Abstract
HIV-1 subtype D (HIV-1D) progresses to disease faster and has lower transmissibility than subtype A (HIV-1A). We examined whether these differences could lead to a population level change in the distribution of these subtypes over time. HIV-1 viral RNA was extracted from stored serum samples from HIV-positive subjects participating in a population-based cohort study in Rakai, Uganda in 1994 and 2002. Portions of the viral proteins gag and gp41 were sequenced and subtyped. HIV-1 subtype assignments were generated for 773 subjects in 1994 and 812 subjects in 2002. The change in subtype distribution of the population as a whole as well as quartile age groups were examined for significant changes using a linear model. There was a significant decrease in the proportion of subjects infected with HIV-1D from 70.2% to 62.4% and a significant increase in subjects infected with HIV-1A from 16.7% to 23.3% over the 8-year period (p = 0.005). The most marked changes in proportion of HIV-1D and A were seen in the younger individuals (<25 and 25–30 years; p < 0.05). The percentages of subjects infected with HIV-1C and recombinant subtypes did not change significantly. Over this 8-year period, the overall viral population in this region evolved toward the less virulent HIV-1A strain, most likely as a consequence of the faster disease progression and lower transmissibility of HIV-1D.
Introduction
One of the defining characteristics of the spread of human immunodeficiency virus type 1 (HIV-1) is its high degree of genetic diversity as a consequence of its replication cycle and the high error rate of reverse transcriptase.1 Due to this diversity, HIV-1 is divided into nine subtypes (HIV-1A–D, F–H, J, and K) and multiple intersubtype recombinant strains.1 Genetic variation between subtypes has been estimated to be 25–33% in the amino acid sequences of the envelope protein, whereas variation within a subtype is approximately 15–20%.1,2 This diversity is most evident in sub-Saharan Africa.1
Previous research has demonstrated that infections with HIV-1D are associated with faster disease progression than those with HIV-1A.3–6 In an open cohort of commercial sex workers with known times of HIV infection in Mombasa, Kenya, HIV-1D infection was associated with a greater than two fold increased risk of death compared to HIV-1A infection.5 A study from Rakai, Uganda also found that subjects who were infected with HIV-1D or with recombinant subtypes incorporating subtype D had a significantly faster rate of progression to death than those who were infected with HIV-1A.7 Similarly, a faster rate of progression has been noted for people infected with HIV-1A/E in Thailand compared to individuals infected with HIV-1B.8,9 The faster progression was independent of HIV-1 viral load.
Additionally, studies of monogamous HIV discordant couples in Rakai, Uganda observed that HIV-1D was less transmissible than HIV-1A.4,10 The adjusted risk ratio of HIV transmission was 1.98 (95% CI, 1.17–3.34) for HIV-1A compared to HIV-1D.4
Phylogenetic analysis of the HIV epidemic in Eastern Africa suggests that HIV-1D was introduced into the Rakai District of southwestern Uganda in the 1970s, and experienced exponential growth during the civil war and Tanzanian invasion.11 This led to a high prevalence of HIV-1D in the early stage of the HIV-1 epidemic in this region, and substantial proportion of A/D recombinant infections in the Rakai population.11–13
We hypothesized that the differences in viral subtype transmission and pathogenicity could lead to a population level decline in the proportion of HIV-1D infections relative to HIV-1A over time in the absence of antiretroviral therapy (ART). Moreover, we speculate that these changes in subtype distribution would be more pronounced in the younger population since this age group should have the greatest percentage of recent infections. To assess changes in HIV-1 subtype distribution, we analyzed HIV-1-infected individuals in a general population cohort in the Rakai district of Uganda in 1994 and 2002.
