Abstract
Background
Recent studies of the genetics of alcoholism have focused on a cluster of genes encoding for γ-aminobutyric acid (GABAA) receptor subunits, which is thought to play a role in the expression of addiction phenotypes. This study examined allelic associations between 2 single nucleotide polymorphisms (SNPs) of the GABRG1 gene (rs1391166 and rs1497571) and alcohol phenotypes, namely level of response to alcohol, alcohol use patterns, and alcohol-related problems.
Method
Participants were non-treatment-seeking seeking hazardous drinkers (n = 124) who provided DNA samples, participated in a face-to-face interview for level of response to alcohol, and completed a series of drinking and individual differences measures.
Results
Analyses revealed that a SNP of the GABRG1 gene (rs1497571) was associated with level of response to alcohol and drinking patterns in this subclinical sample. Follow-up mediational analyses were also conducted to examine putative mechanisms underlying these associations.
Discussion
These findings replicate and extend recent research suggesting that genetic variation at the GABRG1 locus may underlie the expression of alcohol phenotypes, including level of response to alcohol.
Keywords: GABRG1, Level of Response, Alcohol, Alcohol Dependence, Genetics
Alcohol abuse and dependence are highly prevalent psychiatric disorders estimated to affect 8.5% of the adult U.S. population within a 1-year period (Grant et al., 2004). The etiology of alcoholism is the result of a complex interplay between genetic and environmental risk factors. Twin and adoption studies indicated that approximately 40% to 60% of the variance in risk for developing alcoholism can be explained by genetic factors (Kendler et al., 1994; Prescott and Kendler, 1999; Schuckit, 2000). Large scale efforts have been undertaken to identify specific genes contributing to the liability to alcoholism. The Collaborative Study on Genetics of Alcoholism (COGA) systematically ascertained families with at least 3 members affected by alcohol dependence (AD) (Edenberg, 2002). Results of whole-genome sibling-pair linkage analyses from the COGA pedigrees revealed several chromosomal regions of interest for which there was evidence of genes contributing to AD (Foroud et al., 2000; Reich et al., 1998). Specifically, linkage results from the COGA project (Reich et al., 1998) and an independent sample of American Indians (Long et al., 1998) have identified a region on chromosome 4p12 where a cluster of genes encoding for γ-aminobutyric acid (GABAA) receptor subunits (i.e., γ-1, α-2, α-4, and β-1) is located.
Linkage results have suggested areas of the genome that may underlie genetic liability for alcoholism and more importantly, have identified areas for further research and identification of specific genes contributing to alcoholism risk. To that end, several studies have focused on the aforementioned cluster of GABAA genes located on chromosome 4p12. Results from the COGA project revealed a strong association of variation in genes encoding the α2 subunit of the GABAA receptor (GABRA2) and both AD as well as the β frequency of the electroencephalogram, a neurobiological endopheno-type (Edenberg et al., 2004). The association between GABRA2 with AD was further replicated in 3 different studies (Covault et al., 2004; Fehr et al., 2006; Lappalainen et al., 2005). The effects of genetic variation in the GABRA2 gene were extended to marijuana and illicit drug dependence phenotypes (Agrawal et al., 2006), conduct disorder and alcohol and drug dependence across developmental stages (Dick et al., 2006b), and gene × environment interactions with marital status (Dick et al., 2006a). Interestingly, GABRA2 genotypes, based on results from the COGA sample, were recently found to moderate psychotherapy outcomes for alcoholism in a reanalysis of Project MATCH (Bauer et al., 2007).
Studies have also examined a cluster of GABAA receptor genes (GABRA1, GABRA6, GABRB2, and GABRG2) in chromosome 5q and while some results suggested significant associations to drinking behaviors, such as level of response to alcohol, history of blackouts, age of first drunkenness, and AD (Dick et al., 2006c; Radel et al., 2005), others found no association between GABAA receptor genes and AD (Dick et al., 2005; Sander et al., 1999). Importantly, a study by Covault and colleagues (2008) suggested that markers in the 5′-region of the GABRG1 gene, which encodes the GABAA receptor γ-1 subunit, are in linkage disequilibrium (LD) with markers in the GABRA2 gene, which is adjacent to the GABRG1 gene and also located in chromosome 4p. Moreover, markers in the 5′-region of the GABRG1 gene showed associations with AD in 2 samples of individuals of European Ancestry (Covault et al., 2008). A recent study of LD patterns of GABRG1 and GABRA2 across various populations showed further support for intergenic LD in 5 different populations, including European American (Ittiwut et al., 2008). These studies highlight the importance of genetic variation in GABAA receptor genes to level of response to alcohol, drinking patterns, and to the alcoholism phenotype more broadly, and suggest that further attention must be paid to the GABRG1 gene.
