Abstract
In less than 20 years, our appreciation for micro-RNA molecules (miRNAs) has grown from an original, curious observation in worms to their current status as incredibly important global regulators of gene expression that play key roles in many transformative biological processes. As our understanding of these small, non-coding transcripts continues to evolve, new approaches for their analysis are emerging. In this critical review we describe recent improvements to classical methods of detection as well as innovative new technologies that are poised to help shape the future landscape of miRNA analysis.
Introduction
MicroRNAs (miRNAs) constitute a critically important class of non-translated, small RNAs which post-transcriptionally regulate gene expression via one of multiple mechanisms.. [1] First reported in 1993 as a curious anomaly in Caenorhabditis elegans, [2] thousands of miRNAs have now been identified and shown to play key roles in many transformative biological processes, including developmental timing, [3–5] stem cell differentiation, [6–8] and disease development. [9–10] Although the complete functional role that miRNAs play still remains to be fully elucidated, their conservation throughout Archaea, [11] bacteria, [12] plants, [13] and animals[14] indicate their importance as key regulatory control elements during both normal and transformative biological processes. In contrast to small interfering RNAs (siRNAs), [15] miRNAs are endogenously encoded into the genome and are initially transcribed as long primary transcripts (≥1 kb; pri-miRNAs), which are then enzymatically processed in the nucleus by Drosha into ~70 nt stem loop structures (pre-miRNAs). Pre-miRNAs are exported into the cytoplasm and processed by the enzyme Dicer into the mature 19–24 nt duplexes.
As opposed to siRNAs, which operate almost exclusively via mRNA cleavage at regions having perfect sequence complementarity, miRNAs can modulate gene expression via one of three distinct mechanisms and do not necessarily require perfect base pairing to act upon a target. [1] In the cytoplasm, the single strands form the mature miRNA duplexes are incorporated into the RNA-induced silencing complex (RISC). Guided by the miRNA, the RISC complex can then act on mRNAs through one of three distinct mechanisms: 1) cleavage of the targeted mRNA, a mechanism commonly observed in plants that often requires perfect complementarity between miRNA and mRNA, 2) translational repression, whereby miRNA/RISCs bind to 3 untranslated regions of mRNAs preventing translation by the ribosome, and 3) the recently discovered enhancement of translation, in which a miRNA binds to the 5′-terminal oligopyrimidine tract (5′-TOP) and relaxes a cis-element in the 5′ UTR that inhibits translation. [16]
There are over 15,000 mature miRNA sequences listed in the recently released miRBase 15.0 database, with ~1000 identified as human miRNAs. [17] Through one or more of the aforementioned mechanisms, each miRNA can potentially regulate the expression of multiple mRNAs, meaning that downstream production of many gene products, ultimately proteins, can be tremendously influenced by alterations in the expression of a single miRNA. [18] In fact, it is known that a majority of human mRNAs are regulated by one (or more) miRNAs. [19] Furthermore, it has recently been experimentally demonstrated that multiple miRNAs, many of which are expressed as clusters that are encoded in close genomic proximity to one another, can target the same mRNA, [20] adding further complexity to the mechanisms through which miRNAs regulate gene expression.
Given the prominent role that miRNAs play in “normal” gene expression and organismal function, it is not surprising that the aberrant expression of miRNAs can lead to a wide range of human diseases and disorders, including: cancer, [21–22] neurodegenerative diseases, [23–24] diabetes, [25] heart diseases, [26] kidney diseases, [27–28] liver diseases, [29] and altered immune system function, [30–31] amongst others. In addition to contributing to the underlying cause of a particular disease, miRNAs can also represent potential therapeutic targets[32–34] and diagnostic biomarkers. [35] Particularly exciting are the discovery of circulating miRNAs, which are promising biomarker candidates since they can be detected from readily attainable blood samples. [36–38]
Almost entirely due to their short size, the analysis of miRNAs is considerably more difficult than it is for much longer mRNAs. In particular, the small size of miRNAs greatly complicates the use of standard molecular biology methods based upon the polymerase chain reaction (PCR), as detailed below. Furthermore, the short size also makes hybridization-based assays difficult as the melting temperature and binding dynamics of complementary probes toward their target miRNAs vary significantly with the identity of the target miRNA. Furthermore, experimental parameters, such as the buffer composition, the hybridization temperature, and incubation time all can contribute to significant assay-to-assay variation. [39–43]
So, what are desirable attributes for existing and emerging miRNA analysis methods? Clearly the most appropriate technique for a given measurement challenge varies tremendously based upon the application and setting. For example, in an academic laboratory setting well-established techniques that rely upon the tools of traditional molecular biology might find favor, whereas emerging micro- or nanotechnology-based methods might eventually be most well-suited for point-of-care diagnostic applications. Two other important considerations when selecting an existing or designing a new method for miRNA analysis include dynamic range and multiplexing capability. The expression level of miRNAs, as determined via intracellular copy number, can vary from sequence to sequence by up to a factor of 105 within a single sample. Furthermore, the recent discoveries of multiple miRNAs targeting a single mRNA and regulated expression amongst entire families of miRNAs provide motivation for global miRNA analyses, which will require methods wherein multiple miRNAs, and perhaps the entire “miRNA-ome”, is simultaneously detected in parallel in order to fully elucidate the important and complex function of these tiny regulators.
On account of the critical biological role that miRNAs play in biological function and the diverse range of applications in which miRNA analysis is of value, significant effort has been invested over the past decade to develop new detection methods. In this critical review we highlight a selection of existing and emerging tools for miRNA analysis, with a particular emphasis on the current state-of-the-art and important developments in this fast moving field, as reported in the primary literature in the past four years.
Computational approaches for miRNA target prediction
While the major focus of the review article lies in existing and emerging miRNA detection methods, it is worthwhile to briefly mention computational methods for predicting miRNA targets. [44] Given that the number of potential mRNA targets and the fact that miRNAs can regulate mRNAs that are not perfectly complementary in sequence, the experimental identification and validation of miRNA regulatory sites is a vast challenge. For this reason, extensive effort has been invested in developing computational methods for predicting the mRNA targets of miRNAs.
One general class of computational methods for the prediction of miRNA targets utilizes perfect or imperfect complementarity via Watson-Crick base-pairing between he miRNA and possible target candidates. [45] Most of these approaches focus on the complimentary at seed sequences, 5–8mers at the 5′ end of an miRNA that are often highly conserved. [46–48] PicTar, utilizes the sequence complementarity to target sites with emphasis on perfect base-pairing in the seed region, [47, 49–50] while TargetScan, one of more established computational tools, accounts for both complementarity as well as evolutionary conservation to provide a relatively likelihood that a given sequence is a miRNA target. [48, 51]
Another general framework for prediction of miRNA targets involves energetic calculations. DIANA-microT, developed by Kiriakidou et al., is an algorithm that identifies miRNA targets based on the binding energies between two imperfectly paired RNAs [52–54] and RNAHybrid predicts miRNA targets by finding the most energetically favorable hybridization sites of a small RNA in a larger RNA sequence. [55–56] The miRNanda prediction algorithm includes contributions from the interaction binding energy, sequence complementarity between a set of mature miRNAs and a given mRNA, and also weights the conservation of the target site across various species. [57–58] In contrast to other energetic calculations, STarMIR, models the secondary structure of an mRNA to determine the likelihood of miRNA binding. [59]
The past few years has seen incredible growth in the area of computational prediction of miRNA targets. However, continued progress remains to be achieved as many of the aforementioned tools offer too many false positive target sites. Furthermore, many of the approaches have been developed using experimentally validated miRNA:mRNA systems, therefore introducing bias against miRNAs having and unusual or uncommon sequence. Nonetheless, the continued evolution of miRNA target prediction methodologies will, along with emerging detection methods, play a key role in fully elucidating the mechanisms by which miRNAs regulate normal and potentiate abnormal organismal function – providing a link between diagnostic insight and potential therapeutic opportunities.
