Abstract
BACKGROUND
Recurrent sinopulmonary and cutaneous viral infections with elevated serum levels of IgE are features of some variants of combined immunodeficiency. The genetic causes of these variants are unknown.
METHODS
We collected longitudinal clinical data on 11 patients from eight families who had recurrent sinopulmonary and cutaneous viral infections. We performed comparative genomic hybridization arrays and targeted gene sequencing. Variants with predicted loss-of-expression mutations were confirmed by means of a quantitative reverse-transcriptase –polymerase-chain-reaction assay and immunoblotting. We evaluated the number and function of lymphocytes with the use of in vitro assays and flow cytometry.
RESULTS
Patients had recurrent otitis media, sinusitis, and pneumonias; recurrent Staphylococcus aureus skin infections with otitis externa; recurrent, severe herpes simplex virus or herpes zoster infections; extensive and persistent infections with molluscum contagiosum; and human papillomavirus infections. Most patients had severe atopy with anaphylaxis; several had squamous-cell carcinomas, and one had T-cell lymphoma –leukemia. Elevated serum IgE levels, hypereosinophilia, low numbers of T cells and B cells, low serum IgM levels, and variable IgG antibody responses were common. Expansion in vitro of activated CD8 T cells was impaired. Novel homozygous or compound heterozygous deletions and point mutations in the gene encoding the dedicator of cytokinesis 8 protein (DOCK8) led to the absence of DOCK8 protein in lymphocytes.
CONCLUSIONS
Autosomal recessive DOCK8 deficiency is associated with a novel variant of combined immunodeficiency.
Inherited susceptibility to viral infections, especially by herpesviruses, occurs in patients who have developmental defects of T cells, such as severe combined immunodeficiency; disorders involving cell killing by cytotoxic lymphocytes, such as hemophagocytic lymphohistiocytosis and X-linked lymphoproliferative disease; and impaired type I interferon production.1,2 Several other conditions confer susceptibility to human papillomavirus infections or molluscum contagiosum.3–6
The hallmarks of severe combined immunodeficiency are impaired T-cell function and a severe deficiency of T cells. In some forms of the disease, there are also low numbers of or dysfunctional B cells and natural killer cells. The molecular defects in severe combined immunodeficiency are diverse,1,7–13 and forms of the disease that are caused by residual gene activity or partial T-cell immunodeficiencies (e.g., the Wiskott–Aldrich syndrome) can feature skin diseases with elevated levels of IgE and hypereosinophilia.14,15
We describe 11 patients who had a combined immunodeficiency with low numbers of T, B, and natural killer cells, extensive cutaneous viral infections, and susceptibility to cancer. These patients had loss-of-function mutations in an uncharacterized gene, dedicator of cytokinesis 8 (DOCK8), which is expressed in lymphocytes.
METHODS
CELLS
We isolated subgroups of T cells from peripheral-blood mononuclear cells by negative selection, using magnetic bead isolation and then stimulation with anti-CD3 plus anti-CD28 antibodies (1 μg per milliliter) and cultured in interleukin-2 (for details, see the Supplementary Appendix, available with the full text of this article at NEJM.org). B cells were immortalized with the Epstein–Barr virus, and T cells with herpesvirus saimiri (HVS), according to standard protocols.
COMPARATIVE GENOMIC HYBRIDIZATION ANALYSES
Comparative genomic hybridization analyses were performed with 244K arrays with the use of the Agilent platform, according to the manufacturer’s instructions. Modifications of the process are described in the Supplementary Appendix.
DNA SEQUENCING
Sequencing of genomic DNA was performed after polymerase-chain-reaction (PCR) amplification of exons with their flanking intronic or untranslated regions. (Primer sequences are listed in Table 1 in the Supplementary Appendix.) Point mutations were confirmed in a second PCR amplification reaction, and frameshift mutations were confirmed after cloning. Novel variants that were identified in the 11 index patients were sought in 6 patients with autosomal dominant hyperimmunoglobulin E syndrome (HIES), in 32 patients with other immunologic diseases, in 15 healthy blood donors of various ethnic backgrounds, and in 100 healthy white control subjects.
IMMUNOBLOTTING
Cytosolic lysates were prepared according to standard methods, followed by separation on 3 to 8% TRIS-acetate gels (Invitrogen) and transfer onto nitrocellulose. Proteins were detected with the use of polyclonal rabbit anti-DOCK8 antibodies (Sigma-Aldrich or donated by Dr. Ruusala, Ludwig Cancer Institute, Uppsala, Sweden16) or mouse anti–β-actin antibodies (Sigma-Aldrich) on the same blot. (For details, see the Methods section in the Supplementary Appendix.)
