Abstract
Semiconducting polymer dots (Pdots) represent a new class of ultrabright fluorescent probes for biological imaging. They exhibit several important characteristics for experimentally demanding in vitro and in vivo fluorescence studies, such as their high brightness, fast emission rate, excellent photostability, non-blinking, and non-toxic feature. However, controlling the surface chemistry and bioconjugation of Pdots has been a challenging problem that prevented their widespread applications in biological studies. Here, we report a facile yet powerful conjugation method that overcomes this challenge. Our strategy for Pdot functionalization is based on entrapping heterogeneous polymer chains into a single dot, driven by hydrophobic interactions during nanoparticle formation. A small amount of amphiphilic polymer bearing functional groups is co-condensed with the majority of semiconducting polymers to modify and functionalize the nanoparticle surface for subsequent covalent conjugation to biomolecules, such as streptavidin and immunoglobulin G (IgG). The Pdot bioconjugates can effectively and specifically label cellular targets, such as cell surface marker in human breast cancer cells, without any detectable non-specific binding. Single-particle imaging, cellular imaging, and flow cytometry experiments indicate a much higher fluorescence brightness of Pdots compared to those of Alexa dye and quantum dot probes. The successful bioconjugation of these ultrabright nanoparticles presents a novel opportunity to apply versatile semiconducting polymers to various fluorescence measurements in modern biology and biomedicine.
Introduction
Advances in understanding biological systems have relied on applications of fluorescence microscopy, flow cytometry, versatile biological assays, and biosensors.1,2 These experimental approaches make extensive use of organic dye molecules as probes. But intrinsic limitations of the conventional dyes, such as low absorptivity and poor photostability, have posed great difficulties in further developments of high-sensitivity imaging techniques and high-throughout assays.3,4 As a result, there has been considerable interest in developing brighter and more photostable fluorescent probes. For example, inorganic semiconducting quantum dots (Qdots) are under active development and now commercially available from Life Technologies (Invitrogen). Qdots are ideal probes for multiplexed target detection because of their broad excitation band and narrow, tunable emission peaks. They exhibit improved brightness and photostability over conventional organic dyes.5–8 However, Qdots are not bright enough for many photon-starved applications because of their low emission rates, blinking, and a significant fraction of non-fluorescent dots.9 There has been recent work to develop non-blinking Qdots,10 but their toxicity, caused by the leaching of heavy metal ions, is still a critical concern for in vivo applications.
Semiconducting polymers are attractive materials for various optoelectronic applications, including light-emitting diodes, field-effect transistors, and photovoltaic devices.11,12 Their appeal is based on the readily-tailored electrical and optical properties of semiconductors combined with the easy processability of polymers. Water-soluble semiconducting polymers have also been demonstrated as highly sensitive biosensors and chemical sensors.13–15 Since our early demonstration of semiconducting polymer nanoparticles (Pdots),16,17 there has been rapid progress in the field, including the characterization of their complex photophysics,18–21 and their development for biological imaging and high resolution single-particle tracking.22–30 Pdots exhibit extraordinarily high fluorescence brightness under both one-photon and two-photon excitations. Their brightness stems from a number of favorable characteristics of semiconducting polymer molecules, including their large absorption cross-sections, fast emission rates, and high fluorescence quantum yields. Recent studies have also shown that Pdots as fluorescent probes were photostable, and not cytotoxic in different cellular assays.23,28,31
However, for a wide range of biological applications, a significant problem of Pdots has yet to be solved — control over their surface chemistry and conjugation to biological molecules. Although research efforts involving silica or phospholipid encapsulation can result in composite particles with surface functional groups,17,30 all the reported results until now on cellular labeling with Pdots are presumably based on endocytosis,23,28,30,31 a far less effective and specific process compared to the established labeling methods for organic fluorophores and Qdots. It is still unclear whether Pdot probes could be made specific enough to recognize cellular targets for effective labeling. This challenge thus far has severely prevented the wide-spread use of Pdots in biological applications.
Here, we describe our results that successfully address the challenge of Pdot bioconjugation and specific cellular targeting. We developed a facile conjugation method that covalently links Pdots to biomolecules for labeling cellular targets by specific antigen-antibody or biotin-streptavidin interactions. This functionalization and bioconjugation strategy can be easily applied to any hydrophobic, fluorescent, semiconducting polymer. We apply the Pdot bioconjugates to single-particle imaging, cellular imaging, and flow cytometry experiments and demonstrate their advantages over conventional organic fluorophores and Qdot probes. This work, therefore, opens up a new and practical pathway for employing a variety of highly fluorescent, photostable, and non-toxic Pdot bioconjugates for biological applications.
