Abstract
DNMT2 is a human protein that displays strong sequence similarities to DNA (cytosine-5)-methyltransferases (m5C MTases) of both prokaryotes and eukaryotes. DNMT2 contains all 10 sequence motifs that are conserved among m5C MTases, including the consensus S-adenosyl-l-methionine-binding motifs and the active site ProCys dipeptide. DNMT2 has close homologs in plants, insects and Schizosaccharomyces pombe, but no related sequence can be found in the genomes of Saccharomyces cerevisiae or Caenorhabditis elegans. The crystal structure of a deletion mutant of DNMT2 complexed with S-adenosyl-l-homocysteine (AdoHcy) has been determined at 1.8 Å resolution. The structure of the large domain that contains the sequence motifs involved in catalysis is remarkably similar to that of M.HhaI, a confirmed bacterial m5C MTase, and the smaller target recognition domains of DNMT2 and M.HhaI are also closely related in overall structure. The small domain of DNMT2 contains three short helices that are not present in M.HhaI. DNMT2 binds AdoHcy in the same conformation as confirmed m5C MTases and, while DNMT2 shares all sequence and structural features with m5C MTases, it has failed to demonstrate detectable transmethylase activity. We show here that homologs of DNMT2, which are present in some organisms that are not known to methylate their genomes, contain a specific target-recognizing sequence motif including an invariant CysPheThr tripeptide. DNMT2 binds DNA to form a denaturant-resistant complex in vitro. While the biological function of DNMT2 is not yet known, the strong binding to DNA suggests that DNMT2 may mark specific sequences in the genome by binding to DNA through the specific target-recognizing motif.
INTRODUCTION
Until recently Dnmt1 was the only known mammalian DNA cytosine methyltransferase (reviewed in 1). However, it had long been speculated that de novo methyltransferases (MTases) distinct from Dnmt1 would be found to act during gametogenesis and the early stages of development. Unequivocal evidence for additional DNA MTases came from the demonstration that Dnmt1 null embryonic stem cells are capable of methylating newly integrated proviral DNA (2). Within the last two years, three DNA (cytosine-5)-methyltransferase (m5C MTase) homologs (Dnmt2, Dnmt3a and Dnmt3b) have been identified from databases of expressed sequence tags based on their content of diagnostic m5C MTase motifs (reviewed in 1). The DNMT1, 2 and 3 families are distantly related to each other and probably diverged early in eukaryotic evolution (1). Dnmt3a and 3b have been shown to have enzymatic activity and are encoded by essential genes (3); inactivation of the catalytic domain of DNMT3B in patients with ICF (immunodeficiency, centromere instability and facial anomalies) syndrome causes demethylation limited almost exclusively to the inactive X chromosome and classical satellite DNA (4). However, no transmethylase activity has been demonstrated for Dnmt2 and embryonic stem cells which lack Dnmt2 appear to have normal de novo and maintenance MTase activities (5).
Dnmt2 is a relatively small protein of 391 amino acids and lacks the large N-terminal domains present in the Dnmt1 and Dnmt3 families (6). The gene appears to be well conserved among eukaryotes, not only in organisms whose genomes are methylated (mammals, Arabidopsis thaliana, Xenopus laevis and Danio rerio), but also in organisms lacking detectable cytosine methylation, such as Schizosaccharomyces pombe and Drosophila melanogaster. Dnmt2 is ubiquitously expressed with multiple mRNA species in most human and mouse adult tissues (5,6). Patterns of Dnmt2 expression in human and mouse tissues are very similar to those of Dnmt1 (6). The expression of D.melanogaster Dnmt2 has been reported to be developmentally regulated (7), although a different pattern of expression in this species has also been reported (8).
We present the X-ray crystal structure of human DNMT2 complexed with S-adenosyl-l-homocysteine (AdoHcy) and report the unusual DNA-binding properties of DNMT2. Although no transmethylase activity has been detected in Dnmt2, our results reveal a two-domain structure of DNMT2 that closely resembles M.HhaI, a prokaryotic m5C MTase. Sequences involved in AdoHcy binding (catalytic domain) and putative DNA recognition (target-recognizing domain, TRD) are structured in the same geometry as M.HhaI. Unlike other m5C MTases, DNMT2 was found to be capable of binding DNA in a denaturant-resistant and probably covalent complex.
MATERIALS AND METHODS
Overexpression and purification
The full-length open reading frame from the human DNMT2 cDNA was cloned into the bacterial expression vector pQE9 (Qiagen) and DNMT2 protein was expressed in Escherichia coli (McrBC-deficient strain ER2488) with a six-histidine tag at the N-terminus. Soluble recombinant proteins were purified from the supernatants on a nickel chelate column (Qiagen), followed by Mono-Q column chromatography.
Limited proteolysis of DNMT2 protein
Protease V8 (100, 10 or 1 ng) was added to 1 µl of 15 mg/ml DNMT2 for 15 min on ice in a 10 µl reaction of 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA and 0.1% β-mercaptoethanol. The reaction was stopped with SDS sample buffer and the products were separated on a 13% SDS–PAGE gel. One large fragment and two closely migrating smaller fragments were observed. Denaturing the cut protein with 4 M urea and passing through a Ni–agarose column demonstrated that only the large fragment contained the His tag and therefore represented the N-terminal cleavage product. The masses of all three fragments were determined with a matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) mass spectrometer. The N-terminal sequence of the smallest fragment was determined using an Applied Biosystems 491A Pulsed-Liquid Sequencer on-line with an Applied Biosystems 140S PTH Analyzer (Procise-HT). Combining these results and the specificity of V8 protease allowed us to deduce that the large fragment represents residues 1–190 and the two smaller fragments represent residues 234–391 and 238–391. The N- and C-terminal fragments remained tightly associated after proteolysis and could be co-purified by chromatography and crystallized.
