Abstract
With many bioactive non-ribosomal peptides and polyketides produced in fungi, studies of their biosyntheses are an active area of research. Practical limitations of working with mega-dalton synthetases including cell lysis and protein extraction to recombinant gene and pathway expression has slowed understanding of many secondary metabolic processes relative to bacterial counterparts. Recent advances in accessing fungal biosynthetic machinery are beginning to change this. Here we describe the successes of some studies of thiotemplate biosynthesis in fungal systems, along with very recent advances in chemical tagging and mass spectrometric strategies to selectively study biosynthetic conveyer belts in isolation, and within a few years, in endogenous fungal proteomes.
Keywords: fungal metabolism, nonribosomal peptide synthetases, polyketide synthases, thiotemplate biosynthesis, secondary metabolism, proteomics, mass spectrometry, Fourier-Transform mass spectrometry
1.0 Introduction
The sustained examination of medicinally relevant natural products and their biosyntheses is driven by the need for new medicines that will selectively inhibit targets implicated in human disease. Drug discovery and synthesis are often inspired by new molecular scaffolds discovered in nature that can be used to probe and provide new information about biological targets. In an effort to maximize the diversity of natural compounds isolated and to minimize the isolation of well-studied compounds, fungal systems offer a significant jump from the `simpler' world of natural product discovery in bacteria. Highly potent fungal molecules such as lovastatin, aflatoxin and fuminosin are reviewed, along with protein-based analysis techniques using mass spectrometry (MS) to evaluate their biosynthetic proteins and/or their small molecule products. Along with facile genome sequencing, such tools promise to increase the rate at which we can access and understand the ultra-large genes involved in production of non-ribosomal peptides and polyketides.
Biosynthetic products span from essential primary metabolites such as fatty acids that make up the cell membrane to polyketides (PKs) and nonribosomal peptides (NRPs) that constitute pigments, antibiotics and siderophores. An often exploited mechanism of natural product genesis is thiotemplate biosynthesis, which allows for indirect encoding of ketide and peptide metabolite biosynthesis by a protein template. Thiotemplating refers to the covalent tethering of intermediates to a unique cofactor, 4'-phosphopantetheine, through a labile thioester bond. These tethered intermediates are arranged along an assembly line of enzymes that are usually a multi-active site, mega-enzyme (type I), a series of interacting enzymes (type II), or (for polyketide synthases (PKSs)) a single active site enzyme that acts repeatedly to form the final product (type III). The assembly lines of nonribosomal peptide synthetases (NRPSs) can be further subdivided into linear (a), iterative (b) and non-linear (c) [1]. The fungal type I iterative PKSs (IPKSs) are classified as either highly-reducing (hr), partially reducing (pr), or non-reducing (nr) [2]. This assembly line style of production lends itself to study by large molecule MS to characterize the proteins involved in the biosynthesis as well as the covalently-attached intermediate at each biosynthetic step, when recombinant proteins are available and activity reconstituted.
Regardless of type, enzymes involved in thiotemplate biosynthesis have enormous primary structures and can form some of the largest protein complexes within cells, sometimes larger than the ribosome [3]. The logic of thiotemplating offers two advantages – 1) substrate/intermediate shuttling through the pathway – intermediates cannot diffuse away, 2) potentially toxic intermediates are not allowed to affect the producing organism – a mechanism of host resistance. In short, thiotemplate biosynthesis enables efficient and safe biosynthesis of highly active secondary metabolites. Substrates are activated for entry into thiotemplate pathways by acyl-CoA ligases, acyl transferases (fatty acids and polyketides), or adenylation enzymes (nonribosomal peptides). The energy of activation of each monomer is conserved throughout the pathway by transthioesterification along the assembly line until the final product is released. Biosynthetic mechanisms of NRPs and PKs share many common elements, most important covalent tethering of intermediates to the assembly line. This commonality allows for techniques specifically targeting this quality to be applied generally to thiotemplate systems. Thiotemplate biosynthesis has been studied extensively in bacteria and through this work many techniques have been developed that are applicable to fungal systems. Fungal systems have historically been difficult to study in vitro due to problems with cloning and expression of the large and sometimes intron containing genes and for this reason, progress in the fungal world has lagged behind that of bacterial counterparts. Progress on that front has been reviewed elsewhere [4]. For this reason we first review the progress made on bacterial thiotemplate systems before moving to the more recent advances in fungal systems.
2.0 Mechanisms of Biosynthesis
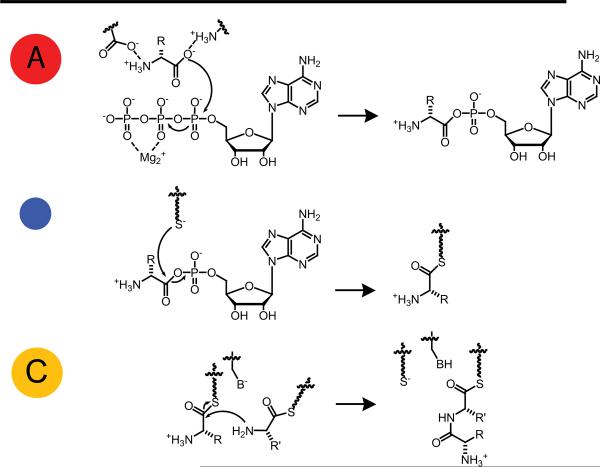
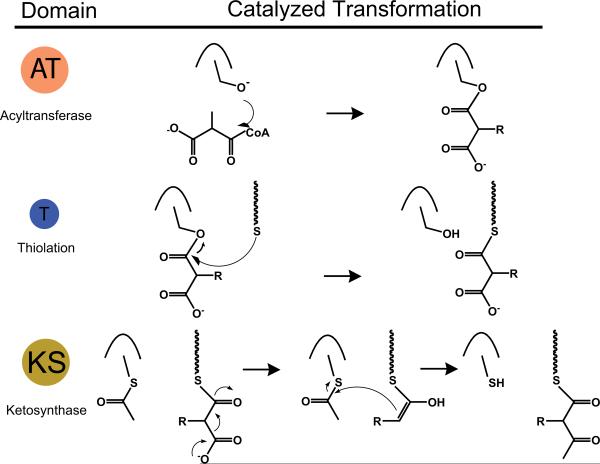
For those familiar with NRPSs and PKSs, we direct you to the text below straight away; for those interested in some background reading, please see Appendix #1. The basic enzymatic functions at the core of NRPSs – adenylation (A), thiolation (T) and condensation (C) are displayed in Fig. 1, the basic functions of PKSs – acyl transferase (AT), thiolation (T) and ketosynthase (KS) are displayed in Fig. 2.
Fig. 1.
Basic enzymatic domains and their reactions for non-ribosomal peptide synthesis; A: adenylation, T: thiolation, and C: condensation. Non-experts can refer to Appendix #1 for detailed descriptions of function.
Fig. 2.
Basic enzymatic domains and their reactions for polyketide synthesis; AT: acyltransferase, T: thiolation, and KS: ketosynthase. Non-experts can refer to Appendix #1 for detailed descriptions of function.
