This study reveals the subcentrosomal distribution of ALMS1, a human protein implicated in primary cilium formation and maintenance, and provides new insight into its centrosome-related functions. The first functional data on two human proteins sharing C-terminal sequence similarity with ALMS1 are also presented.
Abstract
Mutations in the human gene ALMS1 cause Alström syndrome, a rare progressive condition characterized by neurosensory degeneration and metabolic defects. ALMS1 protein localizes to the centrosome and has been implicated in the assembly and/or maintenance of primary cilia; however its precise function, distribution within the centrosome, and mechanism of centrosomal recruitment are unknown. The C-terminus of ALMS1 contains a region with similarity to the uncharacterized human protein C10orf90, termed the ALMS motif. Here, we show that a third human protein, the candidate centrosomal protein KIAA1731, contains an ALMS motif and that exogenously expressed KIAA1731 and C10orf90 localize to the centrosome. However, based on deletion analysis of ALMS1, the ALMS motif appears unlikely to be critical for centrosomal targeting. RNAi analyses suggest that C10orf90 and KIAA1731 have roles in primary cilium assembly and centriole formation/stability, respectively. We also show that ALMS1 localizes specifically to the proximal ends of centrioles and basal bodies, where it colocalizes with the centrosome cohesion protein C-Nap1. RNAi analysis reveals markedly diminished centrosomal levels of C-Nap1 and compromised cohesion of parental centrioles in ALMS1-depleted cells. In summary, these data suggest centrosomal functions for C10orf90 and KIAA1731 and new centriole-related functions for ALMS1.
INTRODUCTION
Mutations in the ALMS1 gene cause Alström syndrome (Collin et al., 2002; Hearn et al., 2002; Marshall et al., 2007), a rare progressive condition characterized by childhood obesity, insulin resistance, retinal degeneration, and sensorineural hearing loss, with other common features including dilated cardiomyopathy, short stature, and hepatic and renal dysfunction (Alstrom et al., 1959; Marshall et al., 2005). The amino acid sequence of ALMS1 contains several notable features, including a large tandem repeat region and a putative leucine-zipper motif, but it bears no significant similarity to proteins of known function (Collin et al., 2002; Hearn et al., 2002). The C-terminus of ALMS1 shares sequence similarity with the C-terminus of an uncharacterized human protein, C10orf90, and this region of similarity has been termed the ALMS motif (Collin et al., 2002). ALMS1 has been implicated in the function, formation, and/or maintenance of primary cilia (Collin et al., 2005; Hearn et al., 2005; Graser et al., 2007a; Li et al., 2007); however, its precise role is unknown.
ALMS1 localizes to the centrosome (Andersen et al., 2003; Hearn et al., 2005), an organelle comprising a pair of centrioles (termed mature and immature parental centrioles) embedded in a matrix of pericentriolar material (PCM). The centrosome is the major microtubule-organizing center in most animal cells and also has roles in, for example, regulating cell cycle transitions and cytokinesis (Doxsey et al., 2005; Azimzadeh and Bornens, 2007). During interphase, the two parental centrioles are thought to be linked by fibers emanating from their proximal ends (Lim et al., 2009). C-Nap1 (centrosomal Nek2-associated protein 1) has a role in maintaining this intercentriolar link (and hence the close association of parental centrioles) most likely by acting as the centriolar docking site for rootletin, a major component of the interconnecting fibers (Bahe et al., 2005; Yang et al., 2006). Several other proteins have been implicated in structural or regulatory roles relating to centrosome cohesion, including Cep215, CPAP, dynamin 2, and β-catenin (Thompson et al., 2004; Graser et al., 2007b; Bahmanyar et al., 2008; Zhao et al., 2010), suggesting the involvement of multiple mechanisms. However quantitative analysis indicates that C-Nap1 and rootletin are particularly important for the maintenance of centrosome cohesion (Graser et al., 2007b).
C-Nap1 and rootletin dissociate from parental centrioles at the onset of mitosis, concomitant with centrosome separation (Fry et al., 1998; Mayor et al., 2002; Bahe et al., 2005). At this stage each centrosome contains one parental and one progeny centriole, centriole duplication having begun during S phase with one procentriole being assembled orthogonally to each parental centriole. The progeny centriole remains attached to its parent until the end of mitosis, at which point the two disengage and concomitantly establish a rootletin-containing link between their proximal ends (Azimzadeh and Bornens, 2007; Lim et al., 2009). The nature and regulation of the link between parental centrioles is therefore of interest in terms of understanding centrosome separation at the G2-M transition as well as centriole dynamics during interphase. Mechanisms controlling centriole duplication are also of considerable interest because strict control of centriole/centrosome number is required to maintain genome stability in cycling cells (Tsou and Stearns, 2006; Cunha-Ferreira et al., 2009).
In most noncycling vertebrate cells the mature parental centriole attaches to the plasma membrane and becomes a basal body, which acts as the template for assembly of the primary cilium, a sensory cell surface organelle (Singla and Reiter, 2006; Dawe et al., 2007). The cilium, whose basic structure comprises a microtubule-based axoneme enclosed by a sheath of plasma membrane, is assembled and maintained by a microtubule motor protein-driven process termed intraflagellar transport (IFT; Silverman and Leroux, 2009). Recent data indicate that ALMS1 is required for the proper formation and/or maintenance of primary cilia (Graser et al., 2007a; Li et al., 2007), strongly suggesting that at least some features of Alström syndrome are due to ciliary dysfunction, as is the case for numerous other human genetic disorders, now collectively termed ciliopathies (Badano et al., 2006; Marshall, 2008; Nigg and Raff, 2009).
Here, we further investigate the localization and function of ALMS1. We use high-resolution immunofluorescence microscopy to determine its subcentrosomal distribution and generate deletion constructs to query which regions of the protein are required for centrosomal-targeting. We use RNA interference (RNAi) in cultured cells to investigate a potential role of ALMS1 suggested by its localization pattern within the centrosome. We also identify an ALMS motif in the candidate centrosomal human protein KIAA1731, analyze the subcellular localization of exogenously expressed KIAA1731 and C10orf90, and use RNAi to investigate the functions of both proteins.
MATERIALS AND METHODS
Plasmid Construction
The coding sequence of ALMS1 was cloned by PCR in pCMV-HA (BD Biosciences, San Jose, CA), providing an N-terminal hemagglutinin (HA) tag. PCRs were performed on human cDNA or BAC DNA (Expand High Fidelity; Roche Diagnostics, Burgess Hill, United Kingdom). Because of its large size, the coding sequence was initially cloned in sections. ALMS1 deletion constructs were generated by restriction enzyme digestion of the full-length clone or by PCR cloning into pCMV-HA. The insert of cDNA clone KIAA1731 (GenBank accession no. AB051518; Kazusa DNA Research Institute, Chiba, Japan) was transferred into pCMV-Myc (BD Biosciences), providing an N-terminal Myc tag. A two-base pair deletion in the open reading frame of the source clone was corrected by replacing a BglII/PmeI fragment with a corresponding fragment amplified by PCR from human cDNA. A MluI/KpnI fragment from IMAGE clone 4823075 (Geneservice, Cambridge, United Kingdom), encoding residues 243–796 of C10orf90 (GenBank accession no. BAG59968), was inserted into pCMV-Myc. The coding sequence of CP110 was amplified by PCR from IMAGE clone 5267904 (Geneservice) using restriction site-tagged primers and inserted into pCMV-HA. All constructs were verified by sequencing.
