Abstract
To identify novel psoriasis susceptibility loci, we carried out a meta-analysis of two recent genome-wide association studies 1,2, yielding a discovery sample of 1,831 cases and 2,546 controls. 102 of the most promising loci in the discovery analysis were followed up in a three-stage replication study using 4,064 cases and 4,685 controls from Michigan, Toronto, Newfoundland, and Germany. Association at a genome-wide level of significance for the combined discovery and replication samples was found for three genomic regions. One contains NOS2 (rs4795067, p = 4 × 10−11), another contains FBXL19 (rs10782001, p = 9 × 10−10), and a third contains PSMA6 and NFKBIA (rs12586317, p = 2 × 10−8). All three loci were also strongly associated with the subphenotypes of psoriatic arthritis and purely cutaneous psoriasis. Finally, we confirmed a recently identified3 association signal near RNF114.
Psoriasis vulgaris (PsV) is one of several immunologically-mediated disorders whose genetic provenance is now coming into focus. While the most evident cellular features of psoriasis are epidermal hyperplasia and altered keratinocyte differentiation, mounting evidence implicates both innate and acquired immunity in disease pathogenesis4. About a third of patients with PsV develop psoriatic arthritis (PsA), an inflammatory arthritis that is usually seronegative for rheumatoid factor5. Herein, the abbreviation PsC refers to individuals with purely cutaneous psoriasis (i.e., PsV without PsA), and the general term “psoriasis” refers to PsV unless otherwise specified. Patients with psoriasis are at significantly greater risk of cardiovascular morbidity and mortality, especially in younger individuals with more severe skin disease6. Psoriasis is also associated with inflammatory bowel disease and other autoimmune disorders7.
Psoriasis is strongly influenced by genetic factors, beginning with long-recognized associations between psoriasis and MHC (major histocompatibility complex) haplotypes bearing HLA-Cw6. More recently the MHC component of susceptibility has been fine-mapped to or very near HLA-Cw6 itself8, and nine additional regions outside the MHC have yielded genome-wide levels of statistical significance in an initial report: IL12B, IL23R, IL23A, TNFAIP3, TNIP1, IL4/IL13, LCE3B/LCE3C, DEFB4, and SPATA2/RNF1143,9,10. In this study, we report the discovery of three novel genetic loci for psoriasis, none of which have been definitively implicated in any complex genetic disorder to date. We also confirm a previous report 3 of association to the SPATA2/RNF114 association (pcomb = 2 × 10−7), expanding the associated region to include two additional genes.
We augmented published genome-wide association study (GWAS) results from the Collaborative Association Study of Psoriasis (CASP)1 by using genotype imputation11 to combine the results with those of an independent GWAS of psoriasis from Kiel, Germany2, using either Phase 2 HapMap or the August 2009 release of the 1000 Genomes Project (1000G) data as reference (Table S1). Only SNPs (simple nucleotide polymorphisms) imputed with relatively high confidence (estimated r2 between imputed and true genotypes >0.3) in both GWAS were considered for meta-analysis. Our discovery sample consisted of up to 1,831 cases and 2,546 controls with imputed genotypes for 2,502,313 (HapMap) or 7,456,344 (1000G) autosomal SNPs. Genomic control inflation factors for the p-values from the meta-analysis of the CASP and Kiel GWAS were low (λ = 1.061 for HapMap and 1.054 for 1000G imputations), and Q-Q plots (Figure S1) also indicated no serious systematic differences between cases and controls due to cryptic relatedness, population substructure, or other biases.
The replication sample consisted of up to 4,064 PsV cases and 4,685 controls from Michigan, Toronto, Newfoundland, and Germany. All participating subjects gave informed consent and protocols were reviewed and approved by local institutional review boards. Ascertainment criteria for psoriasis have been described8. PsA was diagnosed according to the CASPAR criteria12. PsC was defined as the presence of skin lesions in the demonstrated absence of PsA at the time of ascertainment.
We selected 102 SNPs for replication based on their p value rankings either in a subset of 350 CASP PsA cases vs. normal controls (53 SNPs, set A, Table S1) or in the combined CASP-Kiel GWAS (49 SNPs, sets B and C, Table S1). Imputation quality of these SNPs was generally excellent. First stage replication genotyped all selected SNPs in the large Michigan sample; 91 of these SNPs passed all quality control (QC) filters. Second stage replication typed all but one of the 51 QC-filtered SNPs from set A of the PsA GWAS in the samples from Toronto and Newfoundland, which contain most of the PsA cases. SNPs with the most promising association results were then followed-up with third stage typing--8 from sets B and C in the two Canadian samples, and 6 from all three sets in the German sample. Statistical significance of association was assessed with an allelic chi-square test for experimentally genotyped SNPs and an equal variance t-test with allele dosages for imputed SNPs. Meta-analysis of association across cohorts incorporated both imputation quality and asymmetry in the number of cases and controls as well as size of cohort. See Online Methods for details of quality control, imputation quality and meta-analysis.