FLOW CYTOMETRY
Standard flow-cytometric methods were used for staining of cell-surface markers and carboxyfluorescein succinimidyl ester (CFSE) dilution assays. Cells were harvested for CFSE analysis after 3 days of culture under conditions of stimulation or no stimulation with anti-CD3 plus anti-CD28 antibodies. (See the Supplementary Appendix for additional details.)
RESULTS
CLINICAL FINDINGS
The patients and their relatives were enrolled in research protocols that were approved by the institutional review board of the National Institute of Allergy and Infectious Diseases. All study subjects provided written informed consent. The clinical features of Patient 1 in Family 3, Patients 1 and 2 in Family 4, and Patients 1 and 2 in Family 8 have been described previously.5,17,18 We initially studied three patients (the index patients from Families 1, 2, and 3) from a group of patients with undefined combined immunodeficiencies. After finding that the three carried a mutation in the same gene (DOCK8), we recognized that their disorders shared certain clinical features with atypical forms of HIES. We therefore sought out additional patients and included in our study several patients who were described previously, for a total of 11 patients between the ages of 6 and 21 years from eight families5,18 (Table 2 in the Supplementary Appendix). Both male and female patients of various ethnic backgrounds were affected. Two patients presented with rash at birth, and all had atopic dermatitis (Fig. 1A). Of the 11 patients, 9 had severe and extensive food or environmental allergies, including anaphylaxis, and 6 had asthma or reactive airway disease. Eosinophilic esophagitis or lung disease was observed in two patients.
Figure 1. Characteristic Dermatologic Findings.
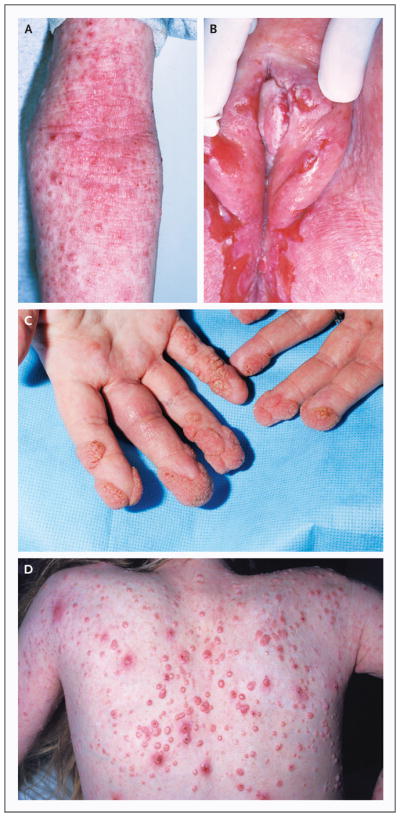
Among the characteristic dermatologic findings in the patients in the study were atopic dermatitis with scattered superimposed molluscum contagiosum lesions on the inside of the arm (Panel A), chronic ulcerative anogenital herpes simplex virus infection (Panel B), periungual and acral warts (Panel C), and molluscum contagiosum on the back (Panel D).
All the patients had recurrent upper and lower respiratory tract infections. Most of the patients had otitis media that had required placement of a tympanostomy tube, two patients had had mastoiditis, and seven patients had recurrent sinusitis. Nine patients had recurrent pneumonias or bronchitis, and bronchiectasis had been diagnosed in two patients. Recovered pulmonary pathogens included Streptococcus pneumoniae, Haemophilus influenzae, Pneumocystis jiroveci, respiratory adenovirus, and respiratory syncytial virus. One patient had recurrent croup.
All the patients had extensive, frequently coexisting, cutaneous viral infections. Superficial, often ulcerating, herpes simplex virus infection occurred in seven patients, manifested as recurrent or persistent orolabial or anogenital involvement, keratitis, or eczema herpeticum (Fig. 1B). Seven patients had persistent flat and verrucous warts (Fig. 1C); five had extensive, disfiguring molluscum contagiosum infections (Fig. 1D); two had recurrent herpes zoster; and one had severe primary varicella–zoster infection.
Eight patients had Staphylococcus aureus skin infections or abscesses, two had S. aureus osteomyelitis, five had mucosal or nail candidiasis, and six had recurrent otitis externa. One patient had had cryptococcal and H. influenzae meningitis. Other infections included several occurrences of salmonella enteritis, giardiasis, and pericarditis.
Except for Patient 1 in Family 8, the 11 patients we studied did not have the neurologic, vasculitic, or autoimmune findings reported in patients with autosomal recessive HIES.5 Nonimmunologic features that are typically seen in autosomal dominant HIES were uncommon.19 Three patients had poor growth, and one had delayed puberty.