Materials and Methods
Study population
The Rakai Community Cohort Study (RCCS) is a rural, population-based open cohort that has conducted annual surveillance since 1994 of persons aged 15–49 years residing in the Rakai district of southwestern Uganda.14 Interviews and venous blood samples were obtained annually from approximately 14,000 consenting adults living in over 50 villages. The same villages were surveyed annually with approximately 85% of subject overlap over subsequent years. These villages were further grouped into northern, central, and southern regions of the district. Data from the 1994 and 2002 surveys were used to assess changes in HIV-1 subtype distributions over time. Both surveys were conducted prior to the introduction of ART in 2004. All subjects provided written consent for their samples to be stored and used for research purposes. The study was approved by Institutional Review Boards (IRBs) in Uganda (the Uganda Virus Research Institute's Science and Ethics Committee and the Uganda National Council for Science and Technology) and from the IRBs of collaborating U.S. institutions (Walter Reed Army Institute of Research, Columbia University, and Johns Hopkins University).
Laboratory analysis
HIV status was assessed by two enzyme immunoassays: Vironostika HV-1 (Organon Teknika, Charlotte, NC) and Cambridge Biotech (Worcester, MA). Discordant enzyme immunoassay (EIA) results were confirmed by Western blot (bioMérieux VITEK, St. Louis, MO). Viral RNA was extracted from all available stored serum samples of HIV-1 antibody-positive subjects from 1994 and 2002 using a QIAmp Viral RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol.15 The eluted RNA was expanded by reverse transcriptase polymerase chain reaction (RT-PCR) and an additional nested PCR in two separate reactions to amplify portions of gag (HXB2 nt 1249–1704) and gp41 (HBX2 nt 7858–8260).16,17 Amplified samples were purified using ExoSAP-IT (USB, Cleveland, OH) and then sequenced using the Applied Biosystems 3730xl DNA Analyzer. Subtype assignments were generated using the NCBI genotyping database, and verified using a phylogenetic analysis of all sequences from each year. Sequence subtype classification for both the gag and gp41 fragments was based on their relation to the closest reference sequence. Subjects with both gag and gp41 sequences that were uniformly subtype A, C, or D were considered infected with that subtype, and subjects with discordant sections were considered infected with a recombinant strain.16,17 Data from 1994 were similarly generated as described previously.18
Statistical analysis
The distributions of subtypes were determined in 1994 and 2002, and changes in subtype distributions were assessed using Chi-square tests. The subtype distributions were also stratified by geographic region and changes in distributions were examined.18 Subtype prevalence rates were estimated by applying the subtype distribution to the overall prevalence of HIV-1 in 1994 and 2002.
For the samples collected at each year, only a subset of samples was able to be successfully amplified and sequenced with valid assignment of HIV-1 subtype information. Therefore, to ensure the generalizability of inference about the subtype distribution change based on the subtyped samples, Pearson Chi-square statistics were used to compare the unsubtyped and subtyped subject characteristics including geographic region, age, gender, current marital status, current nonmarital sexual relationship, number of sex partners during the past year, number of sex partners during the past 5 years, and condom use.
The empirical change between 1994 and 2002 in subtype distribution was initially assessed using Chi-square tests for all subtyped samples. To generalize the result based on the subtyped samples to the HIV-1-infected population, the subtype distribution change was further estimated while adjusting for geographic region. Specifically, a linear model was used to model the proportions of subtypes as functions of year and region. Moreover, age (grouped as <25, 25–30, 30–35, or >35) and its interaction with year were included in the region adjusted model to test the hypothesis about the subtype distribution change in a younger infected population. Model fitness was examined using residuals, and all the modeling was performed using SAS PROC CATMOD.
Results
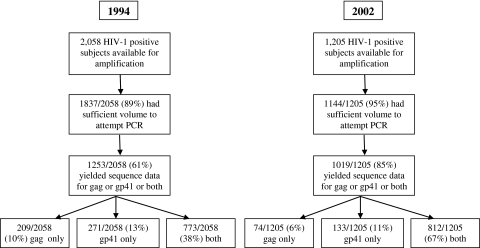
A total of 2058 HIV-positive subjects were identified in 1994, of which 1837 (89.3%) had sufficient volume for PCR, and 773 (37.6% of total) of these samples had sequence data for both gag and gp41 and were assigned a subtype (Fig. 1).18 These regions were examined because of their relative genetic stability over time and distinct separation within the genome. There were 1205 HIV-positive subjects identified in 2002, of whom 1144 (95%) had sufficient volume for PCR, and 812 (67% of total) of these samples yielded sequence data for both the gag and gp41 regions and could be assigned a subtype (Fig. 1). The difference in the total number of HIV-infected subjects between 1994 and 2002 is reflective of a 4% decrease in overall HIV prevalence in the Rakai district during this 8-year period.