Molecular biology-based analysis methods
Early reports featuring miRNA measurements were fueled by what was already available in the laboratories of researchers at the forefront of the field—traditional molecular biology techniques such as cloning and enzymatic ligation assays. As timing would have it, miRNA research began to gather momentum directly on the heels of the genome technology explosion, and thus technologies such as RT-PCR and cDNA microarrays were rapidly adapted to accommodate the needs of the miRNA researcher. This section details the current state-of-the-art for miRNA detection. Based upon well-established methodologies, but with the recent incorporation of several very important innovations, these techniques represent the most commonly utilized methods for miRNA analysis in the research biology laboratory setting.
Cloning
Cloning was one of the first techniques utilized to detect and discover miRNAs. [60–62] Although slow and laborious, cloning is still at times used for miRNA detection. A more recent development that has been developed for the discovery of miRNAs is miRAGE – miRNA serial analysis of gene expression. [63] Similar to cloning, small RNAs are extracted and amplified via the reverse-transcriptase polymerase chain reaction (RT-PCR) into complementary DNAs (cDNAs). In this application, biotinylated primers are utilized in the PCR step allowing the cDNA products to be purified via affinity chromatography with streptavidin-coated beads. The cDNAs are enzymatically cleaved from the beads and the eluted products can be cloned and sequenced. miRAGE is advantageous in that it can identify up to 35 tags in a single iteration, versus about five using conventional cloning. [63] However, this technique is extremely labor intensive, requires hundreds of μg of total RNA, and only provides information as to the presence or absence of a particular miRNA from within a sample. [64] While cloning still remains a powerful technique for the validation and discovery of novel miRNAs, the associated shortcomings of the technique make it impractical for high-throughput miRNA detection and expression profiling. [37]
Northern Blotting
At present, the most standard method for the detection of miRNAs is Northern blotting. [65–67] Northern blotting offers a number of advantages for miRNA analysis including a number of well established protocols and amenability to equipment readily available in most molecular biology laboratories. Additionally, since Northern blotting involves a size-based separation step, it can be used to detect both mature and precursor forms of a miRNA, which is appealing for studies which focus on the mechanisms of miRNA processing.
Common protocols for Northern blotting involve miRNA isolation, polyacrylamide gel electrophoresis, transfer of the separated sample to the blotting membrane, and visualization via hybridization with a radioactively labeled DNA strand complementary to the miRNA of interest. Despite its widespread use, traditional Northern blotting is, in general, plagued by a lack of sensitivity (up to 20 μg of total RNA required per blot) and a laborious and time consuming protocols (often taking several days for complete analysis), which limits its utility in a clinical setting. [68] Furthermore, the technique often displays a limited dynamic range (2–3 orders of magnitude depending on the visualization method) and the reliance on a radioactive tag (typically 32P) can be disadvantageous in some settings. [69] Northern blots do allow for multiple samples to be analyzed in a side-by-side format, but only one miRNA can be assayed for at a given time, a drawback which is of increasing importance as researchers strive towards global analyses for a systems level understanding of miRNA function.
A number of improvements have been made to traditional Northern blotting protocols that help assuage several of the aforementioned problems. Of particular significance is the incorporation of locked nucleic acid (LNA) hybridization probes. [70–72] LNAs are based upon DNA bases but feature the addition of a methylene bridge connecting the 2′-oxygen of the ribose to the 4′-carbon, effectively rigidifying the strand by inducing organization of the phosphate backbone. [73] As a result, oligonucleotide strands that incorporate LNAs have been shown to bind complementary RNA strands with considerably higher affinity and target specificity compared to their DNA-only analogues. Furthermore, RNA:LNA duplexes are unique from RNA:DNA or RNA:RNA duplexes in that they have altered interactions with several nucleic acid recognizing proteins, including some enzymes. In order to avoid the necessity for a radioactive tag, Ramkissoon et al. demonstrated that digoxigenin (DIG), a steroid hapten, could be incorporated into complementary RNA strands used to visualize Northern blots for three different miRNAs. [74] Their incorporation of DIG and the accompanying chemiluminescent readout reduced the time-to-result from days to hours, and increased the shelf-life of the probes, compared to radioactively labeled strands.
Reverse-Transcriptase Polymerase Chain Reaction
Similarly to its use in conventional studies of RNA expression, RT-PCR can also be applied to the analysis of miRNAs. Reverse transcription is first utilized to convert the target RNA into its cDNA, which is then subsequently amplified and quantified via one of several conventional PCR methods. However, the simple translation of these methods to miRNAs is complicated by the short size of the target, as the length of the primers normally used in the PCR step are as long as mature miRNAs themselves. Shorter primers are typically not useful as their low duplex melting temperature with the miRNA can introduce signal bias. To avoid these challenges, researchers have developed an array of creative approaches based upon enzymatic modification of conventional primers or altogether new primers for mature miRNA.
One of the first applications of RT-PCR for the detection of miRNA was reported by Schmittgen et. al, who examined pre-miRNA expression. [75] Because the study did not examine mature miRNAs, shortened primers were not necessary and the researchers were able to successfully detect amplicons using a fluorescent readout. However, the assumption that the amount of pre-miRNA is strictly representative of mature miRNA expression does not rigorously hold and thus the most straight-forward application of RT-PCR is of limited utility.
As a method to analyze mature miRNAs without modifying the target strand itself, Raymond and coworkers utilized miRNA-specific reverse transcription primers that featured an overhanging 5′ tail so that the resulting cDNA was extended in length from that of the original target. [76] Following reverse transcription (RT), a LNA-containing PCR primer was added which, together with a universal primer contained within the 5′ tail, enabled sensitive quantitation of miRNAs.
There has also been significant effort in applying enzymatic methods to the elongation of the miRNA itself by ligation of oligo sequences. The addition of these flanking sequences allows for longer primer sequences to be utilized, increasing the efficiency of RT-PCR. Separate reports by Miska et. al. and Barad et. al. utilized the addition 3′ and 5′ adapter oligos to the target miRNAs via T4 ligase prior to reverse transcription. [77–78] A limitation of many of the ligation based RT-PCR techniques, however, is that the sensitivity and specificity of the method is ultimately dependent on the efficiency of ligation. In particular, the kinetics of T4 ligase has been shown to vary with substrate sequence, and the incorporation of the ligation step can potentially introduce a signal bias into the measurements. [79–80]
An alternative method for RT-PCR analysis was developed by Shi and Chiang, who used poly(A) polymerase to add poly(A) tails to the 3′ end of target miRNAs in solution. [81] The corresponding RT primers included poly(T) tails to increase the Tm of the heteroduplex and promote reverse transcription. This method was further adapted by Andreasen et al. at Exiqon to include two microRNA-specific, LNA primers during the PCR amplification, drastically increasing the specificity and sensitivity of the assay. [82] An advantage of poly(A) polymerase is that the enzyme shows no sequence preference in its activity and thus it should be a useful tool for high throughput miRNA analysis applications. Similar technologies are available commercially from Agilent and Invitrogen.