Results and Discussion
Functionalization and bioconjugation of Pdots
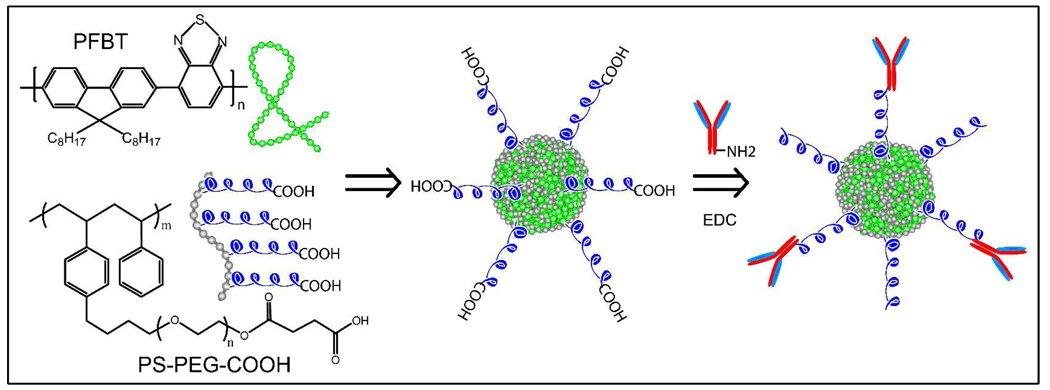
Our strategy for functionalizing the surface of Pdots is based on entrapping heterogeneous polymer chains, driven by hydrophobic interactions during nanoparticle formation, into a single Pdot. A small amount of amphiphilic polymers is co-condensed with the semiconducting polymers of the Pdots to modify and functionalize the nanoparticle surface (Scheme 1). This paper employs a functional, amphiphilic, comb-like, polystyrene polymer PS-PEG-COOH, but other amphiphilic polymers with different functional groups can also be used. PS-PEG-COOH consists of a hydrophobic polystyrene backbone and several hydrophilic side chains of ethylene oxide terminated with carboxylic acid. During nanoparticle formation, the hydrophobic polystyrene backbones are most likely embedded inside the Pdot particles while the hydrophilic PEG chains and functional groups extend outside into the aqueous environment. Unlike physical adsorption, therefore, this method should permanently anchors the PEG and functional groups to the Pdot surface. The PEG chains provide a biocompatible layer that minimizes non-specific absorption. The PEG chains also act as a steric barrier against nanoparticle aggregation, while the functional carboxyl group can be easily covalently-linked to biomolecules using established protocols.32
Scheme 1.
Surface functionalization of semiconducting polymer dots and subsequent bioconjugation via EDC-catalyzed coupling.
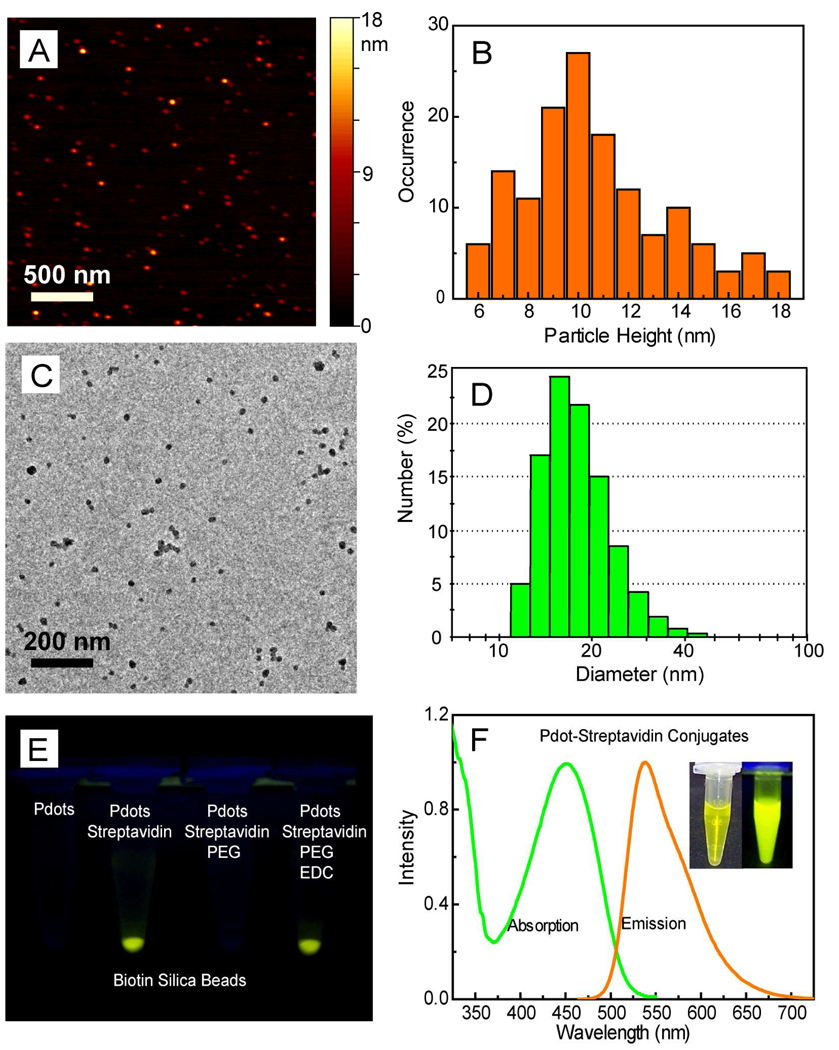
We used the PS-PEG-COOH polymer to functionalize Pdots made from the highly fluorescent semiconducting PFBT (Scheme 1). Functionalized PFBT dots were prepared using a precursor solution mixture with a constant PFBT concentration and PS-PEG-COOH/PFBT fractions ranging from 0 to 20 weight percent. The size and morphology of the functionalized PFBT dots were characterized by atomic force microscopy (AFM, Figure 1A). Particle height histogram obtained from AFM images indicated that the majority of PFBT dots possessed diameters in the range of 10±3 nm (Figure 1B). The small particle size was further validated with transmission electron microscopy (TEM, Figure 1C) and dynamic light scattering (DLS, Figure 1D). Both measurements indicated comparable particle sizes of the functionalized Pdots, with an average diameter of ~15 nm. The sizing measurements are consistent with each other in that the lateral size of collapsed particle from TEM and the hydrodynamic size from DLS should be slightly larger than the particle height obtained from AFM. In comparison with the unfunctionalized Pdots, the presence of a small amount of PS-PEG-COOH polymer (less than 20 wt%) did not cause any noticeable effects on particle size and morphology. The size control of functionalized Pdots exhibited similar trend as that for bare Pdots, that is, the nanoparticles produced using a lower precursor concentration possess smaller particle size, while those prepared from more concentrated precursor solutions exhibited larger diameters. The functionalized Pdots in all the characterizations and bioconjugations described in this paper were prepared by using a precursor solution containing 80 wt% PFBT and 20 wt% PS-PEG-COOH. Absorption and emission spectra of Pdots do not change with the nanoparticle size 23, and thus this size-independent feature of Pdot significantly relaxes the constraint on size control in nanoparticle preparation. Moreover, there does not appear to be an obvious size effect on fluorescence quantum yield for PFBT particles, likely due to the excellent quality and chemical stability of the polymer, which results in minimal quenching by defects. This size-independence feature may be advantageous to obtain brighter probes for certain applications because a larger size merely increases the brightness of the probe. It should be noted that this functionalization strategy led to the most effective nanoparticle probes in terms of fluorophore density: more than 80 percent of the semiconducting polymer nanoparticles were effective fluorophores. In contrast, for Qdots and dye-loaded spheres, the effective fluorophores are limited to a few percent of the particle volume or weight due to the presence of a thick encapsulation layer (for Qdots) or self-quenching of dyes (for dye-doped spheres).
Figure 1.
