lower than that of the wild-type strain, respectively.
Quantification of lipid droplets by fluorescence microscopy
Aliquots of 5 µl of cellular suspension (10 OD 600 nm) and 5 µl of 10 µM BODIPY were mounted onto a polylysine-treated glass slide. Microscopy was performed on Axio Observer.Z1 (Zeiss) or Olympus IX 81 using a 100x oil immersion objective. Images captured were analyzed using ImageJ software and total fluorescence was determined with the aid of the nucleus counter and cell counter function of the Wright Cell Imaging Facility (WCIF) plug-in collection. LDs were quantified in at least 50 cells.
Protein homogenate preparation and western blot
Total protein homogenates were prepared using the rapid procedure described by Yaffe and Schatz [31] with minor modifications. Briefly, 0.25 M NaOH and 1% β-mercaptoethanol (final concentration) were added to culture samples, which were then incubated at room temperature for 10 min, followed by the addition of trichloroacetic acid to a final concentration of 6%. Protein was collected by centrifugation and the pellets were stored at −80°C until use. For western blot analysis, pellets were resuspended directly in electrophoresis Laemlli sample buffer to provide a 10-OD suspension. Aliquots of 10 µl of these extracts, containing 0.1-OD cells, were separated in 7.5% SDS-acrylamide gel using the Mini-Protean II (BioRad) and electrotransferred to Immobilon-P for 30 min at 18 V in 25 mM Tris, 192 mM glycine and 10% methanol, using a trans-blot semi-dry cell (BioRad). Membranes were treated with anti-phospho-AMPKα Thr172 (Cell Signaling) or anti-Snf1p (Santa Cruz) antibodies. Blots were detected using the chemiluminescence ECL Plus kit (GE Healthcare).
Measurement of triacylglycerol
Cultures were grown in YPD medium, harvested by centrifugation at 3000 rpm for 5 minutes at room temperature and washed with distilled water. An aliquot of 15 OD cells was resuspended in 300 µl of extraction buffer (50 mM Tris-HCl, 0.3% Triton X-100, pH 7.5) and lysed with glass beads vortexing for 5 cycles of 30 seconds at 4°C. Lysed cells were separated and the glass beads were washed with 300 µl of extraction buffer. The total lysate was centrifuged at 3,000 rpm for ten minutes. Neutral lipids were extracted from 200 µl of the supernatant as described by Bligh & Dyer [32]. Briefly cells were incubated with shaking for 5 minutes after successive addition of 750 µl chloroform/methanol (1∶2), 250 µl chloroform and 250 µl distilled water. Tubes were centrifuged at 1,000 rpm for 5 minutes and the organic phase was dried under N2 vapor at room temperature. Neutral lipids were resuspended in 150 µl of extraction buffer. Triglycerides were measured with a clinical kit (Doles, Brazil) as indicated by the manufacturer. This kit is based on hydrolysis of triacylglycerol with lipase and further determination of glycerol.
Soraphen A susceptibility tests
Drug susceptibilities were measured by both broth microdilution and spot assays. MICs for the strains were determined using the broth microdilution method. Spot assays were carried out as in [33]. Briefly, yeast strains were grown in YPD to stationary phase and inoculated at serial dilutions of 107, 106 and 105 cells/ml onto YPD plus 2% agar with increasing concentrations of soraphen A and then incubated for two days at 30°C, as previously described [34]. For microdilution tests, cells from stationary culture were diluted to 105 cells/ml in fresh YPD with increasing concentrations of soraphen A in a total volume of 200 µl and then incubated in 96-well microtiter plates at 30°C. Growth was recorded after 24 h by measuring absorbance at 600 nm. The IC50 was defined as the drug concentration that inhibited growth by 50% compared to growth of the drug-free controls.
Results
Liquid fluorescence recovery assay (LFR assay)
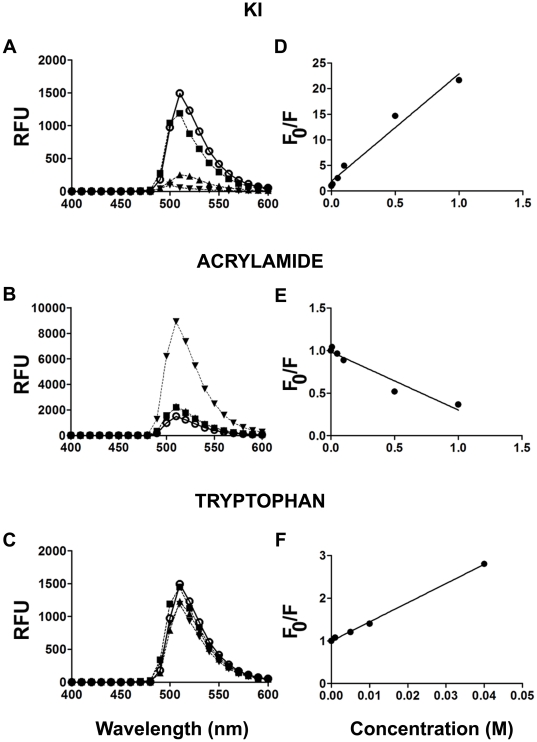
LDs are often visualized by microscopy after staining with Nile red, osmium or BODIPY [35]–[37]. BODIPY has a lipophilic nature and exhibits high specificity for lipid droplets [26]; these qualities render the compound a good candidate to use as a probe for LDs in a fluorescent liquid high-throughput experiment. Based on fluorescence-quenched assays [38], we selected a hydrophilic quencher that would permit BODIPY to fluoresce only when localized inside LDs and not when present in solution (Figure 1). Three classical fluorescence quenchers (KI, acrylamide and tryptophan) were titrated against a 5 µM BODIPY solution. The emission spectra (485 nm excitation) showed that only KI quenched the fluorescence efficiently (Figure 2A–C). From Stern-Volmer plots, where the quenching efficiency is related to the total quencher concentration [39] it can be observed that quenching increased linearly with increasing concentrations of KI and tryptophane (Figure 2D,E) but not acrylamide (Figure 2F). From these plots, a Stern-Volmer constant of 44 and 14.1 was calculated for KI and acrylamide, respectively. KI was selected as the quencher because a higher F0/F ratio was reached with this compound as compared to tryptophan in the conditions tested.
Figure 2. Quenchers of BODIPY fluorescence.
A and B, Emission spectra (ex 485 nm) of a 5 µM aqueous solution of BODIPY in the presence of KI (A), acrylamide (B) and tryptophane (C). The concentrations of KI and acrylamide were: none (○), 0.01 M (▪), 0.1 M (▴) and 1 M (▾), Due to its reduced solubility in the conditions tested, tryptophan was used at concentrations of 0 (○), 0.005 M (▪), 0.01 M (▴) and 0.02 M (▾). RFU = relative fluorescence unit.D–F, Stern-Volmer plots (indicate fluorescence quenching (F0/F) as a function of quencher concentration [39]) for BODIPY with increasing concentrations of KI, tryptophan and acrylamide, respectively. F0 (fluorescence in the absence of quencher) and F (fluorescence in the presence of quencher).
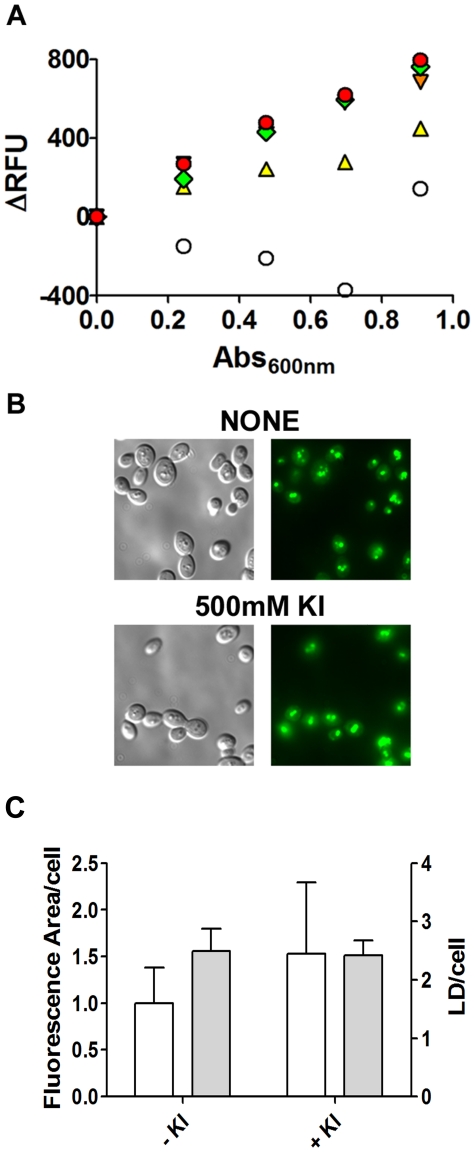
When fixed-formaldehyde cells were added to a BODIPY-KI-quenched solution, fluorescence was recovered (Figure 3A). The best linearity between the number of cells and recovered fluorescence was obtained using KI in the quenching medium at a concentration ranging from 0.25 M to 1.0 M. For subsequent experiments, the reading solution contained 500 mM KI and 5 µM BODIPY. To discard artifacts, we verified by microscopy that KI did not quench the fluorescence of LDs stained with BODIPY. The fluorescence area/cell and the number of LDs did not change significantly when 500 mM KI was added, suggesting that iodide is excluded from LDs (Figure 3B and C). This phenomenon allows researchers to measure the levels of lipid droplets within the cells in an easy and inexpensive manner (Figure 1). Some fluorophores exhibit a red-shift of the fluorescence spectra when in a non-polar environment, but the absorbance and emission spectra of BODIPY were unaltered, showing maximum absorbance at 485 nm and maximum emission at 510 nm with excitation at 485 nm (Figure S3).