FIG. 1.
The trial profile of the subject population from which sequence data were obtained is shown for 1994 and 2002.
In 1994, 70.2% of the subtyped samples were HIV-1D, and the proportion of HIV-1D declined to 62.4% in 2002 (p = 0.005) (Table 1). Conversely, HIV-1A constituted 16.7% of samples in 1994, and this proportion increased to 23.3% in 2002 (p = 0.005) (Table 1). There were no significant changes in the proportion of subtype C or recombinant infections.
Table 1.
Comparison of Overall Subtype Distribution in 1994 and 2002
| HIV-1 subtype | 1994 n = 773, (%) | 2002 n = 812, (%) | Change (%) | Chi-square p-value |
|---|---|---|---|---|
| A | 129 (16.7) | 189 (23.3) | 6.6 | 0.005 |
| C | 7 (0.9) | 7 (0.9) | 0.0 | |
| D | 543 (70.2) | 507 (62.4) | −7.8 | |
| Recombinant | 94 (12.2) | 109 (13.4) | 1.2 |
Previous studies from Rakai have demonstrated that HIV-1 subtypes differed by geographic region.18 Stratifying by geographic region, the subject characteristics of the subtyped and unsubtyped samples were compared at year 1994 and 2002, respectively; it was found that the subtyped and unsubtyped samples were comparable at both years with the only exception that two of the three regions had significant differences in terms of gender in 1994. Therefore, the subtyped samples are representative of the HIV-1-infected population as a whole in Rakai when stratifying by region.
The changes in subtype distribution and other possible confounding factors (age, gender, marriage status, nonmarital relationships, sex partners in the past year and past 5 years, and condom use) were subsequently examined by geographic region using a linear model (Table 2). Stratified by region, the subtype distribution change is not significantly different between the geographic regions, with a consistent decrease in the proportion of infections with HIV-1D and a corresponding increase in HIV-1A (Table 2). Controlling for region and other factors, the model estimated a significant decrease in HIV-1D of –8.8% (95% CI: –4.2–13.4%) and a significant increase in HIV-1A of 7.8% (95% CI: 4.0–11.6%). The proportion of HIV-1C and recombinant infections did not change significantly, with an estimated joint increase of 1.1% (95% CI: –2.4–4.5%).
Table 2.
Comparison of Observed Region Specific Subtype Proportions in 1994 and 2002
| |
North |
Central |
South |
||||||
|---|---|---|---|---|---|---|---|---|---|
| |
1994 |
2002 |
|
1994 |
2002 |
|
1994 |
2002 |
|
| Subtype | n (%) | n (%) | Change (%) | n (%) | n (%) | Change (%) | n (%) | n (%) | Change (%) |
| A | 77 (21.1) | 91 (27.0) | 5.9 | 39 (15.5) | 55 (23.8) | 8.3 | 13 (8.3) | 42 (17.5) | 9.2 |
| C | 3 (0.8) | 1 (0.3) | −0.5 | 1 (0.4) | 1 (0.4) | 0.0 | 3 (1.9) | 5 (2.1) | 0.2 |
| D | 239 (65.5) | 196 (58.2) | −7.4 | 182 (72.2) | 145 (62.8) | −9.4 | 122 (77.7) | 163 (67.9) | −9.8 |
| Recombinant | 46 (12.6) | 49 (14.5) | 1.9 | 30 (11.9) | 30 (13.0) | 1.1 | 19 (12.1) | 30 (12.5) | 0.4 |
| Total | 365 | 337 | 252 | 231 | 157 | 240 | |||
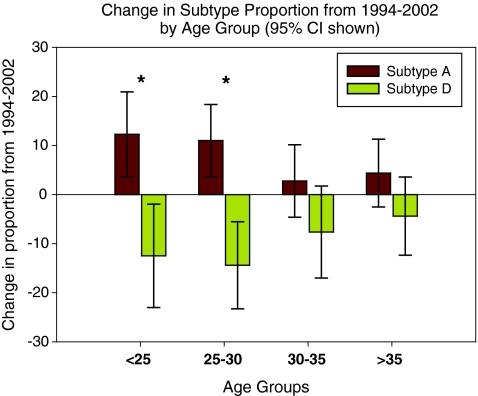
The linear model was used to examine the proportions of the subtypes adjusting for functions of the indicator variables and relevant covariables. We hypothesized that if the proportion of subtypes changed over time, this change would be most prominent in the younger population since they were more likely to be recent seroconverters. Thus, the variable of age (grouped as <25, 25–30, 30–35, or >35) and its interaction with the variable indicating year were included in the model to assess the age group-specific changes of HIV subtype distribution. The most prominent change in subtype distribution was observed in the age group younger than 25 years old (p = 0.017) and the group aged between 25 and 30 years (p = 0.004) (Fig. 2). Specifically, for the group younger than 25 years, HIV-1A increased 12.3% (95% CI: 3.6–20.9%), coupled with a significant drop in HIV-1D of 12.5% (95% CI: 1.9–20.3%). For the group of 25–30 years, HIV-1A increased 11.0% (95% CI: 3.7–18.4%) while HIV-1D decreased 14.4% (95% CI: 5.5–23.3%); HIV-1C and recombinants did not change significantly in either age group.