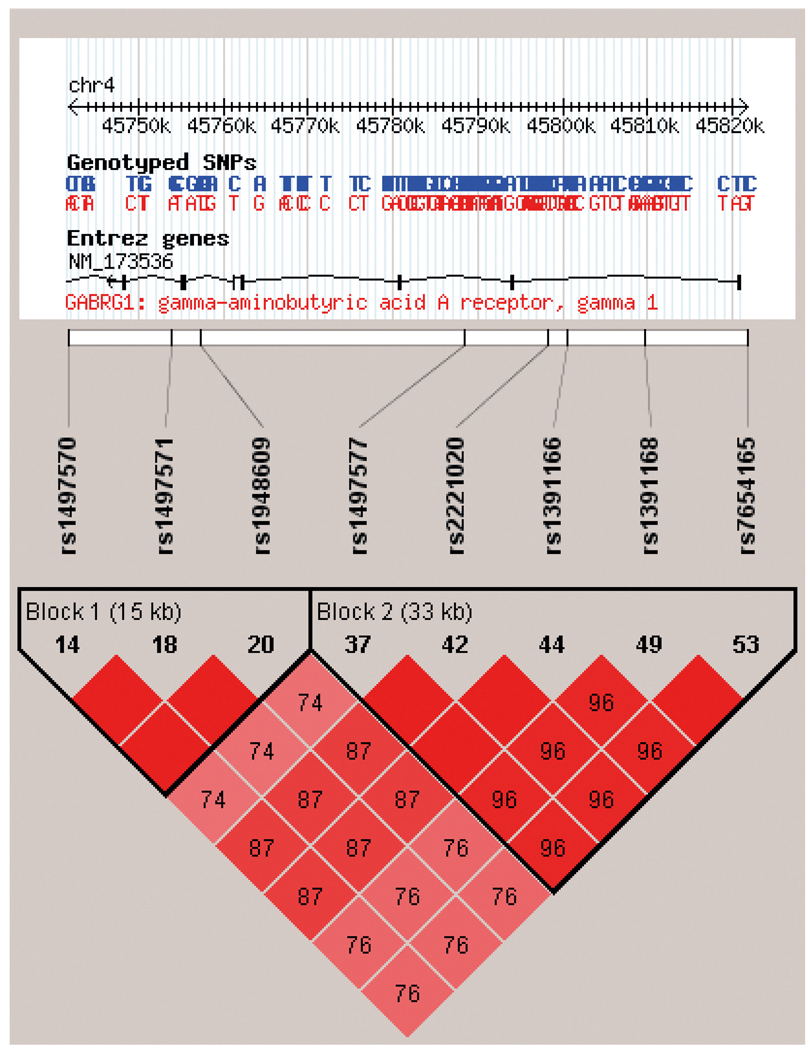
The purpose of this study is to examine allelic associations between 2 single nucleotide polymorphisms (SNPs) of the GABRG1 gene (rs1391166 and rs1497571) and alcohol use phenotypes in a sample of hazardous drinkers. The GABRG1 SNPs were selected on the basis of recent research demonstrating allelic and haplotyic associations between the GABRG1 gene and AD (Covault et al., 2008; Enoch et al., 2009). Specifically, the 2 SNPs selected are representative of the major haplotype blocks in GABRG1 and validated SNP assays are commercially available for both of them. Broadly, the GABRG1 gene is part of a cluster of GABAA genes located in chromosome 4, which appears to harbor genetic variation underlying the expression of alcohol and drug dependence phenotypes (Agrawal et al., 2006; Edenberg et al., 2004). Figure 1 shows the 2 candidate SNPs examined in this study in relation to SNPs examined in previous reports of alcoholism phenotypes along with their patterns of LD. The alcohol phenotypes were chosen in light of the subclinical nature of the sample of hazardous drinkers. These phenotypes include alcohol use patterns, alcohol-related problems, and level of response to alcohol, a well-established biobehavioral marker of risk for AD (Schuckit and Smith, 1996, 2006).
Fig. 1.
Linkage disequilibrium (LD) plot from Haploview 4.1 for EA subjects based on HapMap (phase II) samples of individuals of European Ancestry for the 2 SNPs evaluated in this study (rs1497571 and rs1391166) and SNPs evaluated in previous studies: (1) rs1497570, rs19448609, rs2221020, and rs1391168 (Edenberg et al., 2004; Enoch et al., 2009); and (2) rs1497577, rs1391166, and rs7654165 (Covault et al., 2008). SNPs rs1497577, rs1391166 (examined in this study), and rs7654165, all of which are located in haplotype block 2, showed significant association with AD (Covault et al., 2008). The same was true for SNP rs1497570 (Enoch et al., 2009). The remaining SNPs were not significantly associated with AD (Edenberg et al., 2004). Pair-wise SNP |D′| values (×100) of linkage are shown along with 2 haplotype blocks identified using the 4-gamete rule. Darkened blocks indicate SNP pairs without evidence of extensive recombination (i.e., 4-gamete rule for haplotype block characterization with at least one 2-SNP haplotype having a frequency <0.02).
GABAergic neurotransmission is thought to play a central role in the behavioral effects of alcohol, particularly sedation, tolerance, loss of motor coordination, and withdrawal (Grobin et al., 1998; Koob, 2004; Krystal et al., 2006). Previous studies have implicated GABAA receptor genes with AD, alcohol use patterns, and level of response to alcohol (Dick et al., 2006c; Schuckit et al., 1999, 2004). Therefore, it is hypothesized that the candidate SNPs of the GABRG1 will be associated with lower level of response to alcohol, heavier drinking, and more alcohol-related problems in this sample of hazardous drinkers.
METHOD
Participants and Procedures
This study was approved by the University of Colorado Human Research Committee and all participants provided written informed consent after receiving a full explanation of the study. Inclusion criteria were the following: (1) age between 21 and 65; (2) a score of 8 or higher on the Alcohol Use Disorders Identification Test (AUDIT), indicating a hazardous drinking pattern (Allen et al., 1997); (3) self-reported current drinking of 6 or more drinks (4 or more for females) per week; this item was used to ensure that participants were current drinkers and not abstainers as a result of recent alcohol problems. Participants were recruited primarily from the University through advertisements and E-mails directed towards the University students and employees. A total of 124 participants met the screening criteria and completed an in-person assessment of demographics, drinking behavior, level of response to alcohol, and individual differences measures. Of the 124 participants in this study, 40 participants went on to complete a pharmacogenetic study of naltrexone (Ray and Hutchison, 2007). This study focuses on the sample of 124 (39 females) participants selected for a pattern of hazardous drinking and unselected for genotype for whom the average age was 22.53 (SD = 2.56; range = 21 to 33). The ethnic composition of the sample was the following: Caucasian (87.8%), Asians (4.9%), Latino (4.1%), Native Americans (2.4%), and African Americans (0.8%).