Figure 3. The BODIPY fluorescence signal quenched by iodide is recovered by the addition of cells to the medium.
A. Increasing amounts of formaldehyde-fixed cells were added (subsequent 5 µl additions) to a 96-well plate containing 200 µl of 5 µM BODIPY in the presence of none (○), 0.1 M (▴, yellow), 0.25 M (▾, orange), 0.50 M (♦, green) or 1 M (•, red) KI. Fluorescence intensity (ex/em = 485/510 nm) and absorbance (600 nm) were recorded. A representative experiment is shown. B. Cells grown in YPD to stationary phase were formaldehyde-fixed, washed and resuspended to 10 DO 600 nm/ml. An aliquot of 5 µl of cells was mixed with 5 µl BODIPY solution (10 µM) in the absence (upper panel) or presence of 500 mM KI (lower panel). Stained cells were analyzed by fluorescence microscopy (100x magnification). C. Microscopy images were analyzed. The total fluorescence area/cell was determined and expressed in pixels/cell (white bars). LDs per cell were quantified using the same images (gray bars). Data are for at least 50 individual cells. No statistical difference was found between data collected from images stained with 5 µM Bodipy with or without 500 mM KI.
Sensitivity of the LFR assay
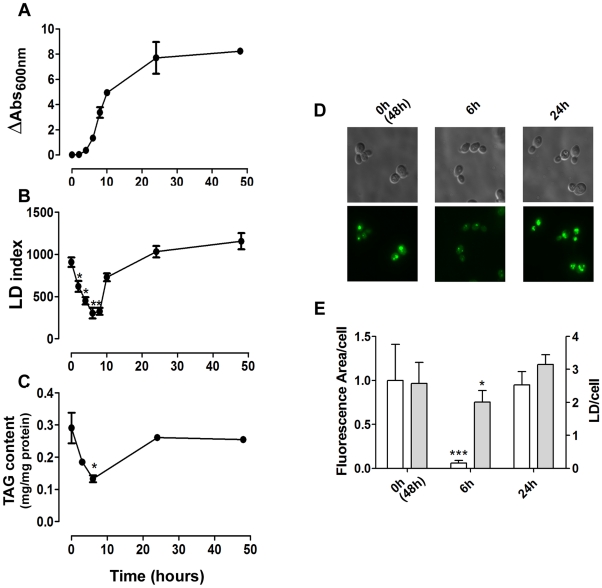
To assess whether our method was able to detect small fluctuations in LD levels, we quantified this parameter during yeast growth as LDs are actively metabolized during growth [23], [40] (Figure 4A, B). Results obtained with the LFR assay (Figure 4B) were similar to those obtained by quantifying the TAG content (Figure 4C) or through microscopic analysis, from which we calculated the total fluorescence area/cell. Furthermore it did not reflect the number of LDs/cell that does not vary as much as the size of LDs along the growth curve (Figure 4D, E). The results were also in agreement with previous results obtained by microscopic analysis [23] and biochemical quantification of cellular TAG [40], which indicated that yeast cells consume LDs when diluted in fresh medium. After six hours, LDs are re-synthesized reaching highest levels at stationary phase [23], [40]. This result demonstrates that our assay was able to detect small physiological lipid stores fluctuations during yeast growth without the need for time-consuming image analysis.
Figure 4. The liquid fluorescence recovery (LFR) assay can detect LD dynamics.
A. Stationary yeast cells pre-grown for 48 hours, were inoculated into fresh YPD medium and cellular growth was recorded by measuring absorbance at 600 nm over the course of 48 hours (n = 3± S.D.). B. Aliquots of cells were withdrawn during growth and fixed in formaldehyde. The LD index was determined (fluorescence/OD cells) using the LFR assay (n = 3± S.D.) C. The triacylglycerol content was measured during growth. D and E. LD content was determined by fluorescence microscopy. Cells were grown for 0 (corresponding to 48 h pre-grown cells), 6 and 24 hours in YPD, incubated with BODIPY and photographed by fluorescence microscopy. The total fluorescence area/cell (white bars) were determined and expressed in pixels/cell (white bars). LDs per cell were quantified using the same images (gray bars). Data are for at least 50 individual cells. *p<0.05, ** p<0.01, ***p<0.001, in comparison to WT values.
High-throughput experiment employing the LFR assay
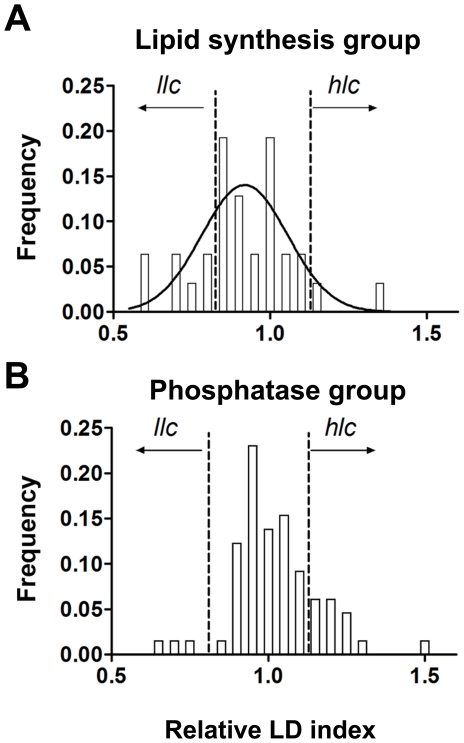
We employed the LFR assay to study the involvement of protein phosphatases in LD metabolism. In this high-throughput experiment, we screened 96 null strains as well as the BY4741 WT strain (14 independent WT clones). We subdivided the strains in two groups. The first was a reference group, referred to as the lipid synthesis group. This reference group included 31 null strains for genes encoding LD-associated enzymes and/or enzymes involved in the metabolism of sterols, the metabolism of triglycerides and the biosynthesis of glycerolipids (Table 1) [41]. The second group, referred to as the phosphatase group, was composed of 65 null strains with all known non-essential protein phosphatases and non-essential phosphatase regulatory subunits (Table 2), according to the annotation in the Yeast Genome Database (www.yeastgenome.org, October 2009). As LD content varies substantially during growth (see Figure 4B) and not all strains have the same growth rate, we decided to asses LD content in strains grown to stationary phase, which was attained for all strains after 48 hours of seeding. A histogram of the distribution of the lipid indexes for all strains was analyzed and an index for cataloguing strains (as containing a higher LD index (hlc) or a lower LD index (llc)) was elaborated using an arbitrary cohort of ±20% in relation to the LD index of WT strains (Figure 5). When the mean LD index of the lipid synthesis group was compared to that of the WT strain, no significant difference was observed (means were 0.9539 and 0.9135, respectively p = 0.276). The same trend was not observed for the mean LD index of the protein phosphatase group, which was significantly higher than the WT sample (1.023, p = 0.0164) and included an over-representation of hlc strains. Interestingly, this result indicates that lipid accumulation is more likely provoked by the random loss of protein phosphatase function as opposed to the random loss of function among proteins involved in lipid metabolism. After analyzing the groups as a whole, individual strains were catalogued as having higher (hlc) or lower LD content (llc) (Tables 1 and 2).
