Inhibition of Golgi fragmentation results in cell cycle arrest at the G2 stage. However, the molecular basis of this G2 block is not known. Here, we show that a block of Golgi partitioning control G2/M progression through the impairment of centrosome recruitment and activation of Aurora-A.
Abstract
At the onset of mitosis, the Golgi complex undergoes a multistep fragmentation process that is required for its correct partitioning into the daughter cells. Inhibition of this Golgi fragmentation results in cell cycle arrest at the G2 stage, suggesting that correct inheritance of the Golgi complex is monitored by a “Golgi mitotic checkpoint.” However, the molecular basis of this G2 block is not known. Here, we show that the G2-specific Golgi fragmentation stage is concomitant with centrosome recruitment and activation of the mitotic kinase Aurora-A, an essential regulator for entry into mitosis. We show that a block of Golgi partitioning impairs centrosome recruitment and activation of Aurora-A, which results in the G2 block of cell cycle progression. Overexpression of Aurora-A overrides this cell cycle block, indicating that Aurora-A is a major effector of the Golgi checkpoint. Our findings provide the basis for further understanding of the signaling pathways that coordinate organelle inheritance and cell duplication.
INTRODUCTION
The Golgi complex has a crucial role in the processing and transport of cellular proteins and lipids. In mammalian cells, the Golgi complex is organized as a continuous membranous system that comprises stacks interconnected by tubules, a structure known as the “Golgi ribbon” (Shorter and Warren, 2002). The mitotic inheritance of the Golgi complex involves progressive and reversible disassembly of this Golgi ribbon into dispersed elements through a multistage process (Shorter and Warren, 2002; Colanzi et al., 2003; Altan-Bonnet et al., 2004). This must be precisely regulated for the optimal distribution of a functional Golgi complex to each of the daughter cells. The first step in this Golgi disassembly is the fragmentation of the noncompact zones of the Golgi ribbon. This occurs in G2 and results in the formation of isolated Golgi stacks (Colanzi et al., 2007; Feinstein and Linstedt, 2007). Importantly, this severing of the Golgi ribbon is necessary for entry into mitosis (Sutterlin et al., 2002; Hidalgo Carcedo et al., 2004; Colanzi and Corda, 2007; Colanzi et al., 2007; Feinstein and Linstedt, 2007).
A functional block of the proteins involved in this Golgi fragmentation step, such as the fissioning protein CtBP1-S/BARS (henceforth referred to as BARS) (Hidalgo Carcedo et al., 2004; Corda et al., 2006) and the Golgi matrix components GRASP65 (Sutterlin et al., 2002) and GRASP55 (Duran et al., 2008), results in inhibition of the severing of the Golgi ribbon and arrest of the cell cycle at the G2 stage (Sutterlin et al., 2002; Hidalgo Carcedo et al., 2004; Yoshimura et al., 2005). Inhibition or depletion of the kinases that control this Golgi fragmentation, such as Raf1, Mek1 and Erk1c, also results in a significant delay in G2/M transition (Acharya et al., 1998; Colanzi et al., 2003; Shaul and Seger, 2006; Feinstein and Linstedt, 2007). Once in mitosis, the Golgi stacks undergo further fragmentation, which is independent of BARS (Colanzi et al., 2007) and Mek1 (Feinstein and Linstedt, 2007) and is controlled by Cdk1 and Polo-like kinase1 (Plk1) (Wang et al., 2005; Colanzi and Corda, 2007).
Therefore, a “Golgi mitotic checkpoint” is dedicated to linking the state of (dis)assembly of the Golgi complex with entry into mitosis (Colanzi and Corda, 2007; Rabouille and Kondylis, 2007). The molecular mechanisms connecting Golgi fragmentation to regulation of mitotic progression are however not known, and their elucidation has the potential for identification of novel proteins/mechanisms that are involved in the overall maintenance of the structure and function of the Golgi complex and in cell cycle regulation.
Here, we show that severing of the Golgi ribbon during G2 is concomitant to centrosome separation and to centrosome recruitment and activation of the mitotic kinase Aurora-A (Aur-A), an essential regulator of entry into mitosis (Hirota et al., 2003; Seki et al., 2008). To identify the cell cycle proteins that are targeted by the Golgi checkpoint, we induced an acute block of Golgi partitioning in cells synchronized for mitotic ingression and analyzed the functional consequences of this inhibition of Golgi fragmentation. We find that a block in Golgi partitioning induces strong impairment of recruitment and activation of Aur-A at the centrosome, and this results in a block of cell entry into mitosis. Overexpression of Aur-A can overcome the cell cycle block induced by inhibition of Golgi fragmentation, indicating that this kinase is a major effector of this Golgi checkpoint.
These findings provide the basis for defining the mechanisms by which the fidelity of Golgi complex partitioning is monitored, and therefore how this controls G2/M transition. This has important physiological and pharmacological consequences, because characterization of this pathway will help in the identification and validation of novel target(s) for antiproliferative therapies and cancer treatment.
MATERIALS AND METHODS
Cell Culture
Normal rat kidney (NRK) and HeLa cells were from the American Type Culture Collection (Manassas, VA) and were respectively cultured in DMEM and minimal essential medium (Invitrogen, Carlsbad, CA), supplemented with 10% fetal calf serum (Biochrom, Berlin, Germany), 1% MEM nonessential amino acids solution, 1% penicillin/streptomycin, and 1% l-glutamine (Invitrogen). All cell lines were grown under controlled atmosphere in the presence of 5% CO2 at 37°C.
Antibodies and Reagents
Aphidicolin and thymidine were from Sigma-Aldrich (Milan, Italy). Dimethylsulfoxide (DMSO) was from Carlo Erba (Italy). RO-3306 and SB203058 were from Calbiochem (Darmstadt, Germany); SB-218078 was from Tocris Bioscience (Bristol, UK); UCN-01, anisomycin, etoposide, and topotecan were from Sigma-Aldrich. MLN8054 was kindly provided by J. Ecsedy (Millennium Pharmaceuticals, Cambridge, MA). Pak kinase inhibitor IPA3 was kindly provided by Dr. Jonathan Chernoff (Fox Chase Cancer Center, Philadelphia, PA). Fluorescein isothiocyanate (FITC)- and tetramethylrhodamine B isothiocyanate (TRITC)-labeled dextran and Hoechst 33342 were from Invitrogen; DRAQ5 was from Enzo Life Sciences (Lausen, Switzerland); and Mowiol was from Calbiochem. Antibodies were from the following sources: Aurora-A, Myt1, phospho-Aurora-A (Thr288), p38 mitogen-activated protein (MAP) kinase, phospho-p38 MAP kinase (Thr180/Tyr182), phospho-Cdc25C (Ser216), and phospho-Pak1(Ser199/204)/Pak2 (Ser192/197) (Cell Signaling Technology, Danvers, MA); Aurora A, green fluorescent protein (GFP), Plk1, and pericentrin (Abcam, Cambridge, MA); phospho-histone H3 (Ser10) (Millipore, Billerica, MA); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ϒ-tubulin, and phospho-Pak1/2/3 (Thr423) (Sigma-Aldrich); GM130 (BD Biosciences, San Jose, CA); CtBP1/BARS and giantin (kindly provided by M. A. De Matteis, Consorzio Mario Negri Sud, Italy); and Alexa 488-, Alexa 633- and Alexa 546-conjugated secondary antibodies (Invitrogen).
Immunofluorescence Microscopy
HeLa cells were grown on 10 μg/ml fibronectin-coated glass coverslips (Sigma-Aldrich) and treated as described above. They were then either fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 min at room temperature or with methanol for 5 min at –20°C. The blocking reagent (0.5% bovine serum albumin, 0.1% saponin, and 50 mM NH4Cl) was then added to the cells for 20 min, followed by a 1-h incubation with the primary antibody in the blocking reagent. The cells were then washed with phosphate-buffered saline and incubated with secondary antibodies (1:400) with 2 μg/ml Hoechst 33342 and/or 5 μM DRAQ5.
Microinjection and Cell Cycle Synchronization
For morphological and mitotic index analysis of synchronized cells, HeLa cells were plated at 60% confluence on 15-mm coverslips. For the double-thymidine cell cycle arrest, the cells were maintained in growth medium plus 2 mM thymidine for 16 h and then rinsed and maintained in growth medium for 8 h. The cells were then maintained in thymidine for an additional 16 h before the final release of the cell cycle arrest. At various times after this release (6–13 h), the cells were fixed and their DNA was labeled with either 2 μg/ml Hoechst 33342 or 5 μM DRAQ5, and with the appropriate antibodies, depending on experimental conditions. The mitotic index was estimated by measuring the number of cells showing clear mitotic (condensed chromosomes) and interphase (diffuse nuclear staining) features, under an Axiophot microscope (Carl Zeiss, Gottingen, Germany) equipped with a 63× objective. For microinjection experiments, the purified recombinant proteins (8–12 mg/ml) were mixed with 0.4 mg/ml FITC- or TRICT-labeled dextran (Invitrogen), as tracers of microinjection, and microinjected into 200–600 HeLa cells 30 min after removal of the S phase block. The cells were then incubated in complete medium for the appropriate times, before fixing. The cells were stained with the DNA dyes Hoechst 33342 or DRAQ5, and with the appropriate antibodies, depending on experimental conditions. For G2 phase cell cycle arrest, HeLa cells were subjected to a single thymidine arrest for 16 h and then rinsed and maintained in growth medium plus 40 μg/ml Hoechst 33258 for additional 18 h. Statistical significance of the measurements was assessed by two-tailed Student's t test.
Cell Transfection and RNA Interference
HeLa cells were transfected with the TransIT-LT1 Transfection Reagent (Mirus, Madison, WI), according to the manufacturer's instructions. The cells were microinjected 24 h after transfection, and processed for immunofluorescence at the mitotic peak. An anti-GFP polyclonal antibody was used to enhance the transfection signal. Small interfering RNA (siRNA) duplexes were transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. The Golgi protein GM130 was targeted using siRNA duplexes directed against the sequence AAGTTAGAGATGACGGAACTC (Dharmacon RNA Technologies, Lafayette, CO). Myt1 protein kinase was targeted using a siGENOME SMARTpool (M-005026-02-0005; Dharmacon RNA Technologies). p38 MAP kinase was targeted using an siRNA pool (SignalSilence Pool p38 MAP kinase siRNA; Cell Signaling Technology). BARS was targeted using siGENOME SMARTpool (M-008609-01; Dharmacon RNA Technologies). Nontargeting siRNA sequences were used as controls (Dharmacon RNA Technologies). After transfection, the intracellular protein contents were assessed by SDS-polyacrylamide gel electrophoresis followed by Western blotting, and the cells were further processed according to the experimental design.
