Abstract
Evolutionary related multisubunit RNA polymerases from all three domains of life, Eukarya, Archaea and Bacteria, have common structural and functional properties. We have recently shown that two RNAP subunits, F/E (RPB4/7)—which are conserved between eukaryotes and Archaea but have no bacterial homologues—interact with the nascent RNA chain and thereby profoundly modulate RNAP activity. Overall F/E increases transcription processivity, but it also stimulates transcription termination in a sequence-dependent manner. In addition to RNA-binding, these two apparently opposed processes are likely to involve an allosteric mechanism of the RNAP clamp. Spt4/5 is the only known RNAP-associated transcription factor that is conserved in all three domains of life, and it stimulates elongation similar to RNAP subunits F/E. Spt4/5 enhances processivity in a fashion that is independent of the nontemplate DNA strand, by interacting with the RNAP clamp. Whereas the molecular mechanism of Spt4/5 is universally conserved in evolution, the added functionality of F/E-like complexes has emerged after the split of the bacterial and archaeo-eukaryotic lineages. Interestingly, bacteriophage-encoded antiterminator proteins could, in theory, fulfil an analogous function in the bacterial RNAP.
Keywords: transcription, RNA polymerase, F/E RPB4/7, Spt4/5, evolution, archaea
Introduction—RNAP Architecture in the Three Domains of Life
RNA polymerases (RNAPs) are among the top five most important enzymes in biology, all cellular life depends on them, and defects in the factors that regulate their activity result in a broad range of pathologies. All multisubunit RNAPs are evolutionary related and have a conserved structural organization represented by the bacterial enzyme, which contains 4 distinct types of subunits. The larger RNAPs from the archaeal and eukaryotic domains contain additional homologous subunits, which are not present in bacteria.1 Two of these, F/E (homologous to RPB4/7 in eukaryotes), form a stalk-like protrusion, which is the most prominent structural feature that distinguishes archaeal and eukaryotic RNAPs from their bacterial counterparts2 (Fig. 1A). All known archaeal and eukaryotic RNAP, including the plant-specific RNAPIV and V have F/E-like subunits.3 How do they contribute to RNAP functionality? And what can we learn about the generic differences between, and the evolution of transcription machineries in the three domains of life?
Figure 1.
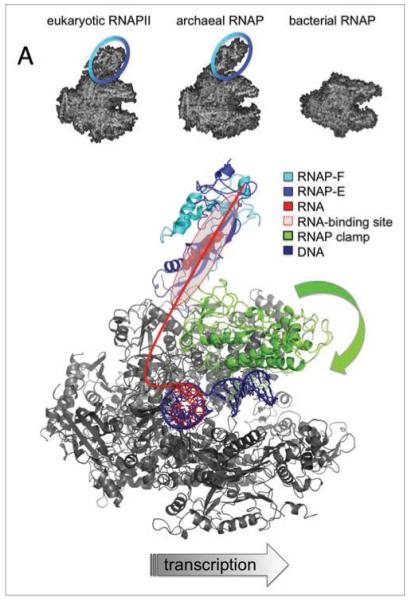
Interactions between RNAP subunits F/E and the transcript RNA modulate transcription elongation and termination. (A) shows the structures of RNAPs in Eukarya (pdb 1NT9), Archaea (pdb 2PMZ) and Bacteria (pdb 1I6V). RNAP subunits F/E form a stalk-like protrusion, which is a signature module of eukaryotic and archaeal RNAPs (highlighted with blue circle). A model of the archaeal transcription elongation complex in top view (S. shibatae RNAP pdb 2WAQ and DNA/RNA scaffold pdb 1Y1W). RNAP subunits, motifs and nucleic acids are colored according to the key in the figure. The path of the RNA transcript is sketched as a red line, and the residues in subunit E that mediate the RNA binding are highlighted as red spheres.12 The closure of the flexible RNAP clamp over the DNA binding channel is indicated with a green block arrow.