Table 1. Lipid synthesis null strains screened by LFR assay.
| NAME | SYSTEMATIC | BIOLOGICAL PROCESS | MEAN | S.D. | P |
| HMG2§ | YLR450W | STE synthesis | 0.58 | 0.19 | <0.001 |
| DGA1§,* | YOR245C | TAG synthesis | 0.59 | 0.16 | <0.001 |
| ERG4§,* | YGL012W | STE synthesis | 0.70 | 0.18 | 0.003 |
| ERG5§,* | YMR015C | STE synthesis | 0.71 | 0.28 | 0.007 |
| ARE2§ | YNR019W | SE synthesis | 0.76 | 0.21 | 0.023 |
| LRO1 | YNR008W | TAG synthesis | 0.81 | 0.09 | 0.069 |
| DPP1 | YDR284C | TAG synthesis | 0.81 | 0.19 | 0.095 |
| LPP1 | YDR503C | TAG synthesis | 0.84 | 0.12 | 0.152 |
| ERG2 | YMR202W | STE synthesis | 0.84 | 0.06 | 0.164 |
| GPT2 | YKR067W | TAG synthesis | 0.85 | 0.06 | 0.178 |
| CSG2 | YBR036C | Ceramide synthesis | 0.85 | 0.26 | 0.233 |
| TGL5 | YOR081C | TAG lipase | 0.86 | 0.17 | 0.265 |
| AYR1 | YIL124W | TAG synthesis | 0.87 | 0.19 | 0.317 |
| LAC1 | YKL008C | Ceramide synthesis | 0.88 | 0.09 | 0.359 |
| HMG1 | YML075C | STE synthesis | 0.88 | 0.15 | 0.380 |
| SUR2 | YDR297W | Ceramide synthesis | 0.91 | 0.01 | 0.048 |
| ARE1 | YCR048W | SE synthesis | 0.91 | 0.10 | 0.626 |
| IPT1 | YDR072C | Ceramide synthesis | 0.93 | 0.09 | 0.782 |
| LAG1 | YHL003C | Ceramide synthesis | 0.94 | 0.01 | 0.577 |
| TGL1 | YKL140W | SE hydrolase | 0.99 | 0.03 | 0.189 |
| YEH2 | YLR020C | SE hydrolase | 1.00 | 0.08 | 0.540 |
| ERG24 | YNL280C | STE synthesis | 1.00 | 0.14 | 0.546 |
| SCS7 | YMR272C | Ceramide synthesis | 1.00 | 0.12 | 0.531 |
| ERG6 | YML008C | STE synthesis | 1.01 | 0.20 | 0.507 |
| SCT1 | YBL011W | TAG synthesis | 1.01 | 0.10 | 0.450 |
| SUR1 | YPL057C | Ceramide synthesis | 1.03 | 0.04 | 0.332 |
| YEH1 | YLL012W | SE hydrolase | 1.05 | 0.03 | 0.002 |
| TGL4 | YKR089C | TAG lipase and SE hydrolase | 1.09 | 0.14 | 0.095 |
| ERG3 | YLR056W | STE synthesis | 1.10 | 0.27 | 0.107 |
| SLC1¥ | YDL052C | TAG synthesis | 1.17 | 0.05 | 0.022 |
| TGL3¥,* | YMR313C | TAG lipase | 1.37 | 0.17 | <0.001 |
7 out of 31 strains showed abnormal LP levels. Statistical analysis was performed against WT values (mean fluorescence = 0.95, S.D. = 0.15, n = 14). It was defined a cutoff using an arbitrary 20% difference between the mutant and WT relative fluorescence means. Phenotypes referred as llc
(§low lipid content,)
and hlc
(¥high lipid content,)
were marked in bold. Gene descriptions based in SGD ( www.yeastgenome.org ).
*strains identified in a previously published screening for LP [25] .
Table 2. Phosphatase null strains screened by LFR assay.
| NAME | SYSTEMATIC | DESCRIPTION | MEAN | S.D. | P |
| SIT4§ | YDL047W | Type 2A-related protein phosphatase | 0.63 | 0.09 | <0.001 |
| REG1§ | YDR028C | Regulatory subunit of type 1 protein phosphatase Glc7p | 0.68 | 0.16 | 0.002 |
| SAP190§ | YKR028W | Regulatory subunit of Sit4 phosphatase | 0.73 | 0.07 | 0.007 |
| SIS2 | YKR072C | Regulatory subunit of type 1 protein phosphatase Ppz1 | 0.86 | 0.07 | 0.223 |
| PIG2 | YIL045W | Subunit for the type-1 protein phosphatase Glc7p | 0.89 | 0.12 | 0.450 |
| LTP1 | YPR073C | Protein tyrosine phosphatase | 0.90 | 0.12 | 0.518 |
| RTS1 | YOR014W | Regulatory subunit of protein phosphatase 2A | 0.91 | 0.07 | 0.598 |
| YBR051W | Dubious ORF, partially overlaps REG2 | 0.92 | 0.29 | 0.809 | |
| PTC6 | YCR079W | Type 2C protein phosphatase | 0.92 | 0.10 | 0.636 |
| GIP2 | YER054C | Subunit for the type-1 protein phosphatase Glc7p | 0.92 | 0.11 | 0.646 |
| YVH1 | YIR026C | Phosphotyrosine-specific protein phosphatase | 0.92 | 0.13 | 0.653 |
| CNA1 | YLR433C | Subunit of calcineurin, a type 2B protein phosphatase | 0.92 | 0.10 | 0.661 |
| PTC3 | YBL056W | Type 2C protein phosphatase | 0.93 | 0.17 | 0.725 |
| PTC5 | YOR090C | Type 2C protein phosphatase | 0.93 | 0.08 | 0.722 |
| GLC8 | YMR311C | Subunit for the type-1 protein phosphatase Glc7p | 0.93 | 0.04 | 0.457 |
| TIP41 | YPR040W | Interacts physically and genetically with Tap42p, regulates Sit4 | 0.93 | 0.09 | 0.781 |
| SIW14 | YNL032W | Phosphotyrosine-specific protein phosphatase | 0.94 | 0.10 | 0.821 |
| CNB1 | YKL190W | Subunit of calcineurin, a type 2B protein phosphatase | 0.94 | 0.08 | 0.897 |
| RTS3 | YGR161C | Regulatory subunit of protein phosphatase 2A | 0.95 | 0.10 | 0.930 |
| PSR2 | YLR019W | Protein phosphatase involved in the general stress response | 0.95 | 0.04 | 0.992 |
| OCA2 | YNL056W | Putative protein tyrosine phosphatase | 0.95 | 0.22 | 0.997 |
| SDP1 | YIL113W | MAP kinase phosphatase | 0.96 | 0.17 | 0.971 |
| YNL010W | Unknown function; similar to phosphoserine phosphatases | 0.96 | 0.08 | 0.948 | |
| PTP1 | YDL230W | Phosphotyrosine-specific protein phosphatase | 0.96 | 0.09 | 0.932 |
| SIP5 | YMR140W | Unknown function; interacts with both the Reg1p/Glc7p | 0.96 | 0.14 | 0.928 |
| CMP2 | YML057W | Subunit of calcineurin, a type 2B protein phosphatase | 0.97 | 0.12 | 0.866 |
| YGR203W | Phosphatase with sequence similarity to Cdc25p, Arr2p and Mih1p | 0.97 | 0.14 | 0.814 | |
| PPH22 | YDL188C | Type 2A protein phosphatase | 0.99 | 0.14 | 0.636 |
| PPT1 | YGR123C | Ser/thr phosphatase with similarity to human phosphatase PP5 | 0.99 | 0.14 | 0.622 |
| PPG1 | YNR032W | Putative serine/threonine protein phosphatase | 1.00 | 0.07 | 0.596 |
| PTC2 | YER089C | Type 2C protein phosphatase | 1.01 | 0.15 | 0.522 |
| RRD2 | YPL152W | Activator of the phosphotyrosyl phosphatase activity of PP2A | 1.01 | 0.09 | 0.474 |
| MIH1 | YMR036C | Protein tyrosine phosphatase | 1.01 | 0.07 | 0.458 |
| PIG1 | YLR273C | Subunit for the type-1 protein phosphatase Glc7p | 1.01 | 0.15 | 0.467 |
| GIP1 | YBR045C | Subunit for the type-1 protein phosphatase Glc7p | 1.02 | 0.04 | 0.048 |
| PHO13 | YDL236W | Alkaline phosphatase, also has protein phosphatase activity | 1.02 | 0.09 | 0.382 |
| SAP4 | YGL229C | Regulatory subunit of Sit4 phosphatase | 1.03 | 0.16 | 0.376 |
| NBP2 | YDR162C | Recruitment of phosphatase Ptc1p to the Pbs2p-Hog1p complex | 1.04 | 0.07 | 0.264 |
| PTP3 | YER075C | Phosphotyrosine-specific protein phosphatase | 1.05 | 0.17 | 0.267 |
| YER121W | Putative protein of unknown function | 1.05 | 0.07 | 0.249 | |
| VHS3 | YOR054C | Regulatory subunit of type 1 protein phosphatase Ppz1 | 1.05 | 0.15 | 0.255 |
| NEM1 | YHR004C | Probable catalytic subunit of Nem1p-Spo7p phosphatase | 1.05 | 0.10 | 0.246 |
| PPZ1 | YML016C | Type 1 protein phosphatase | 1.05 | 0.16 | 0.221 |
| PSR1 | YLL010C | Protein phosphatase involved in the general stress response | 1.05 | 0.11 | 0.210 |
| PSY4 | YBL046W | Regulatory subunit of type 2A protein phosphatase Pph3 | 1.06 | 0.19 | 0.200 |
| SAP155 | YFR040W | Regulatory subunit of Sit4 phosphatase | 1.06 | 0.14 | 0.179 |
| PPQ1 | YPL179W | Putative protein serine/threonine phosphatase | 1.08 | 0.21 | 0.147 |