These experiments yielded four SNPs from three genomic regions for which association p values achieved genome-wide statistical significance (p ≥ 5 × 10−8) in the combined discovery and replication samples. Table 1 presents the results for these four SNPs, along with three additional SNPs yielding pcomb ≥ 1 × 10−6. Genotype counts for these seven SNPs are shown in Table S2. Results for the remaining SNPs passing quality control are summarized in Table S3, and a comparison of association results for PsA vs. PsC is presented in Table 2.
Table 1.
Loci with strongest evidence of association with psoriasis in the combined sample.
| SNPb | Setc | Chr | Pos (Mb) |
Alleles risk/ nonrisk |
Discovery Samples (1831 cases, 2546 controls)a |
Replication Samples (4064 cases, 4685 controls)a |
Combined p-value (meta) |
Notable Nearby Genes (relative position)e |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequencyd |
OR (meta) |
p-value (meta) |
Frequencyd |
OR (meta) |
p-value (meta) |
|||||||||
| Case | Control | Case | Control | |||||||||||
| rs4795067 | B | 17 | 23.13 | G/A | .397 | .354 | 1.20 | 5×10−5 | .389 | .349 | 1.19 | 2×10−7 | 4×10−11 | NOS2 (intronic) |
| rs10782001 | B | 16 | 30.85 | G/A | .394 | .347 | 1.22 | 1×10−5 | .402 | .368 | 1.16 | 1×10−5 | 9×10−10 | FBXL19 (intronic) |
| rs12924903 | A | 16 | 30.84 | A/G | .393 | .343 | 1.24 | 1×10−4 | .384 | .349 | 1.16 | 2×10−6 | 1×10−9 | FBXL19 (−6.9 kb) |
| rs12586317 | C | 14 | 34.75 | T/C | .783 | .742 | 1.25 | 1×10−5 | .777 | .751 | 1.15 | 1×10−4 | 2×10−8 | NFKBIA, PSMA6 |
| rs1008953 | B | 20 | 43.41 | C/T | .809 | .770 | 1.27 | 1×10−5 | .809 | .787 | 1.14 | 8×10−4 | 1×10−7 | SDC4 (−3.7 kb) |
| rs495337 | C | 20 | 47.96 | G/A | .610 | .573 | 1.17 | 7×10−4 | .616 | .570 | 1.21 | 7×10−5 | 2×10−7 | RNF114 (silent) |
| rs12580100 | B | 12 | 54.73 | A/G | .898 | .868 | 1.33 | 2×10−6 | .913 | .896 | 1.17 | 1×10−2 | 1×10−6 | RPS26 (−1.2 kb) |
The number of cases and controls that were genotyped and passed quality control for at least one of the 91 SNPs in the three replication sets. The actual numbers of cases and controls typed for each SNP varies and and can be determined from the genotype counts in Table S2.
SNPs attaining genome-wide significance and their notable nearby genes are indicated by bold font.
Replication set for the SNP (see main text and Table S1 for more details).
Frequency of the risk allele.
Position of each SNP relative to notable nearby genes is given. +/− indicates whether the SNP is upstream (−) or downstream (+) of the transcription start site. SNPs within the gene are labeled as “intronic”, “UTR”, “silent” or “missense”.
Table 2.