2.1 NRP Biosynthesis
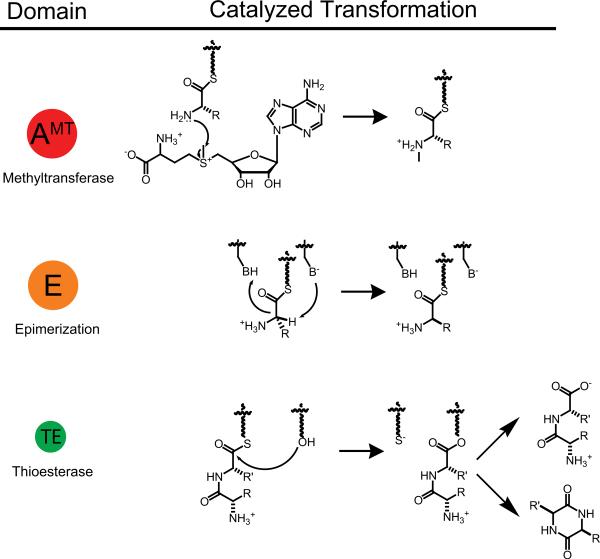
In addition to these three basic functions for peptide elongation in NRPSs, product release can be achieved by hydrolysis of the last thioester with or without macrocyclization [5] as seen in Fig. 3. Also there are several other tailoring activities in NRPS assembly lines, as seen in Fig. 3. These include C- and N-methyltransferases, N-formyltransferases, amino transferases, L-D epimerases, oxidases, reductases, cyclases, and halogenases among others [6]. NRPS pathways may also be integrated into hybrid NRPS-PKS pathways, as seen with emericellamide [7], and the final products may be decorated with lipid and/or carbohydrate moieties. Given these extensive biotransformations in NRPSs, the peptide products exhibit a diverse array of structures and biological effects.
Fig. 3.
Some typical NRPS tailoring domains and their catalyzed reactions; AMT: N-methyl transferase (preferentially found in fungi), E: epimerase, and TE: thioesterase.
2.2 PK Biosynthesis
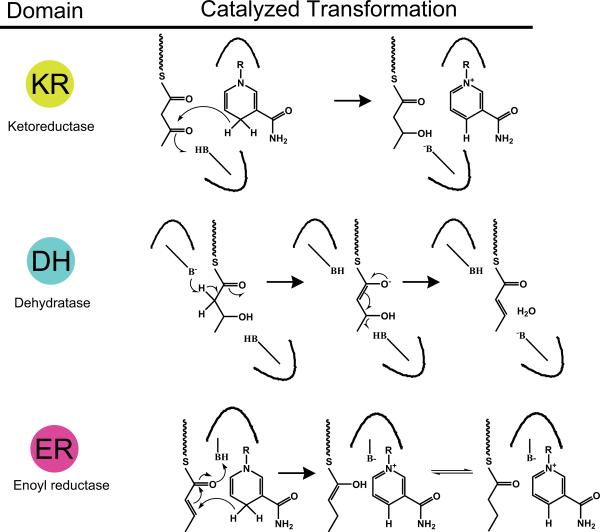
PKs, unlike NRPs must undergo extensive on assembly-line transformations in order to achieve complex structural content. The presence of the optional ketoreductase (KR), dehydratase (DH) and enoyl reductase (ER) functions after initial activation allows for reduced C-C bonds and chemical stability for linear or macrocyclic PKs (Fig. 4). Stereocenters introduced by the action of KRs that are not further reduced by DH and/or ER enzymes contribute to the diversity in structure PK products possess [8]. Absence of theses reducing functions results in highly reactive poly-carbonyl backbones that undergo aromatization and cyclization guided by product template (PT) domains [9,10]. C-methyl transferases (C-MT) and utilization of alternate acyl-CoAs such as methyl-malonyl- CoA, results in addition of branched structures to the backbone. Additionally, like NRPs, PKs can undergo post assembly line tailoring by O-methyl transferases, glycosylases and others [11].
Fig. 4.
Some typical PKS tailoring domains and their catalyzed reactions; KR: ketoreductase, DH: dehydratase, and ER: enoyl reductase. R = ribose-ADP(P). The KR and ER domains are NADPH dependent enzymes.
2.3 Differences Between Fungal and Bacterial Thiotemplate Pathways
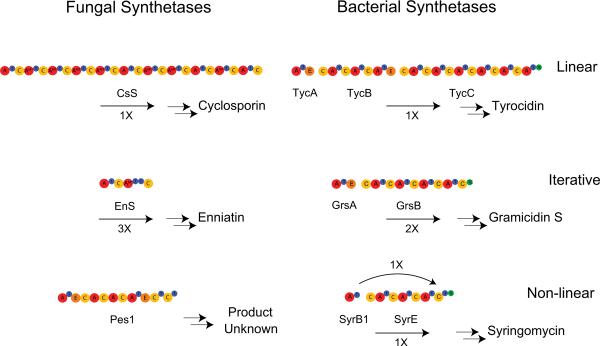
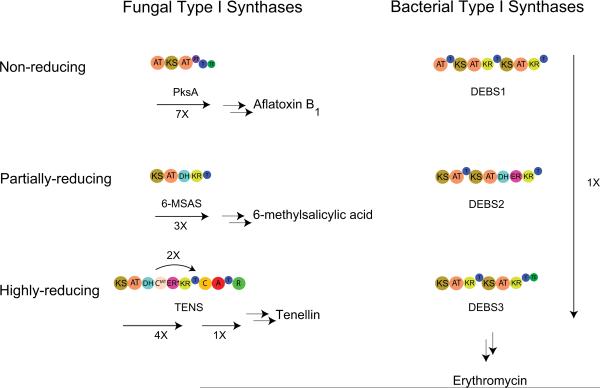
While there are no well-defined rules regarding differences between fungal and bacterial NRPSs and PKSs, there are some general trends that distinguish them (Fig. 5, 6). First, fungal pathways tend to be mostly large megasynthetases (type I) while bacterial pathways tend more toward modularity (type II). Bacterial NRPSs generally terminate by the action of a thioesterase (TE) domain whereas fungal NRPSs tend to terminate by the action of a terminal condensation domain. Another distinguishing characteristic of fungal NRPSs is that many fungal A-domains have an inserted N-methyltransferase domain [12]. Fungal type I PKSs tend to follow the iterative model; each domain acts repeatedly activating and modifying the same monomers. Serial use of the same set of enzymatic functions can result in polyketides dominated by one structural motif; these polyketides are characterized as non-reduced or highly reduced due the presence or absence of PKS reducing functions in the synthase. In addition, presence of KR or KR and DH domains (but not ER domains) results in an intermediate class of partially reduced polyketides. Bacterial type I PKSs act modularly; each domain acts once. Large polyketides require multiple large enzymes for biosynthesis in bacteria, because each domain functions once in the bacterial systems. A benefit of modularity is it allows for mixing of non-reducing and reducing modules. Both bacteria and fungi have numerous hybrid NRPS-PKS pathways. Both fungal and bacterial pathways are often clustered together on the genome, but fungal pathways may be split onto separate chromosomes [13]. Generally, fungal thiotemplate genes do not contain introns or only contain a few introns, however as more fungal genomes are sequenced it has become apparent this is not always the case [14].
Fig. 5.