Microscopy
Cells were imaged with a confocal laser microscope (LSM510 META confocal microscope system, Carl Zeiss; objective: 63 × 1.4 numerical aperture [NA] oil; definition: 512 × 512 pixels; pinhole diameter: 1 Airy unit for each emission channel; acquisition LSM510 software: LSM 510 [3.2]). For quantitative analysis of Aur-A and phospho-Aur-A on centrosomes, the images were acquired using identical confocal settings. Cells also were imaged using a DM5000-B fluorescence microscope and acquisition software FW4000 V 1.2.1. (both Leica, Wetzlar, Germany). Images were cropped and optimized for brightness and contrast with Photoshop and composed using Illustrator (Adobe Systems, Mountain View, CA).
Quantification of Aurora-A Fluorescence Intensity
Cells were imaged with a confocal laser microscope (LSM710, Carl Zeiss; objective: 63 × 1.4 NA oil; definition: 1024 × 1024 pixels). The bright centrosomal regions identified by a centrosome marker were circled, the Aurora-A fluorescence intensity in these regions and in a similarly sized background region were determined using LSM710 software (ZEN 2008 SP1), and the Aurora-A centrosomal fluorescence was calculated from these values.
RESULTS
Severing of the Golgi Ribbon during G2 Is Coincident with Centrosome Separation
The molecular dissection of the signaling pathways connecting Golgi fragmentation to the regulation of mitotic progression requires first the identification of the cell cycle proteins that are targeted by the Golgi checkpoint. For this, we used a microinjection-based experimental approach to induce an acute block of Golgi partitioning in cells synchronized for mitotic ingression, and a single-cell immunofluorescence-based analysis of the functional consequences of this inhibition of Golgi fragmentation. This demanding experimental approach was essential to focus our observations on processes that are precisely regulated and that occur over limited space and time and to reduce the intervention of potential compensatory mechanisms.
To inhibit the G2-specific severing of the Golgi ribbon, HeLa cells were microinjected with recombinant proteins or antibodies aimed at interfering with the function of either BARS, a protein essential for the G2-specific fission of the tubular membranes connecting the Golgi stacks (Hidalgo Carcedo et al., 2004; Colanzi et al., 2007), or GRASP65, a Golgi-associated protein that is involved in the maintenance of Golgi structure and function (Sutterlin et al., 2002) through a BARS-independent mechanism (Colanzi and Corda, 2007). This microinjection-based approach has already been extensively validated as an effective method for inducing specific, potent and persistent Golgi-dependent G2 block (Sutterlin et al., 2002; Hidalgo Carcedo et al., 2004).
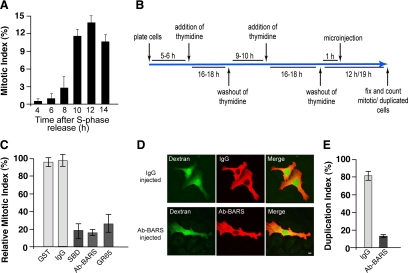
HeLa cells were seeded on glass coverslips and synchronized for cell cycle arrest at the G1/S boundary by double-thymidine S phase block (Colanzi et al., 2007). After the release of this S phase block by thymidine washout, the number of cells in mitosis increased with time, peaking 12 h after the removal of thymidine (mitotic peak) (Figure 1A). During a time frame corresponding to S phase, >200 cells on each coverslip were microinjected with generic immunoglobulin (Ig)Gs as a control or with an anti-BARS blocking antibody to block Golgi fragmentation (Figure 1B). In addition, the cells also were injected with purified recombinant glutathione transferase (GST) as a control, with the recombinant BARS deletion mutant corresponding to the BARS substrate-binding domain (SBD) (Hidalgo Carcedo et al., 2004) or with the recombinant cis-Golgi protein GRASP65 (Sutterlin et al., 2002). These two recombinant proteins acted as dominant-negative mutants and inhibited Golgi fragmentation (Supplemental Figure S1) (Sutterlin et al., 2002; Hidalgo Carcedo et al., 2004). The cells were fixed at the mitotic peak (12 h after S phase release) to evaluate the number of cells in mitosis (Figure 1B), as described previously (Hidalgo Carcedo et al., 2004). The injection of generic IgGs or GST did not alter mitotic entry, whereas microinjection of the anti-BARS antibody, SBD, or GRASP65 resulted in strong inhibition of the mitotic index (Figure 1C), indicating that the HeLa cells used in this study are responsive to the Golgi checkpoint. Furthermore, to test whether inhibition of Golgi fragmentation causes a delay or a block in G2/mitotic progression, we set up a different mitotic assay. Double-thymidine synchronized HeLa cells where injected with generic IgGs or anti-BARS antibodies and fixed 7 h after they had passed their mitotic peak (Figure 1B; 19 h after thymidine washout). Cells that had completed mitosis were identified as cell couplets that shared the injected fluorescent marker (Figure 1D, top). Because the count of cell couplets included all of the mitotic events that had occurred during the experimental time frame, this represents a cumulative mitotic index. In the cell population injected with generic IgGs, >80% of the cells were found as couplets (Figure 1D, top; and E). Conversely, in the cell population injected with the anti-BARS antibody, the majority of the cells were in the form of single cells (not duplicated; Figure 1D, bottom), and <20% were found as cell couplets (Figure 1E). More than 70% of the anti-BARS-antibody–injected cells were recovered at the end of the experiment, suggesting that during the time frame of the experiment, only a minor fraction had been lost as a result of induction of an apoptotic response (data not shown). In line with an essential role of Golgi fragmentation for entry into mitosis, siRNA-mediated depletion of BARS (Supplemental Figure S2A) reduced the levels of BARS by >80% (Supplemental Figure S2B), and this caused a strong reduction of the mitotic index (Supplemental Figure S2C) and of the mitotic-specific phosphorylation of the histone H3 (Supplemental Figure S2B). Therefore, the important role of the Golgi checkpoint is shown by this induction of persistent cell cycle arrest.
Figure 1.
Inhibition of Golgi fragmentation causes a block of cell cycle progression in HeLa cells. (A) Quantification of the mitotic index of cells grown on coverslips and arrested in S phase using the double-thymidine block, as shown schematically in B. The cells were fixed at the indicated times after the S phase block release, the DNA was labeled with Hoechst-33342, and the mitotic indices were calculated under fluorescence microscopy (see Materials and Methods). (C) Quantification of cells treated as illustrated in B. One hour after the S phase block release, the cells were microinjected with fluorescent dextran as injection marker, together with recombinant GST (8 mg/ml), generic IgGs (5 mg/ml), recombinant SBD (8–12 mg/ml), an anti-BARS antibody (Ab-BARS; 5–7 mg/ml), or recombinant GRASP65 (GR65; 8–10 mg/ml). The cells were fixed at the mitotic peak (12 h after S phase release) and processed for immunofluorescence under confocal microscopy, with DRAQ5 (blue) for the cell cycle phase. The relative mitotic indices were calculated as percentages of microinjected cells in mitosis normalized to nonmicroinjected cells on the same coverslip. (D and E) HeLa cells were treated as described in C with microinjections with generic IgGs (5 mg/ml) and the anti-BARS antibody (Ab-BARS; 5–7 mg/ml). The cells were fixed 19 h after the S phase block release and processed for immunofluorescence under confocal microscopy, with fluorescently labeled secondary antibodies to reveal IgG- or anti-BARS-injected cells. (D) Representative images of post-mitotic couplets (top) and premitotic singlets (bottom), microinjected as indicated. Bar, 10 μm. (E) Quantification of duplication index as percentages of postmitotic couplets (see Materials and Methods). Quantification data are means ± SD from two (A and E) and four (C) independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition.
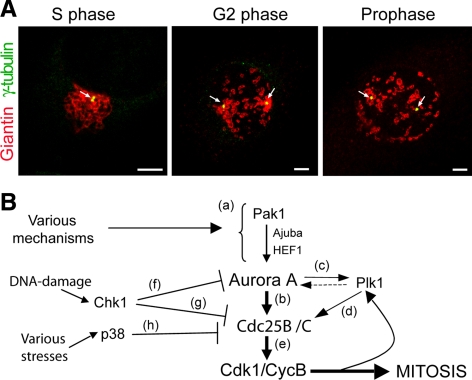
Because the inhibition of the severing of the Golgi ribbon induces a cell cycle block in G2 (Sutterlin et al., 2002; Hidalgo Carcedo et al., 2004), this must affect the signaling processes that govern the activation of the cyclin B–Cdk1 complex (cycB-Cdk1), the master regulator of entry into mitosis (Nigg, 2001). The first activation of cycB-Cdk1 occurs at the centrosome during G2, in coincidence with centrosome separation, and before the hallmarks of mitotic entry become morphologically discernible, such as chromosome condensation and assembly of mitotic spindles (Hirota et al., 2003; Jackman et al., 2003). In HeLa cells, the S phase Golgi ribbon (identified using an anti-giantin antibody; Figure 2A, S phase) was located in close association with the duplicated centrosomes (identified using an anti-γ-tubulin antibody; Figure 2A). During early G2 phase (identified using an anti-phospho-H3 antibody; Figure 6A), the centrosomes began to separate (Figure 2A; G2 phase, arrows), and this coincided with the first evident severing of the Golgi ribbon (i.e., the Golgi fragmentation step essential for mitotic entrance). The Golgi complex thus appeared to be divided into two main groups of isolated membranes, each of which maintained close association with a separated centrosome (Figure 2A, G2 phase). The centrosomes reached their final positions in prophase (Figure 2A, prophase), in preparation for the formation of the mitotic spindles (Nigg, 2001).