Picking the Strands: RNAP-F/E are Involved in DNA Melting during Transcription Initiation
During early transcription initiation RNAP is recruited to the promoter by the basal factors TBP and TFB and the template DNA strand is loaded into the active site. The archaeal RNAP subunits F/E facilitate DNA melting and are required for the function of a third basal factor, TFE4,5 that increases the stability of the complex and assists DNA strand separation. Likewise RPB4/7, the yeast homologues of archaeal RNAP subunits F/E, play an important role during transcription initiation of S. cerevisiae RNAPII.6 The molecular mechanisms of F/E during ‘open complex’ formation are unclear, but structural data have suggested that F/E modulates the position of the RNAP clamp, which in turn leads to DNA strand separation.7 The tip of RPB7 forms a wedge that is inserted at the base of the RNAP clamp and thereby might close it over the main DNA binding channel7 (Fig. 1). In the yeast system it has been argued that RPB4/7 dynamically associates with RNAP during the transcription cycle.8 This hypothesis is mostly based on indirect evidence: RNAPII purified from exponentially growing yeast is subsaturated with respect to RPB4/7, whereas RNAPII isolated from stationary phase cells contain RPB4/7 in stoichiometric amounts.9,10 In order to directly characterise the association of F/E with RNAP we monitored complex formation using fluorescence anisotropy. Our results demonstrated that F/E is stably incorporated into RNAP and not in a dynamic equilibrium with F/E complexes in solution.11 This result implies that F/E is present in the elongating form of RNAP, which prompted us to investigate the functional impact of F/E during the elongation phase of transcription.12
Stability of Elongation Complexes Facilitates Processivity
Transcription elongation complexes have a remarkable processivity; RNAPII molecules routinely transcribe several million basepairs (e.g., the 2.4 Mb dystrophin gene) during which they remain associated with the template DNA for as long as 16 hours.13 This stability is crucial for the elongation process since RNAP cannot re-elongate a partial RNA transcript after the enzyme has disengaged from the template, and consequently RNAP has to initiate transcription from the promoter de novo. Despite the requirement of forming stable elongation complexes, RNAP cannot bind too tightly to its template or its product, since this would impede efficient transcription elongation. Therefore transcribing RNAPs have to interact dynamically with the nucleic acid scaffold of the elongation complex. Crystal structures of elongating RNAPs (e.g., pdb codes 1I6H, 1Y1W) have resolved the downstream duplex DNA template and a 9-basepair (bp) DNA-RNA hybrid but have so far failed to resolve the upstream duplex DNA or the nascent RNA.14,15 An elegant study by the Michaelis group probed the location of the upstream DNA by using a Förster resonance energy transfer system coined the nanopositioning system (NPS).16 However, the same approach failed to determine the path of the nascent RNA transcript beyond 23 nt.17 The nascent RNA emerges from the main body of the RNAP through the RNA exit channel and is directed towards the F/E subunits (Fig. 1B). Indeed, both F/E and RPB4/7 bind RNA in vitro and in vivo.18-20 This interaction is apparently sequence independent, of intermediate affinity (F/E-RNA Kd = 0.34 ± 0.06 μM),21 and covers approximately 15 nt of RNA between position +26 to +41 relative to the active site.20 In order to test whether the F/E-RNA interaction had an impact on transcription elongation we carried out transcription assays using a recombinant archaeal RNAP system, which allowed us to assemble RNAP variants lacking the F/E subunits.5,11,12,22,23 Since we wanted to investigate the effect of F/E on elongation independently of events relating to transcription initiation (see above), we carried out promoter-independent transcription elongation assays with synthetic nucleic acid scaffolds consisting of template DNA and a 14-nucleotide (nt) RNA primer.12 Our experiments demonstrated that RNAPs lacking F/E suffer a severe elongation failure in the absence of the nontemplate strand (NTS), and display a marked reduction in processivity compared to wild type RNAPs when using the more biological relevant scaffold containing the NTS.12 In order to test whether the RNA-binding activity of F/E was vital for its stimulatory activity we produced F/E mutants that were defective in RNA-binding, and assembled RNAPs carrying these mutations. Our data showed a correlation between elongation activity and RNA-binding, which strongly suggest that interactions between F/E and the transcript increase processivity.12 Interestingly, the most severe RNA-binding deficient mutant variant of F/E was still able to stimulate elongation at low levels. This effect could be due to a residual RNA-binding by F/E in the context of the elongation complex, which could not be detected in electrophoretic mobility shift assays (EMSAs) with the isolated F/E complex. Alternatively F/E could stimulate elongation allosterically by inducing a conformational change in RNAP, such as the closure of the RNAP clamp.7 An inward movement of the RNAP clamp would narrow the main DNA binding channel, stabilise the elongation complex and increase processivity (see above and Fig. 1B).