A recently developed approach for RT-PCR-based miRNA expression profiling that eliminates the need for enzymatic extension is based upon the hybridization of stem-loop RT primers. The stem-loops are designed so that they are complementary to the 3′ end of the miRNA while at the same time having a 5′ end that is derived fr om the pre-miRNA sequence that composes the antisense half of a hairpin loop, as shown in Figure 1. These primers offer heightened specificity and sensitivity for miRNAs as compared to linear RT primers, largely on account of the increased base stacking and steric limitations imposed by the stem loop structure. By incorporating stem-loop primers into their assays, Chen and co-workers were able to quantitatively monitor the expression profile of mature miRNAs. [83] This procedure was further adapted by Varkonyi-Gasic et. al., who incorporated an additional 5–7 nucleotide extension of the primer to further increase the melting temperature. [84] Applied Biosystems offers a commercial miRNA analysis method based upon stem-loop primer RT-PCR with TaqMan quantitation.
Figure 1.

Schematic description of a RT-PCR assay for a target miRNA. Stem-loop primers, are first hybridized to the miRNA followed by reverse transcription. The resulting transcript is then quantitated using conventional real-time PCR, using a TaqMan probe. Figure adapted from reference 83.
Li and colleagues developed a clever alternative to this general stem-loop procedure by using T4 ligase to attach two DNA stem-loop probes to one another, using the target miRNA as a template, as shown in Figure 2. [85] The two separate stem loop probes were designed to each contain one half of the miRNA complementary sequence masked within the hairpin structure of the stem-loop. Only in the presence of the target miRNA are the stem-loops extended and accessible to the ligase. The resulting long DNA strand can then be detected via standard PCR techniques. A major advantage of this approach is that increased specificity is achieved compared to methods that only utilize the 3′ specificity of a primer.
Figure 2.

Schematic diagram of the enzymatic ligation-based real-time PCR assay for measurement of mature miRNAs. In the presence of the target miRNA, two stem-loop probes, each of which is partially complementary to the target, brought into close proximity via hybridization with the miRNA. T4 ligase is then used to attach the probes together, forming an extended primer than is amenable to real-time PCR-based quantitation. Figure adapted from reference 85.
A significant limitation of the previously mentioned RT-PCR based methods is a restricted ability to simultaneously quantitate multiple miRNAs from a single sample. While multiple RT-PCR analyses can be run in parallel, the increased sample required for such assays is a motivation for the development of multiplexed miRNA analysis methods. However, there are two factors that generally complicate the application of RT-PCR for monitoring multiple miRNAs within a single volume: 1) multiple, sequence specific primers (or primer sets) will be necessary, placing an impetus on detection specificity, and 2) the presence of each strand must be uniquely encoded by a sequence-specific read-out mechanism, such as an independent fluorophore signal in a qPCR experiment.
To tackle the first issue, Lao et al. proposed a pseudo-multiplexed RT-PCR method for the high-throughput detection of miRNAs in which carefully designed stem-loop primers allowed the simultaneous RT and PCR amplification of all of the target miRNAs. [86] The sequence-specific cDNAs were then split into six aliquots and quantitation was performed in parallel using separate single-plex TaqMan PCR reactions for each target miRNAs. Unfortunately, the many PCR cycles needed between the separate amplification and quantitation steps compromises the quantitative utility of the approach.
In the previous example, multiplexed quantitative PCR (qPCR) cannot be performed because there are a limited number of spectrally unique probes that can encode for cDNAs derived from each of the target miRNAs. Furthermore, spectral overlap is in general a significant challenge in the translation of many single-plex biomolecular techniques/assays multiplexed formats. For these reasons, amongst others, there has been a significant effort invested in demonstrating spatial rather than spectral multiplexing schemes, and several of these approaches will be described in more detail below as they apply to miRNA analysis.
Microarrays
Helping to fuel the enormous growth of genomics, and to some extent proteomics, microarray analysis technologies are well-suited to massively multiplexed biomolecular detection on account of spatial, rather than spectral, multiplexing. Not surprisingly, microarrays have been extensively applied to the high-throughput detection of miRNAs as they are capable of simultaneously screening hundreds of target sequences within a single sample volume. Moreover, with proper design of capture probes, microarrays can be used to identify both precursor and mature miRNAs. In general, microarrays are not particularly well-suited for quantitative detection or copy number determination, but rather are very good tools to examine the relative expression of miRNAs between two different biological samples.
As with all miRNA analysis methods, specificity is of utmost importance for microarray methods as cross hybridization can lead to false positive signals. Similarly to Northern blotting, the incorporation of LNA capture probes significantly increases the specificity of a microarray towards target miRNAs. [87] However, even more importantly, is the ability to normalize the melting temperature across all of the capture probe-target duplexes through selective integration of LNAs, an approach that has been led commercially by Exiqon in their miRCURY line of miRNA analysis products. This adjustment allows for uniform stringency rinses to be used with the microarray, and helps accounts for differences in binding kinetics normally observed for cDNA-only capture probes.
In addition to prudent design of capture probes, conventional microarray analysis methods require the target miRNAs be labeled, most commonly with a fluorescent tag. This labeling is often performed prior to hybridization and can be accomplished via a number of methods including the attachment of a pre-labeled oligo via T4 ligase, [88–91] poly(A) extension from the 3′ end via poly(A) polymerase, [92] and covalent modification with mono-reactive and fluorescently tagged cisplatin derivatives that can complex with guanine nucleotides. [93–94]
Another popular method for labeling a miRNA-containing sample, prior to microarray analysis, involves the incorporation of fluorescent tags (often Cy3 and Cy5) during the process of RT-PCR. [64, 77–78] This approach, which borrows from conventional mRNA transcript profiling, provides a convenient method of labeling the total cDNA derived from the miRNA targets in a sample, but also increases the amount of available target via the PCR amplification. However, many of the same challenges faced by stand alone RT-PCR analysis such as sequence bias and run-to-run reproducibility are still encountered when analyzing on a microarray platform. Furthermore, additional complications can be encountered since the presence of a fluorescent tag can significantly perturb duplex stability, an effect that is particularly significant when considering the short lengths of the strands analyzed in miRNA hybridization assays.
As an alternative to labeling miRNAs prior to hybridization, there have been a number of recently developed techniques that focus on introducing labels to the target miRNA after it has been bound to the microarray surface. This approach may, in some cases, help to avoid label-induced perturbations to the duplex hybridization. Liang et al. developed an interesting hybrid scheme by which the vicinal diol at the 3′ of a hybridized miRNA was converted to two aldehyde groups via oxidation with sodium periodate and subsequently conjugated to biotin in solution. [95] The biotinylated miRNAs were then hybridized to the microarray and detected with streptavidin coated quantum dots, giving a 0.4 fmol limit of detection. While this method does involve pre-labeling of the miRNA, it is thought that biotin represents a very small and thus non-disruptive tag, compared with larger labels, such as the conventional Cy3 and Cy5 dyes.