Figure 2.
Morphology of Golgi membranes and centrosomes through the cell cycle in HeLa cells. (A) Representative images of cells grown on coverslips and processed for immunofluorescence under confocal microscopy 12 h after the S phase block release. The cells were labeled with antibodies against giantin (red; Golgi complex) and γ-tubulin (green) to mark the centrosomes (arrows) at the indicated cell cycle stages. Images were acquired at maximal resolution, with fixed imaging conditions. Bars, 5 μm. (B) Schematic representation of the mechanisms governing activation of cycB-Cdk1 at the centrosome during G2. See text for details.
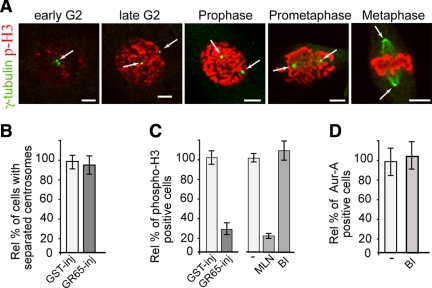
Figure 6.
Block of Golgi fragmentation does not require Plk1 to inhibit Aur-A recruitment. Cells were grown on coverslips and arrested in S phase using the double-thymidine block and processed for immunofluorescence under confocal microscopy. (A) Representative images of cells at the indicated cell cycle stages, labeled with an antibody against γ-tubulin (green, arrow) and with DRAQ5 (data not shown) for cell cycle phase, and labeled with antibodies against phosphorylated histone H3 (p-H3, red). (B and C) The cells were either nonmicroinjected or microinjected 1 h after thymidine washout with recombinant GST (GST-inj, 8 mg/ml), or recombinant GRASP65 (GR65-inj, 8–10 mg/ml), and with FITC-conjugated dextran as microinjection marker. For the noninjected cells, 8 h after thymidine washout they were treated with vehicle (−) or 0.25 μM MLN8054 (MLN), or with 100 nM BI2536 (BI). The cells were then processed 12 h after thymidine washout, for immunofluorescence under confocal microscopy with antibodies against γ-tubulin (B) or phosphorylated histone H3 (C). The relative percentages of cells with separated centrosomes (B) or positive for phosphorylated histone H3 (C) were calculated according to the relevant nonmicroinjected cells on the same coverslip or to cells treated with vehicle (−). (D) Eight hours after S phase block release, the cells were either left in growth medium (−) or treated with 100 nM BI2536 (BI), fixed at the mitotic peak (at 12 h) and labeled with antibodies against T288-phosphorylated Aur-A. The relative percentages of PT288 Aur-A–positive cells were calculated according to the cells treated with dilution buffer (−). Quantification data are means ± SD from at least two independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition. Bar, 5 μm.
Thus, because during G2 the severing of the Golgi ribbon seemed to be coordinated with the movement of the separating centrosomes, we explored the hypothesis that a block in this Golgi partitioning affects the mechanisms governing the centrosomal activation of cycB-Cdk1. The earliest essential event for this activation is the recruitment and activation of the kinase Aur-A at the centrosome (Fu et al., 2007). Aur-A is activated by either autophosphorylation or Pak1-mediated transphosphorylation on T288 in its activation domain (Figure 2Ba; Zhao et al., 2005). This phosphorylation, and thus Aur-A activity, is protected from the action of phosphatases (e.g., PP1) through Aur-A binding to scaffolding proteins, such as HEF1 and Ajuba, which are components of the focal adhesion complexes (Figure 2Ba; Hirota et al., 2003; Pugacheva and Golemis, 2005). Once activated, Aur-A directly phosphorylates Cdc25B phosphatase (Figure 2Bb; Cazales et al., 2005) and Plk1 (Figure 2Bc), which in turn phosphorylates Cdc25C phosphatase (Figure 2Bd; Seki et al., 2008). The two phosphatases then together activate the cycB–Cdk1 complex, which leads to irreversible entry into mitosis (Figure 2Be; Jackman et al., 2003). Plk1 is another important regulator of mitotic entry (Nigg, 2001). However, a possible contribution of Plk1 to Aur-A activation is still under debate (Figure 2Bc, dashed arrow; Chan et al., 2008). Finally, negative regulation of mitotic entry is operated by stress-damage– or DNA-damage–activated checkpoints; by acting through the kinases p38 (Cha et al., 2007) and Chk1 (Loffler et al., 2007), Cdc25B can be phosphorylated, which leads to its inhibition (Figure 2B, g and h). In addition, Chk1 can also inhibit Aur-A (targeting its Ser342) (Figure 2Bf; Krystyniak et al., 2006).
A Block of Golgi Fragmentation Results in Inhibition of Aur-A Recruitment to and Activation at the Centrosomes
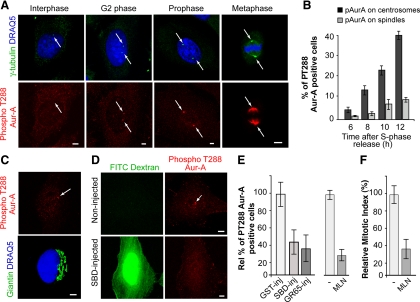
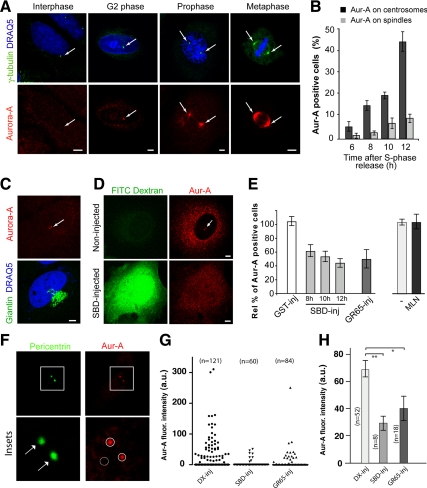
To determine whether and how the block of Golgi fragmentation affects the mechanisms governing centrosomal activation of cycB-Cdk1, we systematically tested whether the Golgi-dependent cell cycle block is mediated by one or more of the mechanisms depicted in Figure 2B. We first focused on the mitotic kinase Aur-A, because it is the first known cell cycle component to be activated at the centrosome during early G2, and it has been shown to have an essential role for entry into mitosis (Hirota et al., 2003; Cazales et al., 2005; Seki et al., 2008). Thus, we determined the kinetics of Aur-A activation at the centrosome in HeLa cells that had been synchronized by the double-thymidine S phase block (Colanzi et al., 2007). Active Aur-A was identified by its phosphorylation on T288, by taking advantage of a well-characterized anti-phospho-T288-Aur-A antibody (Figure 3A; Hirota et al., 2003). Active Aur-A was detected at the centrosome (Figure 3A, G2 phase and prophase) 6 h after release of the S phase block (Figure 3B), and its signal increased gradually and became detectable at the spindle (Figure 3A, metaphase) with progression into the mitotic phase of the cell cycle (Figure 3B). Importantly, cells with active Aur-A detected on unseparated centrosomes (Figure 3C, top), corresponding to the earliest stage of Aur-A recruitment, did not show evident alterations of Golgi structure (Figure 3C, bottom), indicating that Aur-A activation precedes the initial severing of the Golgi ribbon.
Figure 3.
Inhibition of Golgi fragmentation blocks Aur-A activation at the centrosomes in HeLa cells. Cells were grown on coverslips and processed for immunofluorescence under confocal microscopy after the S phase block release. (A) Representative images of cells at the indicated cell cycle stages, labeled with antibodies against γ-tubulin (green, arrow), and with DRAQ5 (blue) for cell cycle phase (top), and labeled with antibodies against T288-phosphorylated Aur-A (red, arrow; bottom). (B) Quantification of cells as described in A, with pT288-Aur-A–positive cells calculated as percentages of cells with active T288-phosphorylated Aur-A at the centrosome (dark gray bars) and the spindle (light gray bars), at the indicated times. (C) Representative images of cells 8 h after thymidine washout, labeled with antibodies against T288-phosphorylated Aur-A (red, arrow), and labeled with antibodies against giantin (green) for the Golgi complex, and with DRAQ5 (blue) for cell cycle phase. (D–F) The cells were either nonmicroinjected or microinjected 1 h after thymidine washout, with recombinant GST (GST-inj; 8 mg/ml), recombinant SBD (SBD-inj; 8–12 mg/ml), or recombinant GRASP65 (GR65-inj; 8–10 mg/ml), and with FITC-conjugated dextran as microinjection marker. For the noninjected cells, 8 h after the S phase block release they were treated with vehicle (−) or 0.25 μM MLN8054 (MLN). The cells were then processed 12 h after the S phase block release for immunofluorescence under confocal microscopy with antibodies against T288-phosphorylated Aur-A and with DRAQ5 (for cell cycle phase). (D) Representative images of noninjected and SBD-injected cells. (E) The relative percentages of PT288 Aur-A–positive cells were calculated according to the relevant nonmicroinjected cells on the same coverslip (see Materials and Methods) or to cells treated with vehicle (−). (F) Quantification of the mitotic index of cells treated with MLN. All images were acquired at maximal resolution, with fixed imaging conditions. Quantification data are means ± SD from two (B and F) and four (E) independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition. Bar, 5 μm.
HeLa cells were subjected to the double-thymidine synchronization protocol and injected with FITC-dextran, as injection marker, together with GST, SBD, or GRASP65. The cells were fixed at the mitotic peak and stained with the antibody against active T288-phosphorylated Aur-A, to determine the percentage of cells with active Aur-A at the centrosome. Injection of GST did not affect Aur-A activation (Figure 3E, GST-inj). Conversely, injection of SBD (Figure 3D) and GRASP65 reduced the proportion of cells with active centrosome-located Aur-A by 60–70% (Figure 3E, SBD-inj, GR65-inj), showing that a block of Golgi fragmentation reduces Aur-A activation. In line with an essential role for Aur-A in G2/mitosis progression, the treatment of synchronized cells for 4 h with 0.25 μM of the Aur-A inhibitor MLN8054 (MLN) reduced both the proportion of cells with active Aur-A (Figure 3E, MNL), and the mitotic index (Figure 3F, MLN).