Cell Culture and DNA Transfection
U2OS cells (ECACC, Porton Down, United Kingdom) and HEK 293 cells (ATCC, Manassas, VA) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37°C and 5% CO2 (reagents from PAA Laboratories, Yeovil, United Kingdom). hTERT-RPE1 cells (ATCC) were maintained in DMEM/Ham's F12 with the same supplements and conditions. Cells to be analyzed by immunofluorescence were seeded in Lab-Tek II chamber slides (VWR International, Lutterworth, United Kingdom). Plasmid transfections were performed with Lipofectamine 2000 (Invitrogen, Paisley, United Kingdom), and cells were processed for immunofluorescence 24 h later.
RNAi
Cells were seeded in chamber slides and transfected with siRNA duplexes (Qiagen, Crawley, West Sussex, United Kingdom) at 50 nM using HiPerFect transfection reagent (Qiagen). Cells were processed for immunofluorescence 96 h after transfection. Small interfering RNA (siRNA) target sequences (5′ to 3′) were as follows: gtgaacatttcagatttcgaa (ALMS1_06), cagagagtaacttaaccgaag (ALMS1_07), cagaactttatacctgatgaa (ALMS1_7966, oligo 343 in Graser et al., 2007a), caggttgacacctcttcctta (KIAA1731_4333), aacaattacttgaatatcaaa (KIAA1731_03), aatgtgggtcatcatatagta (C10orf90_07), cagcgctggaacccaggttta (C10orf90_08), tcagcttcgtgattctcag (PCM1; Dammermann and Merdes, 2002), and ctggaagagcgtctaactgat (C-Nap1; Bahe et al., 2005). AllStars Negative Control siRNA was purchased from Qiagen. Preliminary attempts to rescue the dispersed-PCM1 phenotype observed in cells treated with ALMS1-directed siRNAs were hampered by the finding that transient expression of a control protein, enhanced green fluorescent protein (eGFP), could affect the subcellular distribution of PCM1, and we did not pursue this approach further. To monitor depletion of KIAA1731 and C10orf90 mRNA, siRNA transfections were performed in six-well plates and total RNA extracted 72 h later using TRI reagent (Sigma-Aldrich Poole, Dorset, UK). Oligo-dT–primed reverse transcription was performed with SuperScript III (Invitrogen), and the resulting cDNAs were amplified by PCR using primers for C10orf90 (5′-gagccctggcctgtccgaagac-3′, 5′-gtcctcccaggtccaggtagtgtg-3′) and HPRT (5′-cctggcgtcgtgattagtgatgat-3′, 5′-agcttgcgaccttgacca-3′), or by quantitative PCR (qPCR) using predesigned Taqman Gene Expression Assays for KIAA1731 and HPRT (Applied Biosystems, Warrington, UK). qPCRs were performed in triplicate using an ABI 7900HT Fast Real-Time System (Applied Biosystems) and relative quantification. For immunoblot analysis, HEK 293 cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) and subsequently were incubated on ice for 20 min in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.5% Triton X-100) supplemented with protease inhibitors (Complete Mini; Roche Diagnostics). Cell lysates were cleared by centrifugation at 13,000 rpm for 10 min at 4°C, and SDS-PAGE and immunoblotting were done as described (Hearn et al., 2005).
Immunofluorescence Microscopy
Cells were rinsed in PBS and fixed in chilled methanol for 5 min. When required for clear visualization of centrioles and/or ciliary axonemes, U2OS and hTERT-RPE1 cells were incubated on ice for 50 and 20 min, respectively (before fixation) to depolymerize cytoplasmic microtubules. Immunofluorescence was performed as described (Hearn et al., 2005) using the following primary antibodies: rabbit anti-ALMS1TRD (1 μg/ml; Hearn et al., 2005); mouse anti-HA, mouse anti-α-actinin BM-75.2, mouse anti-γ-tubulin GTU-88, and rabbit anti-γ-tubulin DQ-19 (1:500; Sigma-Aldrich); mouse anti-acetylated α-tubulin 6-11B-1 (1:2000; Sigma-Aldrich); mouse or rabbit anti-pericentrin (1:1000; Abcam, Cambridge, United Kingdom); mouse anti-c-Myc, anti-Cyclin B1 GNS1, and anti-C-Nap1 (1:500; BD Biosciences); and rabbit anti-PCM1 (1:2000; Dammermann and Merdes, 2002). Goat secondary antibodies (1:200) were from Sigma-Aldrich (FITC conjugates) or Invitrogen (Alexa Fluor 350, 488 and 594 conjugates). Cells were mounted in Vectashield (Vector Laboratories, Peterborough, United Kingdom) with or without DAPI. Images were captured using an Axio Observer Z1 microscope equipped with an AxioCam MRm digital camera (Carl Zeiss, Welwyn Garden City, United Kingdom). 2D deconvolution was performed where stated using AxioVision software. For experiments requiring comparison of signal intensities, images were captured using identical settings. Mean pixel intensity, minus background, within an area of 1.5 μm2 encompassing the centrosome was measured using the Photoshop histogram tool (Adobe Systems, San Jose, CA).
4Pi Microscopy
Cells were grown on quartz coverslips in six-well plates and processed for immunofluorescence as above. After antibody incubations and washes, cells were mounted in glycerol (refractive index n = 1.46). Images were collected using a Leica TCS 4Pi microscope with 100× 1.35 NA objectives and Leica LCS software using appropriate bandpass filters for each fluorochrome (Leica MicroSystems Inc, Bannockburn, IL). After linear three-point deconvolution, three-dimensional reconstructions were performed using Imaris software (Bitplane, Saint Paul, MN).
Sequence Analysis
The National Center for Biotechnology Informatics (NCBI) nonredundant protein sequence database was searched using BLASTP with composition-based statistics (http://blast.ncbi.nlm.nih.gov/Blast.cgi/). Position-Specific Iterated (PSI)-BLAST analysis was performed using the default inclusion threshold (0.005) and composition-based statistics. Pattern hit-initiated (PHI)-BLAST analysis of the ALMS motif was performed using the following pattern: Y-x(4)-EV-x(5)-[EQ]-x(10)-[RK]-x(5)-[FY], with the default inclusion threshold. The resulting hits were then used for a single iteration of PSI-BLAST. Multiple sequence alignments were performed using ClustalW (gap penalty 10; gap length penalty 0.2) within Lasergene software (DNASTAR, Madison, WI) and shaded using GeneDoc software (www.nrbsc.org) with default similarity groups.
RESULTS
Mapping Centrosome-targeting Domains of ALMS1
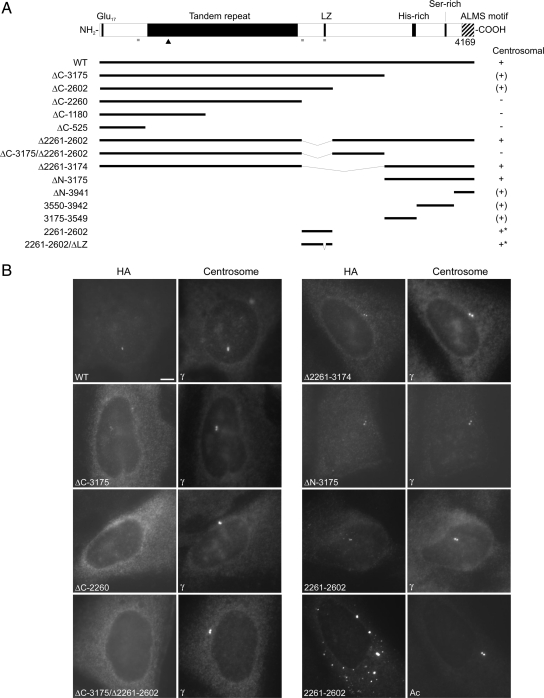
ALMS1 is a large (4169 amino acid) protein with several notable sequence features, including an extensive tandem repeat domain, a putative leucine-zipper motif, and an ALMS motif (Figure 1A). Unlike many other centrosomal proteins, it has limited potential to form coiled-coils (Figure 1A). To query if a particular motif or domain targets ALMS1 to centrosomes, we generated a panel of HA-tagged deletion constructs for immunolocalization analysis. We transiently expressed constructs in U2OS cells and compared their localizations to that of the centrosome marker γ-tubulin. Briefly, this suggested that the C-terminal quarter of ALMS1 (represented in construct ΔN-3175) is important for efficient centrosome-targeting, but also implicated an internal region (residues 2261–2602; Figure 1, A and B, and data not shown). In general, specific and compact centrosomal localization of transfected ALMS1 constructs was observed at low expression levels, whereas additional diffuse intracellular staining was apparent at higher expression levels.