(A) Typical AFM image of functionalized PFBT dots. (B) Histogram of particle height taken on AFM images of functionalized PFBT dots. (C) TEM image of functionalized PFBT dots. (D) Hydrodynamic diameter of functionalized PFBT dots measured by dynamic light scattering. (E) An assay using biotin silica beads to verify bioconjugation through EDC-catalyzed covalent coupling. (F) Absorption and fluorescence spectra of PFBT dot-streptavidin bioconjugates in 1× PBS buffer solution after 6 months of storage, the inset shows photographs of the Pdot-bioconjugate solution under room (left picture) and UV (right picture) illumination.
As a start, we chose to conjugate Pdots with streptavidin because most biological labeling molecules, such as antibodies, can be easily derivatized with biotin. However, because the relatively large surface area of Pdots is intrinsically hydrophobic — although surface modification tends to make it more hydrophilic — there is a concern that biomolecules will be non-specifically adsorbed onto the Pdot surface. We found this concern was indeed valid: carboxyl functionalized Pdots (but lacking the coupling reagent that links carboxyl to amine groups on proteins) were mixed with streptavidin in a buffer solution and then incubated with biotin silica beads. After centrifugation, the Pdots with streptavidin were clearly retained in pellet of the biotin silica beads, and those without streptavidin showed no binding to the beads (Figure 1E), thus indicating severe non-specific adsorption of streptavidin onto the Pdot surface.
To overcome non-specific adsorption, Pdots were mixed with streptavidin in a buffer solution containing 0.1 wt% polyethylene glycol (PEG). The resulting Pdots showed no detectable binding to biotin silica beads, suggesting that the presence of PEG significantly reduced non-specific adsorption (Figure 1E). Accordingly, covalent bioconjugation was successfully performed in a PEG-containing buffer. The peptide bond formation between the carboxyl groups on Pdots and the amine groups of streptavidin was catalyzed by a carbodiimide such as 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC). The EDC-catalyzed, Pdot-streptavidin conjugates showed clear binding to biotin silica beads; binding was not observed for the products obtained in the absence of EDC (Figure 1E). In a separate control, we used identical bioconjugation conditions (i.e. streptavidin and EDC in PEG-containing buffer) but with bare, unfunctionalized Pdots. Binding was not detectable on biotin beads, further confirming that the bioconjugation of streptavidin to Pdots was covalent and that labeling of strepadvidin-Pdots to biotin beads was specific and without any detectable non-specific binding.
The Pdots may be further passivated with additives such as bovine serum albumin (BSA), which can maintain long-term colloidal stability, block hydrophobic surfaces, and reduce non-specific binding in labeling experiments. We found BSA-passivated Pdot bioconjugates are stable for months at physiological pH in HEPES, PBS, Tris, and borate buffers. Figure 1F inset shows two photographs of PFBT-streptavidin conjugates in 1× PBS buffer after 6 months of storage. The suspension of PFBT conjugates was stable, clear (not turbid), and exhibited strong fluorescence under UV lamp illumination (365 nm).
We have achieved successful conjugations of streptavidin or antibodies to different types of Pdots, including the five Pdots described in our previous report.23 In this paper, we focus our results and discussions on PFBT dots for single-particle imaging, cellular labeling, and flow cytometry applications. PFBT dots exhibit a relatively broad absorption peak around 460 nm (Figure 1F), which is a convenient wavelength region for fluorescence microscopy and laser excitations. Analysis of the absorption and fluorescence spectra from ~10 nm-diameter PFBT dots indicated a peak extinction coefficient of 1.5×107 M−1cm−1 and a fluorescence quantum yield of 0.30. The photophysical properties of PFBT dots are summarized in Table 1, together with the properties of two widely used probes purchased from Invitrogen: Qdot 565 and fluorescent IgG-Alexa 488 (~6 dye molecules per IgG). These two commercial probes were selected because they have emissions in a similar wavelength region as that of PFBT dots. It is important to note that Pdots contain multiple emitters, thus, although the lifetimes of Pdots are ~ 50 times shorter than Qdots, the emission rates of Pdots can be three orders of magnitude faster than Qdots.23
Table 1.