Vulvar, facial, and anal squamous-cell dysplasia and carcinomas had occurred in patients with long-standing herpes simplex virus infections, human papillomavirus infections, and molluscum contagiosum. Cancers had developed in three patients during late childhood or early adulthood. Two patients died from metastatic squamous-cell carcinoma. One patient with resected microcystic adenoma died from cutaneous T-cell lymphoma–leukemia.17
IMMUNOLOGIC ASSESSMENT
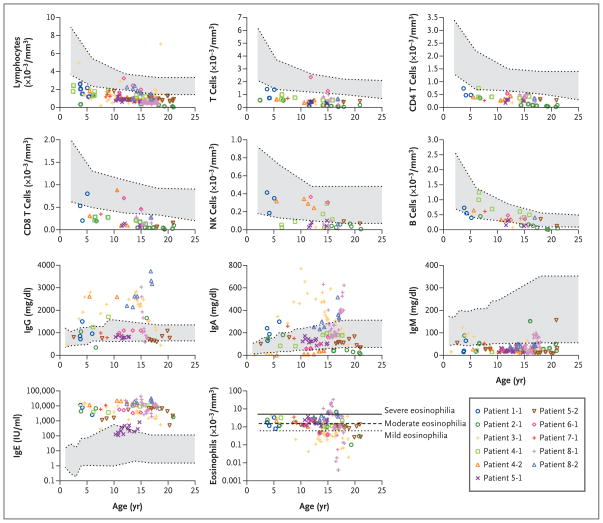
Studies of peripheral-blood mononuclear cells revealed low absolute lymphocyte counts in 9 of the 11 patients, including low counts for total T cells (in 10 of 11 patients), CD4 T cells (in all 11 patients), and CD8 T cells (in 10 of 11 patients) (Fig. 2).20–22 Ratios of T4 cells to T8 cells were within normal ranges. The numbers of regulatory T cells, as assessed by CD4+CD25hiFOXP3+ coexpression, in two patients were decreased because of overall lymphopenia but were proportionally normal (Fig. 1 in the Supplementary Appendix). The number of natural killer cells was decreased in 6 of 10 patients, and the number of B cells was decreased in 5 of 11 patients. Although one patient had a normal number of eosinophils, most patients had mild-to-moderate eosinophilia, with a mean (±SD) of 2.021±1.292×103 cells per cubic millimeter (normal count, <0.600×103). Patient 1 in Family 8 had an eosinophil count as high as 33×103 per cubic millimeter. The numbers of neutrophils and monocytes were normal in all patients (data not shown).
Figure 2. Immunologic Assessment.
Values for blood analyses are shown according to age in the index patients in Families 1, 2, and 3; Patients 1 and 2 in Family 4; Patients 1 and 2 in Family 5; Patient 1 in Family 6; Patient 1 in Family 7; and Patients 1 and 2 in Family 8. In the color key, patients are listed according to their family number, followed by their patient number (Patient 1-1, Patient 2-1, etc.). Normal ranges for age are shown in gray. For lymphocyte subgroups, the normal ranges represent the 10th to 90th percentiles based on published data.20 For leukocyte subgroups and serum immunoglobulin, the normal ranges are shown with 95% confidence intervals.21 For eosinophils, the lines represent arbitrary thresholds for mild, moderate, or severe eosinophilia.22
Despite a decreased number of B cells in some patients, six patients had hypergammaglobulinemia, and five had normal levels of serum IgG. Levels of serum IgA varied, but levels of IgM were consistently low in all patients, with a mean of 35±13 mg per deciliter (normal value, >49). Except for Patient 2 in Family 5, who had high-normal IgE levels (up to 818 IU per milliliter), the patients had very high IgE levels (peak range, 5630 to 43,600 IU per milliliter) (Fig. 2).
Levels of antibodies against a panel of bacterial and viral antigens were variable (Table 3 in the Supplementary Appendix). All patients who were evaluated had protective levels of antibodies to rubella (8 of 8 patients) and varicella–zoster virus (4 of 4 patients). Some patients had a response to vaccines against H. influenzae type B (4 of 9 patients), diphtheria toxoid (5 of 10 patients), and tetanus toxoid (3 of 11 patients). Patients 1 and 2 in Family 4 had impaired T-cell–dependent primary antibody responses after immunization with the neoantigen bacteriophage φX174, with peak rates of bacteriophage inactivation in serum of 0.8 and 0.4 per minute, respectively, at 7 days (normal range at 14 days, 7 to 500). These two siblings also did not have an IgG antibody response after a second challenge with the bacteriophage antigen, with peak rates of inactivation of 0.7 and 0.3 per minute, respectively, on the day of the second challenge with no IgG. (The normal rate of inactivation in serum peaks at 7 days after a second challenge, with a range of 700 to 2000 per minute and with a mean IgG representing 56±24% of the neutralizing immunoglobulins.) Responses to pneumococcal polysaccharide antigens were variable (Table 3 in the Supplementary Appendix). Six of the patients were treated with long-term replacement immune globulin, which seemed to improve the incidence and severity of sinopulmonary infections in three patients without influencing the course of viral infections in any patient.