Figure 1.
Deletion analysis identifies two potential centrosome-targeting regions in ALMS1. (A) ALMS1 primary structure and summary of HA-tagged deletion constructs. All constructs lack exon 2 (not shown), which encodes 42 residues but appears to be rarely expressed in humans (Collin et al., 2002; our unpublished observations). The ability of transiently transfected constructs to localize to the centrosome is summarized to the right: +, compact centrosomal; (+), diffuse or less compact centrosomal; −, no detectable centrosomal staining. Asterisks denote constructs that gave additional prominent punctate/aggregate-like staining throughout the cytoplasm. LZ, putative leucine zipper; Glu17, stretch of consecutive glutamic acid residues. Gray bars denote regions with >0.8 probability of forming coiled coils (COILS program, window size 21; Lupas, 1996); the arrowhead indicates the epitope of the antibody used in subsequent experiments to detect endogenous ALMS1. (B) Immunofluorescence data for selected constructs. Cells were costained with antibodies to HA and either γ-tubulin or acetylated tubulin (Ac), as indicated, to mark the centrosome. Size bar, 5 μm.
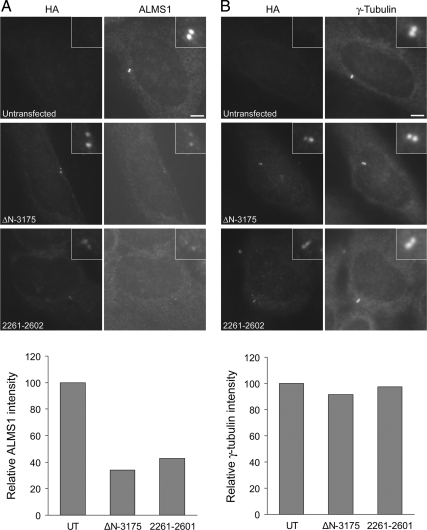
Notably, at moderate-to-high expression levels the construct representing residues 2261–2602 gave punctate staining throughout the cytoplasm and appeared to form aggregates, one of which typically coincided with the centrosome (Figure 1B). Misfolded proteins can accumulate in the vicinity of the centrosome via the aggresome pathway (Kopito, 2000). However, at low expression levels, construct 2261–2602 (and construct ΔN-3175) colocalized specifically with endogenous ALMS1 (Supplemental Figure S1), suggesting that at least a fraction of this construct assembles at the centrosome similarly to the endogenous protein. The antibody used to detect endogenous ALMS1 recognizes an epitope within the tandem repeat domain, which is not present in either construct 2261–2602 or ΔN-3175 (Figure 1A). Deletion of the putative leucine-zipper motif in construct 2261–2602, which eliminated its short predicted coiled-coil, did not appear to alter its intracellular distribution (Figure 1A and data not shown). We noted that immunostaining of endogenous ALMS1 was diminished in cells expressing either construct ΔN-3175 or construct 2261–2602 (Figure 2A). At low-to-moderate levels of construct expression this effect appeared to be specific because centrosomal γ-tubulin immunostaining was not appreciably altered (Figure 2B). These data imply that transiently expressed ALMS1 deletion constructs compete with endogenous ALMS1 for access to the centrosome.
Figure 2.
Transiently expressed ALMS1 deletion constructs appear to displace endogenous ALMS1 from the centrosome. U2OS cells were transfected with HA-tagged constructs ΔN-3175 or 2261–2602, representing two putative centrosome-targeting domains in ALMS1, and costained with antibodies to HA and either endogenous ALMS1 (A) or γ-tubulin (B). The epitope recognized by the ALMS1 antibody is not present in either deletion construct, allowing constructs to be distinguished from endogenous ALMS1. The mean intensity of ALMS1 and γ-tubulin immunofluorescence in cells expressing each construct, relative to that in untransfected (UT) cells, is shown; 11–20 cells were analyzed for each condition.
We attempted to narrow the part of construct ΔN-3175 required for centrosome-targeting by generating three shorter, nonoverlapping constructs. However, each of these showed some evidence of centrosomal localization in a proportion of cells (Figure 1, A and B, and data not shown). Specifically, constructs 3175–3549, 3550–3942, and ΔN-3941 gave detectable centrosomal staining in 58, 93, and 10% of cells, respectively (n = 40). Construct ΔN-3941 preferentially localized to the nucleus (data not shown). These data suggest that all or a large part of construct ΔN-3175 contributes to centrosome targeting, but that the extreme C-terminal portion including the ALMS motif may be less important.
In summary, these findings suggest that two regions of ALMS1 have roles in targeting it to centrosomes: a relatively small internal region (residues 2261–2602) and a larger C-terminal region (residues 3176–4169).
Human KIAA1731 Contains an ALMS Motif and Tagged C10orf90 and KIAA1731 Localize to the Centrosome
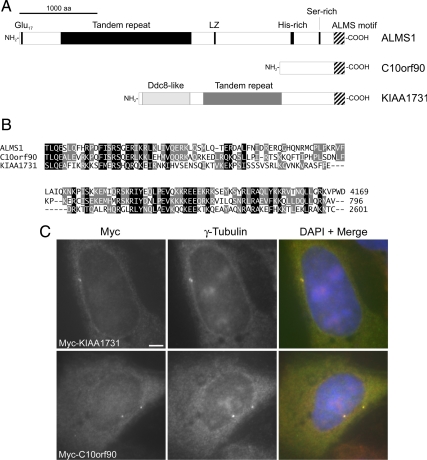
The ALMS motif (residues 4037–4169) appears to be the only element of either putative centrosome-targeting domain of ALMS1 that bears significant similarity to another human protein. Notably, PSI-BLAST analysis identified an ALMS motif-like sequence at the C-terminus of the human protein KIAA1731 (Figure 3, A and B; Supplemental Table S1), in addition to that previously identified at the C-terminus of C10orf90 (Collin et al., 2002). Interestingly, KIAA1731 was previously identified as a candidate centrosomal protein by proteomic analysis (Andersen et al., 2003) and contains an N-terminal region with similarity to Ddc8, a protein that is highly expressed in testis and localizes to the tails of elongated spermatids and spermatozoa (Catalano et al., 1997; Jaworski et al., 2007; Shi et al., 2009; Figure 3A). The Ddc8-like region of KIAA1731 also shares limited similarity with Drosophila Ana1, a protein implicated in centriole formation (Goshima et al., 2007; Dobbelaere et al., 2008; Blachon et al., 2009).
Figure 3.
The candidate centrosomal protein KIAA1731 contains a C-terminal ALMS motif, and tagged KIAA1731 and C10orf90 localize to the centrosome. (A) Primary structures of human ALMS1, C10orf90 (GenBank accession no. BAG59968) and KIAA1731 (GenBank accession no. NP_203753); the ALMS motif is denoted by striped shading. In addition to its C-terminal ALMS motif, KIAA1731 contains a region with similarity to Ddc8 and a degenerate tandem repeat apparently unrelated to that in ALMS1. (B) Multiple sequence alignment of human ALMS motifs. The C-terminus of each protein is shown; numbers denote amino acid positions. Black and gray shading denote positions conserved in all or 2/3 sequences, respectively, with conservative substitutions permitted. (C) Immunolocalization of Myc-tagged KIAA1731 (full-length) and C10orf90 (residues 243–796). U2OS cells were transiently transfected and coimmunostained for Myc and γ-tubulin, as indicated. In this and subsequent figures, DNA was visualized by DAPI staining (blue). Size bar, 5 μm.