Photophysical Properties of PFBT dots, IgG-Alexa 488, and Qdot 565.
| Probe (Size) |
PFBT dot (~10 nm) |
Alexa 488 (~1 nm) |
Qdot 565 (~15 nm) |
|---|---|---|---|
| Abs. / Fluores. Max | 460 nm / 540 nm | 496 nm / 519 nm | UV/565 nm |
| Extinction Coeffocient at 488 nm | 1.0×107 M−1 cm−1 | 5.3×104 M−1 cm−1 | 2.9×105 M−1 cm−1 |
| Quantum Yield | 0.3 | 0.9 | 0.3~0.5 |
| Fluorescence Lifetime | 0.6 ns | 4.2 ns | ~2.0 ns |
Note: The data for Alexa 488 and Qdot 565 are according to the probe specification provided by Invitrogen. The parameters of Alexa 488 are for single-dye molecules. An IgG-Alexa 488 probe has a hydrodynamic diameter of 12 nm, contains an average of 6 dye molecules, but its brightness corresponds to ~ 2–4 dye molecules due to self-quenching. Fluorescence lifetime of PFBT dots was measured by a TCSPC setup (Supplementary Figure S5). Also note that single PFBT dots contain multiple emitters, which results in photon emission rates that are higher than those predicted from fluorescence lifetime alone.
Single-particle fluorescence brightness
A useful estimate of fluorescence brightness is given by the product of the peak absorption cross section and the fluorescence quantum yield. Photophysical data indicate that PFBT dots of ~10 nm diameter are about 30 times brighter than IgG-Alexa 488, and Qdot 565 probes under a typical laser excitation (488 nm). We note that the absorption cross section of Qdots increases as excitation wavelength is shifted to the blue (e.g. 405 nm), and that the absorption cross section of PFBT dots at 488 nm is ~2× less than that at ~460 nm. However, excitation wavelengths above 450 nm are typically preferred in biological imaging, due to increased autofluorescence and phototoxicity at bluer wavelengths. Therefore, while multicolored Pdots have been developed,23 there remains a need to further improve the brightness of red-emitting Pdots. Nevertheless, a side-by-side brightness comparison of green-emitting PFBT dots with the IgG-Alexa 488, and Qdot 565 probes should serve as a useful reference. Therfore, we carried out single-particle imaging to experimentally evaluate and compare the brightness and photostability of the three probes.
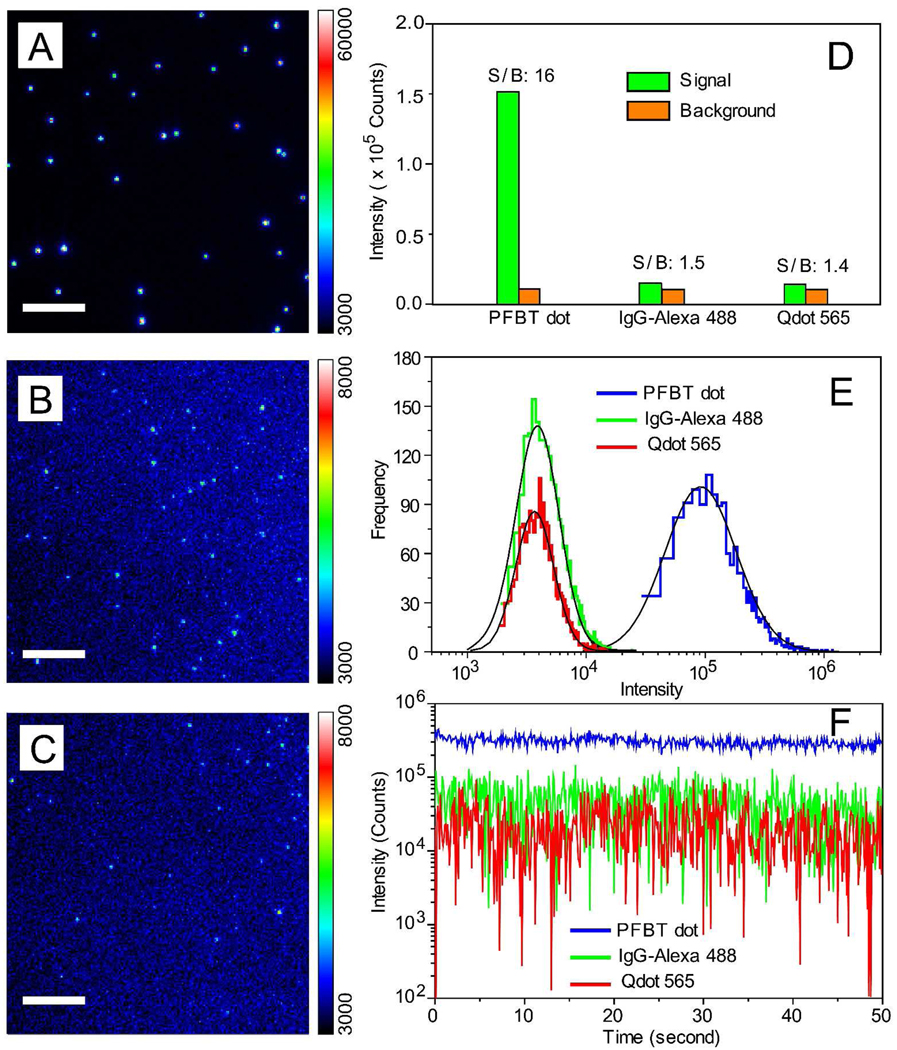
Figure 2A, 2B, and 2C show typical single-particle epi-fluorescence images of PFBT dots, IgG-Alexa 488, and Qdot 565, respectively, obtained under identical acquisition and laser excitation conditions. With a relative low excitation power (1 mW) from a 488 nm laser, very bright, near-diffraction-limited spots were clearly observed for individual PFBT dots. Some Pdots actually saturated the detector (Figure 2A), whereas the IgG-Alexa 488 and Qdots exhibited much lower intensity levels, barely detected by the camera at the low excitation power we used (Figure 2B, 2C). The PFBT dots exhibited an order-of-magnitude improvement in signal-to-background ratio compared to those of Qdot 565 and IgG-Alexa 488 (Figure 2D). Such a prominent contrast is primarily due to the high per-particle absorption cross section of Pdots, which would be particularly suitable for fluorescence detection requiring low excitation conditions. For