GENETIC ANALYSES
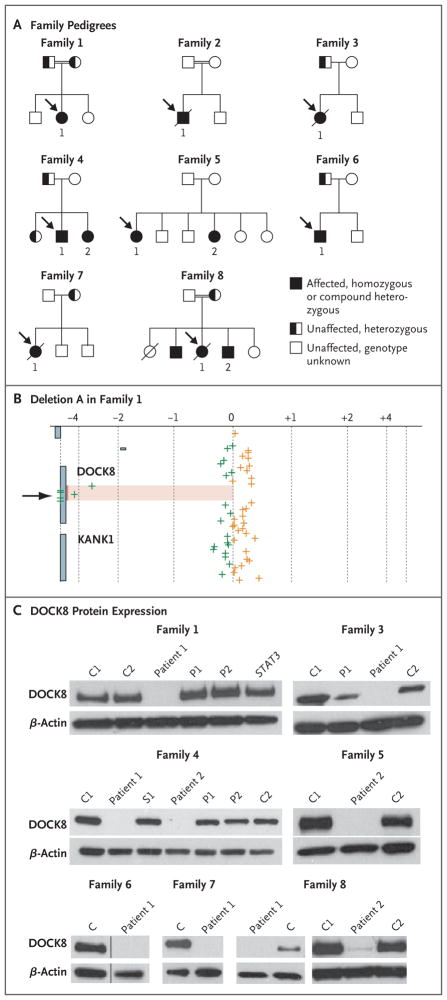
Figure 3A shows the pedigrees of the eight families. Families 1, 2, and 8 were consanguineous. Comparative genomic hybridization arrays of genomic DNA from the index patients in Families 1 and 2 revealed homozygous deletions in the DOCK8 gene (Fig. 3B, and Fig. 2A in the Supplementary Appendix). Deletion A in Family 1 spanned exons 10 through 23, and deletion B in Family 2 spanned exons 5 through 24 (Fig. 3B, and Fig. 2A and 2C in the Supplementary Appendix). Deletions A and B were confirmed by the failure of PCR to amplify deleted exons; furthermore, the juxtaposition of normally distant exons resulted in PCR amplification across the deleted region, allowing precise identification of the sequence breakpoint (Fig. 3 and 4 in the Supplementary Appendix). The deletion-spanning PCR products were not amplified from the 38 patients with other immune disorders, including the 6 patients with autosomal dominant HIES, or from the 115 healthy control subjects.
Figure 3. Patient Pedigrees and DOCK8 Molecular Analyses.
Panel A shows the pedigrees of the eight families in the study. Probands are designated by arrows. Circles represent female family members, and squares male family members. A slash through a symbol represents a deceased person. Panel B shows log2 ratios of DNA from Patient 1 of Family 1 with deletion A (spanning exons 10 through 23) to reference DNA (on the x axis) after hybridization to probes at or near the DOCK8 locus, according to the genomic position (on the y axis). The arrow pointing to the shaded region indicates the homozygous DOCK8 deletion. The green crosses indicate probes where the signal of the log2 ratio was less than zero, and the orange crosses more than zero. Panel C shows immunoblots for DOCK8 protein expression in seven families that were included in the study. Lysates are from primary T cells (for all members of Family 1 and Patient 2 of Family 8), Epstein–Barr virus–transformed B-cell lines (for Families 3, 4, 6, and 7 and Patient 1 of Family 8), and herpesvirus saimiri–transformed T-cell lines (for Family 5). The data for Family 6 show two lanes (samples from a control [C] and from Patient 1) that are aligned from the same gel. Immunoblots for Patient 1 in Family 2 and for Patient 1 in Family 5, which were analyzed after the completion of the study, are shown in Figure 7A in the Supplementary Appendix. No DOCK8 protein was found in samples from any of the 11 patients who were tested. Included in the samples for Family 1 is an unrelated STAT3-mutant control with autosomal dominant hyperimmunoglobulin E syndrome. β-actin is included as a loading control in all the analyses. P denotes parent, and S a heterozygous member of Family 4.
Heterozygous deletions in the DOCK8 gene were found in patients from Families 3, 4, 5, and 6: deletions C, D, E, and F, respectively (Fig. 2A and 2C in the Supplementary Appendix). As expected, the heterozygous deletions resulted in apparent homozygosity of single-nucleotide polymorphisms (SNPs) within the corresponding sequenced regions (data not shown). Deletions A through F were novel structural variants that were not found in the Database of Genomic Variants, and no homozygous losses in this area of copy-number variation have been reported previously.
Patients with heterozygous deletions in one allele of DOCK8 had compound heterozygosity for deleterious mutations (Fig. 2B and 2C in the Supplementary Appendix). None of these mutations were reported in SNP databases, and they were not identified in the 38 patients with other immune disorders, including the 6 patients with autosomal dominant HIES, or in the 115 control subjects.