Construction and purification of DNMT2Δ47
DNMT2Δ47 was generated by oligonucleotide-directed mutagenesis that removed the sequences encoding residues 191–237 and was expressed in E.coli as a glutathione S-transferase (GST) fusion. A thrombin cleavage site was introduced between the DNMT2Δ47 and GST moieties. The fusion protein, GST–DNMT2Δ47, was purified on a glutathione–agarose column (Pharmacia) and DNMT2Δ47 was cleaved from the GST moiety by on-column thrombin digestion, resulting in the final DNMT2Δ47 protein, which contains three additional N-terminal amino acids (GlySerArg). The DNMT2Δ47 protein was separated from thrombin by Mono-Q column chromatography. Binary complexes of DNMT2Δ47 and AdoHcy were formed by incubation of DNMT2Δ47 (in 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA and 0.1% β-mercaptoethanol) and AdoHcy (in water) at a protein:AdoHcy molar ratio of 1:1.5, either before or after protein concentration.
Crystallization and X-ray data collection
Crystals were obtained in hanging drops containing ∼20 mg/ml DNMT2Δ47–AdoHcy complex in 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.1% 2-mercaptoethanol and 150 mM NaCl, with concentration by vapor diffusion against 1.3–1.4 M ammonium sulfate, 8% glycerol, 100 mM glycine–NaOH pH 9.0, at 16°C. Glycerol and 2-mercaptoethanol were found to be essential for successful crystallization. One molecule of glycerol bound in a surface cavity on the small domain by making six hydrogen bonds (three proton acceptors and three proton donors) to the main chain nitrogen and oxygen atoms. One molecule of 2-mercaptoethanol was found to react with the sulfur atom of Cys24 and to occupy a surface cavity of the crystallographically related neighboring protein molecule.
Crystals were of space group I41, determined based on the systematic absence of reflections along the z-axis, with unit cell dimensions a = b = 116.5 Å and c = 69.8 Å. The crystals diffracted from 2.3 to 1.8 Å resolution (Tables 1 and 2) under cryogenic conditions. Crystals were transferred to a drop containing the cryobuffer (∼1.4 M ammonium sulfate, 100 mM glycine–NaOH pH 9.0, 24% glycerol) and quenched into and then maintained at 95 K in a stream of liquid nitrogen gas. X-rays were produced by a Rigaku rotating anode generator (50 kV, 100 mA) equipped with a RAXIS-IV imaging plate detector and synchrotron radiation at the National Synchrotron Light Source beamline X12-C equipped with a Brandeis B2 (1 × 1) CCD-based detector. The resulting images were processed using the program HKL (9).
Table 1. X-ray data collection statistics at intensity cut-off I/σ(I) ≥ 0.
| Crystal |
Native |
(CH3)3PbCOOCH3 |
|
Xenon |
| Wavelength (Å) | 0.980 | 0.95189 | 0.950051 | 1.5418 |
| Energy (keV) | 12.651 | 13.023 | 13.050 | 8.042 |
| Resolution (Å) | 20–1.8/1.83–1.8 | 20–1.9/1.93–1.9 | 20–1.9/1.93–1.9 | 20–2.3/2.38–2.3 |
| Completeness (%) | 96.8/93.2 | 84.6/50.4 | 84.8/50.1 | 96.1/83.1 |
| R linear | 0.051/0.206 | 0.050/0.229 | 0.049/0.209 | 0.057/0.196 |
| <I/σ(I)> | 16.4/3.74 | 13.4/2.78 | 15.3/3.16 | 18.1/4.92 |
| Unique reflections | 41 963/1976 | 31 250/927 | 31 376/925 | 20 017/1702 |
| Total reflections | 146 987 | 89 660 | 92 108 | 82 748 |
| Twin fraction | 0.4144 | 0.2647 | 0.2647 | 0.3588 |
| Completeness after detwinning (%) | 94.5 | 72.0 | 74.5 | 89.4 |
| Unique reflections | 40 953 | 26 590 | 27 553 | 18 622 |
| Phasing statistics at 2.3 Å resolution | ||||
| Heavy atom sites | – | 1 | 1 | 3 |
| Phasing power | – | 1.55 | 1.52 | 1.41 |
| R Cullis | – | 0.719 | 0.688 | 0.570 |
| R Kraut | – | 0.070 | 0.069 | 0.126 |
| Figure of merit | – | 0.311 | 0.321 | 0.273 |
| Overall FOM | 0.499 |
Table 2. Structural refinement statistics at 1.8 Å resolution.
| Resolution (Å) |
Completeness (%)/reflections
(no.) |
R value/Rfree (%) |
| 20.00–3.59 | 91/5012 | 17.74/22.49 |
| 3.59–2.86 | 93/5084 | 18.05/22.55 |
| 2.86–2.50 | 91/4910 | 20.47/27.06 |
| 2.50–2.27 | 90/4884 | 22.15/24.65 |
| 2.27–2.10 | 88/4749 | 24.36/24.87 |
| 2.10–1.98 | 84/4512 | 27.03/27.36 |
| 1.98–1.88 | 77/4155 | 30.85/32.85 |
| 1.88–1.80 | 71/3815 | 35.16/37.48 |
| 20.00–1.80 | 86/37 121 | 21.21/25.04 |
| Ramachandran plot | ||
| Most favored regions (%) | 93.4 | |
| Additionally allowed regions (%) | 6.6 | |
| Root mean square deviation from ideality | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.5 | |
| Dihedrals (°) | 23.4 | |
| Improper (°) | 1.4 | |
| G factor | ||
| Dihedrals | 0.21 | |
| Covalent | 0.45 | |
| Overall | 0.31 | |
| Average thermal factor (Å2) | ||
| Main chain | 19.7 | |
| Side chain | 20.9 |
Xenon and trimethyllead derivatives
A three-site xenon derivative was obtained by exposing a single crystal (after transferring to the cryobuffer) to xenon gas (Nova Gas Technologies, Cryogenic Rare Gas Laboratories Inc.) at 270 p.s.i. for 20 min and flash freezing in liquid carbon tetrafluoride without depressurization, using a Cryo-Xe-Siter (Molecular Structure Corp.). Three xenon molecules locate in the large domain, in the hydrophobic interfaces between the β sheet and the helices on each side.