Pause and Terminate—Allostery at the Core of Transcription
Transcription termination is one of the least understood mechanisms of RNAP. In the archaeal system transcription of short poly-U stretches (5–8 U-residues) leads to termination in vitro and in vivo, independently of secondary structures or exogenous transcription factors.24-26 The transcription of a poly-U stretch serves as a universal pause signal for all RNAPs. In the bacterial RNAP additional events are required for termination, either a stable RNA hairpin (intrinsic terminator) or the action of the rho termination factor triggers conformational changes in the RNAP active site (trigger and bridge helices, Fig. 2A) and the RNAP clamp, which result in the dissociation of the elongation complex and termination.27,28 Based on this model and two of our working hypotheses that involve F/E-induced allostery during initiation and elongation (see above), we investigated whether F/E had any influence on transcription termination.12 Our results showed that F/E is not strictly required for termination, however, the termination efficiency at weak termination signals (comprised of 5 rather than 8 U-residues) is significantly increased by F/E in a manner that is largely dependent on F/E-RNA interactions.12
Figure 2.
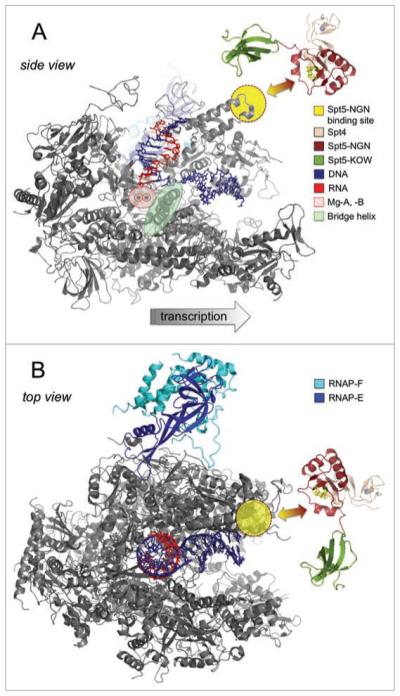
Stimulation of transcription elongation through the RNAP clamp coiled coil by Spt4/5. (A) shows a side view of the RNAPII elongation complex (pdb 1Y1W) with the RPB2 subunit removed for clarity, and a model of archaeal Spt4/5.31 The Spt5 NGN binding site and the active centre of RNAP are highlighted according to the key in the figure. The hydrophobic residues (A4, Y42 and L44) in the Spt5 NGN domain that interact with the RNAP coiled coil are highlighted as yellow sticks. (B) shows a top view of the elongation complex including all RNAP subunits and Spt4/5 in an alternative orientation to (A). The spatial arrangement of Spt4/5 relative to the RNAP is unknown. The Spt5 KOW domain (highlighted in pea green) could project towards RNAP subunits F/E (A) or across the major DNA binding channel (B).
How does F/E enhance transcription termination? In the archaeal system the pausing induced by transcription of a poly-U tract appears to be sufficient to trigger termination, since neither RNA hairpins nor exogenous factors are required. The efficiency of termination correlates with the duration of the pause,29 which in turn is likely to be affected by the stability of the elongation complex—that could be increased by F/E-RNA interactions. In addition F/E could directly affect the conformational changes within RNAP that lead to the dissociation of the elongation complex, possibly via the RNAP clamp or the trigger/bridge helices (Fig. 2A). The role and distribution of termination signals at the 3′ end of archaeal transcription units, and their relative strength in vivo (mainly determined by the number of U residues) is far from clear.23,24 The effect of F/E on termination is likely to have an even greater impact in vivo than in vitro, since efficient and accurate transcription termination is of crucial importance in Archaea that typically have small genomes, very short intergenic regions and transcription-translation are coupled.24,25,30
Converging on the RNAP Clamp—Regulation of Transcription Elongation by Spt4/5
The transcription elongation properties of RNAPs are influenced by their subunit composition (i.e., ±F/E) and regulated by exogenous transcription factors including Spt5, the only known RNAP-associated transcription factor that is universally conserved in evolution. We have recently solved the X-ray structure of M. jannaschii Spt4/5 and characterized its function.31 Our results demonstrate an astonishing degree of conservation in terms of Spt4/5 structure, its interaction with RNAP and its stimulatory properties on transcription elongation.31 Archaeal Spt5 is, like its bacterial homologue NusG, comprised of two domains, the NGN domain (NusG N-terminal domain) and a KOW domain (Kyrpidis, Ouzounis and Woese domain). We carried out a deletion analysis, which revealed that Spt5-NGN is the effector domain of Spt5 that mediates the dimerisation with Spt4, the binding to RNAP, and is required for the elongation activity of Spt4/5. The last two features are reliant on an interaction between a deep hydrophobic cavity in the Spt5-NGN domain and the tip of the RNAP clamp coiled coil, a surface exposed structural feature that is conserved in all multisubunit RNAPs (Fig. 2). A very similar interaction between the Spt5 homologue NusG and its cognate RNAP has been proposed in the bacterial system.32,33