Association of strongest replicated loci with psoriatic arthritis (PsA) and cutaneous psoriasis (PsC)
| SNP | Chr | Pos (Mb) |
Alleles risk/ nonrisk |
PsA vs. control (1747 PsA, 7231 controls)a |
PsC vs. control (3394 PsC, 7231 controls)a |
PsA vs. PsC (1747 PsA, 3394 PsC)a |
Notable Nearby Genes |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequenciesb | OR | p-value | Frequenciesb | OR | p-value | Frequenciesb | OR | p-value | |||||
| rs4795067 | 17 | 23.13 | G/A | .389 / .341 | 1.23 | 1x10−6 | .391 / .348 | 1.20 | 1×10−8 | .396 / .382 | 1.06 | .25 | NOS2 |
| rs10782001 | 16 | 30.85 | G/A | .412 / .358 | 1.26 | 4×10−8 | .388 / .353 | 1.16 | 2×10−6 | .412 / .385 | 1.12 | .022 | FBXL19 |
| rs12924903 | 16 | 30.84 | A/G | .412 / .352 | 1.29 | 4×10−9 | .380 / .347 | 1.15 | 4×10−5 | .403 / .378 | 1.11 | .058 | FBXL19 |
| rs12586317 | 14 | 34.75 | T/C | .786 / .751 | 1.22 | 6×10−5 | .778 / .747 | 1.19 | 1×10−6 | .785 / .780 | 1.03 | .60 | NFKBIA |
| rs495337 | 20 | 47.96 | G/A | .615 / .573 | 1.20 | 2×10−3 | .614 / .572 | 1.19 | 1×10−6 | .615 / .618 | 0.99 | 0.86 | RNF114 |
| rs1008953 | 20 | 43.41 | C/T | .803 / .772 | 1.21 | 2×10−4 | .792 / .769 | 1.15 | 2×10−4 | .803 / .795 | 1.05 | .37 | SDC4 |
| rs12580100 | 12 | 54.73 | A/G | .882 / .867 | 1.15 | .029 | .893 / .866 | 1.29 | 1×10−6 | .888 / .893 | 0.95 | .54 | RPS26 |
The number of cases and controls for the specified phenotype comparison that were genotyped and passed quality control for at least one of the 91 SNPs in the three replication sets. The actual numbers of cases and controls typed for each SNP varies and can be determined from the genotype counts in Table S2.
Frequencies of the PsV risk allele for the two phenotypes being compared. Allele frequencies for the same phenotype (PsA, PsC, or control) and the same SNP often differ among comparisons because of differing number of qualifying cohorts (e.g., some cohorts have PsA but no PsC cases so qualify for PsA vs. control but not PsA vs. PsC or PsC vs. control comparisons) and because variations in the proportions of each phenotype per cohort alter the relative cohort weights in the meta-analyses.
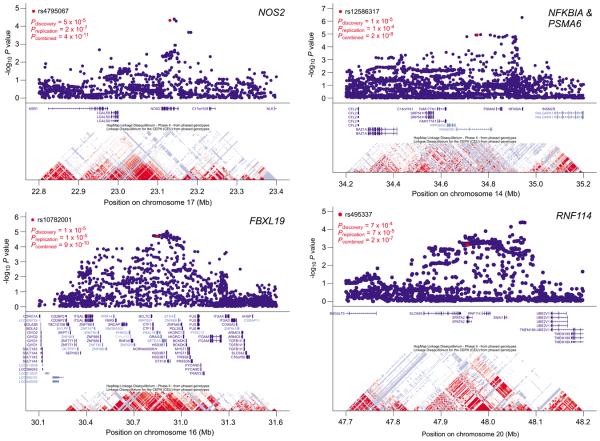
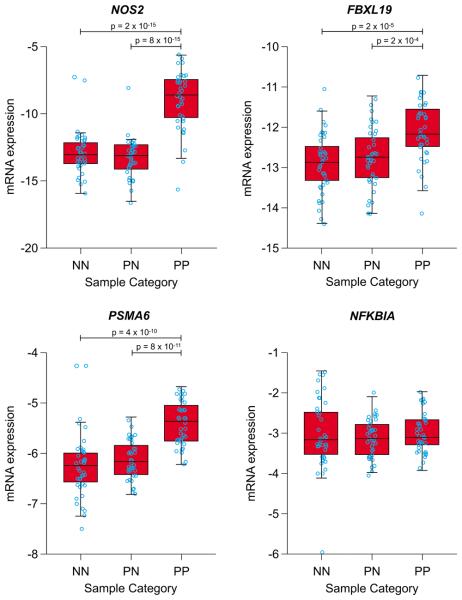
One of the associated SNPs (rs4795067, pcomb = 4 × 10−11, ORrepl = 1.19) mapped to an intron of the NOS2 gene (Table 1). As shown in Figure 1, the strongest imputed association signals from the CASP-Kiel GWAS clustered in a region of low linkage disequilibrium (LD) containing no other genes. NOS2 encodes iNOS (inducible nitric oxide synthase), which is expressed by a subset of CD11c-positive, CD1c-negative, TNF-α (tumor necrosis factor-α)-producing inflammatory dendritic cells (DC) that is markedly expanded (30-fold) in psoriatic lesions13. Consistent with this, we found marked overexpression of NOS2 mRNA in lesional psoriatic skin by microarray analysis (Table S4) and especially by QRT-PCR (Table S5, Figure 2). However, neither rs4795067 nor any other SNP in this region was found to significantly regulate NOS2 mRNA expression (i.e., to function as cis-acting expression quantitative trait loci (cis-eQTL), as assessed by either microarray (Table S6) or qRT-PCR assays (data not shown). While this SNP attained genome-wide significance in subphenotype analysis only for PsC, it also yielded a strong association with PsA (OR = 1.23, p = 1 × 10−6, Table 2). Notably similar to inflammatory DC in cutaneous psoriasis, cells infiltrating the sub-lining layer of diseased synovium express markedly higher levels of iNOS and TNF-α in PsA than in osteoarthritis or traumatic arthritis14. While Online Mendelian Inheritance in Man (OMIM 163730) lists unconfirmed associations between NOS2 variants and malaria, mycetoma, preterm delivery, and Parkinson's disease, this is the first description of a genome-wide significant association between NOS2 and any complex genetic disorder.