Overview of fungal vs. bacterial NRPSs. NRPSs can be divided into three types, linear (a), iterative (b) and non-linear (c). Fungal NRPSs generally follow a single megasynthetase logic, whereas bacterial NRPS pathways are generally spread over a greater number of polypeptides. Fungal NRPSs are also rich in A-MT domains, a feature rarely seen in bacterial NRPSs. CsS: cyclosporine synthetase; EnS: enniatin synthetase; Pes1: synthetase encoded by pes1, TycA, B, C: tyrocidine synthetase A, B, C; GrsA, B: gramicidin S synthetase A, B; SyrB1, E: syringomycin synthetase B1, F.
Fig. 6.
Overview of fungal vs. bacterial type I PKSs. Type I PKSs can be divided into three types, non-reducing (nr), partially-reducing (pr) and highly-reducing (hr). Fungal type I PKSs generally follow an iterative logic using a single AT, KS, and T iteratively, whereas bacterial type I PKS use modules of PKS machinery sequentially. PksA, polyketide synthase A from aflatoxin biosynthesis; 6-MSAS, 6-methyl salicylic acid cynthase; TENS, tenellin synthase; DEBS, 6-deoxyerythronolide B synthase.
2.4 Fungal PKs
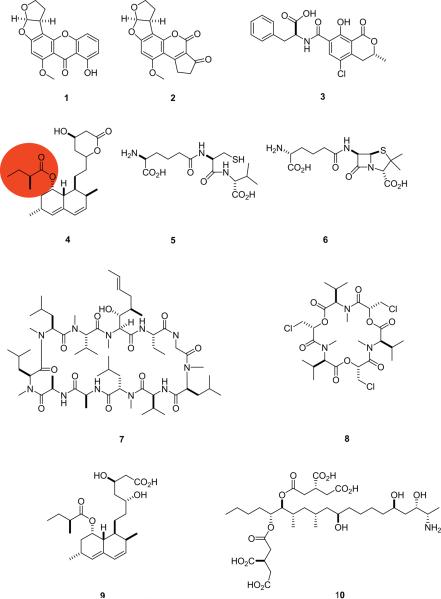
As described above, polyketides are synthesized by acetyl building blocks, with common variation established by methylation, oxidation, and reduction. Commonality between NRPs and PKs is the compelling range of bioactivities they exhibit. One of the most broadly studied, and most potent mycotoxic compound classes are the aflatoxins. Aflatoxins are isolated from A. nomius, A. tamari, A. flavus and A. parasiticus and share an analogous biosynthetic route with A. nidulans derived sterigmatocystin (1) (Fig. 7). The potent liver carcinogenic properties exhibited by 1 and the aflatoxins have impelled the review and publication of a significant body of literature [15–19]. In terms of the tailoring potential of fungal PKs, both aflatoxin B1 (2) and compound 1 utilize the above mentioned tailoring domains to derivatize the starter, fatty acid synthase derived, hexanoic acid. Significant protein study has characterized the role of a pathway-specific Zn2Cys6 transcription activator that is common to both 1 and 2, but not a necessary characteristic of these biosynthetic pathways [20]. This Zn2Cys6 transcription factor (AflR) was the first pathway-specific fungal natural product formation factor identified. A second mycotoxic compound of mixed NRPS/PKS biogenesis is ochratoxin A (3), produced by both Penicilli and Aspergilli generas. This unique dihydroisocoumarin phenylalanine compound has been studied relative to its B analogue (Cl in 3 is replaced with H in ochratoxin B) and determined that chlorination is the penultimate biosynthetic step [21]. In addition to the negative effects induced by ochratoxin and aflatoxin compound classes, numerous other PKs including T-toxins [22], fumonisins [23], citrinin [24], aurofusarin [25], patulin [26], and dothistromin [27] are closely regulated and actively studied to limit exposure and to prevent severe mycotoxic effects. In addition to their negative mycotoxic properties, fungal PKs have been flagged as a notable source of medicinally applicable natural products. Most noteworthy is the lovastatin/compactin compound class. Lovastatin (4) has been commercially developed as a hypolipidemic drug via HMG-CoA reductase inhibition. In summary, fungal-derived PKs induce a broad range of harmful and beneficial biological responses and continue to drive the future studies of this principal compound class.
Fig. 7.
Some fungal- and bacterial-derived compounds, 1 –10.
2.5 Fungal NRPs
Fungal-derived NRPs exhibit a number of pharmacologically relevant properties including both antibiotic (penicillins) and possible anticancer agents such as gliotoxin [28] as well as those like HC-toxin exhibiting destructive mycotoxic properties [29]. The significant bioactivity of these compounds prompts elucidation of their biosynthetic mechanisms. Prominent NRPS systems in fungi are now well-understood including the β-lactam antibiotics: penicillins and cephalosporins. Both compound classes share initial linear tripeptide biosynthesis of ACV (5) from L-α-aminoadipic acid, L-cysteine and L-valine (the latter epimerized to D-valine). Mechanistic studies have confirmed a two-step β-lactam formation which precedes thiazolidine ring formation as seen in penicillin N (6, [30]). Diverse biosynthetic routes ensue from compound 6 to the resulting penicillin or cephalosporin analogues. An equally significant fungal NRP that spurred extensive biosynthetic study was cyclosporin A (7), an immunosuppressant used to prevent allotransplantation rejection under the trademarked name Sandimmune®. Early evaluation of the undecapeptide gene cluster of 7 led to isolation of one enzyme responsible for adenylation and N-methylation of seven out of 11 amino acids, later designated cyclosporin synthetase [31]. This was not only one of the earliest fungal NRPS breakthroughs, but also one of the largest synthetases identified (1.6 MDa) and demonstrated the extensive degree of N-methylation exhibited in fungal NRPs. Beyond peptide diversity induced by tailoring activities, the use of domains or modules more than once in an assembly line adds variation to linear fungal derived NRPs. One example of this that led to an early understanding of iterative biosynthesis was ennaitin A (8), a cyclohexadepsipeptide consisting of D-2-hydroxyisovaleryl – N-methyl-L-leucine [32]. Enniatin A (8) is biosynthesized in three iterative condensations of dipeptidol building blocks [33,34] with an intramolecular reaction mechanism [35] forming 8. Together, compounds 1 –10 (Fig. 7) represent variations in the potential of fungi to produce pharmacologically relevant and biosynthetically diverse natural products.
Recent advances in mass spectrometry for protein characterization have propelled biochemical analysis of thiotemplate systems. Mass spectrometry and activity-based profiling approaches have been primarily applied to the investigation of bacterial systems, but there have been three recent and high profile examples of mass spectrometry being used to study important fungal systems, lovastatin, aflatoxin and fumonisin.
3. Protein Level Analysis of Thiotemplate Enzymes
3.1 Thiotemplate protein studies in bacteria
The path to implementation of proteomic-like techniques with fungi builds from bioinformatic, genomic and mass spectrometric methods already established for bacteria. Bioinformatic analyses often provide successful prediction of adenylation domain specificity; however, in vitro analysis should always be performed to fully characterize a biosynthetic pathway (particularly for orphan gene clusters where the metabolite produced is unknown). The traditional methods used to analyze substrate specificity of adenylation domains have relied on the pyrophosphate exchange assay [36–38]. While these methods are sensitive, they cannot be used to determine substrate specificity of adenylation domains in a complex or undefined mixture such as the native metabolome. In addition, analysis may be hampered by unavailability of radiolabeled substrates. Radiolabeling also has a deficit in that it does not report on any further transformations to the substrate once tethered to the carrier protein by the activating module or other enzymes present in a reaction mixture. The ability to rapidly and accurately analyze the intermediates of a thiotemplate pathway is of particular importance for fungal systems because the current models of adenylation domain specificity do not include a well-defined fungal nonribosomal code [39].