A notable purely post-hybridization strand modification scheme that actually allows read out without any covalent modification of the bound miRNA is the RNA-Assisted-Klenow-Enzyme (RAKE) assay, developed by Nelson and co-workers and illustrated in Figure 3. [96] In this methodology, DNA capture probes, which are linked to the surface via its 5′ end, are carefully designed to have a spacer sequence presenting three thymidine bases directly adjacent to the region complementary to specific miRNA targets. Following hybridization, the entire microarray is exposed to DNA exonuclease I, which enzymatically degrades the capture probes that are not duplexed with miRNA. The Klenow fragment of DNA polymerase I, an enzyme that can act as an RNA-primed DNA polymerase, is then added with biotinylated dATP, which is incorporated complementary to the three thymidines in the capture probe template. The amount of bound target miRNA can then be determined after incubation with fluorescently labeled streptavidin. Because both polymerase I and the Klenow enzyme fragment are sequence independent, the assay is not susceptible to any intrinsic signal bias and a detection limit of 10 pg was reported. However, one limitation of the technique is that the Klenow enzyme is specific only towards the 3′ end of the bound miRNA and thus certain isoforms may elicit unwanted cross-hybridization. Nevertheless, similar approaches have been successfully adapted by a number of other researchers. [97–99]
Figure 3.

Schematic of the RNA-primer, array-based Klenow enzyme (RAKE) assay. Hybridized miRNA bound to specially designed capture probes both shields the capture probe from enzymatic degradation, but also serves as a primer for strand extension, during which a biotinylated nucleotide is introduced. Following extension, the microarray is stained with fluorescent streptavidin and imaged to determine the relative amount of miRNA present in the original sample. Figure adapted from reference 96.
Emerging methods of miRNA analysis
While the previously described techniques were based upon more conventional tools and methods in molecular biology, there is increasing interest in developing completely new analytical approaches to analyzing miRNA expression. Many of these emerging methods take advantage of micro or nanotechnologies and aim to address one or more of the shortcomings associated with the previously mentioned techniques including a minimization of sample size, increases in measurement sensitivity, precision, and dynamic range, and reduction in sequence dependent bias, cost, and time-to-result. Furthermore, a goal of many of these new technologies is to allow very high levels of multiplexing, ideally without sacrificing other key performance metrics, with cost and assay simplicity being a major driver for clinical diagnostic applications. Among the many miRNA analysis methods currently under investigation for miRNA biomarker based diagnostics, some of the most promising advances have involved new detection schemes based on electronic and optical signal transduction, and many already excel in key performance benchmarks. Given their current rapid rate of development, these techniques appear to be promising candidates to provide solutions for emerging miRNA analysis applications.
Electrical Detection
Electrical detection methods are based on changes in circuit properties that occur upon target miRNA hybridization. Signal amplification, often made possible through redox reporters and chemical ligation, can confer ultra-high sensitivity to these devices. However, sometimes this increase in sensitivity is accompanied by a loss of dynamic range. Here we discuss a selection of recently described methodologies, categorized broadly as either direct or indirect based according to their reliance on chemical modification of the target miRNA. Indirect methods usually involve a chemical ligation step which provides an amplified electrical signal following specific target miRNA-DNA hybridization. Though successful, these approaches are being challenged by label free technologies which offer equivalent or superior performance with a simpler assay, amongst other advantages[100]. At first glance, direct methods appear to be the most attractive owing to a reduced number of error-introducing sample preparation steps and thus the potential for faster analysis times, providing that they are able to provide adequate sensitivities for the given bioanalytical challenge.
A good example of a direct miRNA detection method is the use of nanoscale field effect transistors to monitor binding in a completely label free assay motif. Peptide nucleic acid (PNA) functionalized silicon nanowires can be incubated with complementary miRNA targets and changes in the resistivity of the nanowires is monitored before and after the binding events. PNAs are DNA analogues in which the deoxyribose and phosphate backbone is replaced by a peptide bonding motif. The resulting oligomer is devoid of charge and displays increased specificity and sensitivity for hybridization assays, similarly to LNAs. [101–102] Using an array of PNA-functionalized silicon nanowires, Zhang et. al. demonstrated a 1 fM detection limit and single base pair mismatch discrimination capability in the detection of let-7b. [103] In this scheme, the negative charges brought to the surface upon miRNA hybridization (phosphate groups in the backbone) act as a gate and locally deplete charge carriers in the semiconducting nanowire, resulting in a decrease in conductivity. One of the most promising aspects of this technology is the ability to fabricate sensor arrays, as shown in Figure 4, via conventional semiconductor processing techniques, which might enable multiplexed miRNA detection. However, this technology still requires further refinement as field effect transistor based biosensor are notoriously prone to variations in sample ionic strength, and cost and fabrication challenges might complicate the use of PNAs and silicon nanowires, respectively, for high throughput miRNA detection applications.
Figure 4.
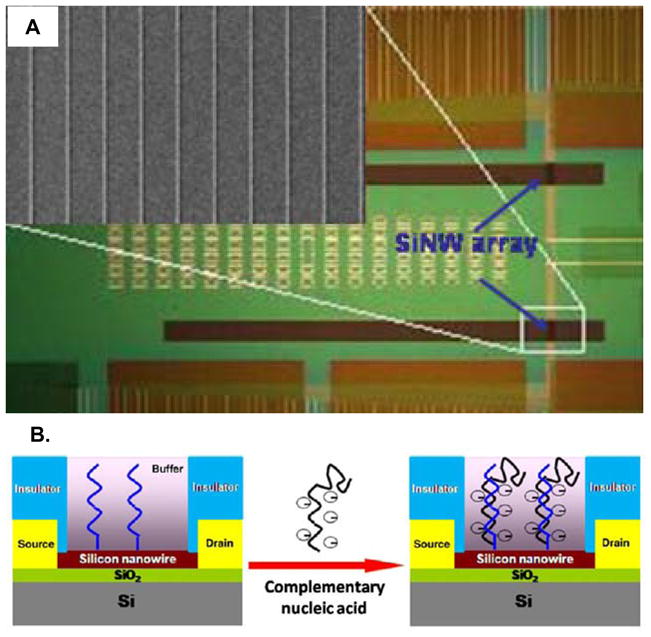
A. Optical and scanning electron micrograph (inset) showing an array of ten silicon nanowire field effect transistors. B. Schematic showing the interaction between a charged nucleic acid and a nanowire field effect transistor. When functionalized with peptide nucleic acids (PNAs) the nanowires can be used to sensitively detect miRNAs as the charge accompanying miRNA hybridization modulates the current flowing through the nanowire due to a gating effect. Figure adapted from references 103–104.
Fan and coworkers reported a method for detecting miRNA based upon changes in conductance accompanying hybridization to PNA-functionalized gaps between a CMOS-based array of microelectrodes. [105] After hybridization, a solution containing aniline, horseradish peroxidase (HRP), and hydrogen peroxide were added, which led to polymerization of the aniline that had associated with the phosphate backbone of the miRNAs via electrostatic interactions. The amount of conductive polyaniline deposited was proportional to the amount of hybridized target and thus the conduction across the microelectrode gap, which drops significantly as the target concentration is increased, could be used for quantitation over a dynamic range of 20 pM to 10 fM, as shown in Figure 5.
Figure 5.
Fan and co-workers developed a miRNA detection scheme based upon polymerization of a conductive polymer across a microscale electrode gap. Aniline selectively interacts with the negatively charged backbone of the miRNA hybridized to PNA capture probes, which have uncharged backbones. The addition of oxidative reagents then leads to the formation of conductive polyaniline and the resistance drop across the electrode gap is proportional to the amount of hybridized miRNA. Figure adapted from reference 105.