Figure 1.
Evidence for psoriasis association in four genomic regions, including the three novel loci attaining genome-wide significance and the confirmed RNF114 region. The upper portion of each plot depicts association p-values for a meta-analysis of the CASP and Kiel discovery GWAS using 1000G-based imputation, while the lower portion depicts RefSeq genes and LD plots from the phase 2 HapMap-CEU sample. For each region, the most strongly associated replicated SNP is highlighted in red, as are its p-values for the discovery, replication, and combined samples.
Figure 2.
Expression data for notable candidate genes within the three psoriasis-associated regions. Log2-transformed levels of mRNA assayed by qRT-PCR are shown for normal (NN), uninvolved (PN), and lesional psoriatic (PP) skin. Figures include both individual measurements (jittered scatterplots) and box-plots summarizing the distribution. Bars and significance levels are shown for all pairwise comparisons of mean mRNA levels that are significant at a nominal (p ≤ 0.05) level.
The next associated SNP (rs10782001, pcomb = 9×10−10, ORrepl = 1.16), mapped to a large region of strong LD on chromosome 16p11.2 containing at least 31 genes (Table 1, Figure 1). Both rs10782001 and rs12924903, which are in strong LD with each other (r2 = 1.0 in Phase 2 HapMap-CEU), attained genome-wide significance for PsA but not for PsC, despite the greater numbers of patients with PsC (n = 2,113) relative to PsA (n = 1,361) (Table 2). Moreover, SNP rs10782001 was significantly more strongly associated with PsA than with PsC (OR = 1.12, p = 0.02). While any of the genes in the associated region could be responsible for the observed association signal, POL3S and FBXL19 are the most differentially expressed among the 16 genes in the most highly associated interval (involved vs. control: 2.66 fold for POL3S, 1.43-fold for FBXL19, Figure 1, Table S4). POL3S encodes a secreted serine protease about which little is known15. FBXL19 is structurally related to FBXL11, an F-box family member recently shown to inhibit NF-κB (nuclear factor kappa light chain enhancer of activated B cells) activity by lysine demethylation16. Of the 68 human members of the F-box protein family, only three (FBXL10, FBXL11, and FBXL19) have zinc finger, plant homology domain, and leucine-rich repeats in addition to the F-box17. FBXL10 and FBXL11 contain jumonji C domains known to be required for demethylase activity, but FBLX19 does not17. Thus, FBXL19 might act as a dominant negative inhibitor of demethylase activity, thereby serving to activate NF-κB.
The third associated SNP (rs12586317, pcomb = 2×10−8, ORrepl = 1.15, Table 1) mapped to the vicinity of the NFKBIA, PSMA6, and KIAA0391 genes. 1000G-based imputation led us to select this SNP for typing, and revealed that the associated region extended to encompass NFKBIA, an observation that was not evident from HapMap-based imputation (Figure 1 and data not shown). Of these three genes, only PSMA6 is overexpressed in psoriatic lesions (Table S4, Figure 2). NFKBIA and PSMA6 are both attractive candidate genes for psoriasis susceptibility, as NFKBIA encodes IκB-α, an inhibitor of NF-κB signaling, and PSMA6 encodes a proteosomal subunit involved in MHC Class I antigen processing. Polymorphisms in both genes (OMIM 164008 and 602855) have been suggestively associated with susceptibility to various disorders including myocardial infarction, Graves disease and inflammatory bowel disease. However, this is the first demonstration of a genome-wide significant association in this region for any complex genetic disorder.