In the absence of antibodies to C10orf90 and KIAA1731, we investigated their subcellular localizations using tagged constructs. Immunofluorescence analysis of transiently expressed constructs showed that both localized to the centrosome (Figure 3C); tagged C10orf90 also localized to the actin cytoskeleton in a proportion of cells (Supplemental Figure S2). These data support centrosomal localization of KIAA1731 and provide the first evidence that C10orf90 also localizes to this organelle. The ALMS motif appears to be the only region of sequence similarity shared by ALMS1, C10orf90, and KIAA1731. However, based on our deletion analysis of ALMS1, it seems unlikely that this motif acts as a discrete centrosome-targeting domain. C10orf90 and KIAA1731 may have additional structural homologies to ALMS1 that we have been unable to detect at the sequence level or may possess unrelated centrosome-targeting domains. Notably, we found that a construct representing the N-terminus of KIAA1731 was able to localize to the centrosome, providing further evidence that the ALMS motif is not critical for centrosome targeting (Supplemental Figure S5A).
Evolutionary Conservation of the ALMS Motif
BLAST analysis also detected C-terminal ALMS motif-like sequences in predicted proteins from numerous metazoan phyla and from two unicellular ciliated eukaryotes (Supplemental Figure S3 and Table S1). These data suggest that the ALMS motif evolved earlier than other regions of ALMS1, similarities to which we have been able to detect only in chordates and vertebrates, and pinpoint residues likely to be critical for the structure and/or function of this motif. The sea urchin and lancelet proteins identified in this analysis may represent orthologues of human KIAA1731 and ALMS1, respectively, based on additional sequence similarity outside the ALMS motif (not shown; similarity to KIAA1731 is noted in the database entry for the sea urchin protein). The remaining predicted proteins identified do not appear to share additional regions of similarity with human ALMS motif proteins.
siRNA-mediated Depletion of KIAA1731 and C10orf90 Suggests that the Encoded Proteins Have Centrosomal Functions
Next we investigated the functions of KIAA1731 and C10orf90 by RNAi, targeting each with two different siRNA duplexes. In the absence of antibodies to the encoded proteins, siRNA-mediated knockdown was monitored at the mRNA level (Figures 4A and 6A). RT-PCR analysis confirmed expression of each gene but suggested that C10orf90 is expressed at a significantly lower level than KIAA1731 and ALMS1 in hTERT-RPE1 cells (our unpublished observation). Strikingly, a proportion of hTERT-RPE1 cells treated with KIAA1731-directed siRNAs displayed markedly reduced or undetectable centrosomal immunostaining for acetylated tubulin, ALMS1, C-Nap1, pericentrin, and γ-tubulin (Figure 4, B–D, and data not shown). Similar effects were observed in U2OS cells (data not shown). Loss of centriole (acetylated tubulin), centriole-associated (C-Nap1 and ALMS1; see below), and PCM (γ-tubulin and pericentrin) markers strongly suggests the absence of centrosomes. Because loss of centrioles can cause dispersal of the PCM (Bobinnec et al., 1998) and presumably also of centriole-associated proteins, these observations may be explained by a defect in centriole formation or stability.
Figure 4.
siRNA-mediated depletion of KIAA1731 leads to loss of centrosome markers in hTERT-RPE1 cells. (A) qRT-PCR analysis showing depletion of KIAA1731 mRNA by two different siRNA duplexes. Results are expressed relative to the negative control siRNA and represent the mean ± SD of triplicate assays. (B) Quantification of cells in which immunostaining of centriolar acetylated tubulin and either ALMS1 or γ-tubulin was undetectable. The mean of three independent experiments is shown; in each experiment 100–300 cells were counted for each condition. Error bars, SE. (C) Examples of siRNA-treated cells costained with antibodies to ALMS1 and acetylated tubulin. The middle and bottom panels show cells with markedly diminished and undetectable immunostaining, respectively. (D) siRNA-treated cells costained with antibodies against C-Nap1 and pericentrin, showing loss of both markers in a cell treated with KIAA1731-directed siRNA. Size bars, 5 μm.
Figure 6.
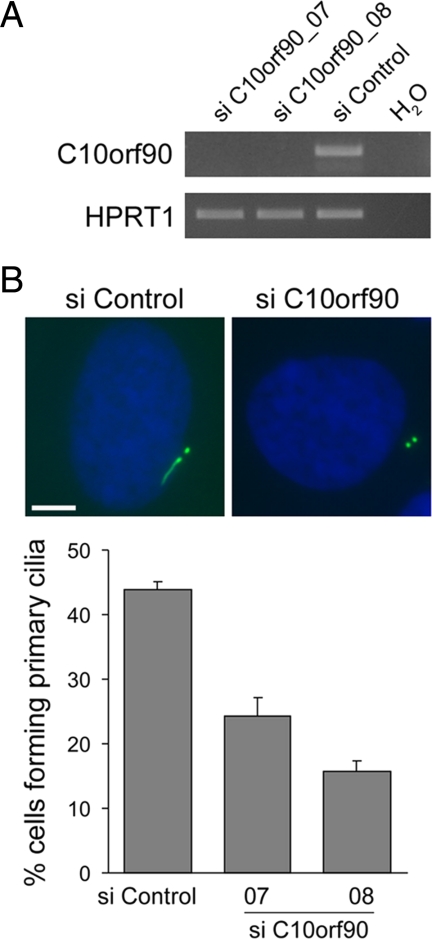
Ciliation is decreased in C10orf90-depleted hTERT-RPE1 cells. (A) RT-PCR analysis showing depletion of C10orf90 by two different siRNA duplexes, using HPRT1 as control. (B) Examples of siRNA-treated cells stained with an antibody to acetylated tubulin (green) to visualize ciliary axonemes and centrioles. Cells were transfected with siRNAs for 72 h and then incubated in serum-free medium for a further 24 h to induce primary cilia formation. The mean percentage of ciliated cells is shown; data are from three independent experiments in which 150–300 cells were counted for each siRNA. Error bars, SE. Size bar, 5 μm.
Failure of centriole biogenesis should result in “dilution” of preexisting centrioles during successive cell divisions (assuming that at least a proportion of cells continue to divide normally). We therefore examined centriole numbers at different time points after siRNA treatment, using acetylated tubulin as a centriole marker. We seeded fewer cells in this experiment in an attempt to increase the level of KIAA1731 knockdown, because in previous experiments <25% of cells displayed complete loss of centriolar acetylated tubulin staining after 96-h siRNA treatment (Figure 4B). As shown in Figure 5A, cells treated with a negative control siRNA had either two or more than two clearly visible centrioles at all time points. In contrast, cells treated with a KIAA1731-directed siRNA showed evidence of centriole loss at all time points (Figure 5A). At 48 h after treatment, the number of cells containing a single centriole was almost double that of cells completely lacking centrioles. The latter increased with time, exceeding the number of cells containing one centriole at 96 h after siRNA treatment. The number of cells containing two or more than two centrioles decreased with time. These data are therefore consistent with a role for KIAA1731 in centriole biogenesis, although they do not exclude the possibility that it is required for centriole stability.