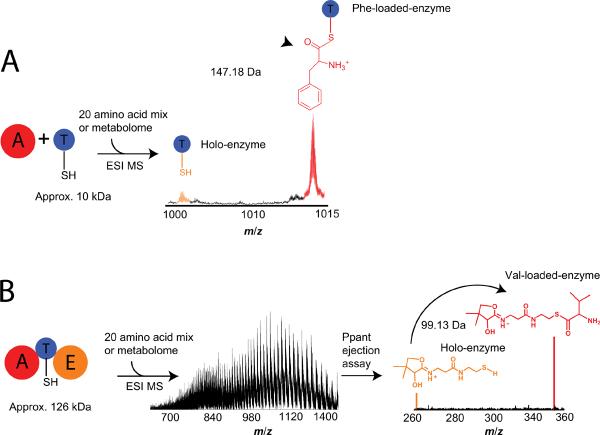
A bacterial example of domain specificity investigation by Dorrestein, et al. [40] included the presentation of nine adenylation-carrier protein di-domain proteins with a pool of substrates of defined content in addition to a full metabolome, followed by identification of intermediates in seven pathways. An example of the workflow is shown in Fig. 8A. Interpretation of specificity was accomplished using high resolution mass spectrometry for proteins or peptides with covalently tethered phosphopantetheine (Ppant) bound intermediates and discrimination between potential intermediates differing in mass by as little as two daltons. Due to the labile nature of the Ppant arm, selective ejection of the complex from the peptide or enzyme containing it in the mass spectrometer is used in a technique known as the “Ppant ejection assay” [41]. Using MS, gentle fragmentation of a mixture containing phosphopantheinylated peptides leads to identification of T-domain active sites and the identity of the intermediate(s) bound to them. Since the focus of analysis is now the small molecule cofactor and bound intermediate(s), accurate precursor mass assignment is no longer a prerequisite for accurately determining the identity of the bound intermediate (Fig. 8B). Dorrestein, et al. pioneered this technique for in vitro characterization by partially elucidating seven systems and extending the analysis to a 126 kDa protein and its bound intermediate. When performed on a Fourier-Transform mass spectrometer (FTMS), the Ppant ejection assay discerns between intermediates differing by one dalton as exemplified by the discrimination between acetoacetate and β-aminobutyrate loaded carrier proteins from the mycosubtilin pathway of B. subtilis [42].
Fig. 8.
Overview of mass spectrometric techniques for analyzing activity in vitro. A) The activity based screening method is an in vitro assay for determining substrate specificity of NRPSs using a complex substrate pool and an accurate mass shift (Δm) as the readout. FTMS enables resolution and accurate assignment of covalently-bound intermediates. B) The Ppant ejection assay simplifies complex mass spectra of peptides or intact proteins by selectively ejecting phosphopantetheine and phosphopantetheine bound intermediates allowing for correct intermediate assignment even at high intact mass.
In addition to determining substrate specificity of activating domains, the Ppant ejection assay can be used to monitor transformations occurring to the covalently bound substrate. Dorrestein, et al. were able to monitor the dehydration and subsequent rehydration of a polyketide intermediate in vitro. Notable was the use of a bench-top mass spectrometer, demonstrating how this type of analysis can be performed in laboratories with access to standard mass spectrometers. The ability of the Ppant ejection assay to shift analysis towards low mass is significant because at masses above 70 kDa or so, intact mass alone cannot discriminate between intermediates that are close (<2 Da) in mass due to complex isotoptic distributions and low signals at high MW. Lee, et al. extended analysis in this high mass regime to determine the substrate specificity of a 108 kDa synthetase in a multiplexed assay [43]. Since 2006, about a dozen labs have employed the Ppant ejection assay using a variety of mass spectrometers to characterize thiotemplate systems in vitro [44–49].
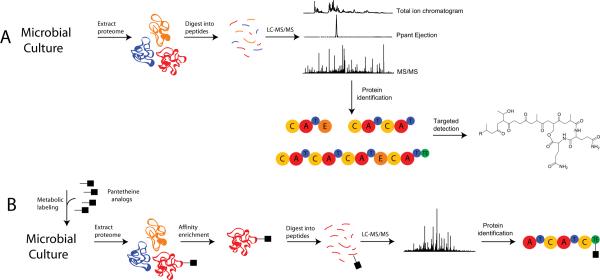
Mass spectrometric tools for the in vitro characterization of recombinantly produced enzymes are now well developed. The advent of a technique called PrISM (for Proteomic Investigation of Secondary Metabolism, see Fig. 9A) now enables the investigation of thiotemplate systems closer to the in vivo level [50]. Bumpus, et al. have shown that identification of T-domain active site peptides can be carried out on crude lysates of recombinantly produced enzymes in addition to full proteomic mixtures of bacterial enzymes at the native expression level. Indeed, the Ppant ejection assay can be used to accurately identify phosphopantetheinylated peptides in full proteome-scale digests. The Ppant ejection assay is very robust because the mass of the Ppant ion is measured so accurately (i.e., <2 part-per-million error) that the detection of thiotemplate biosynthetic pathways can be highly selective. These features allow for the Ppant ejection assay to be used in the PrISM workflow for monitoring expression of targeted systems and for the discovery of thiotemplate systems (even with no genome sequence). PrISM has been demonstrated in the proteome of an unsequenced Bacillus for the identification of a NRPS and hybrid NRPS-PKS gene clusters and association of the small molecule products to their biosynthetic enzymes. This study led to the identification of a new kurstakin lipopeptide.
Fig. 9.
Overview of proteomic techniques for detecting the expression of thiotemplate enzymes in vivo. A) The PrISM workflow incorporates the “Ppant ejection assay” for the selective identification of thiotemplate enzymes. The workflow utilizes tandem mass spectrometric data of thiotemplate enzymes as a starting point for the cloning and analysis of thiotemplate pathways. B) OASIS utilizes thiotemplate-specific metabolic labeling with affinity tags to allow for enrichment and identification of thiotemplate pathways.