| PPH3 | YDR075W | Type 2A protein phosphatase | 1.09 | 0.04 | 0.091 |
| PSY2 | YNL201C | Regulatory subunit of type 2A protein phosphatase Pph3 | 1.09 | 0.13 | 0.098 |
| REG2 | YBR050C | Regulatory subunit of type 1 protein phosphatase Glc7p | 1.09 | 0.05 | 0.086 |
| PTP2 | YOR208W | Phosphotyrosine-specific protein phosphatase | 1.12 | 0.22 | 0.057 |
| OCA1 | YNL099C | Putative protein tyrosine phosphatase | 1.12 | 0.25 | 0.058 |
| PAM1 | YDR251W | Essential protein of unknown function | 1.13 | 0.15 | 0.028 |
| PTC7¥ | YHR076W | Type 2C protein phosphatase | 1.14 | 0.22 | 0.028 |
| PPS1¥ | YBR276C | Protein phosphatase with specificity for Ser, Tthr and Tyr residues | 1.15 | 0.10 | 0.017 |
| SAP185¥ | YJL098W | Regulatory subunit of Sit4 phosphatase | 1.16 | 0.18 | 0.015 |
| PPZ2¥ | YDR436W | Type 1 protein phosphatase | 1.18 | 0.12 | 0.007 |
| MSG5¥ | YNL053W | Dual-specificity protein phosphatase | 1.18 | 0.21 | 0.009 |
| PTC4¥ | YBR125C | Type 2C protein phosphatase | 1.20 | 0.19 | 0.004 |
| SPO7 | YAL009W | Regulatory subunit of Nem1p-Spo7p phosphatase | 1.22 | 0.30 | 0.172 |
| PPH21¥ | YDL134C | Type 2A protein phosphatase | 1.23 | 0.17 | 0.002 |
| GAC1¥ | YOR178C | Subunit for the type-1 protein phosphatase Glc7p | 1.25 | 0.14 | 0.001 |
| BNI4¥ | YNL233W | Subunit for the type-1 protein phosphatase Glc7p | 1.26 | 0.11 | 0.002 |
| PTC1¥ | YDL006W | Type 2C protein phosphatase | 1.28 | 0.26 | <0.001 |
| TPD3 | YAL016W | Regulatory subunit of protein phosphatase 2A | 1.52 | 0.39 | 0.127 |
13 out of 65 strains showed abnormal LP levels. Statistical analysis was performed against WT values (mean fluorescence = 0.95, S.D. = 0.15, from 14 WT samples). It was defined a cutoff using an arbitrary 20% difference between the mutant and WT relative fluorescence means. Phenotypes referred as llc
(§low lipid content,)
and hlc
(¥high lipid content,)
were marked in bold. Gene descriptions based in SGD (www.yeastgenome.org).
Figure 5. Statistical analysis of a high-throughput experiment employing the LFR assay.
LDs were quantified by the LFR assay. The frequency distribution of the relative LD index of strains from the lipid synthesis group (A, n = 31) and protein phosphatase group (B, n = 65) was calculated. Data were collected from four independent experiments as described in the Materials and Methods section. The D'Agostino & Pearson normality test was employed at P<0.05. The lipid synthesis group was normally distributed (p = 0.281), while the protein phosphatase group failed to pass the normality test (p = 0.016). The arbitrary cohort (±20% in relation to WT values) is indicated in the graphs by a dashed line (llc, low lipid droplet content and hlc, high lipid droplet content).
Most of the results obtained in the lipid synthesis group corroborated results previously reported in the literature. Five strains were identified as llc strains (hmg2, dga1, erg4, erg5, are2) and two were identified as hlc strains (slc1 and tgl3). Interestingly, among the llc strains we identified were dga1, which encodes an enzyme responsible for the last step in TAG synthesis (acyl-CoA-dependent pathway); hmg2, which encodes HMG-CoA reductase, which catalyzes the rate-limiting step in ergosterol synthesis; and are2, which esterifies sterols. Deletion of Dga1p and Are2p would lead to diminished levels of TAG and sterol/steryl esters levels [42], respectively. In the hlc group, we identified tgl3 which encodes a TAG lipase. Deletion of this enzyme increases TAG levels [12]. In the same group, we also identified slc1, which encodes a 1-acylglycerol-3-phosphate acyltransferase involved in phosphatidic acid biosynthesis. The exact role of this protein in LD formation has not been demonstrated, but its deletion led to higher TAG levels in cells [43]. Reinforcing our results, in a previously published screening, dga1 and tgl3 were identified as having diminished and increased number of LDs, respectively [44]. Although we classified the erg4 and erg5 mutants as llc strains, these strains have been reported to have more LDs than WT strains when analyzed by fluorescent microscopy [44]. Microscopic analysis confirmed that these strains have more LDs than WT strains but show diminished total fluorescence area (Figure S4), thus indicating a reduction in LD size. These results are interesting because they suggest that fluctuations measured by LFR assay are due to increased total fluorescence area. However the above results do not show if differences assayed in the LFR assay are due to different levels of SE or TAG, or both. To test this we have compared the fluorescence lipid droplet index and fluorescence area/cell of are1are2 and dga1lro1 cells, lacking SE and TAG, respectively (Figure S5). The mean fluorescence area/cell was 40% lower in are1are2 compared to WT while the LFR assay showed an 8% reduction. The dga1lro1 presented a diffuse pattern without distinguishable LDs when stained with BODIPY using the same exposure times as WT being not valid to compare the fluorescence area/cell, however with longer exposure times it was possible to visualize a few LDs/cell. The LFR assay detected a 40% reduction in dga1lro1 LD index. These results show that the LFR assay detects the total area/cell of lipid droplets composed of mainly TAG or SE and that it is more sensitive than fluorescence microscopy studies.
The Ser/Thr protein phosphatase Sit4p regulates Snf1 kinase phosphorylation in association with its regulatory subunit Sap190p
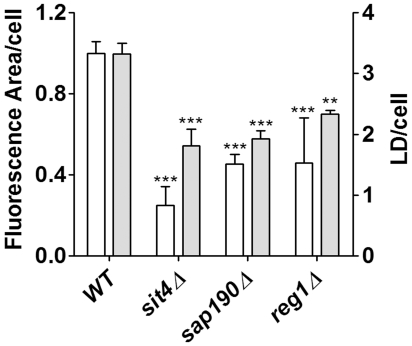
Among the protein phosphatase group, three strains were marked as llc (sit4, sap190 and reg1). Two of these genes are related: SIT4 encodes the catalytic subunit of a type 2A-like Ser-Thr phosphatase and SAP190 encodes one of its regulatory subunits. REG1 codes for a regulatory subunit of Glc7 phosphatase, an essential type-1 protein phosphatase. On the other hand, 10 strains were marked as hlc. Three of these were type 2C protein phosphatases (ptc1, ptc4, ptc7); three were related to type 1 protein phosphatases, a catalytic subunit (ppz2) and Glc7 regulatory subunits (gac1 and bni4). Pph21p is a type 2A protein phosphatase and Sap185p is a regulatory subunit of Sit4p. Other null strains marked as hlc were pps1 and msg5. These results call for a new perspective on the topic at hand and indicate that an intricate regulation of protein phosphorylation is involved in the regulation of LD metabolism by protein phosphatases. Since previous screens have missed sit4, sap190 and reg1 as having less LDs we counted LDs and measured the fluorescent area/cell (Figure 6). Results show that both parameters are significantly lower compared to WT.