FIG. 2.
The amount of change from 1994 to 2002 in the proportion of HIV-1A and D is shown for the four age groups examined. The linear model's 95% confidence intervals are shown with significance indicated (*p < 0.05). The model fitness was examined based on the significance of the residuals. (Color image can be found at www.liebertonline.com/aid).
Over this 8-year period it has been shown that HIV prevalence in the Rakai cohort declined from 17% to 13%.11 The observed subtype proportions for 1994 and 2002 were multiplied by the overall HIV-1 prevalence rates to estimate subtype-specific prevalence. The estimated prevalence of HIV-1D decreased from 11.9% to 8.1% over time, and the prevalence of HIV-1A increased from 2.8% to 3.0%. This represents an absolute decrease of 3.8% in the estimated prevalence of HIV-1D and a 0.2% increase in the estimated prevalence of HIV-1A.
Discussion
We observed a significant decrease in the proportion of HIV-1D infections and an increase in the proportion of HIV-1A infections in Rakai, Uganda between 1994 and 2002. The greatest increase in HIV-1A and decrease in HIV-1D occurred in the younger age groups, which should reflect recent infections. These findings are consistent with a previous report from Kenya that also observed a decrease in the proportion of HIV-1D, but did not see a significant increase in HIV-1A.19 Additionally, in both studies there was no significant increase in the proportion of HIV-1C, which differs from other studies performed in Tanzania.20 Previous studies have demonstrated HIV-1D progresses to disease and death faster than HIV-1A and has a lower transmission rate relative to HIV-1A.4,5 It is not yet known what causes these differences in transmissibility and pathogenicity, but it is most likely virological in nature since these differences have been described in similar populations in the same location.1,5,7,21
This study is somewhat limited since subtype assignment was dependent on viral amplification, and a lower proportion of samples had amplifiable samples in 1994 (61%) than in 2002 (85%). In addition, among all HIV-positive subjects the samples that were successfully sequenced for both the gag and g41 genes was lower in 1994 (38%) than in 2002 (67%). This discrepancy is likely due in part to lower serum volumes found in the older samples and decreased amplification caused by extended storage. If viral loads differed by subtype, this could have led to selective amplification of the two target genes between the two time periods. However, several studies, including one previous study in Rakai, have demonstrated that HIV-1D and HIV-1A have comparable median viral loads, which suggests that amplification should not be differential between subtypes.5,7 It is also possible that certain subtypes were not amplifiable with the primer sets used, although these primers were found to expand HIV-1A, B, D, and C. Only two regions were sequenced so it is possible that some individuals may be infected with undetected recombinant strains. However, this should be equivalent for both subtypes and therefore should not significantly affect the overall findings. Identical extraction, amplification, and sequencing techniques were used for the two time points to eliminate possible technical bias. Moreover, within each geographic area, the participants with subtyped and unsubtyped samples had comparable characteristics at both time points, suggesting that after adjusting for geographic region, the inference based on the subtyped samples is generalizable to the HIV-1-infected population in Rakai.