further comparing the probe performance, we increased laser excitation power to 4 mW so that Qdot 565 and IgG-488 probes can be sufficiently detected by the camera. Because saturation of the detector was observed for Pdot particles, a neutral density filter (optical density of 1.5, which blocks 97% of the emitted fluorescence) was placed together with the emission filter when imaging Pdot samples, and their fluorescence intensities were back-calculated according to the attenuation factor. For all the three probes, background was subtracted. Fluorescence intensity distribution of several thousands particles indicated that PFBT dots were ~30 times brighter than IgG-Alexa 488 and Qdot 565 (Figure 2E), consistent with the brightness comparison based on the photophysical parameters.
Figure 2.
Single-particle fluorescence images of (A) PFBT dot, (B) IgG-Alexa 488, and (C) Qdot 565, obtained under identical excitation conditions. Note the color bar for IgG-Alexa and Qdot 565 has to be set to a lower value (8000 counts rather than 60,000 counts) because they are significantly dimmer than PFBT dots. Scale bar represents 5 µm. (D) Signal and background for single Pdots as compared to single IgG-Alexa 488 and single Qdots, observed under identical excitation power of 1 mW. (E) Intensity distributions of single particle fluorescence for the three probes under the excitation power of 4 mW. Pdots are ~30 brighter than either IgG-Alexa 488 or Qdots. (F) Single-particle photobleaching trajectories. Blinking was not observed for PFBT dots (blue), while frequent blinking was observed for Qdots (red).
Single-particle photobleaching measurements indicated excellent photostability of PFBT dots (Figure 2F). Statistical analyses of multiple photobleaching trajectories showed that over 109 photons per Pdot were emitted prior to photobleaching, two or three orders of magnitude larger than those emitted by individual Qdot 565 and IgG-Alexa 488 particles. Furthermore, a large number of photons could be obtained from individual Pdots at high acquisition rates (~200,000 photons detected per Pdot per 20 ms exposure) because of their high brightness, short fluorescence lifetime, and the presence of multiple emitters per particle. This feature was recently exploited to yield a particle tracking uncertainty of ~1 nm,29 which makes Pdots far superior in high-speed single-particle tracking experiments than conventional fluorescent dyes and Qdots. It is worth noting that most PFBT dots exhibit continuous emission behavior without any obvious fluorescence blinking while most Qdots exhibit pronounced blinking (Figure 2F). This non-blinking feature of Pdots is particularly valuable in single-molecule applications.
Specific labeling of cellular targets with Pdots
It was previously demonstrated that bare Pdots could be delivered into cultured cells, presumably by endocytosis when Pdots non-specifically bound to the cell surface; specific cellular targets were not labeled.23,28,30,31 Therefore, it was unclear from these studies whether Pdot probes could be made specific enough to recognize cellular targets for effective labeling in real applications.