While not attaining genome-wide significance, SNPs rs1008953 (pcomb = 1 × 10−7, ORrepl = 1.14) and rs12580100 (pcomb = 1 × 10−6, ORrepl = 1.17) were also of interest. SNP rs1008953 resides 3.7 kb upstream of SDC4, which encodes syndecan-4, a cell-surface heparin sulfate proteoglycan that inhibits T-cell activation18. Expression of SDC4 mRNA was markedly reduced in lesional psoriatic skin relative to both uninvolved and normal skin (Tables S4 and S5). However, there are several other genes under the association peak, and several SNPs in the region function as cis-eQTLs for SYS1, DBNDD2, and PIGT gene expression (Table S6, Figure S2). SNP rs12580100 resides 1.2 kb upstream of RPS26, which resides in a region that has been implicated in susceptibility to type I diabetes19 and that encodes a ribosomal protein subunit whose expression is under cis-acting genetic control in the liver20. SNP rs12580100 lies only 300 kb from a confirmed association signal at rs2066808 near the IL23A gene1, but these association signals appear to be independent since the two SNPs reside in different LD blocks (Figure S2) and consequently show low pairwise LD (r2 = 0.23). Of note, we found that RPS26 is overexpressed in lesional psoriatic skin (Tables S4 and S5) and confirm in skin that this region contains several highly significant eQTLs that clearly co-localize with the disease association signal for RPS26, but not for IL23A (Table S6, Figure S2).
While also not reaching genome-wide significance, SNP rs495337 (pcomb = 2 × 10−7, ORrepl = 1.21), confirms previous reports of association between psoriasis and the SPATA2/RNF114 region3. However, the 1000G-based imputation demonstrates that the region of association extends farther than previously reported, encompassing the SLC9A8 and SNAI1 genes (Figure 1). Of these four genes, RNF114 was the most strongly expressed in skin (Table S4). None of the transcripts encoded by these four genes were increased in psoriatic lesions; one SPATA2 probe detected a 23% decrease (Table S4). We were unable to confirm a previous report of a cis-eQTL in this region3, either by microarray (Tables S6) or qRT-PCR assays (data not shown); however, our gene expression sample was only about half the size of that used in the previous study.
Clearly, fine mapping studies are needed to identify causal variants in the three novel susceptibility regions we have identified. Nevertheless, genes contained in these novel regions fit well with emerging concepts of psoriasis pathogenesis10. Given genetic, therapeutic, and immunological results indicating that IL-23 acts on DC to promote the survival and expansion of pathogenic T-cells in psoriasis10, it is noteworthy that one of our novel regions contains NOS2, which is expressed by a massively expanded subset of TNF-α producing inflammatory DC in psoriatic lesions13. Moreover, given that TNFAIP3 and TNIP1 regulate NF-κB signaling and have been implicated in psoriasis susceptibility1, the fact that FBXL19 and NFKBIA regulate the same pathway further suggests the central importance of NF-κB signaling in psoriasis. Finally, when combined with the profound response of PsA to TNF-α blockers4 and prior genetic results implicating IL12B and TNIP1 in PsA susceptibility1, the identification of a novel PsA susceptibility region containing FBXL19 suggests that this multicellular IL-23/IL-17/NF-κB signaling circuit may also be of critical importance to the development of joint disease in psoriatic patients.
URLs
MACH software, http://www.sph.umich.edu/csg/abecasis/mach/ HapMap 2 CEU phased haplotypes, http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2006-07_phaseII/phased/ August 2009 release of 1000G phased data, http://www.sph.umich.edu/csg/abecasis/mach/download/1000G-Sanger-0908.html
Online Methods
Genotyping
We determined genotypes for the discovery phase of the Collaborative Association Study of Psoriasis (CASP) on a Perlegen platform1, and genotypes for the discovery phase of the Kiel GWAS on an Illumina HumanHap 550k array2. In Ann Arbor, follow-up genotyping was performed utilizing either the TaqMan (Applied Biosystems, Foster City, CA) or the Sequenom MassArray (Sequenom, San Diego, CA) platforms. Additional follow-up genotyping was performed using the TaqMan platform in Kiel and the Sequenom platform in Newfoundland.
Quality control filters
For the CASP and Kiel GWAS analyses, multiple quality control filters were applied to both samples and genotypes1,2. For the replication samples, we excluded SNPs with < 95% genotyping call rates, with a Bonferonni-corrected Hardy Weinberg p-value < 0.05, or with cluster plots that did not show clear separation of the three genotype clusters. We also excluded samples with < 95% genotyping call rate; for genotyping performed on the Sequenom platform the sample filter was applied separately to each typing group (ranging from 15-23 SNPs in size). Sample and SNP filters were applied separately to each replication cohort.