Figure 5.
siRNA-mediated depletion of KIAA1731 leads to progressive loss of centrioles and spindle pole abnormalities. (A) Representative images of siRNA-treated cells stained with an antibody to acetylated tubulin (green) to label centrioles. The number of clearly visible centrioles is indicated. The chart shows the percentage of cells containing >2, 2, 1, and 0 centrioles after 48-, 72-, and 96-h siRNA treatment. Approximately 120 cells were counted for each experimental condition. (B) Examples of mitotic siRNA-treated cells costained with antibodies to acetylated tubulin and γ-tubulin. Size bars, 5 μm.
Consistent with loss of centrioles/centrosomes in KIAA1731-depleted cells, we observed mitotic cells with only one detectable focus of acetylated tubulin/γ-tubulin (Figure 5B). Metaphase cells with just one focus of γ-tubulin frequently displayed abnormal arrangements of chromosomes, suggesting failure to assemble a bipolar spindle (Figure 5B). We also observed mitotic cells with highly asymmetric distributions of γ-tubulin between spindle poles (Figure 5B). Similar asymmetry is seen in a proportion of cells depleted of CPAP, a human protein known to be required for centriole formation (Kohlmaier et al., 2009).
In contrast to KIAA1731-depleted cells, C10orf90-depleted cells did not exhibit appreciably reduced staining intensity of centrosome markers (Figure 6B and data not shown). To query if C10orf90 has a role in cilium formation or maintenance, similarly to ALMS1, we incubated siRNA-treated cells in serum-free medium to induce cilium formation and visualized ciliary axonemes by immunofluorescence. We observed decreased ciliation in cells treated with C10orf90-directed siRNAs, compared with cells treated with a negative control siRNA (Figure 6B), suggesting that C10orf90 is required for primary cilium formation.
In summary, these findings provide evidence of centrosomal functions for the ALMS motif–containing proteins KIAA1731 and C10orf90, consistent with our data showing centrosomal targeting of tagged versions of these proteins (Figure 3C) and with the previous mass spectrometry-based classification of KIAA1731 as a candidate centrosomal protein (Andersen et al., 2003).
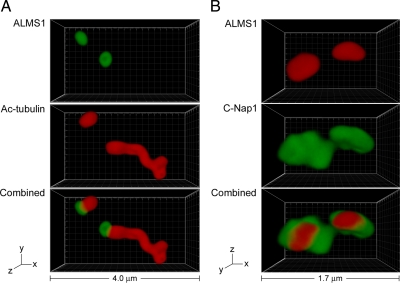
ALMS1 Localizes to the Proximal Ends of Centrioles and Basal Bodies
Immunofluorescence microscopy analysis suggested that ALMS1 localizes adjacent to centrioles (Supplemental Figure S1). To examine its distribution in more detail, we analyzed immunostained cells by 4Pi microscopy. To visualize ALMS1 in relation to the immature parental centriole and basal body of ciliated cells, we used acetylated tubulin as a marker for centrioles, basal bodies, and ciliary axonemes (Piperno et al., 1987). This analysis revealed that ALMS1 specifically caps one end of both the immature parental centriole and the basal body (Figure 7A). The basal body was unequivocally distinguished from the distal tip of the cilium by additionally labeling cells for γ-tubulin (analyzed by standard microscopy; data not shown). Because the ciliary axoneme is known to extend from the distal end of the basal body, these data demonstrate that ALMS1 localizes specifically to the proximal end of the basal body. This localization was confirmed by costaining cells for C-Nap1, a marker for the proximal ends of centrioles (Fry et al., 1998), which revealed similar spatial distributions of ALMS1 and C-Nap1, with ALMS1 having a slightly more compact distribution (Figure 7B). Colocalization of ALMS1 and C-Nap1 was also observed in nonciliated cells (analyzed by standard microscopy; Supplemental Figure S4). In summary, these data establish that ALMS1 is targeted specifically to the proximal ends of centrioles and basal bodies, where it partially colocalizes with C-Nap1. This localization pattern suggests that ALMS1 may have a role in maintaining centrosome cohesion.
Figure 7.
ALMS1 localizes specifically to the proximal ends of centrioles and basal bodies and colocalizes with the centrosome cohesion protein C-Nap1. (A) 4Pi microscopy analysis of a ciliated hTERT-RPE1 cell costained with antibodies to ALMS1 (green) and acetylated tubulin (red), which marks the immature parental centriole, basal body, and ciliary axoneme. (B) 4Pi microscopy analysis of a cell costained with antibodies to ALMS1 (red) and C-Nap1 (green), a marker for the proximal ends of centrioles. The axes and total width of images are indicated.
Next we asked if tagged versions of the ALMS motif–containing proteins C10orf90 and KIAA1731 localize similarly to ALMS1. Using standard microscopy we found that, in contrast to ALMS1, the bulk of centrosomal Myc-KIAA1731 immunostaining colocalized with centriolar acetylated tubulin (Supplemental Figure S5A). Notably, a construct representing the N-terminus of KIAA1731, containing most of the Ddc8-like region of this protein, localized similarly to the full-length construct (Supplemental Figure S5A). Interestingly, this localization pattern resembles that of the Drosophila protein Ana1, which localizes along the lengths of centrioles and is known to share sequence similarity with the N-terminus of KIAA1731 (Blachon et al., 2009). Global sequence alignment of KIAA1731 and Ana1 indicated only 15% amino acid identity (27% similarity) over their entire lengths, but did identify an additional conserved motif in the C-terminal half of each protein (Supplemental Figure S5B and data not shown). Taken together with the N-terminal sequence similarity and our RNAi data, these findings suggest that KIAA1731 is evolutionarily related to Ana1. Tagged C10orf90 also colocalized with centriolar acetylated tubulin, but appeared slightly more diffuse (Supplemental Figure S5A). In summary, these data suggest that, in contrast to ALMS1, C10orf90 and KIAA1731 are not specifically targeted to the proximal ends of centrioles.
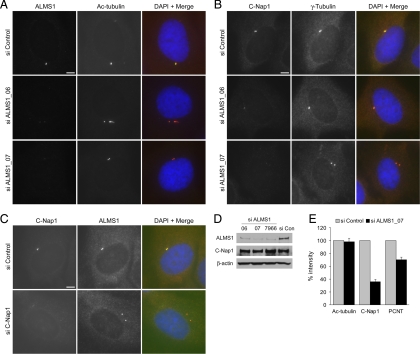
siRNA-mediated Depletion of ALMS1 Diminishes Centriolar Levels of C-Nap1
C-Nap1 dissociates from parental centrioles at the onset of mitosis, concomitant with centrosome separation. At the end of cell division, after disengagement of parent and progeny centrioles, C-Nap1 reassociates with the parent centriole and associates, for the first time, with the progeny centriole (Fry et al., 1998; Mayor et al., 2000). Centriolar association/dissociation of C-Nap1 is regulated by the balance in activities of a NIMA-related kinase (Nek2) and protein phosphatase 1 (Fry et al., 1998; Helps et al., 2000; Mayor et al., 2002). Little is known about how C-Nap1 is tethered to the proximal ends of centrioles during interphase, although Cep135 has recently been implicated in this role (Kim et al., 2008b). The highly similar localizations of ALMS1 and C-Nap1 prompted us to investigate if ALMS1 is also involved in retaining C-Nap1 at centrioles. We abrogated ALMS1 expression by RNAi (Figure 8A) and examined centrosomal levels of C-Nap1 by immunofluorescence. This revealed marked reductions in the intensity of C-Nap1 staining at the centrosome, compared with cells treated with a negative control siRNA (Figure 8, B and E). Immunoblot analysis indicated that total cellular levels of C-Nap1 were not substantially altered by treatment of cells with ALMS1-directed siRNAs (Figure 8D). Similar effects were observed with two independent ALMS1-directed siRNAs and in U2OS and HEK 293 cells (Figure 8B; Supplemental Figure S6 and data not shown). U2OS cells do not normally form primary cilia, indicating that the effect is not cilium-dependent. In the reciprocal experiment, depletion of C-Nap1 by RNAi did not appreciably alter the level of ALMS1 immunostaining at the centrosome (Figure 8C), suggesting that centrosomal assembly of ALMS1 does not depend on C-Nap1.