The migration rate in and out of the Rakai cohort is approximately 15% annually, but has been shown to be equivalent in the HIV-negative and -positive population.22 We did not observe a significant increase in recombinant strains, which are found at higher levels northeast of Rakai, or in HIV-1C.20 Therefore, differential migration is unlikely to affect our findings. The lack of increase in proportion of recombinants probably reflects the low number of lifetime sex partners among this rural population. This may partly explain why other studies of more urban settings have found consistent increases in the proportion of recombinant strains.20,23–25
Phylogenetic analysis of HIV-1 in Uganda suggests that HIV-1A and D were introduced into this population as two distinct founder events, and subsequently HIV-1D prevalence appears to have increased during the social disruption caused by the civil wars and Tanzanian invasion of the 1970s leading to a predominance of HIV-1D in the early 1990s.11 The present study suggests that after political stabilization in 1986, the overall HIV-1 viral population in this area evolved toward a less virulent strain, HIV-1A, and that this was caused in part by the selective advantage of decreased pathogenicity and increased infectivity of HIV-1A over D.
Acknowledgments
The authors would like to thank all the participants of the Rakai cohort and the staff of the Rakai health science program. We would also like to thank Susanna Lamers for her technical expertise. This study was supported in part by funding from the Division of Intramural Research, NIAID, NIH; NIAID (Grants R01 A134826 and R01 A134265); NICHD (Grant 5P30HD06826); the World Bank STI Project, Uganda; the Henry M. Jackson Foundation; the Fogarty Foundation (Grant 5D43TW00010); and the Bill and Melinda Gates Institute for Population and Reproductive Health at J.H.U. All sequence data have been submitted to GenBank (ascension numbers for the gp41 sequences: HM114622–HM115320 and for the p24 sequences: HM115321–HM115893).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Taylor BS. Sobieszczyk ME. McCutchan FE. Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 3.Sankale JL. Hamel D. Woolsey A, et al. Molecular evolution of human immunodeficiency virus type 1 subtype A in Senegal: 1988–1997. J Hum Virol. 2000;3:157–164. [PubMed] [Google Scholar]
- 4.Kiwanuka N. Laeyendecker O. Gray RH, et al. HIV-1 subtypes and differences in heterosexual transmission of HIV among HIV-1 discordant couples in Rakai, Uganda. AIDS. 2009;23:2479–2484. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baeten JM. Chohan B. Lavreys L, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195:1177–1180. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 6.Vasan A. Renjifo B. Hertzmark E, et al. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42:843–852. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- 7.Kiwanuka N. Laeyendecker O. Robb M, et al. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197:707–713. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 8.Nelson KE. Costello C. Suriyanon V. Sennun S. Duerr A. Survival of blood donors and their spouses with HIV-1 subtype E (CRF01 A_E) infection in northern Thailand, 1992–2007. AIDS. 2007;21(Suppl 6):S47–54. doi: 10.1097/01.aids.0000299410.37152.17. [DOI] [PubMed] [Google Scholar]
- 9.Rangsin R. Chiu J. Khamboonruang C, et al. The natural history of HIV-1 infection in young Thai men after seroconversion. J Acquir Immune Defic Syndr. 2004;36:622–629. doi: 10.1097/00126334-200405010-00011. [DOI] [PubMed] [Google Scholar]
- 10.Shepherd JC. Laeyendecker O. Serwadda D, et al. 42nd Annual Meeting of IDSA; Boston, MA: 2004. Reduced transmission of HIV-1 subtype D compared with subtype A in Rakai, Uganda. [Google Scholar]