Next, we tested whether the block of Golgi fragmentation also affects recruitment of Aur-A to the centrosome, which is the step that precedes its activation (Hirota et al., 2003). Aur-A became detectable at the centrosomes of synchronized HeLa cells (Figure 4A) 6 h after S phase block release (Figure 4B). The percentage of cells with Aur-A detectable at the centrosomes (Figure 4A, G2 phase and prophase) and the spindle (Figure 4A, metaphase) increased with progression into the mitotic phases (Figure 4B). In line with the timing of Aur-A activation, cells with Aur-A detected at the duplicated centrosomes did not show structural alterations of the Golgi complex (Figure 4C). Thus, HeLa cells were synchronized by the double-thymidine block and microinjected with GST, SBD, or GRASP65 after thymidine washout. The cells were again fixed at the mitotic peak and stained with antibodies to Aur-A to determine the percentages of cells with Aur-A on the centrosome. In addition, SBD-injected cells were also fixed 8 and 10 h after thymidine washout. As a control, injection of GST did not affect Aur-A recruitment (Figure 4E, GST-inj), whereas microinjection of either of these inhibitors of Golgi fragmentation (SBD, GRASP65) induced >50% reduction in the proportion of cells with detectable levels of Aur-A on the centrosomes (Figure 4, D and E). This Golgi-dependent inhibition of Aur-A recruitment was already evident at early time points after thymidine washout (Figure 4E), indicating that inhibition of Golgi fragmentation resulted in a persistent and early reduction in Aur-A recruitment to the centrosomes. Treatment of synchronized cells for 4 h with the Aur-A inhibitor (0.25 μM MLN8054) did not affect Aur-A recruitment to the centrosome (Figure 4E, MLN), indicating that the kinase activity of Aur-A is not necessary for its localization. Next, we assessed the effect of the inhibition of Golgi fragmentation on the fluorescence intensity of Aur-A in a region of interest defined by pericentrin-stained centrosomes (Figure 4F, circle). As expected, the injection of either SBD or GRASP65 reduced the fraction of cells with above-background levels of Aur-A on the centrosomes, compared with dextran-injected cells (Figure 4G). In addition, this quantitative analysis of Aur-A recruitment showed that the injection of the blockers of Golgi fragmentation also significantly reduced the total Aur-A fluorescence intensity measured in the Aur-A–positive centrosomes (Figure 4H). Therefore, our findings also indicate that the Aur-A regulators HEF1 and Ajuba (Figure 2Ba) cannot be targets of the Golgi checkpoint, because they affect the activation status of Aur-A rather than its recruitment (Hirota et al., 2003; Pugacheva and Golemis, 2005). Although Aur-A is recruited to the centrosome before the Golgi ribbon is fragmented (Figure 4C), our data are compatible with mechanisms that either reduce the rate of recruitment or accelerate the rate of release from the centrosomes when Golgi fragmentation is blocked, because centrosomal Aur-A has been shown to undergo fast dynamic exchange with the Aur-A cytoplasmic pool, with a half-life of ∼3 s (Stenoien et al., 2003). In addition, cold-induced microtubule depolymerization suppressed Aur-A localization at the spindle, but did not affect the G2-specific centrosome localization of Aur-A, indicating that this centrosome localization is microtubule independent (data not shown). Thus, these findings also exclude that the Golgi checkpoint is mediated by TPX2, which is the best-known Aur-A regulator, and which is required for recruiting Aur-A to the spindle (Kufer et al., 2002).
Figure 4.
Inhibition of Golgi fragmentation blocks Aur-A recruitment to the centrosomes in HeLa cells. Cells were grown on coverslips and arrested in S phase using the double-thymidine block and processed for immunofluorescence under confocal microscopy. (A) Representative images of cells at the indicated cell cycle stages, labeled with antibodies against γ-tubulin (green, arrow) and with DRAQ5 (blue) for cell cycle phase, and labeled with antibodies against Aur-A (red, arrow). (B) Quantification of cells as described in A, with percentages of Aur-A positive cells calculated as percentages of cells with Aur-A at the centrosome (dark gray bars) and the spindle (light gray bars), at the indicated times. (C) Representative images of cells 8 h after thymidine washout, labeled with antibodies against giantin (green) for the Golgi complex, and with DRAQ5 (blue) for cell cycle phase, and labeled with antibodies against Aur-A (red, arrow). (D and E) The cells were either nonmicroinjected or microinjected 1 h after thymidine washout with recombinant GST (GST-inj; 8 mg/ml), recombinant SBD (SBD-inj; 8–12 mg/ml), or recombinant GRASP65 (GR65-inj; 8–10 mg/ml), and with FITC-conjugated dextran as microinjection marker. For the noninjected cells, 8 h after thymidine washout they were treated with vehicle (−) or 0.25 μM MLN8054 (MLN). The cells were then processed 8 h, 10 h (SBD-injected cells only) and 12 h (all of the cells) after thymidine washout, for immunofluorescence under confocal microscopy with antibodies against Aur-A and with DRAQ5 (for cell cycle phase). (D) Representative images of noninjected and SBD-injected cells. (E) The relative percentages of Aur-A–positive cells were calculated according to the relevant nonmicroinjected cells on the same coverslip (see Materials and Methods), or to cells treated with vehicle (−). All images were acquired at maximal resolution, with fixed imaging conditions. Bars, 5 μm. Quantification data are means ± SD from two (B) and three (E) independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition. (F) Representative images of cells 12 h after thymidine washout, labeled with antibodies against pericentrin (green) as a centrosome marker, and with Aurora-A (red). Insets are higher magnification views of representative centrosomes. Equal areas were used to select the centrosome regions (circle) and a noncentrosome region with a similar background (dashed circle). (G) Fluorescence intensity (a.u., arbitrary units) of centrosome-associated Aur-A per cell was quantified by using LSM 710 ZEN 2008 SP1 (see Materials and Methods) in cells that had been treated as described in A and injected with FITC-dextran alone (Dx-inj) or in the presence of SBD (SBD-inj) or GRASP65 (GR65-inj). One data set representative of three independent experiments is shown as a scatter plot. More than 250 cells where injected and analyzed. (H) Fluorescence intensity ±SEM of the samples reported in (G) that showed above-background fluorescence levels of centrosome-associated Aur-A. Two-tailed Student's t tests were applied to the data (*p < 0.005; **p < 0.001). Bar, 5 μm.
A Block of Golgi Fragmentation Inhibits Aur-A Recruitment and Activation through a Novel Mechanism
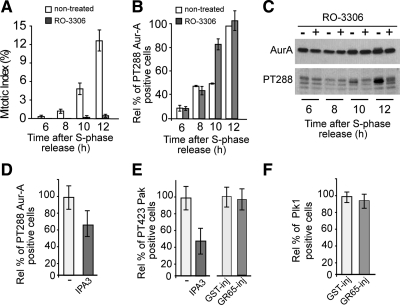
Because many signaling networks are composed of elements that are functionally connected by feedback loops, it was important to determine whether reduction of Aur-A recruitment to and activation at the centrosome is a secondary consequence of a block of the essential mitotic complex cycB-Cdk1. Indeed, although according to some reports Aur-A activation is independent of Cdk1 activity (Hirota et al., 2003), other reports have proposed that Aur-A is activated downstream of cycB-Cdk1 (Marumoto et al., 2002). HeLa cells were subjected to the double-thymidine synchronization protocol, and 4 h after thymidine washout they were incubated with 9 μM RO-3306 (RO), a specific inhibitor of Cdk1 kinase (Vassilev, 2006). The cells were fixed at various times after thymidine washout and stained with Hoechst to determine the mitotic index and with antibodies against the active T288-phosphorylated Aur-A. A similar set of samples was treated for immunoblotting, to reveal the levels of total and active Aur-A. Despite a complete block of entry into mitosis in the RO-treated cells (Figure 5A), activation of Aur-A at the centrosome was not affected by RO addition (Figure 5B). Conversely, this treatment strongly reduced the global levels of active Aur-A (Figure 5C). Thus, through this single-cell analysis and in our experimental setting, early activation of Aur-A at the centrosome (i.e., during the G2 phase) is independent of cycB-Cdk1. This centrosome pool of active Aur-A represents a minimal yet functionally significant fraction that is below the detection limits of Western blotting. On the contrary, the global levels of active Aur-A become detectable by Western blotting only during the mitotic phase, and they were influenced by a positive feedback loop that includes cycB-Cdk1. More importantly, these data show that Golgi-dependent inhibition of Aur-A activation cannot be mediated by a signaling pathway that primarily affects cycB-Cdk1.
Figure 5.
Block of Golgi fragmentation inhibits Aur-A recruitment through a novel mechanism. HeLa cells were grown on coverslips and arrested in S phase by using the double-thymidine block. (A–C) Four hours after S phase block release, the cells were either left in growth medium (nontreated) or treated with 9 μM RO-3306 (a Cdk1 inhibitor) and fixed and processed for immunofluorescence under confocal microscopy or immunoblotting at the indicated times after thymidine removal. Cells were labeled with Hoechst 33342 to determine the mitotic indices up to 12 h after S phase release (A) and with antibodies against T288-phosphorylated Aur-A to determine the relative percentages of pT288 Aur-A–positive cells calculated according to the nontreated cells fixed up to 12 h after S phase release (B). A similar set of samples was processed for immunoblotting to reveal total Aur-A and active Aur-A (PT288) (C). (D–F) Cells were either nonmicroinjected (D and E) or microinjected 1 h after thymidine washout with recombinant GST (GST-inj; 8 mg/ml) or recombinant GRASP65 (GR65-inj, 8–10 mg/ml; E and F), and with FITC-conjugated dextran as microinjection marker (E and F). Eight hours after S phase block release, the nonmicroinjected cells were either left in growth medium (D and E, −) or treated with 30 μM IPA3 (D and E, a Pak inhibitor) and fixed 12 h after the S-phase block release for immunofluorescence under confocal microscopy with antibodies against T288-phosphorylated Aur-A (D), against T423-phosphorylated Pak (E), or against Plk1 (F). The relative percentages of PT288 Aur-A-positive (D), T423 Pak-positive (E) were calculated according to the relevant cells treated with vehicle (−). The relative percentages of Plk1-positive cells (F) were calculated according the noninjected cells on the same coverslip. Quantification data are means ± SD from at least two independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition.