In addition, a lead derivative was obtained by soaking a crystal in 10 mM trimethyllead acetate for 6 days. The lead atom did not directly interact with the protein side chains, but instead interacted with a SO4–2 molecule (from the crystallization procedure) which was held in position by the side chains of Arg108, His101 and the symmetry-related Lys334.
Crystal twinning
Several lines of evidence indicated possible twinning in the I41 crystal. Two lead sites were found in the Patterson Harker sections, but their positions could not be reconciled by cross-difference Fourier analysis and no cross-peaks were found between them. A self-rotation function indicated two molecules present in one asymmetry unit, which would suggest that the solvent content is only ∼16%. A twinning server (http:// www.doe-mbi.u cla.edu/services/twinning) operated by Yeates (10) detected the presence of twinning in the crystal (with the twinning fraction ranging from 0.26 to 0.44; Table 1); space group I41 with a = b permits twinning. The twinned intensity data were thus detwinned according to Yeates (10) between the two related reflections (h,k,l) and (h,–k,–l). The detwinned data contain one DNMT2Δ47–AdoHcy complex per asymmetric unit. Although the twinned data from lead- and xenon-containing crystals showed strong anomalous signals, the detwinned anomalous data produced a much worse Patterson signal and thus were not included in the phasing calculations.
Phasing, model building and refinement
Using detwinned data the positions of the lead and xenon atoms were determined from isomorphous difference Patterson syntheses and confirmed by difference Fourier methods. The multiple isomorphous replacement method was used to generate initial protein phases, using PHASES (11), and subsequent phasing was done using SOLOMON (12). The resulting initial map was of sufficient quality to place the amino acids of DNMT2 in recognizable densities at 2.8 Å resolution using O (13). The resultant model was refined to 1.8 Å resolution (Table 2) using X-PLOR (14). The Protein Data Bank code is 1G55.
Molecular replacement
A different crystal form was obtained, under similar conditions as for the I41 crystal, in the presence of p-chloromercuribenzene sulfate. The crystal belongs to space group P43 with cell dimensions a = b = 114 Å and c = 71 Å. The crystal diffracted to 3.5 Å resolution (11 644 unique reflections out of 36 142 observations with a completeness of 98.5% and Rsym = 0.075 and <I/σ> = 8.5). The twinning server also detected the presence of twinning in the P43 crystal with a twinning fraction of 0.206. The structure was solved by molecular replacement using the detwinned diffraction data (each asymmetric unit contains two molecules) and the refined DNMT2Δ47 structure as the search model. Two solutions were found using AmoRe (15), with a correlation coefficient of 0.66 and R factor of 0.36. The non-crystallographic 2-fold axis was only ∼5° tilted away from the z-axis, resulting in the lower symmetry space group.
Preparation of embryonic stem (ES) cell lysates and analysis of denaturant-resistant DNMT2–DNA interactions
ES cells of strain R1 (kindly provided by Andras Nagy) were grown to mid-log phase and lysed in 5 vol of 20 mM Tris–HCl pH 7.4, 0.32 M sucrose, 0.3% Triton X-100, 0.4 M NaCl, 3 mM MgCl2, 0.5 mM DTT and 0.2 mM PMSF on ice. An equal volume of DEAE–Sephacel (50% v/v slurry equilibrated with 20 mM Tris–HCl pH 7.4) was added and removed by centrifugation after 5 min on ice to deplete the extract of nucleic acids. The details of the binding reactions and their preparation for denaturing gel electrophoresis were as described (16), except that heating was at 65°C for 10 min after addition of SDS to 2%. Electrophoresis on 6% polyacrylamide–0.1% SDS gels, transfer to nitrocellulose membranes and autoradiography were as described (16). Denaturant-resistant binding was defined as resistance to SDS at 65°C and denaturing gel electrophoresis in the presence of SDS.
RESULTS
The widely distributed and well-conserved Dnmt2 family
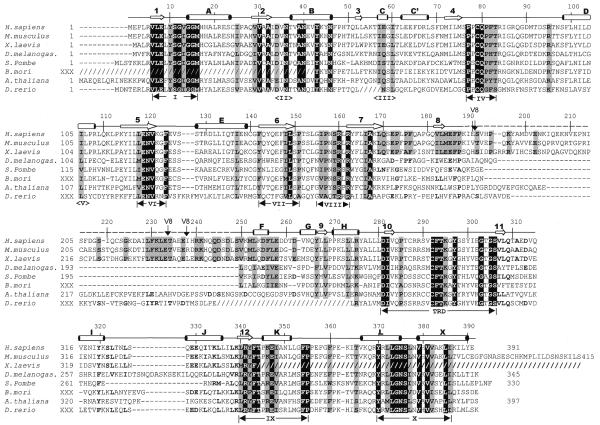
Dnmt2 had been found in mouse and human (6), S.pombe (pmt1p; 17) and D.melanogaster (dDnmt2; 7). In addition, ESTs with strong similarity to DNMT2 were found in X.laevis, A.thaliana, the silkworm Bombyx mori and the zebrafish D.rerio (Fig. 1). DNMT2 homologs are highly conserved in those eukaryotes in which they occur, but no related sequences could be found in the genomes of Caenorhabditis elegans or Saccharomyces cerevisiae. The m5C MTases (EC 2.1.1.73 for bacterial enzymes and EC 2.1.1.37 for animal and plant versions) share six strongly conserved and four weakly conserved sequence motifs that are diagnostic of this class of enzymes (1,18,19). All members of the DNMT2 family contain all 10 MTase motifs in the typical order, with motif I occurring within a few amino acids of the N-terminus (Fig. 1). In addition to the 10 motifs, all DNMT2 homologs share a distinctive conserved stretch of 41 amino acids (266–306 of DNMT2), including an invariant CysPheThr tripeptide and an AspIle dipeptide between motifs VIII and IX, in a region corresponding to the TRD of the bacterial MTases (18). The characteristic CysPheThr tripeptide conserved in the DNMT2 family was not found in other eukaryotic enzymes (DNMT1, DNMT3A, DNMT3B, masc1 and masc2) or in approximately 90 bacterial MTases (20). This high degree of internal conservation in the region surrounding the CysPheThr motif suggests that DNMT2 homologs are members of a subfamily of m5C MTases that may recognize a specific kind of target through the specific TRD, although the nature of the target is not yet known.