Genotype imputation
We performed genotype imputation using a hidden Markov model algorithm implemented in MACH software version 1.011, as previously described11,21. For replication sets A and B we used HapMap 2 CEU phased haplotypes22, which serve as a good reference for populations of European descent11,23. For replication set C, we used phased haplotypes from the August 2009 release of the 1000 Genomes Project (1000G) as the reference. Parameters for the hidden Markov model for the 1000G imputation were first estimated using a subset of 250 randomly sampled individuals, and then all individuals were imputed based on those parameters. For both imputations, a dosage score (ranging from 0 to 2) was computed at each SNP for each individual, which is the expected number of copies of a given allele conditional on the genotypes of directly assayed SNPs and integrating over all possible configurations of the phased reference haplotypes. Imputed dosages for the two GWAS were analyzed separately, and the association test results were then combined by meta-analysis using the weighting scheme described in the next section.
Predicted imputation quality of the 91 SNPs selected for replication was excellent: median MACH-r2 values were 0.9858 and 0.9688, respectively, for the HapMap and 1000G imputations of the CASP GWAS, and 0.9887 and 0.9841 for the two imputations of the Kiel GWAS. We also tested the accuracy of MACH's predicted estimates of r2 between imputed and true genotypes. All 91 replication SNPs were genotyped for a large subset of the CASP GWAS samples (456 of 1359 cases and 670 of 1400 controls), and six of these SNPs yielding significance at or near genome-wide levels were also typed for a substantial subset of the Kiel GWAS samples (443 of 472 cases). A strong correspondence was seen between the observed and predicted squared correlation of imputed and experimentally determined genotypes for both the CASP GWAS (Pearson r = 0.914 for HapMap and 0.937 for 1000G imputations) and the Kiel GWAS (r = 1.000 for HapMap and 0.999 for 1000G).
Meta-analysis of association
Evidence for association was combined across cohorts using a method adapted from published recommendations24. Signed z-scores were calculated directly from association test statistics, with the sign of each z-score indicating the direction of effect in that cohort. Z-scores were then summed across multiple cohorts using weights equal to the square root of the effective sample size in each cohort divided by the sum of effective sample sizes in all cohorts, which ensures that the squared weights sum to one. Effective sample size (Ne) was determined for each combination of cohort and SNP by adjusting the raw sample size for asymmetry in the number of cases and controls, and by scaling with the imputation uncertainty when applicable, as follows:
where Na and Nu are the number of affecteds (cases) and unaffecteds (controls) typed or imputed for the given SNP in the given cohort, and MACH r̂2 is an estimate of the squared Pearson correlation between the imputed genotype scores and the true genotypes for the given SNP. The first term on the right-hand side of the equation calculates the symmetric case/control sample size yielding the same non-centrality parameter (NCP) and hence power, under the null hypothesis for an allelic association test of a biallelic marker, as does the given asymmetric case/control sample size. Under the alternative hypothesis of association, both NCP and Ne depend upon not only Na and Nu but also upon the genetic model (i.e, the risk allele frequencies in cases and controls). However, for the effect sizes normally seen for complex disease loci (OR < 2.0), estimates of Ne under the null and under the alternative differ only modestly for a wide variety of genetic models, total sample size, and Na: Nu sample allocation. The second term on the right-hand side of the equation down-weights the contribution of a GWAS when a particular SNP was poorly imputed.
Estimates of odds ratios and allele frequencies were summed across multiple cohorts using the square of the weights described above. Odds ratios were first log-transformed and then back-transformed after summation across cohorts.
Q-Q plots and genomic control for meta-analysis of CASP and Kiel GWAS
Quantile-quantile (Q-Q) plots and genomic control inflation factors for the p-values from a meta-analysis of the CASP and Kiel GWAS were restricted to those SNPs with a MACH-r2 imputation quality of at least 0.3 for both studies. The genomic control variance inflation factor (λ) was computed using the bounded median method 25. Q-Q plots and genomic control λ values were determined both for all reliably imputed SNPs, and after exclusion of SNPs within nine previously published regions of genome-wide significant association with psoriasis. For the MHC, the excluded interval was the 7.7 Mb extended region defined by Horton et al. 26. For regions harboring candidate loci IL12B, IL23R, IL23A, IL13, TNIP1, and TNFAIP3, boundaries of the excluded intervals were chosen as ± 500 kb relative to the most strongly associated SNP as determined by Nair et al.1. Boundaries were also chosen to be ± 500 kb relative to the most strongly associated SNP in the RNF114 region 3 and ± 500 kb relative to the 32 kb deletion spanning the LCE3B and LCE3C genes on chromosome 1 27. The published psoriasis locus that maps to a highly variable copy number variation (CNV) near the β-defensin cluster on chromosome 8 was not excluded, because there are no known SNPs in strong LD with this CNV 28.