What are the molecular mechanisms by which Spt4/5 stimulates elongation? The Spt5-NGN binding site on RNAP is approximately 70 Å distant from the active site, which suggests an allosteric mechanism of stimulation (Fig. 2A). We find it significant that both RNAP subunits F/E and Spt4/5 are in close proximity of the RNAP clamp and that both affect transcription elongation in a similar manner: by increasing the processivity in a fashion that is not dependent on the NTS. The latter result makes it unlikely that F/E and Spt4/5 function solely by interacting with the NTS, or act by affecting downstream DNA-strand separation, or upstream DNA-strand joining. Rather we propose that Spt4/5 induces a conformational change in the RNAP clamp that is translated into the RNAP interior and results in increased translocation efficiency. This mechanism is reminiscent of NusG and its paralogue RfaH, which have been proposed to stimulate elongation by stabilizing the forward translocated state of the RNAP active site.32 Alternatively, Spt4/5 could bridge the gap over the main DNA-binding channel of RNAP and ‘lock’ the RNAP clamp in a closed position, which would result in an increased elongation complex stability and enhanced processivity (Fig. 2B).
Evolutionary Implications
The F/E [RPB4/7] complex is the most prominent structural feature that distinguishes archaeal and eukaryotic from bacterial RNAPs. What can be learned from their function about the generic differences between the molecular mechanisms and evolution of transcription systems in the three domains of life? The proteins that are required for transcription initiation in archaea/eukaryotes (TBP/TF[II] B/TF[II]E) and bacteria (sigma factors) are not homologous, despite carrying out almost identical functions. F/E acts during transcription initiation in concert with the basal initiation factor TFE;5 they are likely to have coevolved since F/E [RPB4/7] and TF[II]E always occur together, and bacterial genomes encode neither. Our experiments show that F/E stimulates processivity during the elongation phase of transcription, but also that it enhances transcription termination in a sequence dependent manner—in effect F/E influences the elongation versus termination decisions of RNAP.12 The increased processivity could be crucial for the transcription of eukaryotic genes, which due to their intron-exon structure are much larger (up to Mbs) than even the longest bacterial multicistronic mRNAs (tens of kb). This prediction has been validated by a systems biology approach in yeast. The whole genome occupancy of wild type RNAPII compared to RNAPII lacking RPB4 (and possibly RPB7) showed that the latter RNAP was significantly depleted from the 3′ end of several mRNA genes.34 However, archaeal and bacterial genes are comparable in size, which suggests that the presence of F/E-like RNAP subunits does not strictly correlate with larger genes, even though F/E may have ‘allowed’ the increase in gene size in eukaryotes. The regulation of gene expression by elongation versus termination decisions of RNAPs is a ubiquitous modus operandi in Bacteria. The transcription regulation of ribosomal RNA operons as well as the genetic programmes of bacteriophage crucially depends on antitermination mechanisms.35,36 The switch from ‘early’ to ‘late’ genes in the lambda phage depends on the Q antiterminator, which reversibly associates with RNAP and results in a termination resistant form of the transcription elongation complex. In the absence of Q transcription terminates in a sequence dependent manner upstream of the ‘late’ genes, whereas in its presence RNAP reads through the terminator signals, and the downstream ‘late’ genes are expressed. There is no apparent homology between F/E and Q, but it is worth noting that both interact with their cognate RNAPs on similar positions in proximity to the RNA exit channel.37 Our results provide evidence that F/E is permanently associated with RNAP,11 which implies that F/E alters the general properties of RNAP and not in a gene-specific manner. In contrast, Spt4/5 is likely to reversibly interact with RNAP and thereby genuinely regulate the elongation phase of transcription. Recently ‘promoter proximal stalling’ has been discovered as a novel ubiquitous mode of transcription regulation of RNAPII in especially metazoans, but possibly all eukaryotes.38,39 During this process RNAPII initiates at the promoter but stalls approximately 30–40 bp downstream of the transcription start site. Phosphorylation of the RNAPII C-terminal domain (CTD) and transcription factors including Spt5 releases the paused RNAP and induces transcription. Even though the mechanisms of promoter-proximal stalling are not completely understood, the phenomenon critically depends on Spt4/5. The bacterial RNAP was also found enriched in positions offset from the promoter in the direction of transcription—reminiscent of