Figure 1.
Alignment of Dnmt2 homologs. The slashed lines indicate sequences missing from the available ESTs and dashed lines indicate gaps introduced to optimize alignments. The human DNMT2 residue numbering is shown above the sequence alignment. Amino acids highlighted are either invariant (white against black) among the proteins or similar (shaded), as defined by the following groupings: V, L, I and M; F, Y and W; K and R; E and D; Q and N; S and T; A, G and P. The secondary structural elements of DNMT2 (helices A–X and strands 1–11) and conserved sequence motifs are labeled. Complete Dnmt2 sequences are available in GenBank for human (AAC39764), mouse (AAC53529) and S.pombe (CAA57824). The two Dnmt2 sequences of D.melanogaster currently deposited in GenBank, AAF03835 and AAF53163, contain a single exon and lack the consensus motif I. We used the sequence reported by Tweedie et al. (24), which includes an additional upstream exon and contains motif I. The A.thaliana sequence was assembled from genomic sequence AC006601 between nucleotides 29989 and 27859. Two of the eight splicing junctions are confirmed by EST sequences. The X.laevis Dnmt2 sequence was assembled from two overlapping ESTs (AW639976 and AW636415) and lacks the C-terminus of the protein. The D.rerio sequence was assembled by jointing four ESTs (AI331437, AI618465, AI331180 and AI331323). The B.mori sequence was assembled from three ESTs (AU005443, AV399933 and AV400288). Shown in dashed overline and labeled V8 are the sequences (from 191 to 237) deleted to make a version of DNMT2 that could be crystallized. Note that these sequences are normally absent from the D.melanogaster, S.pombe and B.mori Dnmt2 homologs.
Structure determination
Structure determination involved production of a recombinant deleted form (DNMT2Δ47) in which residue 190 was fused to residue 238 (see Materials and Methods). The deleted sequences are poorly conserved within the DNMT2 family and are completely absent from the S.pombe and D.melanogaster DNMT2 homologs (Fig. 1). DNMT2Δ47 was found to bind DNA in electrophoretic mobility shift assays as efficiently as the wild-type protein (data not shown). The deletion, however, allowed crystal formation, which had not occurred in many crystallization trials with wild-type DNMT2 protein. We therefore chose to determine the structure of DNMT2Δ47 complexed with AdoHcy.
We calculated electron density maps in space group I41 by isomorphous replacement of xenon and trimethyllead acetate derivatives (Table 1) and were able to build a total of 313 of the 344 residues of the DNMT2Δ47 protein into a model. Two residues before (amino acids 189–190) and 10 residues after (amino acids 238–247) the Δ47 deletion were disordered; amino acids 79–97 (a 19 residue loop including the active site Cys79) had some limited mobility within the crystals and were modeled only as alanine. The model was refined to 1.8 Å resolution with a crystallographic R factor of 0.21 and Rfree value of 0.25 (Table 2). In addition, a lower resolution structure was solved by molecular replacement in space group P43 (see Materials and Methods) in the presence of a mercuric compound. In this case the side chain of Cys79 is clearly visible as a result of interaction with a mercury atom.
Structured segments
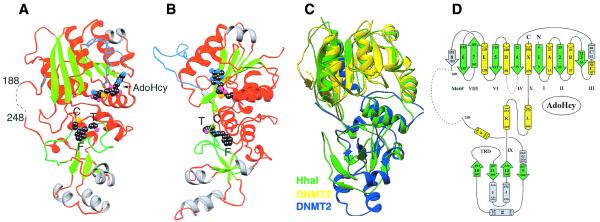
Figure 2A and B presents two views of the DNMT2Δ47 structure; the location of structural elements with respect to the amino acid sequence of DNMT2 is shown in Figure 1. In the orientation shown in Figure 2A the missing segment (residues 189–247) would be located on the left, as indicated by the dotted line. The overall structures of DNMT2Δ47 and M.HhaI MTase are nearly superimposable (Fig. 2C). As in M.HhaI, the DNMT2 structure can be divided into three parts: the large domain (strands β1–β8 and helices αA–αE and αX), the small domain (strands β9–β12 and helices αF–αJ) and the two parallel helices (αK and αL) hinge region (Fig. 2D). With respect to the primary sequence, the small domain and the hinge consists of contiguous sequences, whereas the large domain is composed of elements from the N-terminal region and C-terminal helix αX.
Figure 2.
Ribbon (42) diagrams of the DNMT2Δ47–AdoHcy complex. (A) Front view. (B) Right side view (the point of view of an observer from the right side of Fig. 2A). Helices and loops are colored red (unique helices of DNMT2 gray), strands in green, the PC loop in light blue and the TRD loop in green. The bound AdoHcy and CFT of the TRD are shown as balls, with carbon atoms in black, nitrogen atoms in light blue, oxygen atoms in magenta and sulfur atoms in orange. (C) Superimposition of DNMT2Δ47, colored yellow (residues 1–188) and blue (residues 248–391), and M.HhaI (pdb 1HMY), colored green. (D) Topology of DNMT2.