Overall, the genetic variants consisted of large missing portions of the DOCK8 coding sequence, including a conserved DOCK homology region 1 (DHR1) domain, or had other expected harmful effects in the gene (Fig. 2C in the Supplementary Appendix). DOCK8 messenger RNA (mRNA) is present in tissues from lung, kidney, pancreas, and placenta, but it is unknown whether hematopoietic cells express DOCK8.16 We found that monocytes, B cells, and T cells from healthy blood donors contain DOCK8 mRNA, as assessed by quantitative reverse-transcriptase–PCR (qRTPCR) assay; high levels were found in activated, expanded primary T-cell cultures and transformed lymphocyte lines (Fig. 6A in the Supplementary Appendix). Immunoblotting showed DOCK8 protein in lymphocytes from an unrelated patient with autosomal dominant HIES (Fig. 3C). By contrast, DOCK8 protein was not detected in primary T-cell cultures or transformed lymphocyte lines from all 11 patients who were tested (Fig. 3C, and Fig. 7A in the Supplementary Appendix). Residual levels of full-length protein in lymphocytes from Patient 2 in Family 8 were detected on overexposure of the immunoblot. Lymphocytes from Patient 1 in Family 1 contained minimal levels of a truncated protein that had been generated from the remaining exons (Fig. 7B in the Supplementary Appendix). These results are consistent with the finding of minimal amounts of transcripts or none in the patients’ lymphocytes (Fig. 6B in the Supplementary Appendix). In the case of premature stop codons, this finding is most likely due to nonsense-mediated decay.23
FUNCTION OF CD8 T CELLS
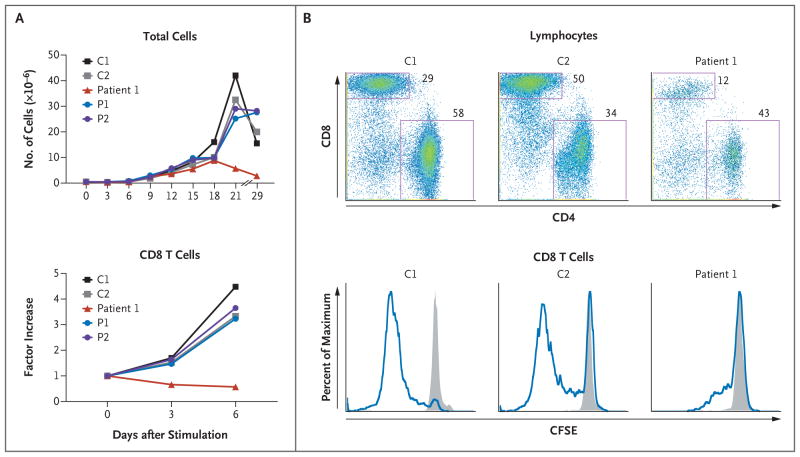
CD8 T cells did not expand well from activated DOCK8-deficient peripheral-blood mononuclear cells from Patient 1 in Family 1 (Fig. 4A) or from three additional patients (Fig. 8A in the Supplementary Appendix). This result was not due to a low starting number of CD8 T cells, since the number of cells did not increase when they were normalized to the starting number. Moreover, CFSE dilution, a marker of cell division, was impaired in DOCK8-deficient CD8 T cells after stimulation of T-cell receptors with anti-CD3 plus anti-CD28 antibodies (Fig. 4B, and Fig. 8C in the Supplementary Appendix). By contrast, we observed no defect in the proliferation of CD4 T cells (Fig. 8B, 8C, and 8D in the Supplementary Appendix).
Figure 4. Impaired CD8 T-Cell Activation and Proliferation in Association with DOCK8 Deficiency in Family 1.
Panels A and B show the results of two independent experiments in Family 1. In Panel A, after activation of peripheral-blood mononuclear cells (PBMCs) with anti-CD3 plus anti-CD28 antibodies for 3 days, T cells were expanded in culture with interleukin-2. Absolute numbers of CD8 T cells were calculated on the basis of flow cytometric determinations of their percentages; CD8 T-cell numbers over time are shown as factor increases normalized to the prestimulated cell population. In Panel B, PBMCs that were labeled with carboxyfluorescein succinimidyl ester (CFSE) were either unstimulated or stimulated, as described for Panel A. After 3 days, cells were stained with the use of fluorescence-labeled antibodies against T-cell subgroup markers and then analyzed on flow cytometry. The top row shows profiles for stimulated cells, with the location of each dot reflecting an individual cell’s intensity for CD4 staining (on the x axis) versus CD8 staining (on the y axis). The boxes delineate the CD8 and CD4 T-cell subgroups. As compared with control samples, samples from Patient 1 had a lower proportion of CD8 T cells (12% of lymphocytes) after activation. Gated CD8 T cells were further analyzed for the fluorescence intensity of CFSE, which is successively diluted with each cell division. The bottom row shows histograms with the height (y axis) reflecting a percent of maximum of gated CD8 T cells that display any given fluorescence intensity of CFSE (on the x axis). The gray shading indicates unstimulated cells, and the blue line indicates stimulation. C denotes healthy control subject, and P parent.