Next, we tested whether the block of Golgi fragmentation affects the centrosomal activation of Pak1 (Figure 2Ba), which has been shown to be the upstream activating kinase that is involved in the first activation of Aur-A at the centrosome (Zhao et al., 2005). HeLa cells were again subjected to the double-thymidine synchronization protocol and injected with GRASP65 or incubated for 4 h with 30 μM of the Pak inhibitor IPA3 (Viaud and Peterson, 2009), until they were fixed at the mitotic peak. In line with the established role of Pak, IPA3 reduced the fraction of cells with detectable levels of active Aur-A by ∼35% (Figure 5D, IPA3). Pak activation was monitored by confocal microscopy using an antibody against a Pak autophosphorylation site, phospho-Pak1/2/3 (T423) (Supplemental Figure S3A; Zhao et al., 2005). The effectiveness of IPA3 was seen by its 50% reduction in the number of cells with active Pak at the centrosome (Figure 5E and Supplemental Figure S3B). However, inhibition of Golgi fragmentation did not reduce the number of cells with detectable levels of active Pak on the centrosome (Figure 5E and Supplemental Figure S3C, GR65-inj). Similar results were observed using another antibody raised against different autophosphorylation sites, anti-phospho-Pak1/2(S199/204) (data not shown). The lack of effect on Pak activation indicates that a block of Golgi fragmentation does not cause a generic inhibition of centrosome-located mitotic kinases and that Pak is not targeted by the Golgi checkpoint.
Aur-A has recently been shown to be the upstream regulatory kinase of Plk1 (Figure 2Bc), another centrosome-localized kinase (Supplemental Figure S3D) with essential roles in mitosis (Seki et al., 2008). This phosphorylation of Plk1 is enhanced by the interaction between Plk1 and Bora, a G2/M-expressed protein that controls the accessibility of the Plk1 activation loop for phosphorylation and activation by Aur-A (Seki et al., 2008). During mitosis, Plk1-dependent Bora degradation promotes Aur-A localization to the centrosome and/or spindle, suggesting that Plk1 can also, in turn, control Aur-A localization by regulating the levels of Bora (Chan et al., 2008). Thus, the synergistic actions of Aur-A, Bora, and Plk1 might, in principle, create a complex regulatory circuit that controls G2/M transition (Figure 2Bc). Moreover, Plk1 has been implicated in mitotic Golgi fragmentation (Sutterlin et al., 2001), suggesting that a Plk1-dependent signaling mechanism might link Golgi partitioning and cell cycle control (Preisinger et al., 2005). Thus, we tested the possible involvement of Plk1 in the Golgi checkpoint by injecting GRASP65 in synchronized cells, to measure the effects of this on Plk1. Our analysis revealed that inhibition of Golgi fragmentation did not impede the recruitment of Plk1 to the centrosome (Figure 5F). However, we could not test whether the block of Golgi fragmentation affects the activation of Plk1, because the available anti-phospho-Plk1 antibody was not sufficiently sensitive to detect by immunofluorescence the active form of Plk1 during G2 (data not shown).
Thus, to further address a possible involvement of Plk1 in the Golgi checkpoint, we followed a different strategy. We first monitored whether a block of Golgi fragmentation can influence other typical mitotic features, such as the separation of the centrosomes and the percentages of cells with nuclear phosphorylation of histone H3. The cells were subjected to the double-thymidine synchronization protocol, and 1 h after thymidine washout, they were microinjected with recombinant GRASP65, or 8 h after the S block release, they were incubated with the Aur-A inhibitor (0.25 μM MLN8054; Hoar et al., 2007), or with a Plk1 inhibitor (100 nM BI2536; Steegmaier et al., 2007), until they were fixed at the mitotic peak. The cells were treated for immunofluorescence to reveal γ-tubulin and phosphorylated histone H3 (Figure 6A). The injection of GRASP65 did not reduce the proportion of cells with disconnected centrosomes (separated >1 μm; Figure 6B), indicating that the cells had passed S phase (Jackman et al., 2003). However, this treatment reduced the proportion of G2 cells (cells with uncondensed chromatin) with detectable levels of nuclear phosphorylation of histone H3 (Figure 6C, GR65-inj). It has been suggested that Aur-A is not directly involved in this phosphorylation, because 24 h of treatment of HCT-116 cells with 0.25 μM MLN did not reduce the fraction of mitotic cells positive to phosphorylated histone H3 (Manfredi et al., 2007). Thus, we tested the effect of an “acute” inhibition of Aur-A on the phosphorylation of histone H3 seen in a G2 population of HeLa cells. The cells were subjected to the double-thymidine synchronization protocol and incubated for 4 h with 0.25 μM MLN8054 or with 100 nM BI2536 until they were fixed at the mitotic peak. This treatment with MLN8054 caused an 80% reduction of cells with phosphorylated histone H3 seen in the G2 phase (i.e., cells with uncondensed chromatin; Figure 6C, MLN), suggesting that similar to the Golgi checkpoint, acute inhibition of Aur-A reduced, although perhaps indirectly, the fraction of cells in G2 with detectable levels of phosphorylation of histone H3. Conversely, the Plk1 inhibitor BI2536 did not affect this phosphorylation (Figure 6C, BI), indicating that Plk1 activity does not correlate with the mitotic features associated with the Golgi checkpoint. Finally, treatment of double-thymidine synchronized cells for 4 h with 100 nM BI2536 did not affect recruitment of Aur-A to the centrosome (Figure 6D), further supporting the concept that the Golgi checkpoint does not reduce Aur-A recruitment through inhibition of Plk1. In line with this conclusion, Plk1 seems to be involved in the disassembly of the Golgi stacks (Wang et al., 2005), which is a fragmentation step that follows the Golgi checkpoint (Colanzi et al., 2007; Feinstein and Linstedt, 2007). Together, our findings further indicate that Aur-A is a major target of G2 arrest induced by a block of Golgi fragmentation.
Injection of Golgi Fragmentation Blockers Reduces Aur-A Recruitment Only in Cells with an Intact Golgi Ribbon
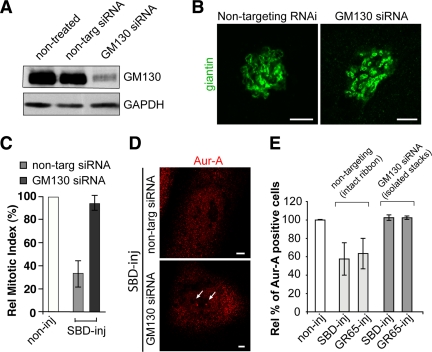
Although we microinjected two unrelated proteins to inhibit Golgi fragmentation, the possibility that SBD and GRASP65 affect Aur-A localization in ways that are not related to Golgi fragmentation was not formally ruled out by these data. To address this point, we exploited the finding that cells with their Golgi complex divided into isolated stacks at all cell cycle stages bypass the need for BARS and GRASP65 for Golgi fragmentation and mitotic entrance (Colanzi et al., 2007; Feinstein and Linstedt, 2007). As a result, such cells are not blocked in G2 when microinjected with SBD (Colanzi et al., 2007) or GRASP65 (Sutterlin et al., 2002). Accordingly, microinjection of SBD or GRASP65 should inhibit Aur-A recruitment to the centrosome only in cells that have an intact Golgi ribbon, whereas this should not affect Aur-A recruitment to the centrosomes when the organization of the Golgi ribbon has already been disrupted. Therefore, here we used siRNA-mediated depletion of the Golgi matrix protein GM130, because GM130-depleted cells show Golgi membranes in the form of isolated, but fully functional, ministacks that are clustered around the nucleus (Marra et al., 2007). According to our experimental conditions, we reduced GM130 expression to levels that were low enough (Figure 7A) to cause the severing of the Golgi ribbon (Figure 7B), whereas not affecting the centrosome morphology (Kodani et al., 2009). Importantly, the microinjection of SBD or GRASP65 into these GM130-depleted cells failed to inhibit entry into mitosis (Figure 7C, GM130 siRNA) and centrosome recruitment of Aur-A (Figure 7, D and E, GM130 siRNA); the same treatments with nontargeting-siRNA-treated cells were still able to reduce entry into mitosis and recruitment of Aur-A (Figure 7, C–E, nontargeting). These data show that injection of SBD or GRASP65 blocks Aur-A activation only in cells that have an intact Golgi ribbon. This excludes the concept that these microinjected proteins per se affect Aur-A directly or through some other nonspecific effects, and more importantly, it establishes a causal link between inhibition of the severing of the Golgi ribbon in G2 and block of Aur-A recruitment to and activation at the centrosomes.
Figure 7.
Blockers of Golgi fragmentation do not affect Aur-A recruitment to the centrosomes in HeLa cells without an intact Golgi ribbon. Cells were grown on coverslips and transfected for 48 h with 100 nM nontargeting siRNAs (i.e., with intact Golgi ribbon) or 100 nM GM130-directed siRNAs (i.e., with isolated Golgi stacks). (A) Representative experiment of cells processed for immunoblotting of GM130 knockdown, as revealed with antibodies against GM130 and against GAPDH. (B) Representative images of cells from A labeled with an anti-giantin antibody (green) for the structure of the Golgi complex. (C–E) Alternatively, the cells were arrested in S phase using the double-thymidine block. One hour after thymidine washout, the cells were either nonmicroinjected (noninj) or microinjected with recombinant SBD (SBD-inj, 8–12 mg/ml) or recombinant GRASP65 (GR65-inj, 8–10 mg/ml), and with FITC-conjugated dextran as microinjection marker. (C) Relative mitotic indices of cells nonmicroinjected (noninj) or microinjected with recombinant SBD (SBD-inj). (D) Representative images of cells fixed 12 h after thymidine washout and processed for immunofluorescence under confocal microscopy with antibodies against Aur-A (red; for cells positive for Aur-A on centrosomes; arrow) and with Hoechst 33342 (for cell cycle phase). (E) Quantification of cells as described in D, with Aur-A–positive cells calculated as percentages of microinjected cells with Aur-A on the centrosomes normalized to nonmicroinjected cells on the same coverslip. Data are means ± SD from three independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition. Bar, 5 μm.