eukaryotic promoter proximally stalled RNAPII. However, the genome distribution of RNAP and NusG did not support the notion that NusG facilitated promoter proximal stalling in bacteria.40 In addition to the domains present in bacterial NusG, the eukaryotic complex contains multiple KOW domains, two C-terminal heptad repeat (CTR) regions which phosphorylation has been implicated in promoter proximal stalling, and Spt4.31 The archaeal complex contains Spt4, but no CTRs. The whole genome distribution of RNAP and Spt4/5 in the Archaea has not been investigated yet. Our results, which demonstrate an exigent structural and functional conservation of the Spt4/5 ‘core’, raise the interesting possibility that promoter-proximal pausing might occur in the archaeal domain, a hypothesis that awaits experimental verification.
Future Directions
Our working hypotheses about the molecular mechanisms of RNAP subunits F/E and the elongation factor Spt4/5 involve conformational changes of transcription complexes. Large molecular machines such as RNAPs are dynamic and consist of rigid cores combined with flexible elements (including the RNAP clamp) that together achieve versatile ‘induced fits’ for a plethora of molecules that functionally interact with RNAP. Conformational changes of RNAP have been predicted by e.g., comparing the X-ray structures of RNAP with varying subunit composition (e.g., ±RPB4/7,7), nucleic acid components (e.g., DNA-RNA scaffolds15 or RNA aptamers41), transcription factors (e.g., TFIIB42 or TFIIS15), or small molecular weight effectors (e.g., ppGpp43). But the inherent limitation in theses predictions is the fact that X-ray structural information is static, even though normal mode analysis of B-factors provide a measure of thermal motions for each structure.44,45 Conformational dynamics of RNAPs in solution have rarely been probed directly due to the complexity of RNAPs, and the challenging nature of the techniques. However, with the advent of recombinant forms of multisubunit RNAPs such as the archaeal RNAP, and the development of biophysical approaches including the FRET-based NPS system46 we are opening up new and promising avenues of research that will address these important and hitherto unresolved questions.
Acknowledgements
We would like to thank Evgeny Nudler (NYU Medical School) for inspiring discussion about transcription termination, and Kristine Bourke Arnvig for critical reading of this manuscript. Research at the RNAP laboratory was funded by project grants from the Wellcome Trust (079351/Z/06/Z) and BBSRC (BB/E008232/1) to Finn Werner.
References
- 1.Werner F. Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 2008;16:247–50. doi: 10.1016/j.tim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Grohmann D, Hirtreiter A, Werner F. Molecular mechanisms of archaeal RNA polymerase. Biochem Soc Trans. 2009;37:12–7. doi: 10.1042/BST0370012. [DOI] [PubMed] [Google Scholar]
- 3.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naji S, Grunberg S, Thomm M. The RPB7 orthologue E’ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J Biol Chem. 2007;282:11047–57. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- 5.Werner F, Weinzierl RO. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol Cell Biol. 2005;25:8344–55. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards AM, Kane CM, Young RA, Kornberg RD. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–5. [PubMed] [Google Scholar]
- 7.Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2005;280:7131–4. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 8.Sampath V, Sadhale P. Rpb4 and Rpb7: a sub-complex integral to multi-subunit RNA polymerases performs a multitude of functions. IUBMB Life. 2005;57:93–102. doi: 10.1080/15216540500078905. [DOI] [PubMed] [Google Scholar]
- 9.Kolodziej PA, Woychik N, Liao SM, Young RA. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol Cell Biol. 1990;10:1915–20. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woychik NA, Young RA. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol. 1989;9:2854–9. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grohmann D, Hirtreiter A, Werner F. The RNAP subunits F/E (RPB4/7) are stably associated with archaeal RNA polymerase—using fluorescence anisotropy to monitor RNAP assembly in vitro. Biochem J. 2009;421:339–43. doi: 10.1042/BJ20090782. [DOI] [PubMed] [Google Scholar]