The induction of cell-surface activation marker CD25 on CD8 T cells was impaired in two of three patients who were tested (Fig. 9A in the Supplementary Appendix). The expression of the antiviral cytokines interferon-γ and tumor necrosis factor α, as assessed by intracellular flow cytometry, was moderately decreased in CD8 T cells in two patients who were tested (Fig. 9B in the Supplementary Appendix). Measurements of cytotoxicity in CD8 T cells on a per-cell basis in four patients were similar to those in control subjects. Both intracellular perforin and degranulation, as assessed by cell-surface extravasation of lysosomal-associated membrane protein 1 (LAMP1, also called CD107a), were normal (Fig. 9C and 9D in the Supplementary Appendix).
DISCUSSION
DOCK8 is a member of the DOCK180 superfamily of guanine nucleotide exchange factors, which interact with Rho GTPases.16 The function of DOCK8 is unknown, but by analogy to related molecules, the protein probably regulates cytoskeletal rearrangements that are required for cellular structure, migration, adhesion, and other functions.24 We found germ-line loss-of-function DOCK8 mutations in patients with a variant of combined immunodeficiency characterized by unusual susceptibility to cutaneous viral infections and cancers. In addition, DOCK8 deficiency was found in a subgroup of patients who were previously thought to have autosomal recessive HIES with severe allergic manifestations. Allergy is not characteristic of other forms of primary immunodeficiency diseases associated with elevated IgE levels, including autosomal dominant HIES.15
The immunologic phenotype in patients with DOCK8 deficiency resembles certain features of mice that have been rendered genetically deficient in a related gene, dock2, by homologous recombination. Such mice have a decreased number of T cells, reduced responsiveness of the T-cell antigen receptor, and allergic disease with high levels of serum IgE.25–27 However, unlike patients with DOCK8 deficiency, dock2-deficient mice have no reported susceptibility to experimental viral infections. Although abnormal neutrophil chemotaxis was found in two of four patients who were tested (data not shown), it has not yet been determined whether the patients had the types of impaired lymphocyte chemotaxis and abnormal lymphoid organ architecture that have been seen in mice.25,28 We evaluated the patients with immunodeficiency for a deficiency of the human orthologue of dock2 but found no evidence of such a deficiency (data not shown). We cannot rule out cooperative effects, but our findings suggest that DOCK2 and DOCK8 do not serve redundant functions in humans. Moreover, the independent discovery of dock8 mutations as a cause of immunodeficiency in ethylnitrosourea-mutagenized mice complements our findings (Cornall R, Goodnow C: personal communication).
HIES consists of primary immunodeficiencies that are characterized by severe eczema, recurrent skin infections (often from S. aureus), mucocutaneous candidiasis, recurrent sinopulmonary infections, elevated serum IgE levels, and eosinophilia. 6 Autosomal dominant HIES results from dominant interfering mutations in the gene encoding transcription factor STAT3. Such mutations lead to failed differentiation of a specialized subgroup of helper T cells that produce interleukin-17 for defense against fungal and extracellular bacterial infections.18,29–32 By contrast, the autosomal recessive form features recurrent cutaneous viral infections but not pneumatocele formation or connective-tissue and skeletal abnormalities. 5 A homozygous loss-of-function mutation in the tyrosine kinase 2 gene, which encodes the TYK2 transcription factor, resulted in impaired cytokine signaling in multiple pathways in a single patient with elevated IgE levels but not in other patients.33,34 Patients with these forms of HIES characteristically have a normal number of lymphocytes.5,6,33
The DOCK8 immunodeficiency syndrome accounts for subgroups of patients who have undefined combined immunodeficiencies or who were previously misclassified as having autosomal recessive HIES. Besides lymphopenia, DOCK8 deficiency has manifestations that have not previously been associated with HIES, including cancers related to cutaneous viral infections. Squamous-cell cancers occurred in three of our patients who had extensive human papillomavirus infections, and one patient also had cutaneous T-cell lymphoma–leukemia. Impaired CD8 T cells in DOCK8 deficiency suggests that these cancers may be due to impaired tumor surveillance. It is also possible that DOCK8 has tumor-suppressor functions. Indeed, DOCK8 deletions in primary lung cancers, gastric- and breast-cancer lines, and gliomas suggest a role of DOCK8 in tumor suppression.35,36 Heterozygous DOCK8 deletions have been reported in several cases of mental retardation, developmental delay, and autisticspectrum disorder.37,38 However, most of the DOCK8-deficient patients and their heterozygous relatives in our study had normal intelligence, suggesting that DOCK8 is not essential for cognitive function.