The large domain is composed of an eight-stranded β sheet with three helices (αA, αB and αX) on one side and four helices (αC, αC′, αD and αE) on the opposite side. Strands β1–β7 adopt a well-characterized AdoMet-dependent MTase fold (21). M.HhaI does not have an equivalent of the short strand β8 in DNMT2. The large domain contains nine of 10 conserved motifs, including the proposed active site nucleophile (Cys79, which reacted with a mercury atom in one of the crystals) and other catalytic residues, as well as the cofactor-binding pocket. The loop containing Cys79 between strand β4 and helix αD is in an open conformation in the absence of DNA substrate (indicated by the red loop in Fig. 2A and B). The structure presented here has AdoHcy bound in a way that is identical to the primed catalytically competent conformation observed in M.HhaI (22).
The small domain has a short four-stranded antiparallel propeller (β9–β12) and five surrounding helices (αF–αJ). M.HhaI structure has a similar propeller, but does not have the equivalent of the three helices αH, αI and αJ (Fig. 2C).
Deleted segment
The dashed line in Figure 2A identifies the non-conserved segment (residues 189–247) of the protein, part of which (Δ47) was deleted to allow crystallization. This segment is clearly present in mammalian and X.laevis Dnmt2, but absent in the DNMT2 homologs of S.pombe, D.melanogaster and B.mori (Fig. 1). In A.thaliana and D.rerio sequences of similar length are present but share little or no sequence similarity with the mammalian and X.laevis sequences.
The segment that had to be deleted to allow crystallization is inserted into the connecting loop between the two structured domains and could fold into an independent domain (Fig. 2A). BLAST searches did not reveal any significant homology between this domain and any known protein. This 58 residue segment (7 kDa) was expressed, purified and eluted as a 24 kDa globular protein on a Superdex 200 column (Pharmacia). Reconstitution of DNMT2Δ47 and this segment did not form a complex that survived chromatography, which indicated that residues 189–247 represent an independent protein domain that does not interact strongly with the remainder of the protein.
Conserved motifs
Inspection of the DNMT2Δ47–AdoHcy structure showed that DNMT2 contains perfect matches to all 10 consensus motifs (Fig. 1), with three exceptions. First, S.pombe pmt1p contains the sequence ProSerCys instead of ProProCys in motif IV, the proposed active site. Deleting the serine to partially restore the consensus ProCys sequence was reported to create an active enzyme that methylates dcm (CCWGG) sites (23), which are normally methylated in E.coli. This was not true of DNMT2, which normally bears the canonical ProProCys motif but does not display this activity. Second, motif I (with the consensus Phe-X-Gly-X-Gly) of DNMT2 contains a tyrosine in place of the canonical phenylalanine, except for the D.melanogaster homolog. The Tyr10 phenol ring forms an edge-to-face van der Waals contact with the adenine moiety of AdoHcy (as does the Phe benzene ring observed in M.HhaI) and its hydroxyl group forms two hydrogen bonds with the Ser98 hydroxyl oxygen atom and the main chain nitrogen atom of Phe99 (Fig. 3A). Ser98 and Phe99 are located at the N-terminus of helix αD and mark the C-terminal boundary of the proposed active site loop containing Cys79. The D.melanogaster homolog contains a Phe at the same position but was catalytically inactive in vitro (24). Interestingly, a Phe→Tyr mutation in motif I of M.MspI, a bacterial MTase, caused an 8-fold increase in the time required for covalent complex formation when assayed with fluorinated target DNA but did not abolish catalytic activity (25). These data suggest that the presence of a Tyr in motif I is unlikely to prevent methyl transfer by DNMT2. Third, DNMT2 motif VIII (with consensus Gln-X-Arg-X-Arg) contains an Asn in place of the canonical Gln, except for A.thaliana and D.rerio, which contain a Tyr and Gln, respectively, at this position. The Asn158 side chain makes several hydrogen bonds to the TRD loop (Fig. 3B): some of these contacts are mediated by a water molecule due to the shorter side chain of Asn, whereas in M.HhaI the equivalent Gln makes direct contacts. A single point mutation that restores the consensus sequence (Asn158→Gln) in DNMT2 did not restore methyl transfer activity (P.Kearney and X.Cheng, data not shown). Therefore, it is unlikely that the Gln→Asn substitution causes DNMT2 to be inactive.
Figure 3.
Detailed plots of interactions centered on (A) Tyr10 of motif I and (B) Asn158 of motif VIII. (C) Superimposition of the TRD loop containing Cys292-Phe293-Thr294 of DNMT2 (blue) and Thr250-Leu251 of M.HhaI (green).
Conserved TRD
m5C MTases usually contain a TRD between conserved motifs VIII and IX (18). The variable TRD sequences determine the substrate specificity of individual enzymes. Although enzymes that have identical specificity often have related TRDs, TRDs of enzymes with different specificity share little sequence conservation, except for a ThrLeu dipeptide diagnostic of most TRDs (20,26,27). In the co-crystal structure of M.HhaI with DNA Thr250 of the dipeptide is located at the interface between the DNA backbone and the enzyme (28). Its side chain hydroxyl oxygen interacts with one of the phosphate oxygen atoms of the nucleotide immediately 5′ to the target cytosine. When the M.HhaI and DNMT2 structures are superimposed (with a root mean square deviation of <1 Å for the corresponding Cα positions) the backbone atoms for residues 289–295 in DNMT2 and 247–253 in M.HhaI overlay well (Fig. 3C). In M.HhaI (28) as well as in M.HaeIII (29) this structurally conserved loop forms part of the scaffold that supports the residues interacting with DNA (27). The dipeptide in DNMT2 that corresponds to Thr250-Leu251 of M.HhaI is Cys292-F293. It should be noted that a Thr250→Cys mutant of M.HhaI behaves very similarly to the wild-type enzyme (20). However, it would be interesting to see whether swapping the TRD between DNMT2 and M.HhaI will generate an active hybrid MTase.