Initial assessment of the inclusion criteria was conducted through a telephone interview. Eligible participants were invited to the laboratory for an in-person assessment session. Upon arrival at the lab, participants read and signed an informed consent form, provided a saliva sample for DNA analyses, and completed a series of face-to-face assessments, as described below.
Assessments
Alcohol Use
Alcohol use was evaluated with a variation of the measure used by White and Labouvie (1989). The instructions defined 1 alcoholic drink as “one beer, one glass of wine, or one serving of hard liquor either by itself or in a mixed drink” (White and Labouvie, 1989). The following 3 questions about drinking patterns in the last year were asked as part of the Alcohol Consumption Questionnaire (ACQ): (1) When you drank alcohol, how many drinks did you consume on average on 1 occasion? (ACQ1); (2) What is the largest number of drinks you consumed on 1 occasion? (ACQ2); and (3) How often you drank alcohol on average? (ACQ3). The last question was answered on a 10-point scale ranging from “never” to “every day” and where 6 = twice per week. Average scores on drinking measures are presented in Table 1.
Table 1.
Average Scores [M ± (SD)] and Intercorrelations Among the SRE and Drinking Variables
| Measure | M | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| 1. First 5 Times (SRE-5) | 4.58 (1.88) | – | |||||||
| 2. Reg 3 Months (SRE-3) | 6.47 (2.05) | 0.71*** | – | ||||||
| 3. Heaviest Drink (SRE-H) | 8.59 (2.78) | 0.55*** | 0.73*** | – | |||||
| 4. Total Score (SRE-T) | 6.43 (2.16) | a | a | a | – | ||||
| 5. Average Drink / Epi (ACQ1) | 5.32 (2.39) | 0.20* | 0.30** | 0.48*** | 0.39*** | – | |||
| 6. High Drink / Epi (ACQ2) | 14.52 (5.87) | 0.39*** | 0.54*** | 0.72*** | 0.65*** | 0.52*** | – | ||
| 7. Often Drink (ACQ3) | 6.69 (1.38) | −0.01 | 0.03 | 0.19* | 0.10 | 0.19* | 0.35*** | – | |
| 8. RAPI Score | 20.26 (13.21) | 0.13 | 0.20* | 0.30** | 0.25** | 0.17 | 0.37*** | 0.32** | – |
| 9. AUDIT Score | 13.62 (4.78) | 0.13 | 0.19* | 0.32** | 0.26** | 0.39*** | 0.41*** | 0.38*** | 0.57*** |
ACQ, Alcohol Consumption Questionnaire; AUDIT, Alcohol Use Disorders Identification Test; RAPI, Rutgers Alcohol Problem Index; SRE. Self-Rating of the Effects of Alcohol; n = 124
p < 0.05
p < 0.01
p < 0.001.
Correlations between SRE-T and the other 3 subscales of the SRE (i.e., SRE-5; SRE-3; SRE-H) were not computed given that these scales are components of the total score (SRE-T).
The Rutgers Alcohol Problem Index
The Rutgers Alcohol Problem Index (RAPI) was used to assess alcohol-related problems. This scale consists of 23 items examining the impact of alcohol on social and health functioning over the past year. The observed Cron-bach a for the RAPI in this study was 0.90, suggesting high internal consistency.
The Self-Rating of the Effects of Alcohol
The Self-Rating of the Effects of Alcohol (SRE) form, delivered in a face-to-face interview format, was used to assess level of response to alcohol (Schuckit et al., 1997a). The SRE is a retrospective measure of responses to alcohol and consists of estimating the number of drinks required to obtain a given intoxication effect, such as “begin to feel different,” “feel a bit dizzy or begin to slur your speech,” “begin stumbling, or walking in an uncoordinated manner,” and “pass out, or fall asleep when you did not want to.” Number of drinks estimates were obtained for the first 5 times participants ever drank (SRE-5), for the period of heaviest drinking (SRE-H). In addition, as recommended by Schuckit and colleagues (1997a), a total score was computed (SRE-T). SRE average scores are reported in Table 1 and are consistent with a previous report on a sample of young nondependent drinking men (Schuckit et al., 1997b). The SRE showed high internal consistency in this sample, Cronbach α = 0.91.
Geno typing
DNA was collected following published procedures (Freeman et al., 1997; Walker et al., 1999). Participants swabbed their cheeks with 3 cotton swabs, followed by a rinse of the mouth with 10 ml of sucrose solution (4% in tap water). All genotyping was conducted at the Neurogenetics Laboratory at theMind ResearchNetwork (Albuquerque, NM). Genomic DNA was isolated from buccal cells using a modification of published procedures (Lench et al., 1988; Spitz et al., 1996). An ABI PRISM 7500 instrument was used to conduct 5′-nuclease (TaqMan) assays of the GABRG1 SNP using assays commercially available from Applied Biosystems (Foster City, CA). This method relies on allele-specific hybridization of oligonucleotide probes (Livak, 1999). Polymerase chain reaction amplifications failed to provide genotypes for rs1391166 in 12 samples and for rs1497571 in 2 samples, despite repeated attempts. For quality control purposes, 34 samples (27%) were genotyped twice for rs1391166 and 20 samples (16%) were genotype twice for rs1497571. Analyses revealed 100% agreement in genotype calls for both SNPs. As an additional check on allele frequencies and genotyping we have reanalyzed every sample for the rs1497571 SNP and again found 100% agreement in the genotype calls for this SNP, which increases our confidence in the analyses reported herein.