Another scheme utilizes a four-component hybridization for sensitive and specific miRNA detection. [106] A capture probe is designed with a gap complementary to the miRNA target of interest. Only upon target binding can a reporter enzyme linked to a further DNA complement then hybridize to the end of the probe. This is due to the additional stabilization conferred by continuous base pair stacking. A hydrolysable substrate is then added and the resulting current monitored. This method benefits from the amplification inherent to enzyme-substrate turnover, as well as electrochemical recycling of the substrate product, p-aminophenol. This system was shown capable of a 2 attomole detection limit and diagnostic capabilities in total RNA extracts from human breast adenocarcinoma MCF-7 cells. Like other direct electrochemical approaches this method does not require chemical modification of the target miRNA.
Ultrasensitive detection down to 10 aM concentration of miRNA was recently demonstrated by Yang et. al. who utilized a Fe-Ru redox pair as a reporter and amplification scheme on a novel nanostructured electrode platform, as shown in Figure 6. [107] Ru3+ accumulates and is reduced at the nanoelectrode surface after miRNA binding to complementary PNA capture probes and a ferricyanide solution phase redox couple chemically regenerates Ru3+ from Ru2+ leading to incredible signal amplification—hundreds of electrons can be generated from a single binding event. [108] In addition to high sensitivity, the sensor shows specificity for mature miRNA over pre-miRNA, and is capable of single base pair mismatch detection. Even more significant, the sensor was used to detect the upregulation of miR-21 and miR-205 in total RNA samples from three human head and neck cancer cell lines. The high surface area of the nanoelectrode is extremely important in this approach as it increases target binding and retention, which is essential to reaching the attomolar regime where there may be only hundreds or thousands of molecules in a sample.
Figure 6.

Schematic diagram illustrating the fabrication and operatin of arrays of novel nanostructured electrodes useful for ultrasensitive miRNA detection. The high surface area of the electrode structure allows sensitive detection of miRNAs via a novel redox reporter system that provides tremendous gain for each target binding event. Figure adapted from reference 107.
A direct approach to miRNA quantitation based on guanine oxidation was demonstrated by Lusi and co-workers based upon the oxidation of guanine bases in the hybridized target strands. [109] While this technique does not require any additional reagents and utilizes less expensive DNA capture probes, as opposed to PNAs, it does require that all of the guanine bases in the capture probe be replaced with inosines. Furthermore, the amount of oxidation current observed is proportional to the number of guanines in the target sequence, complicating the application of this technique for highly multiplexed analyses.
A common type of indirect electrical detection method for miRNAs involves the ligation of an electrocatalytic tag or other nanoparticle to the target, which upon hybridization provides a sequence specific signal. [110–113] The strength of this amplified chemical ligation strategy is its generality, as an extensive number of catalytic or enzymatic moieties can be exploited for improved sensor performance. Several examples of this approach have been reported by Gao and coworkers, who have used inorganic nanoparticle catalysts. [111–113] In one such example, the 3′ ends of target miRNAs were first oxidized with sodium periodate and then hybridized to DNA capture probes on an electrode surface. Amine modified OsO2 nanoparticles were then attached to the 3′ aldehydes of the immobilized miRNA and the current measured from the catalytic degradation of hydrazine, which had been added to the solution. This approach allowed detection of miRNA over a 0.3 pM to 200 pM dynamic range. Notably, a five-fold difference in signal was observed between sequences that had only a single base pair mismatch.
Optical Detection
In addition to electrical signals, optical transduction methods have recently been successfully applied for miRNA detection. Several different classes of optical biosensors have been used to detect miRNAs and here we highlight several innovative examples of fluorescence, bioluminescence, spectroscopic, and refractive index based detection platforms. Optical fluorescence from labeled oligomers (miRNA or cDNA) is the basis for most of the microarray measurements mentioned earlier. However, novel approaches and materials have recently been developed that hold promise to significantly improve fluorescence based miRNA analysis methods.
For example, Li et. al. demonstrated a very sensitive method for miRNA analysis using hairpin probes, T4 ligase, and the fluorescent detection of Cd2+ ions. [114] Target miRNAs bind to carefully designed stem hairpin probes which are then subsequently hybridized with complementary CdSe nanoparticle-labeled DNA. T4 ligase is then added to stabilize the extended duplex structure before Ag+ is added in order to cation exchange the Cd2+ ions out of the nanocrystals and into solution. The authors state that thousands of Cd2+ ions can be liberated from each nanocrystal; a mechanism that provides signal amplification when using a fluorescent assay for Cd2+, allowing miRNA detection down to 35 fM. Sequence specificity is achieved by the use of T4 ligase in two ways: 1) the ligation has a much lower yield if the two strands are not bound with perfect complementarity, and 2) the resulting long duplex has a higher Tm, which allows aggressive stringency washes to be utilized. However, several potential limitations still exist, including the use of CdSe nanoparticles that present an unknown toxicity risk, significant cross reactivity of the Cd2+-sensitive fluorescent dye with Ca2+, meaning that the sample must be rigorously purified prior to analysis, and assay complexity, since multiple reagents and incubation steps are required.
Neely et. al. employed a single molecule fluorescence detection method and dual tagged miRNA-DNA duplexes to detect down to 500 fM miRNA. [115] Importantly, this work established the robust nature of this technique as the authors impressively demonstrated the expression profiling of 45 different miRNA targets in 16 different human tissues, including detection of the key cancer biomarkers mir-16, mir-22, mir-145, and mir-191 from as little as 50 ng of total RNA.
Cissell and coworkers developed a hybridization assay for miRNA detection based on the displacement of the bioluminescent enzyme, Renilla luciferase (Rluc). [116] The Rluc enzyme was conjugated to a synthetic oligonucleotide with a miR-21 sequence was hybridized to an appropriate capture probe and used in a competitive assay. miR-21 in the sample displaced the Rluc-conjugated strand resulting in a decrease in fluorescence that was used to achieve a detection limit of 40 pM with a greater than 3-order of magnitude dynamic range. An assay time of just 90 minutes and potential for integration into a 96 or 384 well plate format makes this an attractive technology for high throughput miRNA analyses.
Surface enhanced Raman spectroscopy (SERS) has been extensively used in the detection of biomolecules, [117–119] but has not generally not achieved widespread use due to poor substrate reproducibility. Using the method of oblique angle vapor deposition to generate sufficiently reproducible substrates, Driskell et. al. were able to detect and differentiate between miRNAs of unrelated sequence based upon the different spectral fingerprints with an incredibly short acquisition time of only 10 seconds![120] However, due to the subtle differences in peak intensity as a function of distinct, but related, sequence composition, identification of specific sequences requires extensive multivariate analysis. Furthermore, the chemical specificity of SERS may complicate detection in complex samples due to high background signals. Nevertheless, this methodology is intriguing for applications in multiplexed miRNA detection.