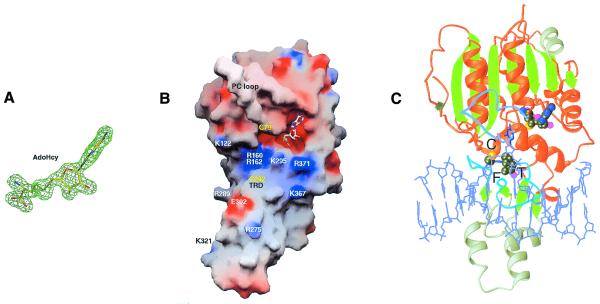
Among DNMT2 family members the 41 residue stretch surrounding the TRD (266–306) contains many highly conserved residues (Fig. 1). The structure of DNMT2 offers a rationale for their conservation. Most of the conserved hydrophobic residues (Tyr266, Leu268, Leu273, Leu282, Phe293 and Tyr297), along with Leu340, Tyr342 and Phe343 of motif IX and Tyr370 of motif X, are involved in structural packing of the small domain and intramolecular interactions (Asp281…Ser306). The arginines (Arg275, Arg288 and Arg289) and Lys295 are located on the potential DNA-binding surface (Fig. 4A).
Figure 4.
(A) AdoHcy is shown in stick form and is superimposed on a Fo – Fc omit electron density map. (B) Solvent-accessible molecular GRASP surface (30) of the electrostatic potential of DNMT2Δ47. The surface is colored blue for positive (10 kT), red for negative (–10 kT) and white for neutral, where k is the Boltzmann constant and T is the temperature. (C) Model of DNMT2Δ47–AdoHcy bound to DNA. The DNA model is from the structure of the M.HhaI–AdoHcy–DNA complex (pdb 3MHT; 31).
Model of the DNMT2–DNA complex
No protein that contains the conserved DNA MTase motifs has previously failed to show methyl transfer activity in biochemical assays or genetic tests. However, abnormalities of methylation patterns were not detected in Dnmt2-deficient mouse ES cells (5) and no detectable methyl transfer activity was observed for wild-type DNMT2 in assays that measured incorporation of methyl groups from [3H-methyl]-AdoMet into a variety of DNA substrates in vitro [including mammalian and E.coli genomic DNA, λ DNA, plasmid DNA, poly(dI·dC), poly(dC·dG), random sequence oligonucleotides and RNA; J.A.Yoder and T.H.Bestor, data not shown]. The DNMT2Δ47–AdoHcy structure shows that the cofactor analog AdoHcy binds normally (Fig. 4A and B) and the protein bears the consensus active site ProCys sequence in motif IV (Fig. 1).
A GRASP (30) representation of the electrostatic surface potential of DNMT2Δ47 shows that AdoHcy binds in an acidic (red) pocket next to a surface enriched in basic (blue) residues (Fig. 4B), which indicates a potential DNA-binding surface. The high degree of structural similarity between DNMT2 and M.HhaI allows us to create a model of DNMT2 bound to DNA. Using the coordinates for the ternary structure of M.HhaI–DNA–AdoHcy (31), we superimposed the protein components and then positioned DNA over the basic surface of DNMT2Δ47. The resulting model showed that DNMT2Δ47 could contact B-form DNA without physical distortion of either the protein or DNA component (Fig. 4C). It is possible that upon association with DNA the catalytic loop (colored light blue in Fig. 4C) would adopt a closed conformation similar to that of M.HhaI (28). From the structure there is no apparent reason why DNMT2Δ47, which is strikingly similar to M.HhaI in size and structure (Fig. 2C) and in AdoHcy binding (Figs 3A and 4A), fails to demonstrate transmethylation activity in vitro and why it should be so strongly conserved in S.pombe and D.melanogaster, organisms whose DNA is not methylated and whose genomes do not contain homologs of Dnmt1 or the Dnmt3 family. We set out to investigate the biochemical properties of DNA binding by DNMT2.
Formation of DNMT2–DNA complexes is resistant to denaturation and is inhibited by ES cell extracts and nucleoside triphosphates in vitro
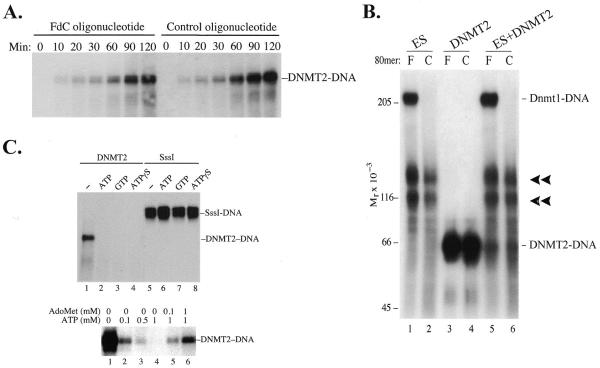
It has been well established that m5C MTases can form an irreversible covalent bond with 5-fluoro-2′-deoxycytosine (FdC) within an oligonucleotide (16,28,32,33). Surprisingly, DNMT2 was found to form denaturant-resistant complexes with oligonucleotides independent of FdC, i.e. DNMT2 forms complexes with both FdC and control oligonucleotides in which FdC has been replaced with dC (Fig. 5A). The time course of product accumulation was linear for >2 h.
Figure 5.