The following genotype frequencies were observed for rs1391166: AA (n = 31), AT (n = 66), and TT (n = 15). For rs1497571, the observed genotype frequencies were: CC (n = 33), CT (n = 71), and TT (n = 18). The first SNP, rs1391166, is located in intron 1 and the estimated heterozygosity according to publicly available databases (http://www.ncbi.nlm.nih.gov/projects/SNP/) is 0.49 (SE = 0.065), while the observed heterozygosity in this sample was 0.59. The second SNP, rs1497571 is located in intron 7 with expected heterozygosity of 0.50 (SE = 0.036) and observed was 0.58. These departures were approximately within 2 SE of the mean. Data available on the SNP database using HapMap samples indicated that allele frequencies for both SNPs differ by ethnicity, such that the major allele was less frequent in individuals of Asian descent compared with individuals of European or African ancestry. For both alleles, tests for conformity to Hardy–Weinberg Equilibrium indicated departure from expected values, χ2(1) = 4.62, p = 0.032 for rs1391166 and χ2(l) = 4.03, p= 0.045 for rs1497571.
Power Analysis
Power analysis was conducted using the continuous outcome design option in Quanto (Gauderman, 2002a,b, 2003). Tests estimated the power to detect genetic effects between each of the SNPs (i.e., rs1391166 and rs1497571) and a continuous outcome (i.e., drinking and level of response to alcohol) in this sample of 124 unrelated individuals. Specifically, we allowed for the following in the power calculations for this study: (1) allele frequency of 0.50 (consistent with the observed minor allele frequencies in this sample); (2) the candidate locus to account for at least 1 % of the variance in the dependent variable with the estimated R2 ranging between 0.01 and 0.08; and (3) dominant gene action. We estimated power at 2 α-levels, 0.05 and 0.01, to assess the changes in statistical power resulting from possible corrections for Type I error. As shown in Table 2, at an α-level of 0.05, a dominant locus accounting for 6% or more of the overall variance would be detectable with better than 79% power. Conversely, at an α-level of 0.01, none of the power estimates for R2 ranging between 0.01 and 0.08 were ≥0.80, which is the recommended threshold (Cohen, 1988,1992).
Table 2.
Power to Detect Genetic Main Effects of Varying Magnitude for Continuous Outcomes at α-Levels of 0.05 and 0.01
| Percent variance (R2) |
Power for genetic effect α = 0.05 |
Power for genetic effect α = 0.01 |
|---|---|---|
| 0.01 | 0.20 | 0.07 |
| 0.02 | 0.35 | 0.16 |
| 0.03 | 0.49 | 0.26 |
| 0.04 | 0.61 | 0.37 |
| 0.05 | 0.71 | 0.48 |
| 0.06 | 0.79 | 0.58 |
| 0.07 | 0.85 | 0.66 |
| 0.08 | 0.89 | 0.74 |
Data Analysis
The genotypes for rs1391166 were grouped into AA (n = 31) and AT/TT (n = 81) while the genotypes for rs1497571 were grouped into CC (n = 33) and CT/TT (n = 89). This was carried out to reduce the number of statistical comparisons and to increase power to detect genotype differences by increasing cell sizes. The decision as to how to best combine the heterozygote group was based on the following steps: (1) we plotted the means of the 3 genotype groups on the dependent variables of interest and examined dominance patterns (as assumed in the power analysis described previously) to decide how to build the 2 groups and (2) if dominance patterns were not clear (the case for rs1391166) then the less frequent homozygote group was combined with the heterozygote group as is generally the case in genetic association studies.
Genotype groups were then compared on age using t-tests and on gender using chi-squared tests. In addition, genotype group comparisons were conducted using Student’s t-tests for continuous variables, as t-tests account for potential violations of the assumption of the homogeneity of variance, an important issue when comparing 2 groups with an unequal number of participants (n). Specifically, for variables in which the homogeneity of variance assumption was held, results of t-tests using pooled variance are reported, whereas for variables that violated the homocedasticity assumption in our sample, the individual sample standard deviation was used to calculate the t-tests. Associations among continuous predictors were examined using Person product–moment correlations. Main outcome analyses were performed using SAS statistical software (SAS Institute, Inc., Cary, NC). For all comparisons, statistical significance was set at p < 0.05, and all tests were 2-tailed.
Linkage disequilibrium plots and haplotype blocks for individuals of European Ancestry were generated from publicly available HapMap (phase II) data using the software program Haploview v4.1 (Barrett et al., 2005). Pair-wise SNP |D′| values (× 100) of linkage were computed using the 4-gamete rule. High D′ values indicate SNP pairs without evidence of extensive recombination (i.e., 4-gamete rule for haplotype block characterization with at least one 2-SNP haplotype having a frequency <0.02). In other words, SNPs with high D′ values indicate that markers are good surrogates for each other, likely to be transmitted together, and therefore likely to capture similar genetic variance.
Corrections for Type I error were considered but ultimately rejected on the basis of the following considerations. First, correction for Type I error would result in a significant loss of statistical power, as demonstrated in Table 1. Second, Type I error needs to be considered for each hypothesis separately, not for the number of variables in the whole set of analyses reported (Dar et al., 1994). In the present analyses 4 measures, all derived from the SRE (i.e., SRE-5, SRE-3, SRE-H, and SRE-T), were used to test the first hypothesis that the candidate SNP would be associated with level of response to alcohol, 3 measures (i.e., ACQ1, ACQ2, and ACQ3) were used to test the second hypothesis that these SNPs would be associated with drinking outcomes, and 2 measures (i.e., RAPI and AUDIT) tested the third hypothesis regarding the association of the candidate SNPs to alcohol-related problems. Together, these considerations indicate that corrections for Type I error may not be advised in this study.