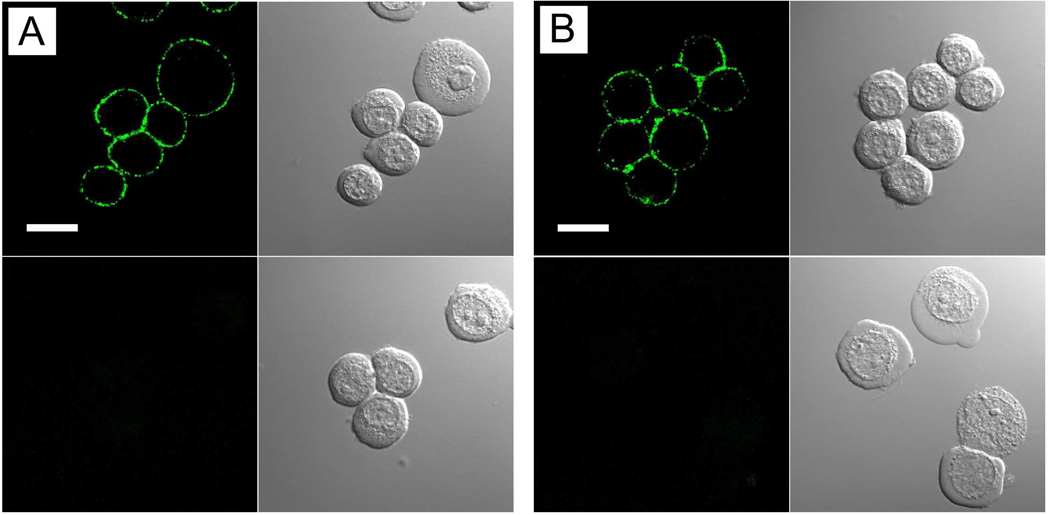
Streptavidin and IgGs are widely used in bioconjugation for immunofluorescent labeling of cellular targets. We created Pdot-IgG and Pdot-streptavidin probes and investigated their ability to label a specific cellular target, EpCAM, an epithelial cell-surface marker currently used for the detection of circulating tumor cells. Figure 3A shows the Pdot-IgG probes successfully labeled EpCAM receptors on the surface of live MCF-7 human breast cancer cells after the cells were incubated with a monoclonal primary anti-EpCAM antibody. When the cells were incubated with just the Pdot-IgG alone, in the absence of the primary antibody, cell-labeling was not detected (Figure 3A, bottom), indicating that the Pdot-IgG conjugates are highly specific for the target.
Figure 3.
Fluorescence imaging of cell-surface marker (EpCAM) in human breast cancer cells labeled with Pdot bioconjugates. (A) Imaging of live MCF-7 cells incubated sequentially with anti-EpCAM primary antibody and Pdot-IgG conjugates. The bottom panels show control samples in which the cells were incubated with Pdot-IgG alone (no primary antibody). The Nomarski (DIC) images are shown to the right of the confocal fluorescence images. Scale bar represents 20 µm. (B) Imaging of live MCF-7 cells incubated sequentially with anti-EpCAM primary antibody, biotinylated goat anti-mouse IgG secondary antibody, and Pdot-streptavidin conjugates. The bottom panels show control samples where the cells were incubated with anti-EpCAM antibody and Pdot-strepavidin (no secondary antibody). The Nomarski (DIC) images are shown to the right of the confocal fluorescence images. Scale bar represents 20 µm.
Next, we used Pdot-streptavidin conjugates as an alternative probe to detect EpCAM. The Pdot-streptavidin probes, together with the primary anti-EpCAM antibody and biotinylated goat anti-mouse IgG secondary antibody, also effectively labeled EpCAM on the surface of live MCF-7 cells (Figure 3B). When the cells were incubated with primary antibody and Pdot-streptavidin in the absence of biotin anti-mouse IgG, no fluorescence was observed on the cell surface (Figure 3B, bottom), thus again demonstrating the highly specific binding of Pdot-strepavidin. The lack of signal also indicated the absence of nonspecific binding in this biotin-streptavidin labeling system. We further employed Pdot bioconjugates to label another cell-surface marker, Her2 (target of the anti breast cancer drug, Heceptin), on a different cell line SK-BR-3 (Supplementary Figure S1), as well as subcellular structures such as microtubules in fixed MCF-7 cells (Supplementary Figure S2). Pdot bioconjugates in both cases labeled the targets specifically and effectively, demonstrating their comprehensive application to cell labeling.
Cell-labeling brightness comparisons
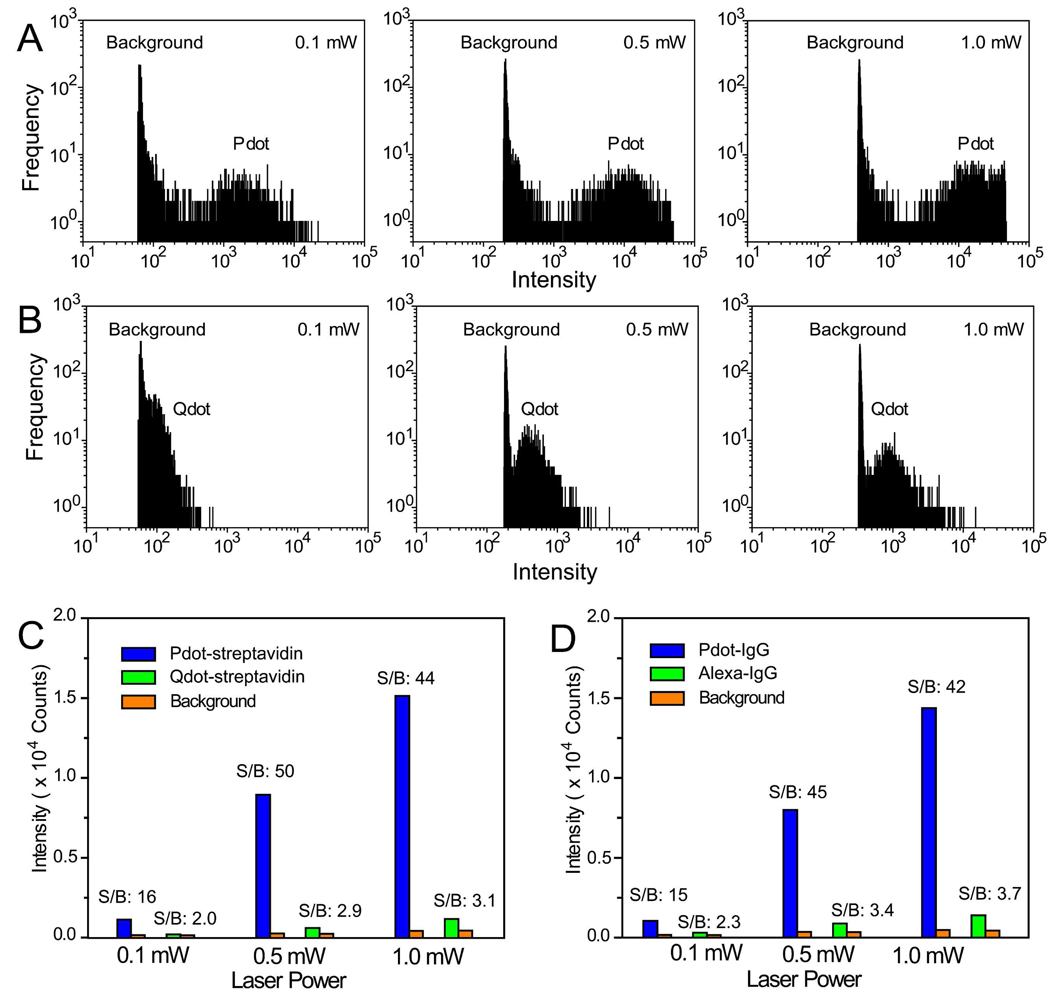
Besides fluorescence imaging, flow cytometry is another area where the brightness of probes is important. We compared the labeling brightness of Pdot bioconjugates with those of commercially available Qdot-streptavidin and Alexa-IgG probes using a microfluidic flow cytometer. Figure 4A shows the flow-through detection of MCF-7 cells labeled with Pdot-streptavidin. At the lowest excitation intensity we used (0.1 mW), a well-defined intensity peak for the Pdot-labeled cells appeared far above the background. In contrast, the peak for Qdot-labeled ones was not clearly separated from the background (Figure 4B). The Pdot peak moved to higher intensity with increasing excitation intensity and started to saturate the detector at a laser power of 0.5 mW. In all excitation conditions, MCF-7 cells labeled with Pdot-streptavidin exhibited much higher intensity levels compared to the results of Qdot-labeled cells using the same labeling concentration as Pdot-streptavidin. The Pdot probes could provide significantly higher signal level at low excitation conditions, a very useful benefit for biological detection in optically turbid media such as blood or thick tissues.
Figure 4.