Other heterozygous deletions and duplications, reported in the Database of Genomic Variants, are consistent with the copy-number variation of DOCK8 within the normal population. Our findings support the increasing recognition that these and other structural variants, which have been estimated to account for a least 5% of the human genome and are enriched in genes of the immune system, represent sources of variation that can contribute to mendelian human diseases and influence immune function with respect to susceptibility to infections.39
Supplementary Material
Acknowledgments
Supported by the Intramural Research Program of the National Institutes of Health, the National Institute of Allergy and Infectious Diseases, and the National Cancer Institute, and by a grant from the Howard Hughes Medical Institute–National Institutes of Health Research Scholar Program (to Mr. Davis). Sequencing was performed by the Genomics Unit of the Rocky Mountain Laboratories Research Technologies Section of the National Institute of Allergy and Infectious Diseases.
We thank Drs. John O’Shea, Pamela Schwartzberg, Andrew Snow, Harry Malech, and Michael Lenardo for helpful discussions or critical reading of the manuscript; Dr. Steve Porcella and Kent Barbian of the Rocky Mountain Laboratories Genomics Unit for sequencing support; Dr. Jennifer Johnston and Amy Hsu-Lin for technical advice; Christopher Dove and Timothy Lenardo for experimental assistance; Drs. Aino Ruusala and Pontus Aspenström for reagents; Christine Spalding for clinical support; Drs. Prescott Atkinson, Michael Cooperstock, Carolyn Fein-Levy, Robin La-Croix, Joe LaRussa, Harold Lischner, and Swayam Sadanandan for patient referrals; Drs. Jennifer Puck and Bodo Grimbacher for their past contributions in assembling several of the case reports of HIES; Andy Johnson and Drs. Michael Lenardo, Richard Cornall, and Christopher Goodnow for sharing information about their independent and complementary findings with respect to the consequences of dock8 mutations in mice; and the patients and their families for participating in the study.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Fischer A. Human primary immunodeficiency diseases. Immunity. 2007;27:835–45. doi: 10.1016/j.immuni.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante J, Zhang SY, von Bernuth H, Abel L, Casanova JL. From infectious diseases to primary immunodeficiencies. Immunol Allergy Clin North Am. 2008;28:235–58. doi: 10.1016/j.iac.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Malech HL. WHIM syndrome: congenital immune deficiency disease. Curr Opin Hematol. 2009;16:20–6. doi: 10.1097/MOH.0b013e32831ac557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prawer SE, Pass F, Vance JC, Greenberg LJ, Yunis EJ, Zelickson AS. Depressed immune function in epidermodysplasia verruciformis. Arch Dermatol. 1977;113:495–9. [PubMed] [Google Scholar]
- 5.Renner ED, Puck JM, Holland SM, et al. Autosomal recessive hyperimmunoglobulin E syndrome: a distinct disease entity. J Pediatr. 2004;144:93–9. doi: 10.1016/S0022-3476(03)00449-9. [DOI] [PubMed] [Google Scholar]
- 6.Freeman AF, Holland SM. The hyper-IgE syndromes. Immunol Allergy Clin North Am. 2008;28:277–91. doi: 10.1016/j.iac.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalman L, Lindegren ML, Kobrynski L, et al. Mutations in genes required for T-cell development: IL7R, CD45, IL2RG, JAK3, RAG1, RAG2, ARTEMIS, and ADA and severe combined immunodeficiency: HuGE review. Genet Med. 2004;6:16–26. doi: 10.1097/01.GIM.0000105752.80592.A3. [DOI] [PubMed] [Google Scholar]
- 8.van der Burg M, van Veelen LR, Verkaik NS, et al. A new type of radiosensitive T-B-NK+ severe combined immunodeficiency caused by a LIG4 mutation. J Clin Invest. 2006;116:137–45. doi: 10.1172/JCI26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Burg M, Ijspeert H, Verkaik NS, et al. A DNA-PKcs mutation in a radiosensitive T-B-SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–8. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Markert ML, Hershfield MS, Schiff RI, Buckley RH. Adenosine deaminase and purine nucleoside phosphorylase deficiencies: evaluation of therapeutic interventions in eight patients. J Clin Immunol. 1987;7:389–99. doi: 10.1007/BF00917017. [DOI] [PubMed] [Google Scholar]
- 12.Pignata C, Fiore M, Guzzetta V, et al. Congenital alopecia and nail dystrophy associated with severe functional T-cell immunodeficiency in two sibs. Am J Med Genet. 1996;65:167–70. doi: 10.1002/(SICI)1096-8628(19961016)65:2<167::AID-AJMG17>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 13.Shiow LR, Roadcap DW, Paris K, et al. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat Immunol. 2008;9:1307–15. doi: 10.1038/ni.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liston A, Enders A, Siggs OM. Unravelling the association of partial T-cell immunodeficiency and immune dysregulation. Nat Rev Immunol. 2008;8:545–58. doi: 10.1038/nri2336. [DOI] [PubMed] [Google Scholar]