Figure 6. LDs size and content in llc phosphatase mutants.
The total area/cell and the number of LDs were analyzed from microscopic images of WT, sit4, sap190, reg1 grown in YPD medium for 48 hours, as described. ** p<0.01, ***p<0.001, in comparison to WT values.
We further studied llc hits. Together with Tap42p, Sit4p is known for its participation in the TOR pathway, which senses nutrient availability and coordinates cell growth [45]. Sit4p was first identified as a type2A-related phosphatase activated by extracellular ceramide [46]. Sit4p is also regulated by four other subunits thought to act as targeting subunits: Sap4p, Sap155p, Sap185p and Sap190p. These proteins have different or overlapping roles in the regulation of the cell cycle [47] and in translation control [48]. Reg1p directs Glc7p to its targets, participating in the regulation of several processes such as glucose de-repression and glycogen synthesis [49], [50]. Reg1p plays a role in energetic metabolism by interacting with Snf1 kinase, a Ser/Thr kinase homologue to mammalian AMPK responsible for maintaining energy homeostasis. Upon depletion of glucose in the medium, Snf1 is phosphorylated by upstream kinases (called Snf1-activating kinases, SAKs) and becomes active [51]. Its dephosphorylation is promoted by Glc7p phosphatase and is dependent on Reg1p, as the reg1 strain displays hyperphosphorylated Snf1p [52]. However, this finding must be investigated further because greater Reg1p-Snf1p interaction is observed in low-glucose conditions when Snf1p is more active [53]. Snf1p kinase, in turn, phosphorylates a key enzyme in FA biosynthesis, acetyl-CoA carboxylase (Acc1), which inhibits the activity of Acc1. It has been shown that deletion of SNF1 increases Acc1 activity as snf1 strain is more resistant to soraphen A which is a potent inhibitor of acetyl-CoA carboxylase – the more resistant a strain is, the greater is its level of acetyl-CoA carboxylase activity [24], [25], [54].
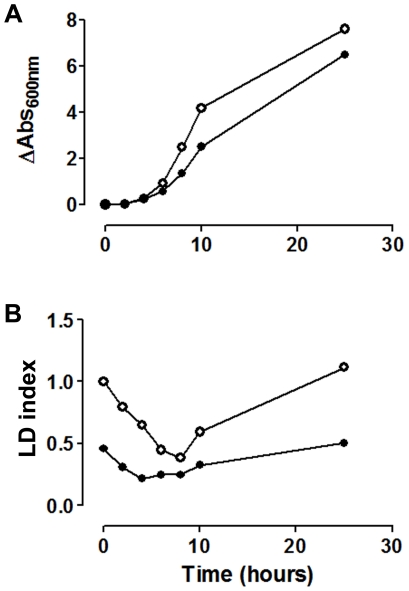
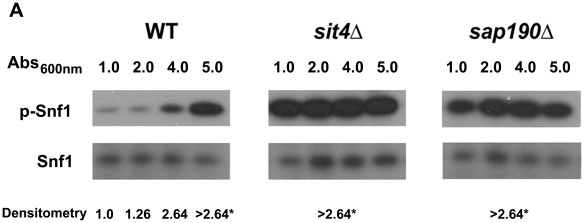
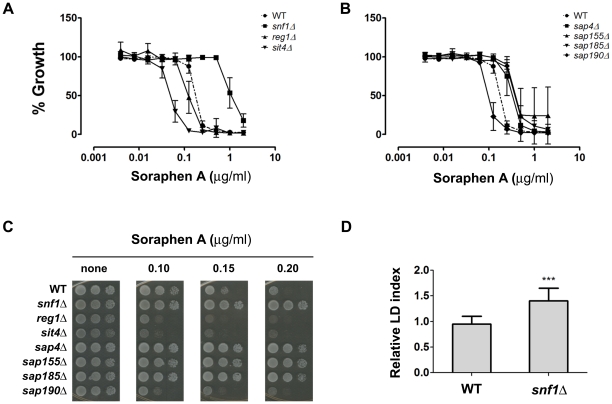
We further investigated whether diminished LD content in sit4 strain was related to a lower rate of synthesis by following its metabolism in a growing culture (Figure 7). Results show that sit4 strain was able to consume LDs after inoculating in fresh medium, during the lag phase of growth, after which the rate of synthesis of LDs was low compared to WT. This result points to a deficient regulation of acetyl-CoA carboxylase activity. In fact, when the phosphorylation state of Snf1p was analyzed during growth, we observed that Snf1p is constitutively phosphorylated in sit4 strain, while it is dephosphorylated in the WT strain during the early log phase. Furthermore, phosphorylation of Snf1p increases throughout culture growth and reaches a maximum during the late exponential phase, when the rate of lipid biosynthesis may have started to slow down (Figure 8). Interestingly Snf1p was also constitutively phosphorylated in sap190 strain during all phases of culture growth. To further confirm that Snf1p is in its active state in sit4 strain (thus inhibiting acetyl-CoA carboxylase activity), we screened the sensitivity of these null mutant strains to soraphen A (Figure 9). Results revealed that sit4 strain is more sensitive to soraphen A. reg1 strain has also been shown to be more sensitive to soraphen A, and it has been shown that in this strain, Snf1p is in its active state [55] and probably leads to an inhibition of acetyl-CoA carboxylase. The results observed were consistent with the classification of sit4 and reg1 as llc strains. As a control, we tested the sensitivity of the snf1 strain. As expected, this resulted in greater resistance to soraphen A due to the activation of acetyl-CoA carboxylase activity [25], [34]. Furthermore, we observed that the snf1 strain presented an LD index that was higher than that of the wild-type (Figure 9D).
Figure 7. LD dynamics is impaired in the sit4 strain.
A. Stationary yeast cells (WT, ○, and sit4Δ, •) grown for 48 in YPD were inoculated into fresh medium. Cellular growth was recorded by measuring absorbance at 600 nm over the course of 25 hours. B. Lipid droplet levels were determined by LFR assay during growth as described in Figure 4. LD index was normalized by stationary WT value. The mean of two independent experiments is shown.
Figure 8. Snf1 kinase phosphorylation levels during growth.
WT, sit4 and sap190 cells were grown in YPD at 30°C while shaking. Aliquots of the culture were harvested at different times during growth (OD 600 nm of 1, 2, 4 or 5), and protein was extracted using the rapid NaOH extraction method [31]. SDS-PAGE gels were loaded with 0.1 OD 600 nm cells per well. Snf1 phosphorylation was analyzed by western blot using anti-phospho AMPKα antibody (P-SNF1, upper panel). As a control for loading, total Snf1 was revealed with anti-SNF1 antibody (lower panel). For autoradiography the films were exposed for the minimum time necessary to detect the phosphorylation signal for WT grown to OD 1.0 which was used as reference. The numbers below the figure represent the densitometry analysis of the P-SNF1/SNF1 ratio (n = 3). Signals were linear unless marked by*.
Figure 9. Soraphen A resistance profiles.
Soraphen A resistance was determined by the microtiter assay and the spot assay. A and B. Serial dilutions of soraphen A were used for the indicated strains (n = 3± S.D.) IC50 were estimated (wt = 0.19, snf1 = 0.95, reg1 = 0.11, sit4 = 0.05, sap4 = 0.36, sap155 = 0.38, sap185 = 0.38 and sap190 = 0.1 µg/ml). C. For the spot assay, serial dilutions (107, 106 and 105 cells/mL, from left to right) of a cellular suspension were seeded onto plates containing YPD solid medium in the presence of increasing concentrations of soraphen A and incubated at 30°C for two days. D. Relative LD index of snf1 cells from a stationary culture in YPD medium was determined by LFR assay, as described. (n = 3,± S.D.) (***p<0.001).