(A) Formation of denaturant-resistant DNMT2–DNA complexes. DNMT2 (4 µg/ml) was incubated at 37°C with oligonucleotides (1 µg/ml) in 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 25 mM NaCl, 0.2 mM PMSF, 0.5 mM DTT and 40 µM AdoMet. Reactions were terminated at the indicated times by addition of SDS to 2% and glycerol to 12% and heating to 65°C for 10 min, subjected to 6% SDS–PAGE, transferred to nitrocellulose and autoradiographed. Oligonucleotides were of sequence 5′-CCTTTACAAATTTCCAATGCNNNNFGNNNNNNNNFNNNNNNNNNFGNNNNCCTGAAAAAAGACTAATTAAATTCATGGTA-3′, where N is a random base. The synthesis, radioactive labeling and purification of FdC oligonucleotides have been described (16). The control oligonucleotide duplexes were not methylated, while FdC duplexes were hemimethylated (16). Methylation status did not affect formation of complexes (data not shown). Lanes headed F contain FdC oligonucleotides and lanes headed C contain control oligonucleotides with C replacing FdC. (B) Effect of ES cell lysates on formation of DNMT2–DNA complexes. Aliquots of 70 µg ES cell lysate were incubated with 0.1 µg internally labeled FdC or control oligonucleotide in 100 µl of 20 mM Tris–HCl pH 7.4, 10% glycerol, 1 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF and 40 µM AdoMet. Where indicated, purified DNMT2 (0.4 µg) was added at the beginning of the reaction. Reactions were terminated after 3 h at 37°C by addition of SDS to 2% and heating to 65°C for 10 min, subjected to 6% SDS–PAGE, transferred to nitrocellulose and autoradiographed. (C) Inhibition of DNMT2–DNA complex formation by nucleoside triphosphates. Incubation of DNMT2 with FdC oligonucleotide and 1 mM ATP, GTP or ATPγS prevented complex formation but had no effect on the reaction of M.SssI with the FdC oligonucleotide. The inhibitory effect could be largely overcome by addition of equimolar amounts of AdoMet (lanes 5 and 6). In all cases electrophoresis was halted just after unbound oligonucleotides migrated off the lower end of the gel.
Figure 5B shows several aspects of the DNMT2–DNA interaction. First, as seen previously (16), Dnmt1 from ES cells is visible as a strictly FdC-dependent protein–DNA complex (lanes 1 and 5), but not with control oligonucleotides that bear cytosines at the positions of the FdC residues (lanes 2 and 6). Second, DNMT2 forms complexes with both FdC and control oligonucleotides (lanes 3 and 4). This indicates that DNMT2 forms a complex with DNA in a manner that differs from that of Dnmt1 and bacterial m5C MTases, which are completely dependent on FdC for stable covalent complex formation. Third, the unidentified protein–DNA complexes (16), indicated with chevrons, are also FdC independent, but have mobilities distinct from that of the DNMT2–DNA complex. Fourth, it was reasoned that recombinant DNMT2 might be lacking accessory factors essential for methyl transfer activity; this seemed especially likely given that DNMT2 lacks the large N-terminal domain of DNMT1 which is necessary for normal function of the latter enzyme (reviewed in 1). ES cells are a likely source of accessory factors as they are capable of de novo methylation and maintain trace levels of m5C when homozygous for null mutations in Dnmt1 (2). Lysates of ES cells were cleared of nucleic acids by absorption to DEAE–Sephacel as described (16) and added to reactions containing purified DNMT2 and DNA. The ES cell extracts did not activate a latent transmethylase activity of DNMT2 but actually inhibited complex formation with both FdC and control oligonucleotides (lanes 5 and 6).
As shown in lanes 1–4 of Figure 5C, the formation of denaturant-resistant complexes by DNMT2 is inhibited by 1 mM ATP, GTP or the non-hydrolyzable ATP analog ATPγS. M.SssI, a CpG-specific bacterial DNA MTase (34), was insensitive to these compounds (lanes 5–8), as was M.HhaI (A.Dong and X.Cheng, data not shown). Inhibition of stable DNMT2–DNA complex formation was largely reversed by addition of equimolar AdoMet (Fig. 5C). Hydrolysis of nucleoside triphosphate was not required for inhibition, as shown by equal inhibition by ATPγS. The competitive inhibition of AdoMet by NTPs suggests that nucleoside triphosphates can occupy the binding sites of both AdoMet and DNA and compete with binding of both. If this is the case, physiological concentrations (low mM) of nucleoside triphosphates would be expected to eliminate AdoMet binding, which is thought to be at cytoplasmic concentrations in the low µM range (35). Therefore, DNMT2 differs from authentic DNA MTases in its formation of a denaturant-resistant complex with DNA and sensitivity of DNA binding to nucleoside triphosphates.
DISCUSSION
Is DNMT2 a DNA MTase? The remarkable isostery with M.HhaI and the presence and correct spatial organization of all of the motifs diagnostic of m5C MTases suggest that DNMT2 is a DNA MTase, but it lacks observable in vitro transmethylase activity. Abnormalities of methylation patterns were not observed in ES cells homozygous for mutations in the Dnmt2 gene and the mutant cells retained the capacity to methylate newly integrated retroviral DNA (5). However, the absence of a phenotype in Dnmt2 null ES cells and in vitro detectable transmethylase activity do not rule out a cellular function for Dnmt2. The lack of Dnmt2 activity could be due to a requirement for a specific post-translational modification or stage-specific cofactor or for a complex DNA target structure not easily mimicked in an in vitro setting. Like Dnmt2, Ascobolus immersus Masc-1 lacks detectable in vitro methylation activity (36). However, genetic data showed that Masc-1 plays an essential role in methylation induced premeiotically, which requires the pairing of duplicated sequences occurring during the premeiotic phase of sexual reproduction (36). If Dnmt2 has a similar role, or plays some role in imprinting, a phenotype may not be appreciated until Dnmt2 null mice are generated and the reproductive phenotypes and the phenotypes of offspring of homozygous parents are determined.