For the 1000G-based imputations, a 210 kb region on chromosome 17p11.2 with multiple SNPs exhibiting strong but spurious association was also excluded for one of the Q-Q plots. These spurious associations are an artifact of a single SNP in the Kiel GWAS (rs4889730), which passed all QC filters but was nevertheless unreliably typed as determined by post hoc inspection of the Illumina genotyping cluster plot. Removal of this SNP from the Kiel GWAS before re-imputation resulted in loss of all evidence of association (data not shown). Notably, this region showed no evidence of association in the CASP GWAS, nor could observed associations in the Kiel GWAS be replicated for SNPs rs1975974 and rs17052344 in the Michigan sample (Table S3) or for SNP rs17052344 in the German sample2.
Measurement and analysis of gene expression
Six millimeter punch biopsies of normal skin (74 subjects) as well as uninvolved and/or involved lesional skin of affected patients (66 subjects) were obtained at the University of Michigan Department of Dermatology, as previously described 29. Involved skin biopsies were taken from psoriasis plaques, and uninvolved and normal skin were sampled from the buttocks, at least 10 cM away from the nearest plaque. Study subjects did not use any systemic anti-psoriatic treatments for 2 weeks prior or topical anti-psoriatic treatments for 1 week prior to biopsy. Informed consent was obtained from all subjects, under protocols approved by the Institutional Review Board of the University of Michigan Medical School and was conducted according to the Declaration of Helsinki Principles. RNA from each biopsy was isolated using the RNeasy kit (Qiagen).
Samples for 64 normal skin biopsies and 58 paired involved and uninvolved skin biopsies were run on Affymetrix U133 Plus 2.0 arrays according to the manufacturer's protocol. The raw data were processed using the Robust Multichip Average (RMA) method, adjusting RMA expression values to account for batch and gender effects 29. Gene expression was further assessed by qRT-PCR, as described previously 29, for 39 normal skin, 38 uninvolved skin, and 37 lesional skin biopsies. These qRT-PCR samples partially overlapped the microarray sample set (29 of normal, 33 of uninvolved, and 29 of involved skin samples common to both).
Two sample t-tests were used to compare mean mRNA expression in skin from normal controls with that in lesional or non-lesional skin from psoriatics. For comparisons of mean expression in involved vs. uninvolved skin from affected individuals, paired t-tests were used for microarray assays since lesional and non-lesional skin was always sampled in pairs from each individual, but two sample t-tests were used for qRT-PCR assays since many of the affected individuals were biopsied for only lesional or non-lesional skin, but not both. Log2-transformed mRNA levels were used for all t-tests.
For microarray data, we tested for cis-acting expression quantitative trait loci (cis-eQTL) separately in normal, uninvolved, and involved skin. We used the score test in Merlin (fastassoc option) to test cis-associations between log2-transformed mRNA levels of each probe (i.e., transcript) and SNPs in its cis-candidate region, mapping from 1 Mb upstream of the transcription start site to 1 Mb downstream of the transcription end site. For genotyped SNPs, which were extracted from the CASP GWAS after QC filtering, the observed number of copies of one allele was modeled. For imputed SNPs, which used HapMap2 CEU phased haplotypes as reference, the dosage of one allele was modeled. 57 normal skin samples and 53 pairs of uninvolved and involved skin samples qualified for eQTL analysis, having both microarray expression profiling and genotype data. To increase power, meta-analysis across independent skin samples (i.e., normal and uninvolved skin or normal and involved skin) was performed by combining sample size-weighted signed z-scores for the individual skin types. Significance thresholds of 9 ×10−7, 2.8 × 10−5, and 1.0 × 10−5, corresponding to FDRs of approximately 0.01, 0.05, and 0.10, were used to assess evidence for cis-eQTLs.
For cis-eQTL analyses using qRT-PCR data, we utilized linear regression of mRNA levels against dosage of the risk allele. Combined analysis across independent skin types was performed by including skin type as an independent indicator variable in the regression model. 37 normal skin, 37 uninvolved skin, and 35 lesional skin samples qualified for analysis, having both qRT-PCR expression and genotype data.