The results of testing the sensitivity of SAP proteins to soraphen A show that sap190 strain leads to a probable inhibition of acetyl-CoA carboxylase activity, which reinforces its classification as an llc mutant. Interestingly, other Sap deletions led to an increase in soraphen A resistance, which is not correlated to an increase in lipid droplets. Such an effect can be explained because it is believed that Saps compete among themselves for Sit4p association and, thus, the deletion of one sap will increase the chances that Sit4 associates with the remaining Saps [47]. These results suggest that the Sit4p-Sap190p complex connects the TOR pathway to the SNF1/AMPK pathway to regulate lipid droplet metabolism (Figure 10).
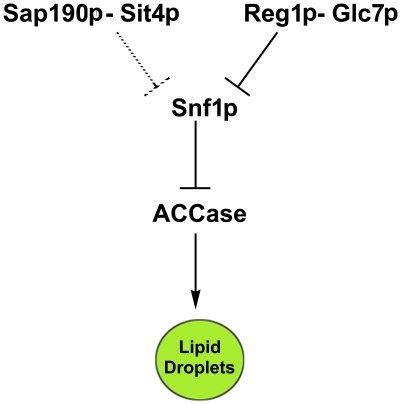
Figure 10. Sit4p orchestrates lipid metabolism in response to nutrients.
Snf1 kinase, a central regulator of energetic homeostasis, when phosphorylated and active, inhibits acetyl CoA carboxylase activity impairing fatty acids synthesis diminishing LD levels. It is dephosphorylated by Reg1/Glc7 phosphatase as glucose levels in the medium increase [50]. Our results suggest that the Sap190-Sit4 pair also participates in Snf1 dephosphorylation and links the TOR pathway to the AMP/ATP-sensitive pathway, mediated by Snf1.
Discussion
There is increasing interest in LD metabolism in the field of biological science as more evidences connect its deregulation to several pathological processes, such as the inflammatory response [56], insulin resistance [57] and even neurodegenerative disorders [58], in addition to the known role of LD in lipid storage. However, there are still fundamental questions to be resolved regarding the signaling pathways involved in LD regulation. The methods employed in lipid studies have not facilitated great advances; such investigations still rely on time-consuming and expensive microscopic studies or on organic solvent extraction and subsequent analysis. Working with such techniques requires well-trained researchers because the steps involved can be tricky and laborious. We propose an alternative, sensitive method for the study of LD dynamics, which can be applied in high-throughput experiments to yield reliable results.
BODIPY 493/503 is a BODIPY fluorophore derivative that, due to its lipophilic nature, accumulates in LDs. It is often employed in microscopic studies of these organelles as an alternative to Nile red. In fact, Nile red acts more as a hydrophobic probe because it only fluoresces in hydrophobic environments in a manner that is proportional to the surrounding hydrophobicity; it has been shown that Nile red stains both membranes and LD [27], [28]. On the other hand, BODIPY is more specific than Nile red for LDs, as revealed by flow cytometry experiments [26], [29]. Furthermore, BODIPY seems to be less sensitive to environmental conditions, such as pH and polarity [59], which could be an advantage when comparing LD staining in different mutant strains. Once optimal conditions were determined, our assay was evaluated by following LD dynamics during culture growth. In this experiment (Figure 4) the size (total area/cell) and number of LDs was assayed by analyzing microscope images of cells stained with BODIPY and TAG levels were measured biochemically. Our fluorescence-based method provided results consistent with the TAG content and with the size of LDs. On the other hand, it did not correlate with the number of LDs which do not vary substantially along the growth curve, when compared to the size of LDs.
Analysis of a reference group comprising strains in which the genes for lipid synthesis and LD dynamics have been knocked out not only validated the entire screening but also reinforced the reliability of our method. In a previous screening in which the entire deletion library was analyzed microscopically to quantify LDs [44], at least two of our hits (dga1 and tgl3) were identified as inducing abnormal phenotypes. However, erg4 and erg5 strains, which were identified as having increased LD number – phenotypes we confirmed microscopically – were identified in our screening as having lower fluorescence levels, which indicates lower lipid content. A plausible explanation is that, although they contain more LDs, these organelles are substantially smaller, which could, in turn, result in reduced BODIPY capture and thus less fluorescence. Interestingly, no relevant protein phosphatases were identified in the previously mentioned screening. In here, among 65 strains in which genes for protein phosphatases or related subunits were knocked out, 10 presented higher lipid droplet content while only 3 (reg1, sit4 and its regulatory subunit sap190) presented lower lipid droplet content. This result suggests that a loss of protein phosphatase function is more likely to result in fluctuations in LDs size as compared to the number of LDs. By analyzing the overall results our attention was drawn to the involvement of three subunits of Glc7 phosphatase. gac1 and bni4 were identified as hlc while reg1 as llc. Glc7p phosphatase interacts with different subunits each leading Glc7p to different substrates regulating several cellular processes as cell cycle, glycogen, sugar and lipid metabolism [60], [61]. Gac1p and Bni4p recruit Glc7p phosphatase to glycogen synthase Gsy2p [62] or the bud neck [63], respectively, and normally they may sequester some Glc7 phosphatase away from Snf1p. Their deletion might make more Glc7p available for Reg1p-Snf1p, thus increasing Snf1p dephosphorylation/inactivation, thereby increasing Acc1p activity and fatty acid synthesis. On the other hand interaction of Reg1p with Glc7p is necessary to dephosphorylate Snf1p. Its deletion leads to phosphorylation/activation of Snf1p, inhibiting the synthesis of LDs.
These three genes, REG1, SIT4 and SAP190, have been identified in a previous screen as necessary for normal growth on oleic acid. The reason how the presence of Sap190p-Sit4p prevents oleate inhibition is not known [64]. Kohlwein et al [17] have shown that TAG synthesis plays an important role to buffer excess oleic acid, as strains lacking the acyltransferases Lro1p, Dga1p, Are1p, and Are2p are sensitive to this fatty acid. Together with our results, these observations suggest that reg1, sit4 and sap190 are necessary to induce synthesis of LDs in the presence of oleate, thus preventing its toxicity.
We further studied the role of Sit4p participation in lipid metabolism. The sit4 strain LD dynamics showed a lower rate of LD synthesis after lag-phase, in comparison to WT (Figure 7). Although we cannot discard defects in LD synthesis or the degradation apparatus, the Sit4p deletion effect is likely due to the diminished availability of construction blocks such as FA. In fact, researchers believe that Sit4 phosphatase might dephosphorylate acetyl-CoA carboxylase, a key enzyme in fatty acid metabolism. However, experimental results have not been published [65]–[67]. Here, we showed that sit4 deletion increases sensitivity to soraphen A, an acetyl-CoA carboxylase inhibitor. The same is observed for the deletion of sap190, a regulatory subunit of SIT4. Moreover, it is important to observe that sap185 deletion induced a phenotype opposite to that elicited by sap190 deletion; the sap185 knockout strain was detected as a high lipid content strain during screening. Although SAPs compete for Sit4 binding [47] and have exhibited opposing functions [68], so far, no antagonistic relationship between Sap185p and Sap190p has been observed. More studies will be necessary to answer whether the opposing LD phenotypes resulting from these SAP genes deletions are related, indicating the existence of a novel antagonistic pair.
To explore a possible connection between TOR and Snf1 signaling pathways (because Snf1 takes part in nitrogen signaling and both are implicated in lifespan extension mechanisms), we investigated Snf1 phosphorylation status in the sit4 strain. Surprisingly, the deletion of sit4 (or its subunit sap190) leads to increased constitutive Snf1 phosphorylation, being more active. This finding would explain an increase in soraphen A sensitivity due to a decrease in acetyl-CoA carboxylase activity. Although we cannot discard the hypothesis that Sit4 directly dephosphorylates acetyl-CoA carboxylase, we provide evidence that Sit4 participation in lipid metabolism occurs through the regulation of Snf1 phosphorylation status and links the TOR pathway to the AMP/ATP-sensitive pathway, which is regulated by Snf1/AMPK.
Supporting Information
Liquid fluorescence recovery assay (LFR assay). Schematic representation of the LFR assay, where cells are first fixed in 3.7% formaldehyde, washed and then added to BODIPY-quenched solution containing 5 µM BODIPY plus 500 mM KI. Fluorescence is recovered and detected with a fluorimeter (ex/em = 485/510 nm).
(1.46 MB TIF)
High-throughput screening of a knockout collection. Individual strains are pre-grown to stationary phase at 30°C and fixed in formaldehyde (3.7%). Cells are washed and added to a 96-well black-wall/clear-bottom plate. Fluorescence (ex/em = 485/510 nm) and absorbance (600 nm) are recorded. Three more subsequent readings were performed after the addition of cells to the wells in order to determine the LD index.