- 15.Ozcan E, Notarangelo LD, Geha RS. Primary immune deficiencies with aberrant IgE production. J Allergy Clin Immunol. 2008;122:1054–62. doi: 10.1016/j.jaci.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Ruusala A, Aspenström P. Isolation and characterisation of DOCK8, a member of the DOCK180-related regulators of cell morphology. FEBS Lett. 2004;572:159–66. doi: 10.1016/j.febslet.2004.06.095. [DOI] [PubMed] [Google Scholar]
- 17.Lei JY, Wang Y, Jaffe ES, et al. Microcystic adnexal carcinoma associated with primary immunodeficiency, recurrent diffuse herpes simplex virus infection, and cutaneous T-cell lymphoma. Am J Dermatopathol. 2000;22:524–9. doi: 10.1097/00000372-200012000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Milner JD, Brenchley JM, Laurence A, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–6. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimbacher B, Schäffer AA, Holland SM, et al. Genetic linkage of hyper-IgE syndrome to chromosome 4. Am J Hum Genet. 1999;65:735–44. doi: 10.1086/302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shearer WT, Rosenblatt HM, Gelman RS, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112:973–80. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Custer JW, Rau RE, editors. The Harriet Lane handbook: a manual for pediatric house officers. 18. Philadelphia: Elsevier Mosby; 2009. [Google Scholar]
- 22.Brito-Babapulle F. The eosinophilias, including the idiopathic hypereosinophilic syndrome. Br J Haematol. 2003;121:203–23. doi: 10.1046/j.1365-2141.2003.04195.x. [DOI] [PubMed] [Google Scholar]
- 23.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 24.Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J Cell Sci. 2005;118:4937–46. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- 25.Fukui Y, Hashimoto O, Sanui T, et al. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–31. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 26.Sanui T, Inayoshi A, Noda M, et al. DOCK2 is essential for antigen-induced translocation of TCR and lipid rafts, but not PKC-theta and LFA-1, in T cells. Immunity. 2003;19:119–29. doi: 10.1016/s1074-7613(03)00169-9. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y, Hamano S, Gotoh K, et al. T helper type 2 differentiation and intracellular trafficking of the interleukin 4 receptor-alpha subunit controlled by the Rac activator Dock2. Nat Immunol. 2007;8:1067–75. doi: 10.1038/ni1506. [DOI] [PubMed] [Google Scholar]
- 28.Nombela-Arrieta C, Mempel TR, Soriano SF, et al. A central role for DOCK2 during interstitial lymphocyte motility and sphingosine-1-phosphate-mediated egress. J Exp Med. 2007;204:497–510. doi: 10.1084/jem.20061780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holland SM, DeLeo FR, Elloumi HZ, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 30.Minegishi Y, Saito M, Tsuchiya S, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 31.Renner ED, Rylaarsdam S, Anover-Sombke S, et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122:181–7. doi: 10.1016/j.jaci.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Beaucoudrey L, Puel A, Filipe-Santos O, et al. Mutations in STAT3 and IL12RB1 impair the development of human IL-17-producing T cells. J Exp Med. 2008;205:1543–50. doi: 10.1084/jem.20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minegishi Y, Saito M, Morio T, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–55. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Woellner C, Schäffer AA, Puck JM, et al. The hyper IgE syndrome and mutations in TYK2. Immunity. 2007;26:535. doi: 10.1016/j.immuni.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi K, Kohno T, Ajima R, et al. Homozygous deletion and reduced expression of the DOCK8 gene in human lung cancer. Int J Oncol. 2006;28:321–8. [PubMed] [Google Scholar]
- 36.Idbaih A, Carvalho Silva R, Crinière E, et al. Genomic changes in progression of low-grade gliomas. J Neurooncol. 2008;90:133–40. doi: 10.1007/s11060-008-9644-z. [DOI] [PubMed] [Google Scholar]
- 37.Griggs BL, Ladd S, Saul RA, DuPont BR, Srivastava AK. Dedicator of cytokinesis 8 is disrupted in two patients with mental retardation and developmental disabilities. Genomics. 2008;91:195–202. doi: 10.1016/j.ygeno.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinci G, Chantot-Bastaraud S, El Houate B, Lortat-Jacob S, Brauner R, McElreavey K. Association of deletion 9p, 46, XY gonadal dysgenesis and autistic spectrum disorder. Mol Hum Reprod. 2007;13:685–9. doi: 10.1093/molehr/gam045. [DOI] [PubMed] [Google Scholar]
- 39.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006;7:407–42. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.