Labeling of carrier proteins by in vivo phosphopantetheinylation or metabolic incorporation of CoA analogues is an approach complementary to PrISM. La Clair et al. synthesized a variety of CoA analogues for chemoenzymatic labeling of carrier proteins using the promiscuous phosphopantetheinyl transferase Sfp from B. subtilis [51]. Chemical conjugation of fluorescent probes to CoA enabled quantitative and qualitative assessment of in vivo phosphopantetheinylation as well as detection of carrier protein containing recombinant enzymes at levels below that detectable by standard SDS-PAGE analysis. Biotinylated CoA analogues were also synthesized and used to pull down nondenatured carrier protein containing enzymes. Meier et al. extended this work by synthesizing a collection of bioorthogonal panthetheine analogues for in vivo labeling of carrier proteins [52]. Advantages of moving towards using smaller panthetheine analogues as opposed to CoA analogues are ease of synthesis, stability of the compounds and increased efficiency of metabolic incorporation due to easier transport across membranes. The panthetheine analogues synthesized were shown to be incorporated into CoA analogues both in the in vitro reconstituted CoA biosynthetic pathway as well as into the endogenous CoA biosynthetic pathway of E. coli. The authors used the in vitro and in vivo synthesized CoA analogues to successfully modify VibB from the vibriobactin pathway of V. cholerae recombinantly expressed in E. coli with purified or co-expressed Sfp. In a follow-up study, Meier et al. synthesized fluorescent acyl transferase, ketosynthase and thioesterase activity based probes and combined them with previously synthesized carrier protein probes [53]. The combined set of probes was shown to be active in vivo for labeling a purified PKS (PikAIII, pikromycin from S. venezuale) and a NRPS TE domain (Tyc-TE, tyrocidine from B. brevis). The probe set was used in vivo to profile fatty acid biosynthesis proteins from cultured human cells and SrfAC, an NRPS involved in surfactin biosynthesis in B. subtilis. These kind of reagents have helped established a new method termed OASIS (for Orthogonal Active Site Identification System, Fig. 9B), which is a new method that couples the metabolic labeling of thiotemplate enzymes with mass spectrometry based proteomics [54]. OASIS was successful in enriching, identifying and profiling proteins of all four NRPS and PKS pathways from B. subtilis, and holds promise for discovery of thiotemplate systems in unsequenced genomes. In conclusion, a number of proteomic techniques have been developed and used to identify mechanisms of bacterial NRPS biosynthesis.
3.2 Thiotemplate protein studies in fungi
The techniques mentioned above have been used to elucidate fungal PKS pathways for two extensively studied compound classes: aflatoxins and lovastatins. Aflatoxins are one of the most studied compound classes produced by A. flavus and A. parasiticus due to their carcinogenic and toxic properties. The biosynthesis of aflatoxins employs a multidomain nonreducing iterative polyketide synthase (IPKS). The biosynthetic pathway has been well studied [55,56], but information regarding individual domains within the aflatoxin IPKS (PksA) has been limited. Most recently, a deconstructive approach to PksA of aflatoxin B1 (2) using heroic efforts for cloning and expression, followed by LC-FTMS resulted in characterization of individual domains and the advanced intermediates assembled within each domain [10]. The relative abundance of seven acylated intermediates, extended in a processive fashion, was established by FTMS in the absence and presence of a product template (PT) domain, which is common among nonreducing IPKSs but functionally ill-defined prior to this report [9]. Specifically, the PT domain unites with ketosynthase and thioesterase domains to irreversibly drive formation of the correct cyclization, aromatization or dehydration product [10]. The use of tandem mass spectrometry (or MSn) for structural evaluation of sub-stoichiometric intermediates also confirmed the architecture and length of ring systems constructed due to the fragmentation of linear units surrounding rings, without intra-ring fragmentation.
Lovastatins from A. terreus and other fungi have been extensively studied and developed for their hypolipidemic properties, producing a great depth of knowledge regarding their biosynthesis. One relatively understudied mechanism was the release of lovastatin, which does not proceed via the expected off-loading TE domain. Therefore, Xie et al. examined the rate of lovastatin acid (9) formation/off-loading by LC/MS to determine the role of two interacting domains LovF and LovD. Previous work by Kennedy et al. determined the biosynthetic product of Lov domains up to LovD was monacolin J, which is the final biosynthetic product in the absence of the final domain, LovF [57]. In the presence of LovF, the α-S-methylbutyrate side chain (highlighted) of 9 was added, which led to completion of 9. To further probe this relationship, Xie et al. used the Ppant ejection assay (vide supra) to confirm the localization of the α-S-methylbutyrate side chain of 6 on LovF (345.1843 Da, Pant arm plus α-methylbutyrate). Next, monacolin J and LovD were added to α-methylbutyrate-loaded LovF and transfer of the diketide side chain from LovF to monacolin J was catalyzed by a dissociated acyltransferase of LovD, resulting in 9. Further, LC-MS was used to evaluate the rate of formation of compound 9 as well as the presence of lovastatin analogues when specific cofactors were excluded [58]. Another thorough study involved the reconstitution of the lovastatin nonaketide synthase LovB and its partner enoyl reductase enzyme LovC and provided the basis for rules dictating highly reducing IPKSs [59]. Ma et al. engineered a S. cerevisiae strain to efficiently express both LovB and LovC domains that were used for complete characterization of their synthetic processes until completion of dihydromonacolin L. MS techniques, including the Ppant ejection assay, were used to determine the identity and level of phosphopantetheinylation of the LovB domain and to monitor and verify intermediate formation [59].
In addition to these lovastatin and aflatoxin studies, a study of fumonisin (10) biosynthesis, from Fusarium also utilized MS product confirmation. Selective alkyl chain addition to the ACP domain was monitored via Ppant ejection, and the mechanism of product release was detrmined to be pyridoxal 5' phosphate (PLP) dependent. Termination was identified to proceed via PLP dependent decarboxylative condensation of L-alanine and acyl-S-ACP with greatest product formation observed for C18-S-ACP. This study by Gerber et al. discloses a new highly reducing IPKS termination mechanism catalyzed by non-standard thioesterase/cyclase chain release enzymes [60]. Utilizing contemporary mass spectrometry, this study contributes to the specific knowledge of fumonisin biosynthesis and the general mechanisms possible for thiotemplate transformations.
4.0 Outlook
These recent studies of fungal PKS proteins are stepping stones towards the implementation of the aforementioned proteomics approaches to natural product elucidation techniques. Mechanism of compound formation as well as early previews of compound structures can be accessed through measurements of biosynthetic proteins directly. Previous challenges including fungal protein purification, isolation, identification and quantitation as well as cell lysis have been recently reviewed with the most efficient techniques to overcome these challenges presented elsewhere [61]. With proteomics maturing rapidly for full proteome coverage, thought now to be defined as detecting over 4000 of the ~6300 genes in the fungal proteomic-benchmark system, S. cerevisiae [62], it is time to apply this expanded tool set to understanding secondary metabolism in fungi.
One example where the reviewed proteomics tools could be put to good use is in the connection of secondary metabolites with their respective biosynthetic genes. For example, A. nidulans is known to produce 22 NRP and PK products, but has the genetic capacity for even more. Analysis of the A. nidulans FGSC A4 genome reveals 26 PKSs and 24 NRPSs [63]. Thus far only 6 PKSs responsible for biosynthesis of monodictyphenone, asperfuranone, orsellinic acid/F9775, asperthicin, napthopyrone, and sterigmatocystin, [64–69] and 4 NRPSs responsible for the biosynthesis of terrequinone, aspyridones, emericellamide and pennicillin [7,70–72] of A. nidulans have been characterized, although it is clear from the detection and characterization of numerous peptide and polyketide products that many more can be expressed. These compounds along with all other compounds covered in this review, their source and bioactivity are listed in Table 1. It is estimated that there are roughly 1.5 million species of fungi [73], but only a small fraction have been sampled by either genomic or proteomic techniques; clearly there is a wealth of fungal biodiversity to be discovered, elucidated and exploited. With the expectation that many if not most of the 1.5 million fungal species will be unculturable, we can expect that metaproteomics [74] will play an eventual role in teasing out the important and complex biosynthetic pathways of fungi.
Table 1.