Surface plasmon resonance imaging (SPRI) has been shown to be an incredibly versatile and effective platform for biomolecule sensing. [121–124] The technique is based on coupling light to the interface of a thin metallic film (typically gold) to excite surface plasmons, which are highly sensitive to changes in the refractive index of the local environment. Properly functionalized with an appropriate capture agent, desired biomolecules can be selectively detected by monitoring changes in reflectivity. While standard SPRI methods would be highly amenable to direct miRNA analysis, an impressive amplification technique incorporating enzymatic strand extension and nanoparticle labeling was developed by Fang and coworkers to achieve an incredible 5 attomole detection limit![125] LNA capture probes immobilized on a gold SPRi substrate were designed so that they were complementary to a targeted miRNA, but left a 6 nucleotide extension of the miRNA beyond the LNA after hybridization. This 3′ overhang can be recognized by poly(A) polymerase, which then enzymatically grows a poly(A) tail at locations where miRNA is localized. Further amplification is achieved by subsequent hybridization of poly(T30) coated Au nanoparticles, which bind to the appended poly(A) tails. The presence of the nanoparticle labels greatly enhances the change in the SPRI reflectivity image, facilitating extremely low limits of detection and a dynamic range from 10–500 fM. Importantly, the dynamic range can be extended to higher concentrations by eliminating the nanoparticle amplification step, if required for the application. Given these developments, and the existing widespread use of this technology for biomolecular measurements, SPRI seems to be a very promising techniques for miRNA expression profiling based on its sensitivity, scalability, dynamic range, and potential for quantitative detection.
Recently, our group has developed a label free and modularly multiplexable biomolecular detection technology based upon arrays of silicon photonic microring resonators. [126–128] These optical structures, which are fabricated via conventional semiconductor processing methods, are incredibly sensitive to binding induced changes in refractive index accompanying the binding of a target analyte to the microring surface, observed as a shift in the resonance wavelength supported by the microcavity. As a demonstration of the applicability of this platform to multiplexed miRNA detection, we recently covalently immobilized DNA capture probes onto the surface of an array of microrings and used it to detect four different disease-relevant miRNAs from a cell line model of brain cancer via a direct hybridization assay. [129] Using this approach we demonstrated a detection limit of 150 fmol after only a 10 minute detection period and a linear dynamic range of over 2 orders of magnitude. We also reported an isothermal method for the discrimination of single base polymorphisms by including stringency-inducing chemical agents directly into the hybridization buffer.
We are currently developing mechanisms for further extending detection limits for the microring resonator technology, and it is worthwhile to point out that many of the enzymatic strand extension or ligation techniques described earlier (poly(A) polymerase, T4 ligase, RAKE, etc.) could be integrated onto the platform in a straightforward fashion. While this technology is still relatively immature in comparison to well-developed methodologies such as RT-PCR and SPRi, the prospects for extremely high level multiplexing and the intrinsic manufacturability of the platform make this an promising technique for many emerging miRNA analysis applications, particularly those related to clinical diagnostics where metrics such as sample size, time to result, and assay cost are of considerable importance.
Conclusions and outlook
Over the past 17 years, our understanding of miRNAs has exploded. As the incredible importance of these small, non-coding transcripts has become increasingly elucidated, the number of tools for their analysis has grown. Still in place today are the original miRNA measurement approaches, many of which are based upon the tried and true tools of molecular biology. More recent adaptations of enabling enzymatic processes have greatly improved many aspects of these classical techniques and allowed higher throughput measurements to be made using RT-PCR or microarray techniques. The introduction of alternative capture probes, incorporating DNA analogues such as LNA and PNA, has been transformative for many of these methods as it in increases the melting temperature for short duplexes.
In the past five years, physical scientists and engineers have become increasingly interested in miRNAs and have intensified efforts to apply emerging detection tools to this important bioanalytical challenge. Some of these approaches incorporate novel materials and reagents, such as metallic nanoparticles, semiconductor quantum dots, and bioluminescent proteins while others utilize the interesting electrical or optical properties of micro- and nanostructures. These emerging approaches all strive to offer one or more advantages over traditional methods, such as of high sensitivity, assay simplicity and reproducibility, multiplexing capability, and device manufacturability.
In the next decade the appetite for enabling miRNA analysis technologies will certainly continue to grow. Recent biological discoveries of correlated expression and action on gene translation have placed impetus on performing global or systems level analyses of miRNAs to uncover the full detail of their regulatory function, and therefore methods that offer high levels of multiplexing will be of great value to these efforts. Furthermore, recent reports describing the value of miRNAs as diagnostic biomarkers for a range of human diseases make the development of point-of-care analysis methods incredibly important. In these applications, metrics such as time-to-result, sample consumption, and assay cost will be key drivers for technology development. As has historically been the demonstrated, transformative biological discoveries are often tied to the development of new technological capabilities. The small size of miRNAs (and other small RNA molecules) challenges conventional biomolecular analysis methodologies and new innovations in miRNA detection will likely play a unique role in enabling future biological breakthroughs.
Figure 7.

Surface plasmon resonance imaging is a promising technique for the detection of miRNAs in an array format. High sensitivity was achieved by Fang and coworkers, who used poly(A) polymerase and poly(T)-coated gold nanoparticles to greatly amplify the SPR response for miRNA binding events. Figure adapted from reference 125.
Figure 8.

Arrays of silicon photonic microring resonators can be used to quantitate miRNAs. A) Schematic illustration of the hybridization of miRNA onto a modified microring, which leads to a shift in the resonance wavelength supported by the integrated microcavity. B) Scanning electron micrograph showing an array of microring resonators. A zoomed in view of a single sensing element is shown in the inset. Figure adapted from reference 129.
Acknowledgments
We gratefully acknowledge financial support for our own efforts in developing a quantitative, multiparameter miRNA analysis method from the National Institutes of Health (NIH) Director’s New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD002190-01; the Camille and Henry Dreyfus Foundation, through a New Faculty Award; and the Eastman Chemical Company (fellowship to AJQ).