(0.46 MB TIF)
Fluorescence properties of BODIPY are not altered in the presence of cells. An aqueous solution of BODIPY (5 µM) was incubated in the presence of the indicated of none (Ο), 0.025 OD (square), 0.05 OD (Δ) and 0.1 OD (∇) of cells and the absorption spectra (upper panel) and emission (excitation at 485 nm) (lower panel) of BODIPY were recorded. Results are in agreement with information provided by the manufacturer (BODIPY Lipid Probes manual, available online at www.invitrogen.com), which states that BODIPY-derivative fluorescence parameters are insensitive to environmental conditions and not quenched by water.
(0.62 MB TIF)
LD content in erg4 and erg5 strains was determined by fluorescence microscopy. Cells were grown to stationary phase and incubated with BODIPY. The total fluorescence area/cell was determined and expressed in pixels/cell (white bars). LDs per cell were quantified using the same images (gray bars). Data provided are for at least 100 individual cells. LD index is indicated below the graph for each strain. *p<0.05, ** p<0.01, ***p<0.001, in comparison to WT values.
(1.59 MB TIF)
LD index correlates with either TAG or SE content. WT, are1Δ are2Δ and dga1Δ lro1Δ strains were grown to stationary phase and incubated with BODIPY. Images were captured for 17msec. A second capture (74msec) for dga1Δ lro1Δ, is also shown (last panel). At the bottom of the figure the LD index (LFR assay) (n = 3) and size (fluorescence area/cell LDs, microscopy images n = 100 cells) for each strain are shown. ** p<0.01, ***p<0.001, in comparison to WT values.
(2.78 MB TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by Conselho Nacional de Ciência e Tecnologia (CNPq, http://www.cnpq.br) and Fundão de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ, http://www.faperj.br) awards to MML and CAM; a Fundão Universitária José Bonifácio/UFRJ (http://www.ufrj.br/) award to CAM; a PEW Charitable Trust (http://www.pewtrusts.org/) award to CAM. JM and BBM were the recipients of an undergraduate fellowship and a doctoral fellowship, respectively, from CNPq. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. S0163-7827(01)00013-3 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Clausen MK, Christiansen K, Jensen PK, Behnke O. Isolation of lipid particles from baker's yeast. FEBS Lett. 1974;43:176–179. doi: 10.1016/0014-5793(74)80994-4. 0014-5793(74)80994-4 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Leber R, Landl K, Zinser E, Ahorn H, Spok A, et al. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, et al. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- 6.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. 10.1074/jbc.M207712200 [doi];M207712200 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. S0968-0004(98)01349-8 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. 10.1074/jbc.M111646200 [doi];M111646200 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, et al. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. 10.1073/pnas.120067297 [doi];120067297 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zweytick D, Leitner E, Kohlwein SD, Yu C, Rothblatt J, Daum G. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur J Biochem. 2000;267:1075–1082. doi: 10.1046/j.1432-1327.2000.01103.x. ejb1103 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Sorger D, Daum G. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J Bacteriol. 2002;184:519–524. doi: 10.1128/JB.184.2.519-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Athenstaedt K, Daum G. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J Biol Chem. 2003;278:23317–23323. doi: 10.1074/jbc.M302577200. 10.1074/jbc.M302577200 [doi];M302577200 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280:37301–37309. doi: 10.1074/jbc.M507261200. M507261200 [pii];10.1074/jbc.M507261200 [doi] [DOI] [PubMed] [Google Scholar]
- 14.Koffel R, Tiwari R, Falquet L, Schneiter R. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol Cell Biol. 2005;25:1655–1668. doi: 10.1128/MCB.25.5.1655-1668.2005. 25/5/1655 [pii];10.1128/MCB.25.5.1655-1668.2005 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullner H, Deutsch G, Leitner E, Ingolic E, Daum G. YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:13321–13328. doi: 10.1074/jbc.M409914200. M409914200 [pii];10.1074/jbc.M409914200 [doi] [DOI] [PubMed] [Google Scholar]
- 16.Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, et al. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. S1097-2765(08)00887-3 [pii];10.1016/j.molcel.2008.12.019 [doi] [DOI] [PubMed] [Google Scholar]
- 17.Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, et al. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. M109.024752 [pii];10.1074/jbc.M109.024752 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garbarino J, Sturley SL. Homoeostatic systems for sterols and other lipids. Biochem Soc Trans. 2005;33:1182–1185. doi: 10.1042/BST20051182. BST20051182 [pii];10.1042/BST20051182 [doi] [DOI] [PubMed] [Google Scholar]
- 19.Garbarino J, Padamsee M, Wilcox L, Oelkers PM, D'Ambrosio D, et al. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J Biol Chem. 2009;284:30994–31005. doi: 10.1074/jbc.M109.050443. M109.050443 [pii];10.1074/jbc.M109.050443 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bozza PT, Viola JP. Lipid droplets in inflammation and cancer. Prostaglandins Leukot Essent Fatty Acids. 2010;82:243–250. doi: 10.1016/j.plefa.2010.02.005. S0952-3278(10)00049-9 [pii];10.1016/j.plefa.2010.02.005 [doi] [DOI] [PubMed] [Google Scholar]
- 21.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000632. e1000632. 10.1371/journal.ppat.1000632 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maya-Monteiro CM, Almeida PE, D'Avila H, Martins AS, Rezende AP, et al. Leptin induces macrophage lipid body formation by a phosphatidylinositol 3-kinase- and mammalian target of rapamycin-dependent mechanism. J Biol Chem. 2008;283:2203–2210. doi: 10.1074/jbc.M706706200. M706706200 [pii];10.1074/jbc.M706706200 [doi] [DOI] [PubMed] [Google Scholar]
- 23.Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, et al. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. M508414200 [pii];10.1074/jbc.M508414200 [doi] [DOI] [PubMed] [Google Scholar]
- 24.Woods A, Munday MR, Scott J, Yang X, Carlson M, et al. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- 25.Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, et al. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gocze PM, Freeman DA. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 1994;17:151–158. doi: 10.1002/cyto.990170207. 10.1002/cyto.990170207 [doi] [DOI] [PubMed] [Google Scholar]
- 27.Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, nile red. J Lipid Res. 1985;26:781–789. [PubMed] [Google Scholar]
- 28.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. 0704154104 [pii];10.1073/pnas.0704154104 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohlwein SD, Petschnigg J. Lipid-induced cell dysfunction and cell death: lessons from yeast. Curr Hypertens Rep. 2007;9:455–461. doi: 10.1007/s11906-007-0084-5. [DOI] [PubMed] [Google Scholar]
- 31.Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 33.Montero-Lomeli M, Morais BL, Figueiredo DL, Neto DC, Martins JR, et al. The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J Biol Chem. 2002;277:21542–21548. doi: 10.1074/jbc.M201977200. 10.1074/jbc.M201977200 [doi];M201977200 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Schneiter R, Guerra CE, Lampl M, Gogg G, Kohlwein SD, et al. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Delta is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol Cell Biol. 1999;19:3415–3422. doi: 10.1128/mcb.19.5.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bozza PT, Payne JL, Morham SG, Langenbach R, Smithies O, et al. Leukocyte lipid body formation and eicosanoid generation: cyclooxygenase-independent inhibition by aspirin. Proc Natl Acad Sci U S A. 1996;93:11091–11096. doi: 10.1073/pnas.93.20.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiDonato D, Brasaemle DL. Fixation methods for the study of lipid droplets by immunofluorescence microscopy. J Histochem Cytochem. 2003;51:773–780. doi: 10.1177/002215540305100608. [DOI] [PubMed] [Google Scholar]
- 37.Fowler SD, Greenspan P. Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. . J Histochem Cytochem. 1985;33:833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- 38.Liao J, Sportsman R, Harris J, Stahl A. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J Lipid Res. 2005;46:597–602. doi: 10.1194/jlr.D400023-JLR200. D400023-JLR200 [pii];10.1194/jlr.D400023-JLR200 [doi] [DOI] [PubMed] [Google Scholar]