Flow-through detection of fluorescently labeled cancer cells. (A) Fluorescence intensity distributions obtained by flowing Pdot-streptavidin labeled MCF-7 cells through a microfluidic flow cytometer; laser excitation was varied from 0.1 to 0.5 to 1 mW. (B) Fluorescence intensity distributions for Qdot 565-streptavidin labeled MCF-7 cells obtained under identical experimental conditions as those used in (A). (C) Comparison of average fluorescence brightness obtained using the microfluidic flow cytometer for cells labeled with Pdot-streptavidin and Qdot-streptavidin. (D) The same experiment and comparison as described in (A–C) was carried using Pdot-IgG and Alexa 488-IgG.
Similar intensity comparisons were performed using Pdot-IgG and Alexa 488-IgG probes using the microfluidic flow cytometer (Supplementary Figure S3). Quantitative analyses of the flow cytometry data showed that the average intensity of Pdot-labeled cells is ~25 times brighter than the Qdot-labeled ones (Figure 4C), and ~18 times brighter than Alexa-IgG labeled cells (Figure 4D). We further quantified the labeling brightness by analyzing fluorescence images of MCF-7 cells labeled with either Pdot-streptavidin or Qdot-streptavidin: The Pdot-labeled cells were ~20 times brighter than the Qdot-labeled ones, consistent with the flow cytometry data (Supplementary Figure S4).
These cell-labeling comparison values are slightly lower than those obtained from single-particle imaging. The lower values may be attributed to several factors, such as discrepancies in collective emission behavior of probe assemblies compared to individual particles; change in binding constants of antibody or streptavidin upon bioconjugation; or variation in emission rate with excitation intensity (saturation). It is also worth noting that cell labeling was performed according to the optimized concentrations for Qdot-streptavidin and Alexa-IgG probes which may not be optimal for Pdot probes. Therefore, we believe the present comparison is a conservative estimate of the advantages provided by Pdot bioconjugates over traditional dye and Qdot bioconjugates. More detailed work is needed for optimizing the bioconjugation reactions, as well as the labeling conditions, for this new class of Pdot-based probes. Nevertheless, the current cell imaging and flow cytometry results clearly indicate that Pdot labeling provides significant improvements in signal level compared to commercially available Alexa-IgG and Qdot probes.
Conclusion
We have developed highly fluorescent semiconducting polymer dots with functional groups that allow for covalent conjugation to biomolecules. The strategy is based on entrapping heterogeneous polymer chains into a Pdot particle, driven by hydrophobic interactions during nanoparticle formation. We have shown that a small amount of amphiphilic polymer bearing functional groups can be co-condensed with the majority of semiconducting polymers to modify and functionalize the nanoparticle surface. Subsequent covalent conjugation to biomolecules such as streptavidin and antibodies were performed using the standard carbodiimide coupling chemistry. These Pdot bioconjugates can effectively and specifically label cell-surface receptors and subcellular structures in both live and fixed cells, without any detectable non-specific binding. We performed single-particle imaging, cellular imaging, and flow cytometry to experimentally evaluate the Pdot performance, and demonstrate their high cellular labeling brightness compared to those of Alexa-IgG and Qdot probes. Our results bring forward a new class of highly fluorescent nanoparticle bioconjugates for a wide range of fluorescence-based biological detection.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Heath (NS062725, CA147831, AG029574). We acknowledge support from the Keck Imaging Center and Center of Nanotechnology at the University of Washington. J.D.M. acknowledges support from the National Institutes of Health (GM 081040) and support from the National Science Foundation (CHE 0547846) for equipment used in this research.