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Poethig RS. Small RNAs and developmental timing in plants. Current Opinion in Genetics & Development. 2009;19(4):374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132(21):4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 5.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64(3):303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69(10):4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin CH, et al. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. Embo Journal. 2009;28(20):3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CZ, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 9.Meola N, V, Gennarino A, Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2(1):7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai LM, Yu D. MicroRNAs in common diseases and potential therapeutic applications. Clinical and Experimental Pharmacology and Physiology. 2010;37(1):102–107. doi: 10.1111/j.1440-1681.2009.05269.x. [DOI] [PubMed] [Google Scholar]
- 11.Dennis PP, Omer A. Small non-coding RNAs in Archaea. Curr Opin Microbiol. 2005;8(6):685–694. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Masse E, Majdalani N, Gottesman S. Regulatory roles for small RNAs in bacteria. Curr Opin Microbiol. 2003;6(2):120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 13.Jones-Rhoades MW, Bartel DP, Bartel B. Annual Review of Plant Biology. Vol. 57. 2006. MicroRNAs and their regulatory roles in plants; pp. 19–53. [DOI] [PubMed] [Google Scholar]
- 14.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′ UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29(15):2302–8. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 21.Cho WC. MicroRNAs in cancer - from research to therapy. Biochim Biophys Acta. 2010;1805(2):209–217. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Ruan K, Fang XG, Ouyang GL. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Letters. 2009;285(2):116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 23.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nature Reviews Neuroscience. 2009;10(12):837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocerha J, Kauppinen S, Wahlestedt C. microRNAs in CNS Disorders. Neuromolecular Medicine. 2009;11(3):162–172. doi: 10.1007/s12017-009-8066-1. [DOI] [PubMed] [Google Scholar]
- 25.Pandey AK, et al. MicroRNAs in Diabetes: Tiny Players in Big Disease. Cellular Physiology and Biochemistry. 2009;23(4–6):221–232. doi: 10.1159/000218169. [DOI] [PubMed] [Google Scholar]
- 26.Cai BZ, Pan ZW, Lu YJ. The Roles of MicroRNAs in Heart Diseases: A Novel Important Regulator. Current Medicinal Chemistry. 2010;17(5):407–411. doi: 10.2174/092986710790226129. [DOI] [PubMed] [Google Scholar]
- 27.Saal S, Harvey SJ. MicroRNAs and the kidney: coming of age. Current Opinion in Nephrology and Hypertension. 2009;18(4):317–323. doi: 10.1097/MNH.0b013e32832c9da2. [DOI] [PubMed] [Google Scholar]
- 28.Liang MY, et al. MicroRNA: a new frontier in kidney and blood pressure research. American Journal of Physiology-Renal Physiology. 2009;297(3):F553–F558. doi: 10.1152/ajprenal.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen XM. MicroRNA signatures in liver diseases. World Journal of Gastroenterology. 2009;15(14):1665–1672. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connell RM, et al. Physiological and pathological roles for microRNAs in the immune system. Nature Reviews Immunology. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 31.Tsitsiou E, Lindsay MA. microRNAs and the immune response. Current Opinion in Pharmacology. 2009;9(4):514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li CS, et al. Therapeutic MicroRNA Strategies in Human Cancer. Aaps Journal. 2009;11(4):747–757. doi: 10.1208/s12248-009-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roshan R, et al. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discovery Today. 2009;14(23–24):1123–1129. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Arenz C. MicroRNAs - Future drug targets.? Angewandte Chemie-International Edition. 2006;45(31):5048–5050. doi: 10.1002/anie.200601537. [DOI] [PubMed] [Google Scholar]
- 35.Fabbri M. miRNAs as Molecular Biomarkers of Cancer. Expert Reviews. 2010;10(4):435–444. doi: 10.1586/erm.10.27. [DOI] [PubMed] [Google Scholar]
- 36.Gilad S, et al. e. Plos One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 39.Dai H, et al. Use of hybridization kinetics for differentiating specific from non-specific binding to oligonucleotide microarrays. Nucl Acids Res. 2002;30(16):e86. doi: 10.1093/nar/gnf085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu WT, Mirzabekov AD, Stahl DA. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environmental Microbiology. 2001;3(10):619–629. doi: 10.1046/j.1462-2920.2001.00233.x. [DOI] [PubMed] [Google Scholar]
- 41.Dorris DR, et al. Oligodeoxyribonucleotide probe accessibility on a three-dimensional DNA microarray surface and the effect of hybridization time on the accuracy of expression ratios. BMC Biotechnol. 2003;3:6. doi: 10.1186/1472-6750-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urakawa H, et al. Optimization of Single-Base-Pair Mismatch Discrimination in Oligonucleotide Microarrays. Appl Environ Microbiol. 2003;69(5):2848–2856. doi: 10.1128/AEM.69.5.2848-2856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guschin D, et al. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl Environ Microbiol. 1997;63(6):2397–2402. doi: 10.1128/aem.63.6.2397-2402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendes ND, Freitas AT, Sagot MF. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Research. 2009;37(8):2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stark A, et al. Identification of Drosophila MicroRNA targets. Plos Biology. 2003;1(3):397–409. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennecke J, et al. Principles of MicroRNA-target recognition. Plos Biology. 2005;3(3):404–418. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krek A, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 48.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flaned by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 49.Grun D, et al. MicroRNA target predictions across seven Drosophila species and comparison to mammalian targets. Plos Computational Biology. 2005;1(1):51–66. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genetics. 2006;38(12):1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 51.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;e(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes & Development. 2004;18(10):1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maragkakis M, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Research. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexiou P, et al. The DIANA-mirExTra Web Server: From Gene Expression Data to MicroRNA Function. Plos One. 2010;5(2):e9171. doi: 10.1371/journal.pone.0009171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rehmsmeier M, et al. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammell M, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nature Methods. 2008;5(9):813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.John B, et al. Human MicroRNA targets. Plos Biology. 2004;2(11):1862–1879. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Betel D, et al. The microRNA.org resource: targets and expression. Nucleic Acids Research. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long D, et al. Potent effect of target structure on microRNA function. Nature Structural & Molecular Biology. 2007;14(4):287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 60.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Current Biology. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 61.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lau NC, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 63.Cummins JM, et al. The colorectal microRNAome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mattie M, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Molecular Cancer. 2006;5(1):24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lagos-Quintana M, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 66.Sempere L, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biology. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Streit S, et al. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat Protocols. 2008;4(1):37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- 69.Fernyhough P. Quantification of mRNA Levels Using Northern Blotting. Neurotrophin Protocols. 2001:53–63. doi: 10.1385/1-59259-060-8:53. [DOI] [PubMed] [Google Scholar]
- 70.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nature Protocols. 2008;3(2):190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 72.Valoczi A, et al. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Research. 2004;32(22):e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chemistry & Biology. 2001;8(1):1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 74.Ramkissoon SH, et al. Nonisotopic detection of microRNA using digoxigenin labeled RNA probes. Molecular and Cellular Probes. 2006;20(1):1–4. doi: 10.1016/j.mcp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Schmittgen TD, et al. A high-throughput method to monitor the expression of microRNA precursors. Nucl Acids Res. 2004;32(4):e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raymond CK, et al. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11(11):1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miska E, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biology. 2004;5(9):R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Research. 2004;14(12):2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohtsuka E, et al. Joining of Synthetic Ribotrinucleotides with Defined Sequences Catalyzed by T4 RNA Ligase. European Journal of Biochemistry. 1977;81(2):285–291. doi: 10.1111/j.1432-1033.1977.tb11950.x. [DOI] [PubMed] [Google Scholar]