- 12.Hirtreiter A, Grohmann D, Werner F. Molecular mechanisms of RNA polymerase—the F/E (RPB4/7) complex is required for high processivity in vitro. Nucleic Acids Res. 2010;38:585–96. doi: 10.1093/nar/gkp928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–90. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 14.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–82. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 15.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–65. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 16.Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, et al. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res. 2009;37:5803–9. doi: 10.1093/nar/gkp601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrecka J, Lewis R, Bruckner F, Lehmann E, Cramer P, Michaelis J. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci USA. 2008;105:135–40. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meka H, Werner F, Cordell SC, Onesti S, Brick P. Crystal structure and RNA binding of the Rpb4/Rpb7 subunits of human RNA polymerase II. Nucleic Acids Res. 2005;33:6435–44. doi: 10.1093/nar/gki945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlicky SM, Tran PT, Sayre MH, Edwards AM. Dissociable Rpb4-Rpb7 subassembly of rna polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J Biol Chem. 2001;276:10097–102. doi: 10.1074/jbc.M003165200. [DOI] [PubMed] [Google Scholar]
- 20.Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- 21.Grohmann D, Klose D, Klare JP, Kay CM, Steinhoff HJ, Werner F. RNA-binding to archaeal RNAP subunits F/E: a DEER and FRET study. J AM Chem Soc. 2010;132:5954–5. doi: 10.1021/ja101663d. [DOI] [PubMed] [Google Scholar]
- 22.Ouhammouch M, Werner F, Weinzierl RO, Geiduschek EP. A fully recombinant system for activator-dependent archaeal transcription. J Biol Chem. 2004;279:51719–21. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- 23.Werner F, Weinzierl RO. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol Cell. 2002;10:635–46. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 24.Santangelo TJ, Cubonova L, Skinner KM, Reeve JN. Archaeal intrinsic transcription termination in vivo. J Bacteriol. 2009;191:7102–8. doi: 10.1128/JB.00982-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santangelo TJ, Reeve JN. Archaeal RNA polymerase is sensitive to intrinsic termination directed by transcribed and remote sequences. J Mol Biol. 2006;355:196–210. doi: 10.1016/j.jmb.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 26.Spitalny P, Thomm M. A polymerase III-like reinitiation mechanism is operating in regulation of histone expression in archaea. Mol Microbiol. 2008;67:958–70. doi: 10.1111/j.1365-2958.2007.06084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–9. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–6. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 30.Santangelo TJ, Cubonova L, Matsumi R, Atomi H, Imanaka T, Reeve JN. Polarity in archaeal operon transcription in Thermococcus kodakaraensis. J Bacteriol. 2008;190:2244–8. doi: 10.1128/JB.01811-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirtreiter A, Damsma F, Cheung A, Klose D, Grohmann D, Vojnic, et al. Spt4/5 Stimulates Transcription Elongation Through The RNA Polymerase Clamp Coiled Coil Motif. NAR. 2010 doi: 10.1093/nar/gkq135. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–29. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mooney RA, Schweimer K, Roesch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Runner VM, Podolny V, Buratowski S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol Cell Biol. 2008;28:1883–91. doi: 10.1128/MCB.01714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkin TM. Control of transcription termination in prokaryotes. Annu Rev Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 36.Torres M, Balada JM, Zellars M, Squires C, Squires CL. In vivo effect of NusB and NusG on rRNA transcription antitermination. J Bacteriol. 2004;186:1304–10. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage lambda Q antiterminator protein contacts the beta-flap domain of RNA polymerase. Proc Natl Acad Sci USA. 2008;105:15305–10. doi: 10.1073/pnas.0805757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–2. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–4. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 40.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–8. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 42.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–30. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 43.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 44.Korkhin Y, Unligil UM, Littlefield O, Nelson PJ, Stuart DI, Sigler PB, et al. Evolution of complex RNA polymerases: The complete Archaeal RNA polymerase structure. PLoS Biol. 2009;7:102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–6. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 46.Muschielok A, Andrecka J, Jawhari A, Bruckner F, Cramer P, Michaelis J. A nano-positioning system for macromolecular structural analysis. Nat Methods. 2008;5:965–71. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]