Therefore, the block of the first stage of the partitioning process, the severing of the Golgi ribbon into separate stacks in G2, specifically inhibits Aur-A recruitment to and activation at the centrosome, causing a general reduction in the mitotic kinases.
Overexpression of Aur-A Can Override the Golgi Checkpoint
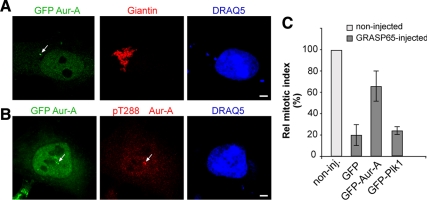
Having established that a block of Golgi fragmentation inhibits activation of Aur-A at the centrosomes, we tested whether inhibition of Aur-A is the major cause of the resulting G2 block. If this is the case, the overexpression of wild-type (wt)-Aur-A should bypass the cell cycle block induced by impaired Golgi fragmentation. Indeed, overexpression of wt-Aur-A does not modify cell cycle progression under normal conditions (Marumoto et al., 2002), although it is an effective approach to override the DNA-damage checkpoint induced by double-strand breaks (Krystyniak et al., 2006). Thus, HeLa cells were transfected with the GFP-tagged forms of Aur-A and Plk1, or with the empty vector as a control, and subjected to the double-thymidine S phase synchronization protocol. AurA overexpression did not induce evident alterations in the structure of the Golgi complex (Figure 8A). After the release from synchronization, the transfected GFP-Aur-A localized to the centrosome and was activated (Figure 8B). Importantly, and in line with a major role for Aur-A in the Golgi checkpoint, the overexpression of GFP-Aur-A induced a substantial overriding of the G2 block that resulted from the inhibition of Golgi fragmentation (Figure 8C, GFP-Aur-A). Conversely, the overexpression of the kinase GFP-Plk1 did not override the Golgi-dependent cell cycle block (Figure 8C, GFP-Plk1), indicating that generic overexpression of a mitotic kinase is not sufficient to nonspecifically force the cell cycle and bypass the Golgi checkpoint.
Figure 8.
Overexpression of Aur-A can override the Golgi checkpoint in HeLa cells. HeLa cells (A–C) were arrested in S phase by using the double-thymidine block and transfected for 24 h with the empty vector (GFP; C only), GFP-Aur-A, or GFP-Plk1 (C only) after the first release from thymidine. (A) Representative images of cells labeled with an antibody against giantin (red) to label the Golgi complex, with DRAQ5 (blue) for the cell cycle phase. (B) Representative images of cells labeled with an antibody against phospho-T288-Aur-A (red) and with DRAQ5 (blue) for the cell cycle phase. (C) One hour after the final thymidine washout, the cells were either nonmicroinjected (noninj) or microinjected with recombinant GRASP65 (8–10 mg/ml), and with TRITC-conjugated dextran as microinjection marker. The cells were fixed 12 h after the final thymidine washout, and processed for immunofluorescence under confocal microscopy, with antibodies against GFP to identify low transfection levels, and with Hoechst 33342 for cell cycle phase. Quantification of the relative mitotic index as percentages of microinjected cells in mitosis were normalized to nonmicroinjected cells on the same coverslip. More than 400 cells were microinjected for each condition. All quantification data are means ± SD from three independent experiments, each carried out in duplicate. Bar, 5 μm.
To test whether Aur-A expression cause Golgi fragmentation, we used a quantitative approach to analyze the state of integrity of the Golgi complex using an immunofluorescence-based measure of the number of “Golgi objects” in interphase and G2-blocked cells (Feinstein and Linstedt, 2007). We found that Aur-A overexpression did not cause any change in the number of Golgi objects, compared with untransfected or mock-transfected cells. Moreover, the number of Golgi objects found in Aur-A–overexpressing cells was significantly lower (<25%) than the objects detectable in G2-blocked cells (Supplemental Figure S4, A and B). In addition, we also tested whether Aur-A inhibition can reduce Golgi fragmentation in a well-characterized assay for Golgi fragmentation (Acharya et al., 1998) and found that the Aur-A inhibitor MLN did not block Golgi fragmentation even if used at high concentration (Supplemental Figure S4, C and D). Collectively, these data indicate that Aur-A is not involved in Golgi fragmentation, and they exclude the possibility that Aur-A overexpression overrides the Golgi checkpoint by inducing Golgi fragmentation.
Known G2 Checkpoints Are Not Involved in the Golgi-dependent Block of the Cell Cycle
Finally, we asked whether other signaling pathways that are known to regulate cycB-Cdk1 are involved in inducing the Golgi-dependent G2-block. In addition to the signaling pathways that provide positive regulation of cycB-Cdk1, entry into mitosis is controlled by negative regulatory signals that are operated by DNA-damage (Nigg, 2001) and stress-related checkpoints (Bulavin et al., 2001). Thus, when damage to a cell is detected, these negative regulatory signals can activate signaling cascades that inhibit key regulatory elements of G2/M transition, such as Cdc25B (Cazales et al., 2005), Aur-A (Krystyniak et al., 2006) and cycB-Cdk1 itself (Nigg, 2001; Figure 2B, f–h). It has already been shown that the kinases ataxia-telangiectasia mutated (ATM) and ATM and Rad 3-related (ATR), which are key players in the DNA-damage checkpoint (Nigg, 2001), are not involved in the Golgi-dependent cell cycle block (Sutterlin et al., 2002). Moreover, induction of damage to DNA by etoposide inhibited entry into mitosis (Supplemental Figure S5, A and B) and Aur-A activation but did not alter Aur-A recruitment (Supplemental Figure S5C), in agreement with previous findings (Cazales et al., 2005). Nevertheless, we also investigated whether the Golgi-dependent block of the cell cycle could be mediated through activation of relevant components of the DNA-damage checkpoint downstream of ATM/ATR, i.e., the centrosome-located Chk1 (Loffler et al., 2007) or the Golgi-localized kinase Myt-1 (Nakajima et al., 2008), all of which can provide negative regulation of cycB-Cdk1 activation (Nigg, 2001; see below). If this is the case, it should be possible to override the Golgi-dependent G2 block by functional inhibition of Chk1 or Myt-1.
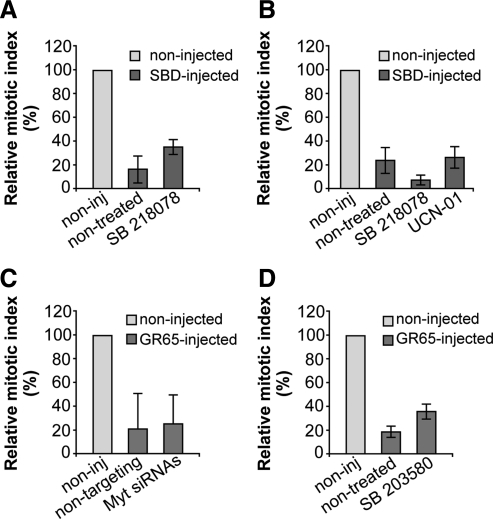
First, Chk1 is a downstream component of the DNA-damage checkpoint that is located on the centrosome, where it has been shown to inhibit Cdc25B (Loffler et al., 2007) and Aur-A (Krystyniak et al., 2006). Chk1 is the only kinase known to date that can negatively regulate Aur-A (Krystyniak et al., 2006); moreover, inhibition or depletion of Chk1 is sufficient to overcome the cell cycle block induced by DNA damage (Loffler et al., 2007). To address the involvement of Chk1, we used two structurally unrelated and well characterized inhibitors of Chk1: SB218078 (Tao and Lin, 2006) and UCN-01 (Cazales et al., 2005), which effectively inhibited the Chk1-mediated phosphorylation of Cdc25C (Supplemental Figure S5D). The cells were subjected to the double-thymidine synchronization protocol, microinjected with the SBD recombinant protein, and 4 h after the S block release, they were incubated with 2.5 μM SB218078 or 300 nM UCN-01, until they were fixed at the mitotic peak. After fixing, the cells were labeled for the cell cycle phase with Hoechst 33342. However, inhibition of Chk1 did not bypass the Golgi checkpoint (Figure 9, A and B), and we also did not see recovery of Aur-A recruitment by inhibition of Chk1 (data not shown).
Figure 9.
The known G2 checkpoints are not involved in the Golgi-dependent block of the cell cycle. HeLa cells (A, C, and D) and NRK cells (B) were grown on coverslips and left untreated (C) or arrested in S phase by using the double-thymidine block. (A and B) One hour after S phase block release, the cells were microinjected (or not; noninj) with recombinant SBD (8–12 mg/ml, SBD-inj) and FITC-conjugated dextran as microinjection marker. Next, 4 h after S phase block release the cells were either left untreated or treated with 2.5 μM SB218078 or 300 nM UCN-01. The cells were fixed 12 h after the final thymidine washout and processed for immunofluorescence under confocal microscopy. (C) The cells were transfected after the first thymidine block of the cell synchronization protocol, for 24 h with 20 nM nontargeting siRNAs or with 20 nM anti-Myt1 siRNAs, or processed without siRNAs transfection. One hour after S phase block release, cells were microinjected (or not; noninj) with recombinant GRASP65 (8–10 mg/ml), and FITC-conjugated dextran as microinjection marker. (D) One hour after S phase block release, cells were microinjected (or not; noninj) with recombinant GRASP65 (8–10 mg/ml), and FITC-conjugated dextran as microinjection marker. After 4 h without thymidine, cells were either left nontreated or treated with 10 μM SB203580 (a p38 kinase inhibitor). For all quantification data (A–D), the cells were fixed 12 h after the second thymidine washout, processed for immunofluorescence, and observed under confocal microscopy, with labeling with Hoechst 33342 for cell cycle phase. Quantification of the relative mitotic indices is shown as percentages of microinjected cells in mitosis normalized to nonmicroinjected cells on the same coverslip. More than 200 cells were microinjected for each condition, and data are means ± SD from two (B) or three (A, C, and D) independent experiments, each carried out in duplicate.