Figure 8.
Centriolar levels of C-Nap1 are diminished in ALMS1-depleted cells. (A) Immunofluorescence microscopy analysis showing depletion of ALMS1 by two different siRNA duplexes (ALMS1_06 and ALMS1_07). hTERT-RPE1 cells were transfected with the indicated siRNAs for 96 h and coimmunostained for ALMS1 and the centriole marker acetylated tubulin. (B) To assess levels of C-Nap1 at the centrosome after siRNA-mediated depletion of ALMS1, cells were costained with antibodies to C-Nap1 and γ-tubulin. (C) In the reciprocal experiment, cells were depleted of C-Nap1 by RNAi and costained with antibodies to ALMS1 and C-Nap1. Size bars, 5 μm. (D) Immunoblot analysis of total cellular levels of C-Nap1 and ALMS1 after siRNA treatment, using β-actin as loading control. ALMS1_7966 is a previously described siRNA duplex (Graser et al., 2007a). HEK293 cells were used due to more efficient extraction of C-Nap1 and ALMS1 compared with RPE1 cells; diminished centrosomal C-Nap1 immunostaining was confirmed in HEK293 cells depleted of ALMS1 (data not shown). (E) Mean intensity of centrosome marker immunostaining in siRNA-treated cells. Mitotic cells were excluded from analysis. Results are expressed relative to the mean intensity in cells treated with a negative control siRNA. Data are from three independent experiments in which 20–50 cells were analyzed for each siRNA; error bars, SE. PCNT, pericentrin.
Centriolar immunostaining of acetylated tubulin was not appreciably lower in ALMS1-depleted cells, indicating that diminished C-Nap1 immunostaining was not due to compromised centriole formation or stability (Figure 8, A and E). Also of note, this phenotype was not due to accumulation of cells in late G2 or M phase, because the majority of ALMS1-depleted cells that had low levels of centriolar C-Nap1 contained just two centrioles (as determined by acetylated tubulin staining) and showed noncondensed DNA staining. Centrosomal immunostaining of the PCM component pericentrin appeared to be slightly diminished in ALMS1-depleted cells (see below; Figure 8E and data not shown).
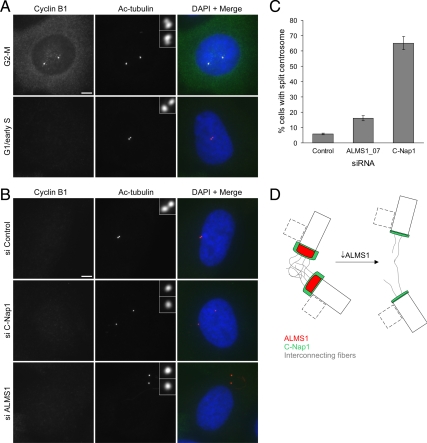
Next we examined the effect of ALMS1 depletion on cohesion between parental centrioles, using direct depletion of C-Nap1 as a positive control. We chose to analyze cells in G1 to avoid including cells undergoing the physiological process of centrosome separation at the G2-M transition and also because siRNA-mediated depletion of C-Nap1 has been reported to induce G1-S arrest (Mikule et al., 2007). We used cyclin B1 as a marker for cell cycle stage (Figure 9A). Cyclin B1 is undetectable in G1, begins to accumulate in the cytoplasm in S phase, and then at the centrosome in late S/early G2 phase, redistributes to the nucleus in prophase and is degraded at the metaphase–anaphase transition (Pines and Hunter, 1991; Bailly et al., 1992). In certain cell lines the immature parental centriole has been reported to be relatively free to move within the cytoplasm in G1 (Piel et al., 2000); however, we found that in ∼95% of interphase cyclin B1–negative hTERT-RPE1 cells the two centrioles were closely associated (Figure 9, A–C). Furthermore, treatment with C-Nap1–directed siRNA led to a dramatic increase in the distance between centrioles in ∼65% of these cells, indicating that these conditions were suitable for analyzing C-Nap1–dependent association of centrioles (Figure 9, B and C). Consistent with reduced levels of C-Nap1 at the centrioles of ALMS1-depleted cells, we noted an increase in the proportion of these cells with centrioles separated by >2 μm (referred to here as split centrosomes; Figure 9, B and C). The increase was modest compared with that observed in cells treated with C-Nap1–directed siRNA, possibly because of higher residual levels of C-Nap1 at the centrosome in ALMS1-depleted cells (Supplemental Figure S7C).
Figure 9.
Depletion of ALMS1 causes centrosome splitting. (A) Cyclin B1 immunostaining was used to determine cell cycle stage, allowing cells entering mitosis to be excluded from analysis. Examples of control cells at the G2-M transition and in G1/early S phase are shown. Centrioles were visualized by costaining with an antibody to acetylated tubulin. Note the separating centrosomes (each containing one parental and one progeny centriole) at G2-M and the close association of the two parental centrioles in G1/early S phase. (B) Immunofluorescence microscopy analysis of centrosome splitting in G1/early S phase cells. Cells were treated with the indicated siRNAs and costained with antibodies to cyclin B1 and acetylated tubulin. Interphase cells with undetectable cyclin B1 and with centrioles >2 μm apart were classed as having a split centrosome. (C) Quantification of centrosome splitting, after siRNA treatment, in interphase cells with undetectable cyclin B1. Depletion of the centrosome cohesion protein C-Nap1 was used as a positive control. The mean ± SE of three independent experiments is shown; in each experiment at least 100 cells were counted for each condition. (D) Model of ALMS1 function in centrosome cohesion, based on the model of C-Nap1/rootletin–dependent centrosome cohesion proposed by Bahe et al. (2005). Depletion of ALMS1 causes, by an unknown mechanism, reductions in the level of C-Nap1 at centrioles. This is predicted to perturb docking of interconnecting or “entangling” fibers with the proximal end of each centriole, leading to centrosome splitting. Procentrioles, which assemble orthogonally to parental centrioles during S and G2 phase, are depicted by dashed lines. Note that C-Nap1 is not thought to localize to the interface between the procentriole and centriole or be involved in maintaining their attachment (Mayor et al., 2000); it is not known if ALMS1 localizes to this interface.
Although these data imply that ALMS1 is required for localization of C-Nap1 to the proximal ends of centrioles, to date we have not been able to demonstrate coimmunoprecipitation of these two proteins, suggesting that ALMS1 might not be directly involved in physically linking C-Nap1 to centrioles. This led us to consider that diminished levels of C-Nap1 at centrioles could also result from disruption of PCM1-based intracellular transport, on which centrosomal recruitment of C-Nap1 has been reported to depend (Hames et al., 2005). PCM1 is a major component of centriolar satellites, nonmembranous 70–100-nm granules that are transported toward the centrosome via the microtubule-based molecular motor dynein/dynactin (Kubo et al., 1999; Kubo and Tsukita, 2003). In support of a functional interaction between ALMS1 and PCM1, we found evidence that the distribution of PCM1 granules was altered in ALMS1-depleted cells, particularly in cells treated with a siRNA duplex previously reported to cause ciliary abnormalities (Supplemental Figure S7A), that centrosomal levels of the PCM1-dependent protein pericentrin (Dammermann and Merdes, 2002) were also reduced (Figure 8E), and that the distribution of a cytoplasmic pool of ALMS1 may be altered in PCM1-depleted cells (Supplemental Figure S7B). However, we also found that, under the conditions used in our experiments, siRNA-mediated depletion of PCM1 did not significantly alter the level of C-Nap1 at the centrosome (Supplemental Figure S7C). These data suggest that although ALMS1-depleted cells may have a defect in PCM1-based trafficking, this defect is unlikely to account for the observed reduction in centriolar levels of C-Nap1.