RESULTS
Pretest Comparisons
There were no differences in allele frequencies by gender [rs1391166, χ2(l) = 0.05, p = 0.83; and rs1497571, χ2(1) = 1.01, p = 0.32] or by ethnicity (rs1391166, Fisher’s exact p = 0.23; and rs1497571, Fisher’s exact p = 0.72) for either SNP. Likewise, the groups did not differ in age for either SNP, t(110) = 1.06, p = 0.29 and t(110) = −1.54, p = 0.13, respectively. Analysis of LD indicated that the D′ for SNPs 1 and 2 was 0.76, suggesting some evidence of recombination; this is also supported by the fact that the 2 SNPs were harbored in different haplotype blocks. As shown in Fig. 1, the 2 SNPs of interest in this study were found to be in high LD with polymorphisms previously studied in the context of alcoholism phenotypes.
GABRG1, Level of Response to Alcohol, and Drinking
As shown in Table 3, there were no significant genotype differences on drinking behavior and level of response to alcohol for SNP rs1391166. However, analyses of SNP rs1497571 revealed that individuals who were homozygous for the C allele, reported requiring a higher number of drinks to obtain alcohol effects at 3 months of regular drinking [SRE-3; t(120) = 2.18, p < 0.05], during a period of heaviest drinking [SRE-H; t(120) = 2.12, p < 0.05], and the SRE total score [SRE-T; t(120) = 2.35, p < 0.05]. Likewise, there was a trend toward a significant genotype effect on SRE scores for the first 5 times participants drank alcohol [SRE-5; t(120) = 1.82, p = 0.071]. These results suggest that carriers of the T allele of SNP rs1497571 of the GABRG1 gene are high responders to alcohol. Follow-up analyses breaking down this genotype group and comparing the TT (n = 18) and CT (n = 71) groups revealed no significant group differences on level of response to alcohol (p > 0.10), which did not support a T allele gene dose effect.
Table 3.
SRE Scores and Drinking Characteristics by GABRG1 Genotype [M ± (SD)]
|
rs1391166 |
rs1497571 |
|||||||
|---|---|---|---|---|---|---|---|---|
| AA (n = 31) | AT (n = 66) | TT (n = 15) | AA versus AT/TT | CC (n = 33) | CT (n = 71) | TT (n = 18) | CC versus CT/TT | |
| First 5 Times (SRE-5) | 4.50 (1.70) | 4.79 (1.84) | 4.52 (1.01) | t(110) < 1.0; p = 0.52 | 5.19(1.69) | 4.51 (1.79) | 4.69(1.60) | t(120) = 1.82; p= 0.071 |
| Reg 3 Months (SRE-3) | 6.45 (2.50) | 6.43 (1.91) | 6.14 (1.56) | t(110) < 1.0; p = 0.89 | 7.14 (1.84) | 6.13 (1.88) | 6.70 (2.78) | t(120) = 2.18; p < 0.05 |
| Heaviest Drink (SRE-H) | 8.66 (3.21) | 8.48 (2.77) | 8.58 (2.42) | t(110) < 1.0; p = 0.79 | 9.46 (2.51) | 8.16 (2.62) | 8.73 (3.49) | t(120) = 2.12; p < 0.05 |
| Total Score (SRE-T) | 6.53 (2.10) | 6.56 (1.98) | 6.42 (1.41) | t(110) < 1.0; p = 0.52 | 7.26 (1.75) | 6.27 (1.85) | 6.71 (2.33) | t(120) = 2.35; p < 0.05 |
| Ave Drink/Epi (ACQ1) | 5.10 (1.76) | 5.29 (2.45) | 5.70 (3.02) | t(110) < 1.0; p = 0.52 | 6.39 (2.66) | 5.10 (2.27) | 4.50 (1.72) | t(120) = 3.00; p < 0.01 |
| High Drink/Epi (ACQ2) | 13.92 (6.41) | 14.83 (6.14) | 13.87 (4.81) | t(110) < 1.0; p = 0.57 | 16.61 (5.09) | 14.03 (5.86) | 12.61 (6.45) | t(120) = 2.44; p < 0.05 |
| Often Drink (ACQ3) | 6.36 (1.28) | 6.71 (1.46) | 7.00 (1.51) | t(110) = −1.38; p = 0.17 | 6.91 (1.49) | 6.73 (1.30) | 6.06 (1.43) | t(120) = 1.11; p= 0.27 |
| RAPI Score | 20.23 (14.26) | 20.48 (13.11) | 23.27 (12.91) | t(110) = −1.84; p = 0.068 | 20.72 (13.95) | 21.82 (13.37) | 13.22 (9.39) | t(120) < 1.0; p= 0.81 |
| AUDIT Score | 12.23 (4.28) | 13.74 (4.73) | 15.60 (5.90) | t(110) = −1.97; p = 0.053 | 15.40 (7.19) | 13.59 (4.60) | 11.17 (3.36) | t(120) = 2.31; p < 0.05 |
ACQ, Alcohol Consumption Questionnaire; AUDIT, Alcohol Use Disorders Identification Test; RAPI, Rutgers Alcohol Problem Index; SRE, Self-Rating of the Effects of Alcohol.
Single nucleotide polymorphism rs1497571 of the GABRG1 gene was also associated with drinking patterns in this sample, such that carriers of the T allele reported a lower average number of drinks per episode, over the past year [ACQ1; t(120) = 3.00, p < 0.01], lower maximum number of drinks per drinking episode over the past year [ACQ2; t(120) = 2.44, p < 0.05] and lower AUDIT Scores [AUDIT; t(120) = 2.31, p < 0.05] when compared with homozygotes for the C allele. However, SNP rs1497571 was unrelated to how often participants drank [ACQ3; t(120) = 1.11, p = 0.27], and their scores on the RAPI [RAPI; t(120) = 0.23, p = 0.81]. As shown by the correlations in Table 1, level of response to alcohol was significantly associated with a heavier drinking pattern and more alcohol-related problems. Finally, restricting the analyses to individual who have valid genotypes for both SNPs of interest (n = 112) did not alter the results reported above.