- 80.Mclaughlin LW, et al. The Effect of Acceptor Oligoribonucleotide Sequence onthe T4 RNA Ligase Reaction. European Journal of Biochemistry. 1982;125(3):639–643. doi: 10.1111/j.1432-1033.1982.tb06730.x. [DOI] [PubMed] [Google Scholar]
- 81.Shi R, V, Chiang L. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39(4):519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 82.Andreasen D, et al. Improved microRNA quantification in total RNA from clinical samples. Methods. 2010;50(4):S6–S9. doi: 10.1016/j.ymeth.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 83.Chen CF, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varkonyi-Gasic E, et al. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J, et al. Real-Time Polymerase Chain Reaction MicroRNA Detection Based on Enzymatic Stem-Loop Probes Ligation. Analytical Chemistry. 2009;81(13):5446–5451. doi: 10.1021/ac900598d. [DOI] [PubMed] [Google Scholar]
- 86.Lao KQ, et al. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochemical and Biophysical Research Communications. 2006;343(1):85–89. doi: 10.1016/j.bbrc.2006.02.106. [DOI] [PubMed] [Google Scholar]
- 87.Castoldi M, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12(5):913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13(1):151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thomson JM, et al. A custom microarray platform for analysis of microRNA gene expression. Nature Methods. 2004;1(1):47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 90.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krichevsky AM, et al. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goff LA, et al. Rational Probe Optimization and Enhanced Detection Strategy for MicroRNAs Using Microarrays. RNA Biology. 2005;2(3):93–100. doi: 10.4161/rna.2.3.2059. [DOI] [PubMed] [Google Scholar]
- 93.Babak T, et al. Probing microRNAs with microarrays: Tissue specificity and functional inference. RNA. 2004;10(11):1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiegant JCAG, et al. ULS: a versatile method of labeling nucleic acids for FISH based on a monofunctional reaction of cisplatin derivatives with guanine moieties. Cytogenetic and Genome Research. 1999;87(1–2):47–52. doi: 10.1159/000015390. [DOI] [PubMed] [Google Scholar]
- 95.Liang RQ, et al. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Research. 2005;33(2):e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nelson PT, et al. Microarray-based, high-throughput gene expression profiling of microRNAs. Nature Methods. 2004;1(2):155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 97.Berezikov E, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Research. 2006;16(10):1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nelson PT, et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;e(2):187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yeung ML, et al. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2(1):81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qavi AJ, et al. Label-Free Technologies for Quantitative Multiparameter Biological Analysis. Analytical and Bioanalytical Chemistry. 2009;394:121–135. doi: 10.1007/s00216-009-2637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Egholm M, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365(6446):566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 102.Nielsen PE, et al. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254(5037):1497–500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 103.Zhang GJ, et al. Label-free direct detection of MiRNAs with silicon nanowire biosensors. Biosensors and Bioelectronics. 2009;24(8):2504–2508. doi: 10.1016/j.bios.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 104.Zhang GJ, et al. Silicon nanowire biosensor for highly sensitive and rapid detection of Dengue virus. Sensors and Actuators B-Chemical. 2010;146(1):138–144. [Google Scholar]
- 105.Fan Y, et al. Detection of microRNAs using target-guided formation of conducting polymer nanowires in nanogaps. Journal of the American Chemical Society. 2007;129(17):5437–5443. doi: 10.1021/ja067477g. [DOI] [PubMed] [Google Scholar]
- 106.Pohlmann C, Sprinzl M. Electrochemical Detection of MicroRNAs via Gap Hybridization Assay. Anal Chem. 2010;82(11):4434–4440. doi: 10.1021/ac100186p. [DOI] [PubMed] [Google Scholar]
- 107.Yang H, et al. Direct, Electronic MicroRNA Detection for the Rapid Determination of Differential Expression Profiles. Angewandte Chemie-International Edition. 2009;48(45):8461–8464. doi: 10.1002/anie.200902577. [DOI] [PubMed] [Google Scholar]
- 108.Soleymani L, et al. Nanostructuring of Patterned Microelectrodes To Enhance the Sensitivity of Electrochemical Nucleic Acids Detection. Angewandte Chemie-International Edition. 2009;48(45):8457–8460. doi: 10.1002/anie.200902439. [DOI] [PubMed] [Google Scholar]
- 109.Lusi EA, et al. Innovative Electrochemical Approach for an Early Detection of microRNAs. Analytical Chemistry. 2009;81(7):2819–2822. doi: 10.1021/ac8026788. [DOI] [PubMed] [Google Scholar]
- 110.Yang WJ, et al. Quantification of microRNA by gold nanoparticle probes. Anal Biochem. 2008;376(2):183–188. doi: 10.1016/j.ab.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 111.Gao ZQ, Yang ZC. Detection of microRNAs using electrocatalytic nanoparticle tags. Analytical Chemistry. 2006;78(5):1470–1477. doi: 10.1021/ac051726m. [DOI] [PubMed] [Google Scholar]
- 112.Gao ZQ, Yu YH. Direct labeling microRNA with an electrocatalytic moiety and its application in ultrasensitive microRNA assays. Biosensors & Bioelectronics. 2007;22(6):933–940. doi: 10.1016/j.bios.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 113.Gao ZQ, Yu YH. A microRNA biosensor based on direct chemical ligation and electrochemically amplified detection. Sensors and Actuators B-Chemical. 2007;121(2):552–559. [Google Scholar]
- 114.Li JS, et al. Detection of MicroRNA by Fluorescence Amplification Based on Cation-Exchange in Nanocrystals. Analytical Chemistry. 2009;81(23):9723–9729. doi: 10.1021/ac901983s. [DOI] [PubMed] [Google Scholar]
- 115.Neely LA, et al. A single-molecule method for the quantitation of microRNA gene expression. Nature Methods. 2006;3(1):41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 116.Cissell KA, et al. Bioluminescence-based detection of MicroRNA, miR21 in breast cancer cells. Analytical Chemistry. 2008;80(7):2319–2325. doi: 10.1021/ac702577a. [DOI] [PubMed] [Google Scholar]
- 117.Cho H, et al. Label-free and highly sensitive biomolecular detection using SERS and electrokinetic preconcentration. Lab Chip. 2009;9(23):3360–3363. doi: 10.1039/b912076a. [DOI] [PubMed] [Google Scholar]
- 118.Huh YS, et al. Enhanced on-chip SERS based biomolecular detection using electrokinetically active microwells. Lab Chip. 2009;9(3):433–439. doi: 10.1039/b809702j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hudson SD, Chumanov G. Bioanalytical applications of SERS (surface-enhanced Raman spectroscopy) Analytical and Bioanalytical Chemistry. 2009;394(3):679–686. doi: 10.1007/s00216-009-2756-2. [DOI] [PubMed] [Google Scholar]
- 120.Driskell JD, et al. Rapid microRNA (miRNA) detection and classification via surface-enhanced Raman spectroscopy (SERS) Biosensors & Bioelectronics. 2008;24(4):917–922. doi: 10.1016/j.bios.2008.07.060. [DOI] [PubMed] [Google Scholar]
- 121.Nelson BP, et al. Surface Plasmon Resonance Imaging Measurements of DNA and RNA Hybridization Adsorption onto DNA Microarrays. Analytical Chemistry. 2000;73(1):1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 122.Wegner GJ, Lee HJ, Corn RM. Characterization and optimization of peptide arrays for the study of epitope-antibody interactions using surface plasmon resonance imaging. Analytical Chemistry. 2002;74(20):5161–5168. doi: 10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 123.Smith EA, et al. Surface plasmon resonance imaging studies of protein-carbohydrate interactions. Journal of the American Chemical Society. 2003;125(20):6140–6148. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 124.Wegner GJ, et al. Real-time surface plasmon resonance imaging measurements for the multiplexed determination of protein adsorption/desorption kinetics and surface enzymatic reactions on peptide microarrays. Analytical Chemistry. 2004;76(19):5677–5684. doi: 10.1021/ac0494275. [DOI] [PubMed] [Google Scholar]
- 125.Fang S, et al. Attomole Microarray Detection of MicroRNAs by Nanoparticle-Amplified SPR Imaging Measurements of Surface Polyadenylation Reactions. Journal of the American Chemical Society. 2006;128(43):14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Washburn AL, Gunn LC, Bailey RC. Label-Free Quantitation of a Cancer Biomarker in Complex Media Using Silicon Photonic Microring Resonators. Analytical Chemistry. 2009;81(22):9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Washburn AL, et al. Quantitative, Label-Free Detection of Five Protein Biomarkers Using Multiplexed Arrays of Silicon Photonic Microring Resonators. Analytical Chemistry. 2010;82(1):69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luchansky MS, Bailey RC. Silicon Photonic Microring Resonators for Quantitative Cytokine Detection and T-Cell Secretion Analysis. Analytical Chemistry. 2010;82(5):1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qavi AJ, Bailey RC. Multiplexed Detection and Label-Free Quantitation of microRNAs using Arrays of Silicon Photonic Microring Resonators. Angewandte Chemie-International Edition. 2010;49(27):4608–4611. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]