In summary, these findings indicate that depletion of ALMS1 leads to a reduction in the level of C-Nap1 at the centrosome, resulting in compromised centrosome cohesion (Figure 9D), and implicate ALMS1 in PCM1-based intracellular transport.
DISCUSSION
ALMS1 Centrosome Targeting
Our analysis of transiently expressed ALMS1 deletion constructs is consistent with and extends the previous finding that a construct representing the C-terminal half (approx. 2000 residues) of ALMS1 localized to the centrosome (Andersen et al., 2003). Although we cannot exclude the possibility that transiently expressed constructs were recruited to the centrosome through interactions with endogenous ALMS1, we believe this to be unlikely because they appeared to displace the endogenous protein from the centrosome. It remains possible that other parts of ALMS1 contribute to centrosome targeting, because failure of some constructs to localize to the centrosome may have been due to misfolding of the relevant regions. Nevertheless, our identification of discrete regions that are capable of centrosome targeting may facilitate the discovery of binding partners involved in this process.
Likely Centriole/Basal Body–related Functions of C10orf90 and KIAA1731
Combined with previous data for ALMS1 and KIAA1731 (Andersen et al., 2003; Hearn et al., 2005), our immunolocalization analysis of transiently expressed C10orf90 and KIAA1731 suggests that all three ALMS motif–containing proteins localize to the centrosome. Based on our deletion analysis of ALMS1, the ALMS motif may contribute to centrosomal targeting but seems unlikely to represent a discrete centrosome-targeting domain. The centrosomal localization of an N-terminal KIAA1731 construct supports the conclusion that the ALMS motif is not required for centrosome targeting. The observation that a short construct encompassing the ALMS motif of ALMS1 localized preferentially to the nucleus is of uncertain significance, because larger constructs containing this region did not show a similar pattern and neither did tagged C10orf90 or KIAA1731. Although confirmation that endogenous C10orf90 and KIAA1731 localize to the centrosome awaits development of suitable antibodies, our RNAi data implicating both proteins in centrosome-related functions provide additional support for such localization.
As noted by Blachon et al. (2009), KIAA1731 shares an N-terminal region of sequence similarity with Drosophila Ana1, a protein implicated in centriole duplication (Goshima et al., 2007; Dobbelaere et al., 2008; Blachon et al., 2009). We have identified further evidence of similarity between KIAA1731 and Ana1, namely, the presence of a conserved motif in the C-terminal half of each protein, the centriolar localization of KIAA1731 suggested by analysis of a tagged construct, and the centriole-loss phenotype observed in KIAA1731-depleted cells. Although their overall level of sequence identity is low, these findings suggest that KIAA1731 is evolutionarily related to Ana1. Much progress has been made recently in understanding the molecular basis of centriole duplication, and a conserved core of essential proteins has been identified in worms, flies, and humans (Kleylein-Sohn et al., 2007; Strnad and Gonczy, 2008; Nigg and Raff, 2009). Ana1 belongs to an additional group of candidate centriole-duplication factors recently identified by RNAi screens in Drosophila cultured cells (Goshima et al., 2007; Dobbelaere et al., 2008). Notably, a recent report suggests that Ana1 is unlikely to be an upstream regulator of centriole duplication (Stevens et al., 2010). Our data are consistent with a role for KIAA1731 in centriole duplication, but this requires clarification. For instance it may be required for the structural integrity of centrioles and basal bodies, similarly to the recently proposed function of Drosophila Ana3 (Stevens et al., 2009). The likely role of KIAA1731 in the formation/stability of basal bodies, structures on which cilium assembly depends, together with our RNAi analysis of C10orf90, suggests that mutations in either KIAA1731 or C10orf90 could represent novel causes of human ciliopathies.
Possible Functional Connections between ALMS1, C-Nap1, and PCM1
The mechanism responsible for the apparent loss of centriolar C-Nap1 in ALMS1-depleted cells remains unknown. Notably, a role for ALMS1 in intracellular transport has been suggested by analysis of a murine model of Alström syndrome (Collin et al., 2005), and we have found evidence that siRNA-mediated depletion of ALMS1 disrupts PCM1-dependent intracellular transport. However, our data also suggest that defective PCM1-dependent recruitment of C-Nap1 is unlikely to account for its diminished intensity in ALMS1-depleted cells. On the basis of this finding and the similar localizations of the two proteins, we speculate that ALMS1 serves a structural role at the proximal ends of centrioles that is important, either directly or indirectly, for the retention of C-Nap1. Interestingly, C-Nap1/rootletin-mediated centrosome cohesion has recently been linked with the Wnt/β-catenin signaling pathway (Bahmanyar et al., 2008; Hadjihannas et al., 2010), a pathway reported to be restrained by the cilium and/or basal body (Gerdes et al., 2007; Corbit et al., 2008; though see also Huang and Schier, 2009; Ocbina et al., 2009). The potential involvement of ALMS1 in this pathway requires further investigation.
Our data raise the question of how a diminished centrosomal level of C-Nap1, or a more generalized perturbation at the proximal ends of centrioles, might contribute to the pathogenesis of Alström syndrome. In terms of causing a ciliary phenotype, it may compromise the ciliary rootlet (a supporting structure mainly composed of the C-Nap1–interacting protein rootletin), which is thought to be important for the long-term stability of sensory cilia (Yang et al., 2002, 2005). However, such a defect is unlikely to underlie the ciliary phenotypes observed over the short-term in ALMS1-depleted cells (Graser et al., 2007a; Li et al., 2007), particularly because siRNA-mediated depletion of C-Nap1 or rootletin does not appear to affect cilium formation or morphology (Graser et al., 2007a).
Defective PCM1-dependent transport could contribute to a ciliary defect in ALMS1-depleted cells, because siRNA-mediated depletion of PCM1 is known to diminish ciliation (Graser et al., 2007a; Mikule et al., 2007; Nachury et al., 2007). A possible functional interaction with PCM1 is of particular interest because PCM1/centriolar satellites associate with Bardet-Biedl syndrome (BBS) proteins (Kim et al., 2004; Nachury et al., 2007), BBS being a ciliopathy with considerable phenotypic overlap with Alström syndrome (Badano et al., 2006). Because we observed a pronounced effect on the cytoplasmic distribution of PCM1 with only one ALMS1-directed siRNA, it will be important to exclude the possibility of an off-target effect by, for example, rescuing the phenotype. Differential targeting of alternatively spliced ALMS1 mRNAs is a possible explanation for the less severe effects seen with two other siRNAs.
In addition to its transport function, PCM1 has been suggested to function as a chaperone, helping proteins to fold and assemble into complexes before transport to the centrosome (Hames et al., 2005). The cytoplasmic ALMS1-positive foci observed in PCM1-depleted cells may therefore represent accumulations of partially folded ALMS1 molecules, arising due to the absence of this chaperone function (though a PCM1-associated fraction of ALMS1 has yet to be demonstrated). The apparently normal levels of ALMS1 at the centrosomes of PCM1-depleted cells indicate either that depletion was not prolonged sufficiently for an effect to be detected or that assembly of centrosome-associated ALMS1 occurs via a different pathway, as suggested for Cep290 (Kim et al., 2008a).
Finally, it is noteworthy that siRNA-mediated depletion of PCM1 does not appear to result in the stunted or morphologically abnormal primary cilia seen in ALMS1-depleted cells (Graser et al., 2007a; Li et al., 2007), which are suggestive of an IFT defect. Localization of ALMS1 to the proximal end of the basal body is intriguing in this regard because, to our knowledge, the role of this part of the basal body in IFT is unknown. It would seem to exclude direct participation in docking of IFT particles, which is thought to occur at “transitional fibers” extending between the distal end of the basal body and the plasma membrane (Deane et al., 2001; Silverman and Leroux, 2009).