Follow-Up Analysis of Mediation
Single nucleotide polymorphism rs1497571 of the GABRG1 gene was associated with both level of response to alcohol and drinking patterns and alcohol problems. Level of response was, in turn, associated with drinking patterns and alcohol problems. This pattern of results led us to test a follow-up mediational model in which the effects of SNP rs1497571 of GABRG1 on drinking behavior are mediated by level of response to alcohol. Analyses used Baron and Kenny’s (1986) conceptual approach and formal tests of the significance of the indirect effects based on Preacher and Hayes’s (2004) bootstrapping estimation approach. This approach is appropriate for small sample sizes because it avoids distributional assumptions and calculates a reliable estimate of the indirect effect and its significance by taking a large number of samples from the data (Preacher and Hayes, 2004).
Bootstrapping estimation using 5,000 resamples was used to test mediational models by which level of response to alcohol (i.e., SRE-T) mediated the effects of SNP rs1497571 of the GABRG1 gene on average number of drinks per episode (ACQ1), maximum number of drinks in a drinking episode (ACQ2), and score on the AUDIT. Results, however, did not support the mediational models for ACQ1 (B = 0.18, SE = 0.13; 95% CI = 0.08 to 0.44, p = 0.17), ACQ2 (B = 0.77, SE = 0.53; 95% CI = −0.27 to 1.82, p = 0.15), or AUDIT Score (B = 0.20, SE = 0.18; 95% CI = 0.16 to 0.56, p = 0.28).
DISCUSSION
This study sought to examine allelic associations between 2 SNPs of the GABRG1 gene (rs1391166 and rs1497571) and alcohol phenotypes, namely level of response to alcohol, alcohol use, and alcohol-related problems. Results revealed that variation in a SNP located on intron 7 of the GABRG1 gene, rs1497571, was associated with level of response to alcohol, drinking behavior, and alcohol problems in a sample of hazardous drinkers. More specifically, individuals who were homozygous for the C allele at this locus reported a lower level of response to alcohol as demonstrated by a higher number of drinks reported to reach certain intoxication effects. These individuals also reported consuming more alcohol per drinking episode, on average, had a higher maximum number of drinks in a drinking episode and scored higher on the AUDIT.
These results are consistent with previous findings implicating genetic variation in the GABRG1 gene with AD (Covault et al., 2008; Edenberg et al., 2004). A very recent study has found significant associations among multiple SNPs and haplotypes of the GABRG1 gene and AD in 2 independent populations: Plain American Indians and Finish Caucasians (Enoch et al., 2009). In the aforementioned study, one of the significant allelic associations was reported for a SNP (rs1497570) which is in complete LD with the SNP for which we found significant effects in this study (rs1497571), as shown in Fig. 1. In short, Enoch and colleagues (2009) found support for GABRG1 haplotype and SNP associations with AD in 2 independent populations and concluded that these are independent genetic contributions from those previously reported for GABRA2 genotypes. Those findings and the ones reported herein suggest that GABRG1 may be an important candidate gene for alcoholism phenotypes, including intermediate phenotypes such as level of response to alcohol and drinking patterns.
From a neurobiological standpoint, genetic variation in the GABRG1 locus may influence the liability to AD through mechanisms of reward and/or drug responsivity. In the rat brain, the γ-1 subunit of the GABAA receptor, encoded by the GABRG1 gene, is expressed selectively in only a few regions such as the amygdala, striatum, and substantia nigra (Pirker et al., 2000); these regions in turn are often implicated in addiction and reward mechanisms. Pharmacological studies have shown that GABAA γ-1 receptors are associated with lower sensitivity to benzodiazepine antagonists (Khom et al., 2006). As such, it is plausible to hypothesize that functional polymorphisms in the GABRG1 leading to alterations γ-1 receptors may ultimately influence the risk to alcoholism by altering tolerance and reward pathways. A great deal of research is needed to more fully ascertain this putative genetic association and its mechanisms, yet from a theoretical and neurobiological point of view, the GABRG1 gene represents a plausible candidate gene for addiction phenotypes.
Follow-up mediational analyses did not support models in which level of response to alcohol mediated the relationship between this SNP (rs1497571) of the GABRG1 gene and drinking behaviors in our sample. It is possible that the effects of this SNP on drinking behavior may be mediated through alternative risk mechanisms, such as conduct disorder, for example, which has also associated with variation in the GABRA2 (Agrawal et al., 2006; Dick et al., 2006b). This study could not directly test the mediating role of conduct disorder, or more broadly, antisocial traits. However, future genetic studies testing such mediational models seem warranted. These models have the potential to advance the field from the identification of genes that are related to psychiatric phenotypes to understanding how specific genes are involved in the development of psychiatric disorders, which represents an important challenge and future direction in the field of psychiatric genetics (Dick et al., 2006d).
While previous studies have found that both haplotype blocks were associated with AD, this study only found significant associations between the SNP in the 3′ haplotype block (rs1497571) and alcohol use and low response to alcohol. Given that the sample size and resulting statistical power is lower in this study compared with previous large-scale trials, as demonstrated by the power analysis reported in Table 2, this discrepancy might suggest that the signal for the 3′ SNP is strong enough to be detected, while power may be simply inadequate for detecting an association for the rs1391166 SNP. Certainly, the lack of association for rs1391166 in this study should not be viewed as a strong contradiction of previous findings of an association between that haplotype block and AD considering the difference in statistical power and phenotypes between this study and previous reports.
Despite a great deal of interest in the cluster of GABAA genes located in chromosome 4, the functional significance of this (rs1497570), or any other, polymorphism of the GABRG1 gene remains unknown. As stated by Enoch et al. (2009), the extensive pattern of LD in the 4-gene cluster located in chromosome 4 suggests the presence of distant enhancers and suppressors; however, basic molecular genetics research is needed to identify a functional locus, or loci, in this gene cluster. As is the case for genetic associations to polymorphisms of unknown functional significance, LD to a functional variant represents an important and plausible, alternative explanation, whereby the signal detected in this study at the rs1497571 locus may be a result of its close proximity to a functional polymorphism.