Footnotes
Supporting Information Available
Materials, experimental details, and supplementary Figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pepperkok R, Ellenberg J. Nat. Rev. Mol. Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat. Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Suarez M, Ting AY. Nat. Rev. Mol. Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 5.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 6.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 7.Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP, Ge NF, Peale F, Bruchez MP. Nat. Biotechnol. 2003;21:452–452. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 8.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao J, Larson DR, Vishwasrao HD, Zipfel WR, Webb WW. Proc. Natl. Acad. Sci. USA. 2005;102:14284–14289. doi: 10.1073/pnas.0506523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XY, Ren XF, Kahen K, Hahn MA, Rajeswaran M, Maccagnano-Zacher S, Silcox J, Cragg GE, Efros AL, Krauss TD. Nature. 2009;459:686–689. doi: 10.1038/nature08072. [DOI] [PubMed] [Google Scholar]
- 11.Friend RH, Gymer RW, Holmes AB, Burroughes JH, Marks RN, Taliani C, Bradley DDC, Dos Santos DA, Bredas JL, Loglund M, Salaneck WR. Nature. 1999;397:121. [Google Scholar]
- 12.Gunes S, Neugebauer H, Sariciftci NS. Chem. Rev. 2007;107:1324–1338. doi: 10.1021/cr050149z. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, McBranch DW, Wang HL, Helgeson R, Wudl F, Whitten DG. Proc. Natl. Acad. Sci. USA. 1999;96:12287–12292. doi: 10.1073/pnas.96.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan CH, Wang S, Hong JW, Bazan GC, Plaxco KW, Heeger AJ. Proc. Natl. Acad. Sci. USA USA. 2003;100:6297–6301. doi: 10.1073/pnas.1132025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas SW, Joly GD, Swager TM. Chem. Rev. 2007;107:1339–1386. doi: 10.1021/cr0501339. [DOI] [PubMed] [Google Scholar]
- 16.Szymanski C, Wu C, Hooper J, Salazar MA, Perdomo A, Dukes A, McNeill JD. J. Phys. Chem. B. 2005;109:8543–8546. doi: 10.1021/jp051062k. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Szymanski C, McNeill J. Langmuir. 2006;22:2956–2960. doi: 10.1021/la060188l. [DOI] [PubMed] [Google Scholar]
- 18.Palacios RE, Fan FRF, Grey JK, Suk J, Bard AJ, Barbara PF. Nat. Mater. 2007;6:680–685. doi: 10.1038/nmat1959. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, Zheng Y, Szymanski C, McNeill J. J. Phys. Chem. C. 2008;112:1772–1781. doi: 10.1021/jp074149+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C, McNeill J. Langmuir. 2008;24:5855–5861. doi: 10.1021/la8000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collini E, Scholes GD. Science. 2009;323:369–373. doi: 10.1126/science.1164016. [DOI] [PubMed] [Google Scholar]
- 22.Wu C, Szymanski C, Cain Z, McNeill J. J. Am. Chem. Soc. 2007;129:12904–12905. doi: 10.1021/ja074590d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. ACS Nano. 2008;2:2415–2423. doi: 10.1021/nn800590n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. Angew. Chem. Int. Ed. 2009;48:2741–2745. doi: 10.1002/anie.200805894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon JH, McDaniel W, MacLean P, Hancock LE. Angew. Chem. Int. Ed. 2007;46:8223–8225. doi: 10.1002/anie.200701991. [DOI] [PubMed] [Google Scholar]
- 26.Baier MC, Huber J, Mecking S. J. Am. Chem. Soc. 2009;131:14267–14273. doi: 10.1021/ja905077c. [DOI] [PubMed] [Google Scholar]
- 27.Abbel R, van der Weegen R, Meijer EW, Schenning APHJ. Chem. Commun. 2009:1697–1699. doi: 10.1039/b822943k. [DOI] [PubMed] [Google Scholar]
- 28.Pu KY, Li K, Shi JB, Liu B. Chem. Mater. 2009;21:3816–3822. [Google Scholar]
- 29.Yu J, Wu C, Sahu S, Fernando L, Szymanski C, McNeill J. J. Am. Chem. Soc. 2009;131:18410–18414. doi: 10.1021/ja907228q. [DOI] [PubMed] [Google Scholar]
- 30.Howes P, Green M, Levitt J, Suhling K, Hughes M. J. Am. Chem. Soc. 2010;132:3989–3996. doi: 10.1021/ja1002179. [DOI] [PubMed] [Google Scholar]
- 31.Rahim NAA, McDaniel W, Bardon K, Srinivasan S, Vickerman V, So PTC, Moon JH. Adv. Mater. 2009;21:3492–3496. [Google Scholar]
- 32.Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, WChung L, Petros JA, O'Regan RM, Yezhelyev MV, Simons JW, Wang MD, Nie S. Nature Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.