- 11.Gray RR. Tatem AJ. Lamers S, et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009;25:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arroyo MA. Sateren WB. Serwadda D, et al. Higher HIV-1 incidence and genetic complexity along main roads in Rakai District, Uganda. J Acquir Immune Defic Syndr. 2006;43:440–445. doi: 10.1097/01.qai.0000243053.80945.f0. [DOI] [PubMed] [Google Scholar]
- 13.Harris ME. Serwadda D. Sewankambo N, et al. Among 46 near full length HIV type 1 genome sequences from Rakai District, Uganda, subtype D and AD recombinants predominate. AIDS Res Hum Retroviruses. 2002;18:1281–1290. doi: 10.1089/088922202320886325. [DOI] [PubMed] [Google Scholar]
- 14.Wawer MJ. Gray RH. Sewankambo NK, et al. A randomized, community trial of intensive sexually transmitted disease control for AIDS prevention, Rakai, Uganda. AIDS. 1998;12:1211–1225. doi: 10.1097/00002030-199810000-00014. [DOI] [PubMed] [Google Scholar]
- 15.Collinson-Streng AN. Redd AD. Sewankambo NK, et al. Geographic HIV type 1 subtype distribution in Rakai district, Uganda. AIDS Res Hum Retroviruses. 2009;25:1045–1048. doi: 10.1089/aid.2009.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C. Dash BC. Simon F, et al. Detection of diverse variants of human immunodeficiency virus-1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primer pairs. J Infect Dis. 2000;181:1791–1795. doi: 10.1086/315439. [DOI] [PubMed] [Google Scholar]
- 17.Yang C. Dash B. Hanna SL, et al. Predominance of HIV type 1 subtype G among commercial sex workers from Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2001;17:361–365. doi: 10.1089/08892220150503726. [DOI] [PubMed] [Google Scholar]
- 18.Collinson-Streng A. Geographic HIV-1 subtype distribution in Rakai District, Uganda. AIDS Res Hum Retroviruses. 2009;25(10):1045–1048. doi: 10.1089/aid.2009.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainwater S. DeVange S. Sagar M, et al. No evidence for rapid subtype C spread within an epidemic in which multiple subtypes and intersubtype recombinants circulate. AIDS Res Hum Retroviruses. 2005;21:1060–1065. doi: 10.1089/aid.2005.21.1060. [DOI] [PubMed] [Google Scholar]
- 20.Renjifo B. Chaplin B. Mwakagile D, et al. Epidemic expansion of HIV type 1 subtype C and recombinant genotypes in Tanzania. AIDS Res Hum Retroviruses. 1998;14:635–638. doi: 10.1089/aid.1998.14.635. [DOI] [PubMed] [Google Scholar]
- 21.Layendecker O. Li X. Arroyo M, et al. the Rakai Health Science Program: The effect of HIV subtype of rapid disease progression in Rakai, Uganda. 13th Conference on Retroviruses and Opportunistic Infections; Denver, CO. 2006. [Google Scholar]
- 22.Wawer MJ. Serwadda D. Gray RH, et al. Trends in HIV-1 prevalence may not reflect trends in incidence in mature epidemics: Data from the Rakai population-based cohort, Uganda. AIDS. 1997;11:1023–1030. doi: 10.1097/00002030-199708000-00011. [DOI] [PubMed] [Google Scholar]
- 23.Herbinger KH. Gerhardt M. Piyasirisilp S, et al. Frequency of HIV type 1 dual infection and HIV diversity: analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res Hum Retroviruses. 2006;22:599–606. doi: 10.1089/aid.2006.22.599. [DOI] [PubMed] [Google Scholar]
- 24.Lihana RW. Khamadi SA. Lwembe RM, et al. The changing trend of HIV type 1 subtypes in Nairobi. AIDS Res Hum Retroviruses. 2009;25:337–342. doi: 10.1089/aid.2008.0228. [DOI] [PubMed] [Google Scholar]
- 25.Vidal N. Mulanga C. Bazepeo SE, et al. Distribution of HIV-1 variants in the Democratic Republic of Congo suggests increase of subtype C in Kinshasa between 1997 and 2002. J Acquir Immune Defic Syndr. 2005;40:456–462. doi: 10.1097/01.qai.0000159670.18326.94. [DOI] [PubMed] [Google Scholar]