- 39.Díaz-García ME, Badía-Laíño R. Fluorescence. In: Townshend A, Worsfold P, Poole C, editors. Encyclopedia of Analytical Science. New York: Academic Press; 2005. pp. 97–106. doi: 10.1016/B0-12-369397-7/00161-8. [Google Scholar]
- 40.Zanghellini J, Natter K, Jungreuthmayer C, Thalhammer A, Kurat CF, et al. Quantitative modeling of triacylglycerol homeostasis in yeast–metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. FEBS J. 2008;275:5552–5563. doi: 10.1111/j.1742-4658.2008.06681.x. EJB6681 [pii];10.1111/j.1742-4658.2008.06681.x [doi] [DOI] [PubMed] [Google Scholar]
- 41.Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, et al. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4:662–672. doi: 10.1074/mcp.M400123-MCP200. M400123-MCP200 [pii];10.1074/mcp.M400123-MCP200 [doi] [DOI] [PubMed] [Google Scholar]
- 42.Lorenz RT, Parks LW. Effects of lovastatin (mevinolin) on sterol levels and on activity of azoles in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1990;34:1660–1665. doi: 10.1128/aac.34.9.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J Bacteriol. 1997;179:7611–7616. doi: 10.1128/jb.179.24.7611-7616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, et al. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. jcb.200711136 [pii];10.1083/jcb.200711136 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohde JR, Bastidas R, Puria R, Cardenas ME. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr Opin Microbiol. 2008;11:153–160. doi: 10.1016/j.mib.2008.02.013. S1369-5274(08)00024-6 [pii];10.1016/j.mib.2008.02.013 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 47.Luke MM, Della SF, Di Como CJ, Sugimoto H, Kobayashi R, et al. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohde JR, Campbell S, Zurita-Martinez SA, Cutler NS, Ashe M, et al. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol Cell Biol. 2004;24:8332–8341. doi: 10.1128/MCB.24.19.8332-8341.2004. 10.1128/MCB.24.19.8332-8341.2004 [doi];24/19/8332 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frederick DL, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol Cell Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Vemuri G, Nielsen J. Systems biology of energy homeostasis in yeast. Curr Opin Microbiol. 2010;13:382–388. doi: 10.1016/j.mib.2010.04.004. S1369-5274(10)00056-1 [pii];10.1016/j.mib.2010.04.004 [doi] [DOI] [PubMed] [Google Scholar]
- 52.McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem . 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. 10.1074/jbc.M104418200 [doi];M104418200 [pii] [DOI] [PubMed] [Google Scholar]
- 53.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vahlensieck HF, Pridzun L, Reichenbach H, Hinnen A. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr Genet. 1994;25:95–100. doi: 10.1007/BF00309532. [DOI] [PubMed] [Google Scholar]
- 55.Hess D, Winston F. Evidence that Spt10 and Spt21 of Saccharomyces cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1. Genetics. 2005;170:87–94. doi: 10.1534/genetics.104.039214. genetics.104.039214 [pii];10.1534/genetics.104.039214 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bozza PT, Melo RC, Bandeira-Melo C. Leukocyte lipid bodies regulation and function: contribution to allergy and host defense. Pharmacol Ther. 2007;113:30–49. doi: 10.1016/j.pharmthera.2006.06.006. S0163-7258(06)00120-3 [pii];10.1016/j.pharmthera.2006.06.006 [doi] [DOI] [PubMed] [Google Scholar]
- 57.Bostrom P, Andersson L, Rutberg M, Perman J, Lidberg U, et al. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. ncb1648 [pii];10.1038/ncb1648 [doi] [DOI] [PubMed] [Google Scholar]
- 58.Cole NB, Murphy DD, Grider T, Rueter S, Brasaemle D, et al. Lipid droplet binding and oligomerization properties of the Parkinson's disease protein alpha-synuclein. J Biol Chem. 2002;277:6344–6352. doi: 10.1074/jbc.M108414200. 10.1074/jbc.M108414200 [doi];M108414200 [pii] [DOI] [PubMed] [Google Scholar]
- 59.Karolin J, Johansson LBA, Strandberg L, Ny T. Fluorescence and Absorption Spectroscopic Properties of Dipyrrometheneboron Difluoride (BODIPY) Derivatives in Liquids, Lipid Membranes, and Proteins. Journal of the American Chemical Society. 2002;116:7801–7806. doi: 10.1021/ja00096a042. [Google Scholar]
- 60.Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. 10.1152/physrev.00013.2003 [doi];84/1/1 [pii] [DOI] [PubMed] [Google Scholar]
- 61.Logan MR, Nguyen T, Szapiel N, Knockleby J, Por H, et al. Genetic interaction network of the Saccharomyces cerevisiae type 1 phosphatase Glc7. BMC Genomics. 2008;9:336. doi: 10.1186/1471-2164-9-336. 1471-2164-9-336 [pii];10.1186/1471-2164-9-336 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X, Hart H, Cheng C, Roach PJ, Tatchell K. Characterization of Gac1p, a regulatory subunit of protein phosphatase type I involved in glycogen accumulation in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:622–635. doi: 10.1007/s004380100455. [DOI] [PubMed] [Google Scholar]
- 63.Kozubowski L, Panek H, Rosenthal A, Bloecher A, DeMarini DJ, et al. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol Biol Cell. 2003;14:26–39. doi: 10.1091/mbc.E02-06-0373. 10.1091/mbc.E02-06-0373 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lockshon D, Surface LE, Kerr EO, Kaeberlein M, Kennedy BK. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics. 2007;175:77–91. doi: 10.1534/genetics.106.064428. 10.1534/genetics.106.064428 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. 10.1038/415180a [doi];415180a [pii] [DOI] [PubMed] [Google Scholar]
- 66.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. 10.1038/415141a [doi];415141a [pii] [DOI] [PubMed] [Google Scholar]
- 67.Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. S1388-1981(06)00199-5 [pii];10.1016/j.bbalip.2006.07.004 [doi] [DOI] [PubMed] [Google Scholar]
- 68.Jablonowski D, Taubert JE, Bar C, Stark MJ, Schaffrath R. Distinct subsets of Sit4 holophosphatases are required for inhibition of Saccharomyces cerevisiae growth by rapamycin and zymocin. Eukaryot Cell. 2009;8:1637–1647. doi: 10.1128/EC.00205-09. EC.00205-09 [pii];10.1128/EC.00205-09 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Liquid fluorescence recovery assay (LFR assay). Schematic representation of the LFR assay, where cells are first fixed in 3.7% formaldehyde, washed and then added to BODIPY-quenched solution containing 5 µM BODIPY plus 500 mM KI. Fluorescence is recovered and detected with a fluorimeter (ex/em = 485/510 nm).
(1.46 MB TIF)
High-throughput screening of a knockout collection. Individual strains are pre-grown to stationary phase at 30°C and fixed in formaldehyde (3.7%). Cells are washed and added to a 96-well black-wall/clear-bottom plate. Fluorescence (ex/em = 485/510 nm) and absorbance (600 nm) are recorded. Three more subsequent readings were performed after the addition of cells to the wells in order to determine the LD index.
(0.46 MB TIF)
Fluorescence properties of BODIPY are not altered in the presence of cells. An aqueous solution of BODIPY (5 µM) was incubated in the presence of the indicated of none (Ο), 0.025 OD (square), 0.05 OD (Δ) and 0.1 OD (∇) of cells and the absorption spectra (upper panel) and emission (excitation at 485 nm) (lower panel) of BODIPY were recorded. Results are in agreement with information provided by the manufacturer (BODIPY Lipid Probes manual, available online at www.invitrogen.com), which states that BODIPY-derivative fluorescence parameters are insensitive to environmental conditions and not quenched by water.
(0.62 MB TIF)
LD content in erg4 and erg5 strains was determined by fluorescence microscopy. Cells were grown to stationary phase and incubated with BODIPY. The total fluorescence area/cell was determined and expressed in pixels/cell (white bars). LDs per cell were quantified using the same images (gray bars). Data provided are for at least 100 individual cells. LD index is indicated below the graph for each strain. *p<0.05, ** p<0.01, ***p<0.001, in comparison to WT values.
(1.59 MB TIF)
LD index correlates with either TAG or SE content. WT, are1Δ are2Δ and dga1Δ lro1Δ strains were grown to stationary phase and incubated with BODIPY. Images were captured for 17msec. A second capture (74msec) for dga1Δ lro1Δ, is also shown (last panel). At the bottom of the figure the LD index (LFR assay) (n = 3) and size (fluorescence area/cell LDs, microscopy images n = 100 cells) for each strain are shown. ** p<0.01, ***p<0.001, in comparison to WT values.
(2.78 MB TIF)