Compounds covered in this review
| Compound | Source | Type | Bioactivity | Reference |
|---|---|---|---|---|
| aflatoxins | fungal | polyketide | toxin | [15–19,55,56] |
| sterigmatocystin | fungal | polyketide | toxin | [69] |
| ochratoxin | fungal | polyketide | toxin | [21] |
| T-toxins | fungal | polyketide | toxin | [22] |
| fumonisins | fungal | polyketide | toxin | [23,60] |
| citrinin | fungal | polyketide | toxin | [24] |
| aurofusarin | fungal | polyketide | toxin | [25] |
| patulin | fungal | polyketide | toxin | [26] |
| dothistromin | fungal | polyketide | toxin | [27] |
| lovastatin | fungal | polyketide | hypolipidemic | [57–59] |
| compactin | fungal | polyketide | hypolipidemic | [75] |
| penicillins | fungal/bacterial | peptide | antibacterial | [30,72,76] |
| gliotoxin | fungal | polyketide | toxin | [28] |
| HC-toxin | fungal | peptide | toxin | [29] |
| cephalosporins | fungal/bacterial | peptide | antibacterial | [76] |
| cyclosporin A | fungal | peptide | immunosuppressant | [31] |
| ennaitin A | fungal | peptide | toxin | [32–35] |
| mycosubtilin | bacterial | peptide | antifungal | [42] |
| kurstakin | bacterial | peptide | antifungal | [50] |
| vibriobactin | bacterial | peptide | siderophore | [52] |
| pikromycin | bacterial | polyketide | antibacterial | [53] |
| tyrocidin | bacterial | peptide | antibacterial | [53] |
| surfactin | bacterial | peptide | antibacterial/antifungal | [53] |
| monodictyphenone | fungal | polyketide | unknown | [64] |
| asperfuranone | fungal | polyketide | antitumor | [65] |
| orsellinic acid/F9775 | fungal | polyketide | protease inhibitor | [66] |
| asperthicin | fungal | polyketide | toxin | [67] |
| napthopyrone | fungal | polyketide | pigment | [68] |
| terrequinone | fungal | peptide | antitumor | [70] |
| aspyridones | fungal | hybrid | toxin | [71] |
| emericellamide | fungal | hybrid | antibacterial | [7] |
5.0 Acknowledgements
The authors thank the National Institutes of Health (GM 067725) for support of our work in natural products discovery and biosynthesis.
Appendix #1
A domain set of A-T-C comprises one module of a NRPS and contains all the functions necessary for incorporation of one monomer [77]. Adenylation domains are the “gatekeepers” of the NRPS assembly line. These enzymes are composed of ~500 amino acids and belong to the AMP-binding super-family of enzymes. Entry of monomers is regulated by specific domains for each monomer and requires ATP for activation. A-domains contain a conserved D-K pair that is responsible for recognition of the amino and carboxyl ends of an amino acid and for coordination of the Mg2+ and ATP cofactors. Substrate specificity of the A-domain is derived from the residues lining the amino acid binding pocket. Well-characterized systems can be used to predict the specificity of uncharacterized A-domains, which is a driving force behind proteomic based NRPS elucidations [1,78,79]. The adenylation domain catalyzes adenylation of the amino acid substrate carboxyl with AMP. The aminoacyl adenylate intermediate is short-lived and is a substrate for a transesterification to the free thiol of the T-domain active site.
The smallest domains, referred to as thiolation or just carrier domains, are ~100 amino acid stretches that carry intermediates and owe their name to the free thiol of their phosphopantetheine (Ppant) cofactor. The T-domain active site is well conserved and contains the GG(H/D)SL phosphopantetheine attachment site motif. The sole function of these domains is to covalently anchor ~20 Å swinging arm that can move from active site to active site shuttling intermediates along the assembly line. The Ppant cofactor is derived from CoA and is attached to the T-domain active site serine through a phosphopantetheinyl transferase most often acting in trans. Condensation domains are composed of ~500 amino acids and catalyze amide bond formation between intermediates on adjacent T-domains. These enzymes carry a signature HHXXXDG motif essential for catalysis. The second H in the strictly conserved motif is proposed to activate the acceptor amino group so that it can act as a nucleophile and attack the donor thioester. The donor T-domain is now available for recharging by its A-domain and the newly formed dipeptide is ready for progression down the assembly line. Peptide bond formation proceeds in this fashion until the end of the last module is reached. Product release can be achieved by hydrolysis of the last thioester with or without macrocyclization [5] as seen in Fig. 2.
PKS core activities of monomer selection (AT), tethering (T, acyl carrier protein (ACP)), and condensation (KS) functions are analogous to the A-, T-, and C- domains of NRPSs and highly related to the AT- ACP- and KS- domains involved in fatty acid synthesis [80]. AT domains are ~45 kDa enzymes that select acyl-CoA starter and extender units for chain initiation and elongation [81]. The AT-domains attack the starter and extender CoA thioester bonds with a conserved serine nucleophile forming a transient covalent intermediate that is the substrate for KSs (for initiation) and ACPs (for elongation) [82]. The T-domains of PKSs are very similar to those of NRPSs and are approximately the same size, 8–10 kDa. The KS is a ~50 kDa enzyme that catalyzes Claisen condensation between a T-domain and KS-domain bound substrates using a conserved cysteine residue [83]. In addition to these minimal functions, PKSs may also include ketoreductase (KR), dehydratase (DH) and enoyl reductase (ER), cyclase (Cyc), thioesterase (TE), C-methyl transferase (C-MT) activities, among others [84]. The diversity in PK structure and activity relies on stereocenters introduced by KRs, degree of reduction, cyclization, aromatization and decoration of the ketide core as opposed to NRPs that owe most of their structural diversity to monomer selection. PKSs can be categorized into three different types, type I, type II and type III, although blurring between the categories has become evident from the growing wealth of DNA sequence [85]. Enzymatic functions are shared between the types; however quaternary structure and chain initiation and termination are different. Type I PKSs are monolithic enzymes comprised of multiple active sites along one polypeptide chain. Type II PKSs are a set of discrete enzymes that act iteratively to produce a polyketide. Type II PKSs utilize stand- alone T-domains. In addition they minimally consist of a KS and chain-length factor (CLF) that are typically encoded by distinct genes. The CLF is an inactive KS enzyme that provides a cavity for the nascent ketide chain to extend into, the depth of which governs ultimate chain length [86]. Type III PKSs use acyl-CoAs but no T-domain and have a single KS which initiates, tethers, extends and terminates the polyketide chain. They function as homodimers and are roughly ~45 kDa in size. Type III PKSs are typically thought of as plant enzymes; however, there are examples of bacterial and fungal type III PKSs [87,88]. For fungi, the most commonly used mode is the type I PKS. In fungi, unlike bacteria, the type I PKSs use a single KS-AT-T module repeatedly and are hence referred to as type I iterative PKSs (IPKSs). These IPKSs can be further subdivided into non-reducing (nr), partially-reducing (pr) and highly-reducing (hr) IPKSs [89]. Theses qualifications reflect the presence or absence of PKS reducing functions, the KR, DH and ER domains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Simpson TJ. Complex enzymes in microbial natural product biosynthesis, part B: polyketides, aminocoumarins and carbohydrates. Chapter 3 Fungal Type I Polyketide Synthases Methods Enzymol. 2009;459:49–98. doi: 10.1016/S0076-6879(09)04625-4. [DOI] [PubMed] [Google Scholar]
- Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J, Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol. 2006;124:690–703. doi: 10.1016/j.jbiotec.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Trauger JW, Schwarzer D, Marahiel MA, Walsh CT. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry. 2001;40:7099–7108. doi: 10.1021/bi010036j. [DOI] [PubMed] [Google Scholar]
- Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: from genes to products. Nat Prod Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CC. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rix U, Fischer C, Remsing LL, Rohr J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat Prod Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- Hornbogen T, Riechers SP, Prinz B, Schultchen J, Lang C, Schmidt S, Mugge C, Turkanovic S, Sussmuth RD, Tauberger E, Zocher R. Functional characterization of the recombinant N-methyltransferase domain from the multienzyme enniatin synthetase. Chembiochem. 2007;8:1048–1054. doi: 10.1002/cbic.200700076. [DOI] [PubMed] [Google Scholar]
- von Dohren H. Biochemistry and general genetics of nonribosomal peptide synthetases in fungi. Adv Biochem Eng Biotechnol. 2004;88:217–264. doi: 10.1007/b99262. [DOI] [PubMed] [Google Scholar]
- Eisfeld K. Non-Ribosomal Peptide Synthetases of Fungi. In: Weber T. A. a. D., editor. Physiology and Genetics: Selected Basic and Applied Aspects. Springer; Heidelberg: 2009. pp. 305–330. [Google Scholar]
- Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Woloshuk CP, Prieto R. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol Lett. 1998;160:169–176. doi: 10.1111/j.1574-6968.1998.tb12907.x. [DOI] [PubMed] [Google Scholar]
- Yabe K, Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2004;64:745–755. doi: 10.1007/s00253-004-1566-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Cary JW, Bhatnagar D, Cleveland TE, Bennett JW, Linz JE, Woloshuk CP, Payne GA. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl Environ Microbiol. 1993;59:3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JP, Mantle PG. Biosynthesis of ochratoxins by Aspergillus ochraceus. Phytochemistry. 2001;58:709–716. doi: 10.1016/s0031-9422(01)00316-8. [DOI] [PubMed] [Google Scholar]
- Baker SE, Kroken S, Inderbitzin P, Asvarak T, Li BY, Shi L, Yoder OC, Turgeon BG. Two polyketide synthase-encoding genes are required for biosynthesis of the polyketide virulence factor, T-toxin, by Cochliobolus heterostrophus. Mol Plant Microbe Interact. 2006;19:139–149. doi: 10.1094/MPMI-19-0139. [DOI] [PubMed] [Google Scholar]
- Desjardins A, Plattner R, Proctor R. Linkage among genes responsible for fumonisin biosynthesis in Gibberella fujikuroi mating population A. Appl. Environ. Microbiol. 1996;62:2571–2576. doi: 10.1128/aem.62.7.2571-2576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjaj H, Klaebe A, Loret MO, Goma G, Blanc PJ, Francois J. Biosynthetic pathway of citrinin in the filamentous fungus monascus ruber as revealed by 13C nuclear magnetic resonance. Appl Environ Microbiol. 1999;65:311–314. doi: 10.1128/aem.65.1.311-314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen RJ, Nielsen NJ, Maolanon N, Sorensen JC, Olsson S, Nielsen J, Giese H. The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol Microbiol. 2006;61:1069–1080. doi: 10.1111/j.1365-2958.2006.05295.x. [DOI] [PubMed] [Google Scholar]
- Puel O, Galtier P, Oswald I. Biosynthesis and Toxicological Effects of Patulin. Toxins. 2010;2:613–631. doi: 10.3390/toxins2040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw RE, Zhang S. Biosynthesis of dothistromin. Mycopathologia. 2006;162:201–213. doi: 10.1007/s11046-006-0054-5. [DOI] [PubMed] [Google Scholar]
- Vigushin DM, Mirsaidi N, Brooke G, Sun C, Pace P, Inman L, Moody CJ, Coombes RC. Gliotoxin is a dual inhibitor of farnesyltransferase and geranylgeranyltransferase I with antitumor activity against breast cancer in vivo. Med Oncol. 2004;21:21–30. doi: 10.1385/MO:21:1:21. [DOI] [PubMed] [Google Scholar]
- Walton JD. HC-toxin. Phytochemistry. 2006;67:1406–1413. doi: 10.1016/j.phytochem.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Hensgens CM, Shibata N, Schofield CJ, Hajdu J, Baldwin JE. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- Weber G, Schorgendorfer K, Schneider-Scherzer E, Leitner E. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr Genet. 1994;26:120–125. doi: 10.1007/BF00313798. [DOI] [PubMed] [Google Scholar]
- Haese A, Schubert M, Herrmann M, Zocher R. Molecular characterization of the enniatin synthetase gene encoding a multifunctional enzyme catalysing N-methyldepsipeptide formation in Fusarium scirpi. Mol Microbiol. 1993;7:905–914. doi: 10.1111/j.1365-2958.1993.tb01181.x. [DOI] [PubMed] [Google Scholar]
- Zocher R, Keller U. Thiol template peptide synthesis systems in bacteria and fungi. Adv Microb Physiol. 1997;38:85–131. doi: 10.1016/s0065-2911(08)60156-3. [DOI] [PubMed] [Google Scholar]
- Zocher R, Keller U, Kleinkauf H. Enniatin synthetase, a novel type of multifunctional enzyme catalyzing depsipeptide synthesis in Fusarium oxysporum. Biochemistry. 1982;21:43–48. doi: 10.1021/bi00530a008. [DOI] [PubMed] [Google Scholar]
- Glinski M, Urbanke C, Hornbogen T, Zocher R. Enniatin synthetase is a monomer with extended structure: evidence for an intramolecular reaction mechanism. Arch Microbiol. 2002;178:267–273. doi: 10.1007/s00203-002-0451-1. [DOI] [PubMed] [Google Scholar]
- Gehring AM, Mori I, Perry RD, Walsh CT. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- Kleinkauf H, Gevers W. Nonribosomal polypeptide synthesis: the biosynthesis of a cyclic peptide antibiotic, gramicidin S. Cold Spring Harb Symp Quant Biol. 1969;34:805–813. doi: 10.1101/sqb.1969.034.01.092. [DOI] [PubMed] [Google Scholar]
- Menkhaus M, Ullrich C, Kluge B, Vater J, Vollenbroich D, Kamp RM. Structural and functional organization of the surfactin synthetase multienzyme system. J Biol Chem. 1993;268:7678–7684. [PubMed] [Google Scholar]
- Schwecke T, Gottling K, Durek P, Duenas I, Kaufer NF, Zock-Emmenthal S, Staub E, Neuhof T, Dieckmann R, von Dohren H. Nonribosomal peptide synthesis in Schizosaccharomyces pombe and the architectures of ferrichrome-type siderophore synthetases in fungi. Chembiochem. 2006;7:612–622. doi: 10.1002/cbic.200500301. [DOI] [PubMed] [Google Scholar]
- Dorrestein PC, Blackhall J, Straight PD, Fischbach MA, Garneau-Tsodikova S, Edwards DJ, McLaughlin S, Lin M, Gerwick WH, Kolter R, Walsh CT, Kelleher NL. Activity screening of carrier domains within nonribosomal peptide synthetases using complex substrate mixtures and large molecule mass spectrometry. Biochemistry. 2006;45:1537–1546. doi: 10.1021/bi052333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Bumpus SB, Calderone CT, Garneau-Tsodikova S, Aron ZD, Straight PD, Kolter R, Walsh CT, Kelleher NL. Facile detection of acyl and peptidyl intermediates on thiotemplate carrier domains via phosphopantetheinyl elimination reactions during tandem mass spectrometry. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DB, Bumpus SB, Aron ZD, Kelleher NL, Walsh CT. The loading module of mycosubtilin: an adenylation domain with fatty acid selectivity. J Am Chem Soc. 2007;129:6366–6367. doi: 10.1021/ja070890j. [DOI] [PubMed] [Google Scholar]