Similarly, population stratification and other unmeasured third variables, both environmental and genetic in nature, may account for these results (Hutchison et al., 2004). This may be especially relevant to GABRG1 genes as Enoch and colleagues (2009) have examined 2 independent populations and found evidence of selection pressure for various alcohol use disorder-linked SNPs. These results may also shed light on the departures from HWE observed for both SNPs in this study. Admixture and selection bias are among the strongest possible explanations for deviations from HWE. To exclude population stratification as an explanation for the associations observed in this study, we reanalyzed the data for individuals of European Ancestry only and doing so did not significantly alter any of the results reported herein. Thus stratification does not appear to account for the results obtained in this study.
Although some studies have used HWE tests to detect genotyping errors, recent research has put forth strong arguments against this practice (Leal, 2005; Zou and Donner, 2006). Additionally, behavioral geneticists have noted that tests for HWE assume that genotypes are at random sample from the general population whereas many behavioral genetic studies ascertain individuals through their disease status, or as is the case in this study, through drinking patterns. As a result, when a marker is associated with a given phenotype, the corresponding genotype may no longer be a random sample, in which case even if the marker is in HWE with the population, departures from HWE may be noted in a selected sample (Li and Li, 2008). In other words, deviations from HWE are thought to be most likely at trait susceptibility loci or polymorphisms that are in LD with the susceptibility locus (Leal, 2005). The departure from HWE observed in this sample may be the result of a selected sample of heavy drinkers, selection pressure as demonstrated by Enoch and colleagues (2009), other unknown factors, or a combination thereof.
In sum, this study provides a replication and extension of previous findings of case-control population-based studies (e.g., Enoch et al., 2009) into a sample of hazardous drinkers. Broadly speaking, consistency in the genetic association literature hinges on the careful methodological replication and extension of findings to various samples, including samples of varying ethnic backgrounds and phenotypic distributions. The fact that this sample is relatively more homogenous than the extreme-groups (e.g., case-control or population-based) would argue for greater difficulty detecting significant genetic effects. Nevertheless, the present results are both statistically significant and potentially relevant to the literature. Future studies seem warranted to more fully characterize genetic variation at the GABRG1 gene, including the replication of these findings in diverse samples, the identification of functional loci through molecular studies, and the characterization of the neurobiological mechanisms underlying these putative associations to alcohol phenotypes.
ACKNOWLEDGMENTS
This research was supported by grants from the National Institute on Alcoholism and Alcohol Abuse to LAR (AA14847) and KEH (AA012238). The authors would like to thank Amy Audette, Stephanie DiCristophoro, Keira Odell, and Jenifer Bishop for their assistance with data collection for this project and Marilee Morgan and Melynda Byers for their assistance with genotyping. The authors are thankful to Dr. Marc Schuckit for his thoughtful comments on an earlier version of this manuscript.
REFERENCES
- Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL, Nurnberger JI, Crowe R, Kuperman S, Schuckit MA, Begleiter H, Porjesz B, Dick DM. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav Genet. 2006;36:640–650. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- Allen JP, Litten RZ, Fertig JB, Babor T. A review of research on the Alcohol Use Disorders Identification Test (AUDIT) Alcohol Clin Exp Res. 1997;21:613–619. [PubMed] [Google Scholar]
- Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bauer LO, Covault J, Harel O, Das S, Gelernter J, Anton R, Kranzler HR. Variation in GABRA2 predicts drinking behavior in project MATCH subjects. Alcohol Clin Exp Res. 2007;31:1780–1787. doi: 10.1111/j.1530-0277.2007.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical Power Analysis for the Social Sciences. Hillsdale, NJ: Erlbaum; 1988. [Google Scholar]
- Cohen J. A power primer. Psychol Bull. 1992;112:155–159. doi: 10.1037//0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar R, Serlin RC, Omer H. Misuse of statistical test in three decades of psychotherapy research. J Consult Clin Psychol. 1994;62:75–82. doi: 10.1037//0022-006x.62.1.75. [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Schuckit MA, Bierut L, Hinrichs A, Fox L, Mullaney J, Cloninger CR, Hesselbrock V, Nurnberger JI, Jr, Almasy L, Foroud T, Porjesz B, Edenberg H, Begleiter H. Maritalstatus, alcohol dependence, and GABRA2: evidence for gene-environment correlation and interaction. J Stud Alcohol. 2006a;67:185–194. doi: 10.15288/jsa.2006.67.185. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, Kuperman S, Hesselbrock V, Schuckit M, Almasy L, Tischfield J, Porjesz B, Begleiter H, Nurnberger J, Jr, Xuei X, Edenberg HJ, Foroud T. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav Genet. 2006b;36:577–590. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Foroud T. No association of the GABAA receptor genes on chromosome 5 with alcoholism in the collaborative study on the genetics of alcoholism sample. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:24–28. doi: 10.1002/ajmg.b.30058. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Edenberg HJ, Foroud T. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcohol Clin Exp Res. 2006c;30:1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Kaprio J. The next challenge for psychiatric genetics: characterizing the risk associated with identified genes. Ann Clin Psychiatry. 2006d;18:223–231. doi: 10.1080/10401230600948407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health. 2002;26:214–218. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34(5):1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Freeman B, Powell J, Ball D, Hill L, Craig I, Plomin R. DNA by mail: an inexpensive and noninvasive method for collecting DNA samples from widely dispersed populations. Behav Genet. 1997;27:251–257. doi: 10.1023/a:1025614231190. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002a;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for matched case–control studies of gene-environment interaction. Stat Med. 2002b;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Candidate gene association analysis for a quantitative trait, using parent–offspring trios. Genet Epidemiol. 2003;25:327–338. doi: 10.1002/gepi.10262. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991–1992 and 2001–2002. Drug Alcohol Depend. 2004;74:223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Grobin AC, Matthews DB, Devaud LL, Morrow AL. The role of GABA(A) receptors in the acute and chronic effects of ethanol. Psychophar-macology (Berl) 1998;139:2–19. doi: 10.1007/s002130050685. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Stallings M, McGeary J, Bryan A. Population stratification in the candidate gene study: fatal threat or red herring? Psychol Bull. 2004;130:66–79. doi: 10.1037/0033-2909.130.1.66. [DOI] [PubMed] [Google Scholar]
- Ittiwut C, Listman J, Mutirangura A, Malison R, Covault J, Kranzler HR, Sughondhabirom A, Thavichachart N, Gelernter J. Interpopulation linkage disequilibrium patterns of GABRA2 and GABRG1 genes at the GABA cluster locus on human chromosome 4. Genomics. 2008;91:61–69. doi: 10.1016/j.ygeno.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Khom S, Baburin I, Timin EN, Hohaus A, Sieghart W, Hering S. Pharmacological properties of GABAA receptors containing gamma1 subunits. Mol Pharmacol. 2006;69:640–649. doi: 10.1124/mol.105.017236. [DOI] [PubMed] [Google Scholar]
- Koob GF. A role for GABA mechanisms in the motivational effects of alcohol. Biochem Pharmacol. 2004;68:1515–1525. doi: 10.1016/j.bcp.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Staley J, Mason G, Petrakis IL, Kaufman J, Harris RA, Gelernter J, Lappalainen J. Gamma-aminobutyric acid type A receptors and alcoholism: intoxication, dependence, vulnerability, and treatment. Arch Gen Psychiatry. 2006;63:957–968. doi: 10.1001/archpsyc.63.9.957. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Leal SM. Detection of genotyping errors and pseudo-SNPs via deviations from Hardy–Weinberg equilibrium. Genet Epidemiol. 2005;29:204–214. doi: 10.1002/gepi.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lench N, Stanier P, Williamson R. Simple non-invasive method to obtain DNA for gene analysis. Lancet. 1988;1:1356–1358. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- Li M, Li C. Assessing departure from Hardy–Weinberg equilibrium in the presence of disease association. Genet Epidemiol. 2008;32:589–599. doi: 10.1002/gepi.20335. [DOI] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, Sperk G. GABA(A) receptors: immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience. 2000;101:815–850. doi: 10.1016/s0306-4522(00)00442-5. [DOI] [PubMed] [Google Scholar]
- Preacher KJ, Hayes AF. SPSS and SAS procedures for estimating indirect effects in simple mediation models. Behav Res Methods Instrum Comput. 2004;36:717–731. doi: 10.3758/bf03206553. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D. Haplotype-based localization of an alcohol dependence gene to the 5q34 γ-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62:47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64:1069–1077. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Sander T, Ball D, Murray R, Patel J, Samochowiec J, Winterer G, Rommelspacher H, Schmidt LG, Loh EW. Association analysis of sequence variants of GABA(A) alpha6, beta2, and gamma2 gene cluster and alcohol dependence. Alcohol Clin Exp Res. 1999;23:427–431. [PubMed] [Google Scholar]
- Schuckit MA. Genetics of the risk for alcoholism. Am J Addict. 2000;9:103–112. doi: 10.1080/10550490050173172. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Mazzanti C, Smith TL, Ahmed U, Radel M, Iwata N, Goldman D. Selective genotyping for the role of 5-HT2A, 5-HT2C, and GABA alpha 6 receptors and the serotonin transporter in the level of response to alcohol: a pilot study. Biol Psychiatry. 1999;45:647–651. doi: 10.1016/s0006-3223(98)00248-0. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. An evaluation of the level of response to alcohol, externalizing symptoms, and depressive symptoms as predictors of alcoholism. J Stud Alcohol. 2006;67:215–227. doi: 10.15288/jsa.2006.67.215. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J. The search for genes contributing to the low level of response to alcohol: patterns of findings across studies. Alcohol Clin Exp Res. 2004;28:1449–1458. doi: 10.1097/01.alc.0000141637.01925.f6. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Tipp JE. The Self-Rating of the Effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction. 1997a;92:979–988. [PubMed] [Google Scholar]
- Schuckit MA, Tipp JE, Smith TL, Wiesbeck GA, Kalmijn J. The relationship between Self-Rating of the Effects of alcohol and alcohol challenge results in ninety-eight young men. J Stud Alcohol. 1997b;58:397–404. doi: 10.15288/jsa.1997.58.397. [DOI] [PubMed] [Google Scholar]
- Spitz E, Moutier R, Reed T, Busnel MC, Marchaland C, Roubertoux PL, Carlier M. Comparative diagnoses of twin zygosity by SSLP variant analysis, questionnaire, and dermatoglyphic analysis. Behav Genet. 1996;26:55–63. doi: 10.1007/BF02361159. [DOI] [PubMed] [Google Scholar]
- Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect. 1999;107:517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HR, Labouvie EW. Towards the assessment of adolescent problem drinking. J Stud Alcohol. 1989;50:30–37. doi: 10.15288/jsa.1989.50.30. [DOI] [PubMed] [Google Scholar]
- Zou GY, Donner A. The merits of testing Hardy–Weinberg equilibrium in the analysis of unmatched case-control data: a cautionary note. Ann Hum Genet. 2006;70(Pt. 6):923–933. doi: 10.1111/j.1469-1809.2006.00267.x. [DOI] [PubMed] [Google Scholar]