In summary, this work shows that ALMS1 associates specifically with the proximal ends of centrioles and basal bodies and implicates it in C-Nap1– and PCM1-related functions. The findings also suggest involvement of the ALMS motif–containing proteins C10orf90 and KIAA1731 in primary cilium assembly and centriole formation/stability, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jürgen Naggert (The Jackson Laboratory) for suggesting and facilitating the use of 4Pi microscopy. We thank Andrew Fry (University of Leicester, UK) and Andreas Merdes (CNRS Toulouse, France) for C-Nap1 and PCM1 antibodies, respectively. We are also grateful for support from Alström syndrome UK, Alström syndrome International and Margaret Miller. This work was supported by the Leverhulme Trust (ECF/2006/0169), the British Heart Foundation (PG/04/020), and the University of Southampton. T.H. also acknowledges support from a Wellcome Trust Value in People Award. G.B.C. was supported by National Institutes of Health Grant HD036878. The Jackson Laboratory core scientific services were supported by an institutional grant (CA034196).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0246) on September 15, 2010.
REFERENCES
- Alstrom C. H., Hallgren B., Nilsson L. B., Asander H. Retinal degeneration combined with obesity, diabetes mellitus and neurogenous deafness: a specific syndrome (not hitherto described) distinct from the Laurence-Moon-Bardet-Biedl syndrome: a clinical, endocrinological and genetic examination based on a large pedigree. Acta Psychiatr. Neurol. Scand. 1959;34:1–35. [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. Structure and duplication of the centrosome. J. Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genom. Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bahe S., Stierhof Y. D., Wilkinson C. J., Leiss F., Nigg E. A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S., et al. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Pines J., Hunter T., Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J. Cell Sci. 1992;101(Pt 3):529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- Blachon S., Cai X., Roberts K. A., Yang K., Polyanovsky A., Church A., Avidor-Reiss T. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182:133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L. M., Rieder C. L., Edde B., Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano R. D., Vlad M., Kennedy R. C. Differential display to identify and isolate novel genes expressed during spermatogenesis. Mol. Hum. Reprod. 1997;3:215–221. doi: 10.1093/molehr/3.3.215. [DOI] [PubMed] [Google Scholar]
- Collin G. B., et al. Alms1-disrupted mice recapitulate human Alstrom syndrome. Hum. Mol. Genet. 2005;14:2323–2333. doi: 10.1093/hmg/ddi235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G. B., et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat. Genet. 2002;31:74–78. doi: 10.1038/ng867. [DOI] [PubMed] [Google Scholar]
- Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., Reiter J. F. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Bento I., Bettencourt-Dias M. From zero to many: control of centriole number in development and disease. Traffic. 2009;10:482–498. doi: 10.1111/j.1600-0854.2009.00905.x. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Fry A. M., Mayor T., Meraldi P., Stierhof Y. D., Tanaka K., Nigg E. A. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J. M., et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Lavoie S. B., Gassner O. S., Lamla S., Le Clech M., Nigg E. A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007a;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Nigg E. A. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 2007b;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- Hadjihannas M. V., Bruckner M., Behrens J. Conductin/axin2 and Wnt signalling regulates centrosome cohesion. EMBO Rep. 2010;11:317–324. doi: 10.1038/embor.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames R. S., Crookes R. E., Straatman K. R., Merdes A., Hayes M. J., Faragher A. J., Fry A. M. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol. Biol. Cell. 2005;16:1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearn T., et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat. Genet. 2002;31:79–83. doi: 10.1038/ng874. [DOI] [PubMed] [Google Scholar]
- Hearn T., Spalluto C., Phillips V. J., Renforth G. L., Copin N., Hanley N. A., Wilson D. I. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]
- Helps N. R., Luo X., Barker H. M., Cohen P. T. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem J. 2000;349:509–518. doi: 10.1042/0264-6021:3490509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Schier A. F. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–3098. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski D. M., Beem-Miller M., Lluri G., Barrantes-Reynolds R. Potential regulatory relationship between the nested gene DDC8 and its host gene tissue inhibitor of metalloproteinase-2. Physiol. Genom. 2007;28:168–178. doi: 10.1152/physiolgenomics.00160.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S. R., Gleeson J. G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 2008a;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. C., et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- Kim K., Lee S., Chang J., Rhee K. A novel function of CEP135 as a platform protein of C-NAP1 for its centriolar localization. Exp. Cell Res. 2008b;314:3692–3700. doi: 10.1016/j.yexcr.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B. F., Mogensen M. M., Spektor A., Dynlacht B. D., Khodjakov A., Gonczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R. R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Tsukita S. Non-membranous granular organelle consisting of PCM-1, subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 2003;116:919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- Li G., Vega R., Nelms K., Gekakis N., Goodnow C., McNamara P., Wu H., Hong N. A., Glynne R. A role for Alstrom syndrome protein, alms1, in kidney ciliogenesis and cellular quiescence. PLoS Genet. 2007;3:e8. doi: 10.1371/journal.pgen.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. H., Zhang T., Surana U. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 2009;19:325–333. doi: 10.1016/j.tcb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Marshall J. D., et al. New Alstrom syndrome phenotypes based on the evaluation of 182 cases. Arch. Intern. Med. 2005;165:675–683. doi: 10.1001/archinte.165.6.675. [DOI] [PubMed] [Google Scholar]
- Marshall J. D., et al. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alstrom syndrome. Hum. Mutat. 2007;28:1114–1123. doi: 10.1002/humu.20577. [DOI] [PubMed] [Google Scholar]
- Marshall W. F. The cell biological basis of ciliary disease. J. Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T., Hacker U., Stierhof Y. D., Nigg E. A. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell Sci. 2002;115:3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- Mayor T., Stierhof Y. D., Tanaka K., Fry A. M., Nigg E. A. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. Loss of centrosome integrity induces p38–p53-p21-dependent G1-S arrest. Nat. Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nigg E. A., Raff J. W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Ocbina P. J., Tuson M., Anderson K. V. Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS One. 2009;4:e6839. doi: 10.1371/journal.pone.0006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C. L., Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., LeDizet M., Chang X. J. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Q., et al. Male germ cell-specific protein Trs4 binds to multiple proteins. Biochem. Biophys. Res. Commun. 2009;388:583–588. doi: 10.1016/j.bbrc.2009.08.053. [DOI] [PubMed] [Google Scholar]
- Silverman M. A., Leroux M. R. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J. F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Stevens N. R., Dobbelaere J., Brunk K., Franz A., Raff J. W. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N. R., Dobbelaere J., Wainman A., Gergely F., Raff J. W. Ana3 is a conserved protein required for the structural integrity of centrioles and basal bodies. J. Cell Biol. 2009;187:355–363. doi: 10.1083/jcb.200905031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Thompson H. M., Cao H., Chen J., Euteneuer U., McNiven M. A. Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat. Cell Biol. 2004;6:335–342. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 2006;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Yang J., Adamian M., Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell. 2006;17:1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Gao J., Adamian M., Wen X. H., Pawlyk B., Zhang L., Sanderson M. J., Zuo J., Makino C. L., Li T. The ciliary rootlet maintains long-term stability of sensory cilia. Mol. Cell. Biol. 2005;25:4129–4137. doi: 10.1128/MCB.25.10.4129-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Liu X., Yue G., Adamian M., Bulgakov O., Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., et al. Dimerization of CPAP orchestrates centrosome cohesion plasticity. J. Biol. Chem. 2010;285:2488–2497. doi: 10.1074/jbc.M109.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.