Next, we examined the role of Myt1, a kinase that inhibits cycB-Cdk1 through phosphorylation of both the T14 and Y15 residues of Cdk1 and that is activated by DNA damage. Importantly, Myt1 is localized at the Golgi complex and the endoplasmic reticulum, and it has been shown to be involved in both mitotic Golgi fragmentation (Cornwell et al., 2002) and Golgi reassembly at telophase (Nakajima et al., 2008). Consequently, Myt1 might have an important role in the Golgi checkpoint and be hyperactivated by a block of Golgi fragmentation. In this case, ablation of Myt1 should bypass the G2 block after the induction of a block of Golgi fragmentation. To test this hypothesis, we knocked down Myt1 and tested whether this could override the Golgi checkpoint. Treatment of cells with siRNA against Myt1 for 24 h was sufficient to achieve an efficient depletion of Myt1 (Supplemental Figure S6A) and override a DNA-damage–mediated cell cycle block in G2 (Supplemental Figure S6, B and C) without affecting the integrity of the Golgi ribbon (data not shown). The cells were subjected to the double-thymidine synchronization protocol, injected with GRASP65, and treated as described above. Again, the depletion of Myt1 did not overcome the Golgi checkpoint (Figure 9C). Together, these data indicate that the components of the DNA-damage checkpoint are not involved in the Golgi-dependent cell cycle block and in the impairment of Aur-A recruitment to the centrosome.
Finally, we examined the involvement of the known stress-related G2 checkpoints. One of these checkpoints is triggered by the ubiquitin ligase CHFR (Yu et al., 2005). Because HeLa cells express a nonfunctional form of CHFR (Yu et al., 2005), this pathway cannot be involved in the Golgi checkpoint. Therefore, we focused on the other stress-related checkpoints that are mediated by p38 kinase (Cha et al., 2007), which is an MAP kinase component localized at the centrosome and known to be activated in response to various environmental stresses and to have an inhibitory role on Cdc25B activity (Cha et al., 2007; see below). Here, we used SB203580, a highly selective inhibitor of p38 that can overcome the stress-dependent G2 checkpoint mediated by p38 itself (Cirillo et al., 2002). To this purpose, HeLa cells were subjected to the double-thymidine synchronization protocol, microinjected with the GRASP65 recombinant protein, and 4 h after S block release, the cells were treated with 10 μM SB203580, until they were fixed at the mitotic peak. The cells were then labeled for the cell cycle phase with Hoechst 33342. This treatment resulted in an effective inhibition of p38 autophosphorylation (Supplemental Figure S7A) but in a negligible recovery of the G2 block in the cells microinjected with GRASP65 (Figure 9D). Similar results were obtained by knocking down p38 by RNA interference (Supplemental Figure S7, B and C). Conversely, stimulation of p38 by anisomycin (Cha et al., 2007) did not cause any alteration of Aur-A recruitment at the centrosome (Supplemental Figure S7, D and E). Together, these data indicate that the p38-mediated checkpoint should only have a minor role, if any, in the Golgi checkpoint.
Thus, the Golgi checkpoint cannot be overridden by treatments that can bypass the known G2 checkpoints. This suggests further that Aur-A recruitment to and activation at the centrosome is connected with the partitioning of the Golgi ribbon through a novel signaling mechanism (Figure 10). Moreover, these last data again indicate that centrosome located and activated Aur-A is a cell cycle component that functions as the major effector of the Golgi checkpoint.
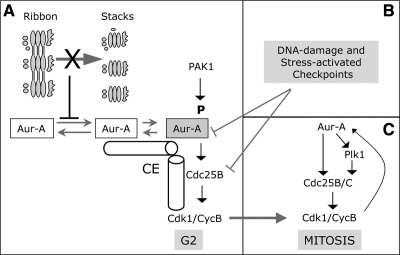
Figure 10.
The block of severing of the Golgi ribbon during G2 inhibits the recruitment and activation of Aur-A at the centrosome trough a novel mechanism. (A) In mammalian cells, the Golgi complex is organized as a continuous membranous system composed of stacks interconnected by tubules, a structure known as the Golgi ribbon (ribbon). During the G2 phase of the cell cycle, the severing of the Golgi ribbon into its constituent stacks (stacks) is a fragmentation step that is essential for mitotic entrance and is concomitant with the initial recruitment and activation of Aur-A kinase at the centrosomes (CE). A block in Golgi fragmentation (X) inhibits Aur-A recruitment and activation at the centrosome, and this in turn impairs the first activation of cycB-Cdk1 at the centrosome. The Golgi-dependent G2 block is not mediated by known Aur-A activators (i.e., Pak1), and neither it is indirectly mediated or Plk1 and the cycB–Cdk1 complex. (B) The Golgi-dependent G2 block is not regulated by known G2 checkpoint mediators (e.g., : Chk1 or p38), and neither it is mediated by Golgi-localized cell cycle regulators (e.g., : Myt-1), which act downstream the Golgi checkpoint. (C) At the onset of mitosis the kinases Aur-A, Plk1 and cycB-Cdk1 become functionally connected by a positive feedback loop, leading to irreversible progression into mitosis.
DISCUSSION
In the present study, we have used microinjection-based assays to induce acute “damage” to Golgi partitioning as a general experimental approach to investigate cell cycle proteins that are “effectors” of the Golgi checkpoint.
Here, we have identified recruitment to and activation at the centrosome of Aur-A kinase as the mitotic signaling step that coordinates the partitioning of Golgi membranes with the regulation of entry into mitosis. This conclusion is based on the following evidence. First, the severing of the Golgi ribbon during G2 (i.e., the fragmentation step that is essential for mitotic entrance) is concomitant with the initial recruitment and activation of Aur-A kinase at the centrosomes. Second, a block in Golgi partitioning leads to reduced recruitment to and impaired activation at the centrosome of Aur-A. The causal link between inhibition of the severing of the Golgi ribbon in G2 and block of Aur-A recruitment and activation is shown by the lack of inhibition of Aur-A recruitment when the SBD and GRASP65 blockers of Golgi fragmentation are microinjected into cells with a fragmented Golgi ribbon. In addition, we have shown that overexpression of Aur-A can overcome the cell cycle block induced by inhibition of Golgi fragmentation, indicating that Aur-A is a major effector of the Golgi checkpoint. Finally, we have shown that the Golgi-dependent G2 block is not influenced by known G2 checkpoint mediators, nor is it mediated by other kinases with essential roles in the activation of the cycB–Cdk1 complex, or by Golgi-localized cell cycle regulators. Overall, our findings reveal the existence of novel mechanisms that upon a block of Golgi fragmentation, lead to inhibition of the recruitment of Aur-A to the centrosome during early G2, by acting either directly on Aur-A or indirectly on an Aur-A activator (Figure 10).
Aur-A is an oncogene that is overexpressed in several human cancers, and it is required for multiple steps as cells progress toward and through mitosis (Marumoto et al., 2005). Aur-A expression increases as cells pass through S phase; then during G2, Aur-A is recruited to the centrosomes where it is activated (Marumoto et al., 2005). Once activated, Aur-A directly phosphorylates Cdc25B phosphatase (Cazales et al., 2005), which itself activates the cycB–Cdk1 complex by removing two inhibitory phosphorylations (Marumoto et al., 2005). This centrosomally activated cycB-Cdk1 then triggers entry into mitosis (Jackman et al., 2003).
Aur-A is activated by either autophosphorylation or transphosphorylation on T288 in its activation domain. The best characterized kinase involved in Aur-A phosphorylation is Pak; however, we show that Pak is not involved in the Golgi checkpoint. This phosphorylation is not sufficient for the maintenance of Aur-A activation, because phosphatases (e.g., PP1) rapidly dephosphorylate the T288 residue (Marumoto et al., 2005). This activity of phosphorylated Aur-A is thus protected by its binding to scaffolding proteins, such as the LIM-domain protein Ajuba (Hirota et al., 2003), the focal adhesion scaffolding protein HEF1 (a member of the p130Cas family; Pugacheva and Golemis, 2005), and the microtubule-associated protein TPX2 (Bayliss et al., 2003). Because Ajuba and HEF1 are focal-adhesion proteins, significant cross-talk between focal adhesions and the mitotic apparatus has been proposed (Pugacheva and Golemis, 2006). It is possible that the disassembly of focal adhesions releases these protein complexes, and leads to the activation of Aur-A, thus coordinating the loss of adhesion to entry into mitosis. However, depletion of HEF1 or Ajuba results in inactivation of Aur-A, without affecting its recruitment to the centrosome (Hirota et al., 2003; Pugacheva and Golemis, 2005); thus, HEF1 and Ajuba cannot be involved in the Golgi checkpoint. The other Aurora-A activator, TPX2, is involved in post-G2 targeting of Aur-A at the mitotic spindle apparatus. Indeed, siRNA-mediated depletion of TPX2 abolishes the association of Aur-A with the spindle, without affecting its association with the centrosomes (Kufer et al., 2002).
The next key question is to understand why and how this severing of the Golgi ribbon controls the recruitment of Aur-A to the centrosome. One simple possibility is that the Golgi membranes “sense” the mechanical tension exerted by the separating centrosomes, and in the case of an unbroken Golgi ribbon, could “emit” a stress signal to turn off recruitment of Aur-A to the centrosomes. Alternatively, Aur-A activation itself might be positively regulated by the severing of the Golgi ribbon. Indeed, even if the severing of the Golgi ribbon results in only a minor acceleration of the timing of mitotic entry and Aur-A recruitment/activation (data not shown), Aur-A might function as a signaling hub. Thus, Aur-A might integrate positive signals from various inputs, among which there are those of the separating Golgi membranes, which would all be required for sustained Aur-A recruitment and activation. In this respect, Golgi fragmentation during G2 might be “signaled” through release of proteins from the severed membranes that would regulate the activation and/or recruitment of Aur-A during early G2. This is analogous to the actions of clathrin or Nir2, which are released from Golgi membranes during prophase and which localize to the mitotic spindle and the midbody, respectively, where they acquire new functions that are related to the formation of the mitotic spindle and cytokinesis (Colanzi and Corda, 2007).