- Lee JH, Evans BS, Li G, Kelleher NL, van der Donk WA. In vitro characterization of a heterologously expressed nonribosomal Peptide synthetase involved in phosphinothricin tripeptide biosynthesis. Biochemistry. 2009;48:5054–5056. doi: 10.1021/bi900164d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone CT, Bumpus SB, Kelleher NL, Walsh CT, Magarvey NA. A ketoreductase domain in the PksJ protein of the bacillaene assembly line carries out both alpha- and beta-ketone reduction during chain growth. Proc Natl Acad Sci U S A. 2008;105:12809–12814. doi: 10.1073/pnas.0806305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Boyne MT, 2nd, Podevels AM, Klimowicz AK, Handelsman J, Kelleher NL, Thomas MG. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc Natl Acad Sci U S A. 2006;103:14349–14354. doi: 10.1073/pnas.0603748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Linne U, Oberthur M, Marahiel MA. Harnessing the chemical activation inherent to carrier protein-bound thioesters for the characterization of lipopeptide fatty acid tailoring enzymes. J Am Chem Soc. 2008;130:2656–2666. doi: 10.1021/ja078081n. [DOI] [PubMed] [Google Scholar]
- Meluzzi D, Zheng WH, Hensler M, Nizet V, Dorrestein PC. Top-down mass spectrometry on low-resolution instruments: characterization of phosphopantetheinylated carrier domains in polyketide and non-ribosomal biosynthetic pathways. Bioorg Med Chem Lett. 2008;18:3107–3111. doi: 10.1016/j.bmcl.2007.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc Natl Acad Sci U S A. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus SB, Evans BS, Thomas PM, Ntai I, Kelleher NL. A proteomics approach to discovering natural products and their biosynthetic pathways. Nat Biotechnol. 2009;27:951–956. doi: 10.1038/nbt.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Manipulation of carrier proteins in antibiotic biosynthesis. Chem Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Meier JL, Mercer AC, Rivera H, Jr., Burkart MD. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. J Am Chem Soc. 2006;128:12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- Meier JL, Mercer AC, Burkart MD. Fluorescent profiling of modular biosynthetic enzymes by complementary metabolic and activity based probes. J Am Chem Soc. 2008;130:5443–5445. doi: 10.1021/ja711263w. [DOI] [PubMed] [Google Scholar]
- Meier JL, Niessen S, Hoover HS, Foley TL, Cravatt BF, Burkart MD. An orthogonal active site identification system (OASIS) for proteomic profiling of natural product biosynthesis. ACS Chem Biol. 2009;4:948–957. doi: 10.1021/cb9002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkersloot JA, Mateles RI, Yang SS. Isolation of averufin from a mutant of Aspergillus parasiticus impaired in aflatoxin biosynthesis. Biochem Biophys Res Commun. 1972;47:1051–1055. doi: 10.1016/0006-291x(72)90939-4. [DOI] [PubMed] [Google Scholar]
- Zhou R, Linz JE. Enzymatic function of the nor-1 protein in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1999;65:5639–5641. doi: 10.1128/aem.65.12.5639-5641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase mediated polyketide release from a fungal megasynthase. J Am Chem Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber R, Lou L, Du L. A PLP-Dependent Polyketide Chain Releasing Mechanism in the Biosynthesis of Mycotoxin Fumonisins in Fusarium verticillioides. J Am Chem Soc. 2009 doi: 10.1021/ja8091054. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nandakumar MP, Marten MR. Proteomics of filamentous fungi. Trends Biotechnol. 2007;25:395–400. doi: 10.1016/j.tibtech.2007.07.008. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- von Dohren H. A survey of nonribosomal peptide synthetase (NRPS) genes in Aspergillus nidulans. Fungal Genet Biol. 2009;46(Suppl 1):S45–52. doi: 10.1016/j.fgb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CC, Oakley BR. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76:2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A Gene Cluster Containing Two Fungal Polyketide Synthases Encodes the Biosynthetic Pathway for a Polyketide, Asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Elizabeth Oakley C, Woo Bok J, Keller N, Oakley BR, Wang CC. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2009;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Fujii I, Sankawa U, Mayorga M, Timberlake W, Ebizuka Y. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Letters. 1999;40:91–94. [Google Scholar]
- Yu JH, Leonard TJ. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol. 1995;177:4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balibar CJ, Howard-Jones AR, Walsh CT. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol. 2007;3:584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- van Liemt H, von Doren H, Klienkauf H. b-(L-a-Aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans. The Journal of Biological Chemistry. 1989;264:3680–3684. [PubMed] [Google Scholar]
- Hawksworth DL. The magnitude of fungal diversity : the 1.5 million species estimate revisited. Mycological Research. 2001;105:1422–1432. [Google Scholar]
- Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, DiBartolo G, Tyson GW, Allen EE, Ram RJ, Detter JC, Richardson P, Thelen MP, Hettich RL, Banfield JF. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- Chakravarti R, Sahai V. Compactin-a review. Appl Microbiol Biotechnol. 2004;64:618–624. doi: 10.1007/s00253-003-1553-7. [DOI] [PubMed] [Google Scholar]
- Aharonowitz Y, Cohen G, Martin JF. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation, and evolution. Annu Rev Microbiol. 1992;46:461–495. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA, Sherman DH. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- Liou GF, Khosla C. Building-block selectivity of polyketide synthases. Curr Opin Chem Biol. 2003;7:279–284. doi: 10.1016/s1367-5931(03)00016-4. [DOI] [PubMed] [Google Scholar]
- Tang Y, Kim CY, Mathews, II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. J Am Chem Soc. 2003;125:12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- Gross F, Luniak N, Perlova O, Gaitatzis N, Jenke-Kodama H, Gerth K, Gottschalk D, Dittmann E, Muller R. Bacterial type III polyketide synthases: phylogenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads. Arch Microbiol. 2006;185:28–38. doi: 10.1007/s00203-005-0059-3. [DOI] [PubMed] [Google Scholar]
- Seshime Y, Juvvadi PR, Fujii I, Kitamoto K. Discovery of a novel superfamily of type III polyketide synthases in Aspergillus oryzae. Biochem Biophys Res Commun. 2005;331:253–260. doi: 10.1016/j.bbrc.2005.03.160. [DOI] [PubMed] [Google Scholar]
- Nicholson TP, Rudd BA, Dawson M, Lazarus CM, Simpson TJ, Cox RJ. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem Biol. 2001;8:157–178. doi: 10.1016/s1074-5521(00)90064-4. [DOI] [PubMed] [Google Scholar]