Supplementary Material
Acknowledgements
The authors wish to thank the many psoriasis and PsA patients and normal controls who participated in this study, and to acknowledge the key contributions of Kristina Callis Duffin, David Goldgar, and Bing Jian Feng of the University of Utah and Cindy Helms of Washington University at St. Louis to the CASP study. This research was supported by grants R01AR42742, R01AR050511, R01AR054966, R01AR050266, R01HG002651, and U01HG005214 from the National Institutes of Health, by the Ann Arbor Veterans Affairs Hospital, by the German Ministry of Education and Research through the National Genome Research Network (BMFT 01GS 0171/ BMBF NUW-S23T10), and by the Krembil Foundation and the Canadian Institutes of Health Research.
Footnotes
Competing Interests Statement:
The authors declare no competing interests.
Author contributions:
R.P.N., P.E.S. and J.T.E. performed SNP selection, data analysis and prepared figures and tables. R.P.N., T.T., P.E.S., P.R., and E.E. performed genotyping. P.E.S., Y.L., and J.D. performed genotype imputation and association analyses, and P.E.S., J.E.G., and J.D. performed the expression analyses. G.R.A. helped with statistical analyses and interpretation of results. R.P.N., T.T., J.J.V., R.I., M.W., S.W., B.E., C.G., H.E.W., H.W.L., P.R., M.K., U.M., and D.D.G. coordinated subject recruitment and collected phenotype data. J.T.E., G.G.K. and A.M.B. contributed genotypes and phenotypes from the CASP discovery GWAS. J.T.E. and P.E.S. drafted the manuscript, R.P.N., G.R.A., E.E., M.W., and A.F. edited the manuscript, and J.T.E. planned and supervised the study. All authors approved the final draft.
References
- 1.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellinghaus E, et al. Genome-wide association study reveals association of psoriasis with TRAF3IP2. Nature Genetics. 2010 doi: 10.1038/ng.689. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capon F, et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum Mol Genet. 2008;17:1938–45. doi: 10.1093/hmg/ddn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–91. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gladman DD. Psoriatic arthritis. In: Koo J, Lee CS, Lebwohl M, Weinstein GD, Gottlieb AB, editors. Moderate to Severe Psoriasis. Informa Health Care; New York: 2009. pp. 239–258. [Google Scholar]
- 6.Gelfand JM, et al. Risk of myocardial infarction in patients with psoriasis. Jama. 2006;296:1735–41. doi: 10.1001/jama.296.14.1735. [DOI] [PubMed] [Google Scholar]
- 7.Makredes M, Robinson D, Jr., Bala M, Kimball AB. The burden of autoimmune disease: a comparison of prevalence ratios in patients with psoriatic arthritis and psoriasis. J Am Acad Dermatol. 2009;61:405–10. doi: 10.1016/j.jaad.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Nair RP, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–51. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elder JT, et al. Molecular dissection of psoriasis: Integrating genetics and biology. J Invest Dermatol. 2009 doi: 10.1038/jid.2009.319. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran V, Schentag CT, Gladman DD. Sensitivity and specificity of the CASPAR criteria for psoriatic arthritis in a family medicine clinic setting. J Rheumatol. 2008;35:2069–70. author reply 2070. [PubMed] [Google Scholar]
- 13.Zaba LC, Krueger JG, Lowes MA. Resident and “inflammatory” dendritic cells in human skin. J Invest Dermatol. 2009;129:302–8. doi: 10.1038/jid.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melchiorri C, et al. Enhanced and coordinated in vivo expression of inflammatory cytokines and nitric oxide synthase by chondrocytes from patients with osteoarthritis. Arthritis Rheum. 1998;41:2165–74. doi: 10.1002/1529-0131(199812)41:12<2165::AID-ART11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 15.Cal S, et al. Identification and characterization of human polyserase-3, a novel protein with tandem serine-protease domains in the same polypeptide chain. BMC Biochem. 2006;7:9. doi: 10.1186/1471-2091-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–80. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung JS, Bonkobara M, Tomihari M, Cruz PD, Jr., Ariizumi K. The DC-HIL/syndecan-4 pathway inhibits human allogeneic T-cell responses. Eur J Immunol. 2009;39:965–74. doi: 10.1002/eji.200838990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–50. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–66. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 26.Horton R, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–99. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 27.de Cid R, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abu Bakar S, Hollox EJ, Armour JA. Allelic recombination between distinct genomic locations generates copy number diversity in human beta-defensins. Proc Natl Acad Sci U S A. 2009;106:853–8. doi: 10.1073/pnas.0809073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gudjonsson JE, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol. 2009;129:2795–804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.