As a further possibility, the basis of the Golgi checkpoint could be that an unbroken Golgi ribbon would impede the Golgi membranes in their correct “positioning” with respect to the separating centrosomes, an aspect that might be connected with mechanisms controlling the geometrical precision of the planar orientation of cell division. This is an essential feature that is critical during development and that remains important for the maintenance of polarized structures in adults (Thery et al., 2006). Recent evidence suggests that the nucleus, centrosome and Golgi complex are localized along the axis of cell polarity, which is determined by spatial cues provided by the geometry of cell–cell and/or cell–extracellular matrix contacts (Thery et al., 2006). These processes are coordinated at the centrosome through the actions of proteins that regulate cytoskeleton organization and function. Moreover, the Golgi/centrosome module is reoriented toward the leading edge of the cell in migrating fibroblasts, and this function involves proteins such as Cdc42 (Nobes and Hall, 1999), Par6 (Palazzo et al., 2001), and GRASP65 (Bisel et al., 2008). Thus, the signaling pathways that form the basis of the coordinated orientation of the Golgi/centrosome module might be essential for a mechanism that checks the correct reciprocal orientation of the Golgi complex with respect to the separating centrosomes during G2. Thus, an unbroken Golgi ribbon would interfere with the correct positioning of the Golgi membranes, and this could in turn interfere with the recruitment of Aur-A.
We have thus here identified the key cell cycle regulatory element that connects the partitioning of the Golgi complex to the molecular events involved in the initiation of mitosis, although how the Golgi checkpoint affects the onset of Aur-A activation remains to be determined. The identification of Aur-A as the cell cycle effector of the Golgi checkpoint provides the basis for defining the mechanisms by which the Golgi complex can monitor its correct partitioning and therefore control G2/M transition. These findings have important physiological and pharmacological consequences, because the definition of this pathway will lead to the identification and validation of novel targets of antiproliferative therapies and cancer treatments.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Luini for insightful discussions and suggestions, I. Ayala and C. Wilson for critical reading of the manuscript, S. Ferrari (University of Zurich, Zurich, Switzerland) and B. Ducommun (University of Toulouse, Toulouse, France) for the generous gift of the Aurora-related constructs, J. Chernoff for the generous gift of IPA3, J. Ecsedy for the generous gift of MLN8054, and J. Pines (Wellcome Trust, Cambridge, United Kingdom) for the generous gift of the Plk1-related constructs. We also thank C. P. Berrie for critical appraisal of the manuscript, E. Fontana for help in preparing part of the figures, C. Cericola for preparing the recombinant proteins, and S. Arbucci for providing assistance with confocal images acquisition. A. C. acknowledges the Italian Association for Cancer Research (AIRC, Milan, Italy; IG 6074) and Telethon (Italy) for financial support. D. C. acknowledges the Italian Association for Cancer Research (AIRC, Milan, Italy; IG 4664), the European Community's Seventh Framework Programme FP7/2007–20013 under grant 202272 (LipidomicNet) for financial support. A. P. was a Fellow of the Italian Foundation for Cancer Research (FIRC, Milan, Italy). R.I.C. was recipient of a “Floriana Dalle Carbonare” Fellowship of the Italian Association for Cancer research (AIRC, Milan, Italy).
Abbreviations used:
- Aur-A
Aurora-A
- GST
glutathione transferase
- SBD
BARS substrate-binding domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0243) on September 15, 2010.
REFERENCES
- Acharya U., Mallabiabarrena A., Acharya J. K., Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. doi: 10.1016/s0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet N., Sougrat R., Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Sardon T., Vernos I., Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- Bisel B., Wang Y., Wei J. H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J. Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin D. V., Higashimoto Y., Popoff I. J., Gaarde W. A., Basrur V., Potapova O., Appella E., Fornace A. J., Jr Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature. 2001;411:102–107. doi: 10.1038/35075107. [DOI] [PubMed] [Google Scholar]
- Cazales M., Schmitt E., Montembault E., Dozier C., Prigent C., Ducommun B. CDC25B phosphorylation by Aurora-A occurs at the G2/M transition and is inhibited by DNA damage. Cell Cycle. 2005;4:1233–1238. doi: 10.4161/cc.4.9.1964. [DOI] [PubMed] [Google Scholar]
- Cha H., Wang X., Li H., Fornace A. J., Jr A functional role for p38 MAPK in modulating mitotic transit in the absence of stress. J. Biol. Chem. 2007;282:22984–22992. doi: 10.1074/jbc.M700735200. [DOI] [PubMed] [Google Scholar]
- Chan E. H., Santamaria A., Sillje H. H., Nigg E. A. Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma. 2008;117:457–469. doi: 10.1007/s00412-008-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo P. F., Pargellis C., Regan J. The non-diaryl heterocycle classes of p38 MAP kinase inhibitors. Curr. Topics Med. Chem. 2002;2:1021–1035. doi: 10.2174/1568026023393390. [DOI] [PubMed] [Google Scholar]
- Colanzi A., Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr. Opin. Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Colanzi A., Hidalgo Carcedo C., Persico A., Cericola C., Turacchio G., Bonazzi M., Luini A., Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Suetterlin C., Malhotra V. Cell-cycle-specific Golgi fragmentation: how and why? Curr. Opin. Cell Biol. 2003;15:462–467. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- Corda D., Colanzi A., Luini A. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 2006;16:167–173. doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Cornwell W. D., Kaminski P. J., Jackson J. R. Identification of Drosophila Myt1 kinase and its role in Golgi during mitosis. Cell Signal. 2002;14:467–476. doi: 10.1016/s0898-6568(01)00276-5. [DOI] [PubMed] [Google Scholar]
- Duran J. M., Kinseth M., Bossard C., Rose D. W., Polishchuk R., Wu C. C., Yates J., Zimmerman T., Malhotra V. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol. Biol. Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T. N., Linstedt A. D. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol. Biol. Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Bian M., Jiang Q., Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- Hidalgo Carcedo C., Bonazzi M., Spano S., Turacchio G., Colanzi A., Luini A., Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- Hoar K., Chakravarty A., Rabino C., Wysong D., Bowman D., Roy N., Ecsedy J. A. MLN8054, a small-molecule inhibitor of Aurora A, causes spindle pole and chromosome congression defects leading to aneuploidy. Mol. Cell. Biol. 2007;27:4513–4525. doi: 10.1128/MCB.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E. A., Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Kodani A., Kristensen I., Huang L., Sutterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol. Biol. Cell. 2009;20:1192–1200. doi: 10.1091/mbc.E08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystyniak A., Garcia-Echeverria C., Prigent C., Ferrari S. Inhibition of Aurora A in response to DNA damage. Oncogene. 2006;25:338–348. doi: 10.1038/sj.onc.1209056. [DOI] [PubMed] [Google Scholar]
- Kufer T. A., Sillje H. H., Korner R., Gruss O. J., Meraldi P., Nigg E. A. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler H., Bochtler T., Fritz B., Tews B., Ho A. D., Lukas J., Bartek J., Kramer A. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle. 2007;6:2541–2548. doi: 10.4161/cc.6.20.4810. [DOI] [PubMed] [Google Scholar]
- Manfredi M. G., et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc. Natl. Acad. Sci. USA. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr, Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M. A. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T., et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- Marumoto T., Zhang D., Saya H. Aurora-A—a guardian of poles. Nat. Rev. Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Yonemura S., Murata M., Nakamura N., Piwnica-Worms H., Nishida E. Myt1 protein kinase is essential for Golgi and ER assembly during mitotic exit. J. Cell Biol. 2008;181:89–103. doi: 10.1083/jcb.200708176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Nobes C. D., Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A. F., Joseph H. L., Chen Y. J., Dujardin D. L., Alberts A. S., Pfister K. K., Vallee R. B., Gundersen G. G. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol. 2001;11:1536–1541. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- Preisinger C., Korner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E. N., Golemis E. A. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat. Cell Biol. 2005;7:937–946. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E. N., Golemis E. A. HEF1-aurora A interactions: points of dialog between the cell cycle and cell attachment signaling networks. Cell Cycle. 2006;5:384–391. doi: 10.4161/cc.5.4.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- Seki A., Coppinger J. A., Jang C. Y., Yates J. R., Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul Y. D., Seger R. ERK1c regulates Golgi fragmentation during mitosis. J. Cell Biol. 2006;172:885–897. doi: 10.1083/jcb.200509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Warren G. Golgi architecture and inheritance. Annu. Rev. Cell. Dev. Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Steegmaier M., et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Stenoien D. L., Sen S., Mancini M. A., Brinkley B. R. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil. Cytoskeleton. 2003;55:134–146. doi: 10.1002/cm.10120. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Hsu P., Mallabiabarrena A., Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Lin C. Y., Feng Y., Ferris D. K., Erikson R. L., Malhotra V. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proc. Natl. Acad. Sci. USA. 2001;98:9128–9132. doi: 10.1073/pnas.161283998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z. F., Lin N. H. Chk1 inhibitors for novel cancer treatment. Anticancer Agents Med. Chem. 2006;6:377–388. doi: 10.2174/187152006777698132. [DOI] [PubMed] [Google Scholar]
- Thery M., Racine V., Piel M., Pepin A., Dimitrov A., Chen Y., Sibarita J. B., Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle. 2006;5:2555–2556. doi: 10.4161/cc.5.22.3463. [DOI] [PubMed] [Google Scholar]
- Viaud J., Peterson J. R. An allosteric kinase inhibitor binds the p21-activated kinase autoregulatory domain covalently. Mol. Cancer Ther. 2009;8:2559–2565. doi: 10.1158/1535-7163.MCT-09-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Satoh A., Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Yoshioka K., Barr F. A., Lowe M., Nakayama K., Ohkuma S., Nakamura N. Convergence of cell cycle regulation and growth factor signals on GRASP65. J. Biol. Chem. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]
- Yu X., Minter-Dykhouse K., Malureanu L., Zhao W. M., Zhang D., Merkle C. J., Ward I. M., Saya H., Fang G., van Deursen J., Chen J. Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- Zhao Z. S., Lim J. P., Ng Y. W., Lim L., Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol. Cell. 2005;20:237–249. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.