It is also possible that DNMT2 is not a DNA MTase. In addition to a lack of detectable in vitro enzymatic activity, DNMT2 homologs are expressed in at least two eukaryotes (S.pombe and D.melanogaster) whose DNA lacks m5C and in these organisms the DNMT2 homologs are the only proteins with similarity to known m5C MTases. All eukaryotes known to methylate their DNA have been found to express at least two DNA MTases, one of which is related to DNMT1 (reviewed in 1). The presence of DNMT2 homologs in organisms not known to methylate their genomes suggests that DNMT2 may not be an active DNA MTase. In addition, Liu and Santi (37) have reported that m5C RNA MTases, which employ the same reaction mechanism and share sequence similarities, including the ProCys sequence, with m5C DNA MTases, use a downstream Cys residue in catalysis. The Cys residue within the CysPheThr tripeptide could have catalytic function within members of the DNMT2 family, although there is at present no reason to believe that DNMT2 is an RNA MTase.
The strong sequence conservation of DNMT2 homologs in diverse eukaryotic taxa suggests an important biological function, but further genetic analysis has thus far failed to identify a role for DNMT2 or its homologs. Human DNMT2 maps very near the SCIDA locus (38), which is involved in a radiation-sensitive form of recessive severe combined immunodeficiency (SCID) common among Athabascan-speaking American Indians. The normal formation of signal joints and defective formation of coding joints during V(D)J recombination in SCIDA patients could involve defects in a DNA-binding protein such as DNMT2. However, mutations in the DNMT2 gene could not be found in DNA from Navajo SCIDA patients (L.Li, M.J.Cowan, J.Russo, X.Qu and T.H.Bestor, data not shown). The D.melanogaster Zucchini (Zuc) gene (http://flybase.bio.indiana.edu/.bin/fbidq.html?FBgn0004056; 39) maps very near dDnmt2 within 33B3–F2. Mutations in the intronless dDnmt2 gene were not observed in three independent ethylmethanesulfonate-induced alleles of Zuc (K.Wehr, G.Schupbach and T.H.Bestor, data not shown).
The denaturant-resistant and probably covalent binding to DNA suggests an evolutionary pathway in which a DNA MTase lost the ability to transfer methyl groups but instead marked sites in the genome by attaching directly to DNA. The catalytic mechanism of m5C MTases normally involves a transient covalent transition state intermediate formed by addition of a cysteine thiolate to the cytosine 6 position. After methyl transfer, abstraction of a proton from the 5 position allows reformation of the 5,6 double bond and release of free enzyme by proton elimination. A mutation that delayed or prevented proton abstraction would cause stable covalent bonding between enzyme and DNA in a manner that does not depend on methyl transfer. Regulated abstraction of a proton from the 6 position would release bound enzyme, making the DNMT2–DNA complex much more reversible than 5-methylcytosine. While a probable role of DNMT2 and its homologs has not yet been determined, it may be significant that centromere structure and function is conserved among those organisms that contain DNMT2 homologs but is divergent in S.cerevisiae and C.elegans, which do not possess DNMT2 homologs. Saccharomyces cerevisiae has very short discrete centromeres that bind a group of proteins that are largely distinct from the well-conserved proteins associated with the centromeres of most eukaryotes and C.elegans has holocentric chromosomes rather than discrete centromeres. These phylogenetic findings suggest the possibility of a role in centromere function, and epigenetic inheritance of the state of centromere function in mammals (reviewed in 40) and S.pombe (41) provides an evolutionary link with epigenetic inheritance of the transcriptionally silent state mediated by DNA MTases. However, if DNMT2 functions via a mechanism that does not involve methyl transfer, the retention by DNMT2 homologs of so much of the character of m5C MTases in terms of sequence and structure becomes an interesting issue, given the strong conservation of DNMT2 homologs in taxa that underwent ancient phylogenetic separation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Goll, P. Warnecke and M. Yanagida for discussions, Paul Kearney for mutagenesis, L. Li and M.J. Cowan for discussions and for providing DNA from SCIDA patients, C. Wehr and G. Schupbach for discussions and for DNA from Zuc mutant flies and A. Nagy for providing R1 ES cells. These studies were funded in part by NIH grant GM49245 and funds from the Georgia Research Alliance (to X.C.) and NIH grants GM59377 and HD37687 and a grant from the Leukemia and Lymphoma Society (to T.H.B.).
PDB no. 1G55
References
- 1.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 2.Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- 3.Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 4.Xu G.-L., Bestor,T.H., Bourc’his,D., Hsieh,C.-L., Tommerup,N., Bugge,M., Hulten,M., Qu,X., Russo,J.J. and Viegas-Pequignot,E. (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 402, 187–191. [DOI] [PubMed] [Google Scholar]
- 5.Okano M., Xie,S. and Li,E. (1998) Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res., 26, 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoder J.A. and Bestor,T.H. (1998) A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet., 7, 279–284. [DOI] [PubMed] [Google Scholar]
- 7.Hung M.-S., Karthikeyan,N., Huang,B., Koo,H.-C., Kiger,J. and Shen,C.-K.J. (1999) Drosophila proteins related to vertebrate DNA (5-cytosine) methyltransferases. Proc. Natl Acad. Sci. USA, 96, 11940–11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyko F., Whittaker,A.J., Orr-Weaver,T.L. and Jaenisch,R. (2000) The putative Drosophila methyltransferase gene dDnmt2 is contained in a transposon-like element and is expressed specifically in ovaries. Mech. Dev., 95, 215–217. [DOI] [PubMed] [Google Scholar]
- 9.Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 10.Yeates T.O. (1997) Detecting and overcoming crystal twinning. Methods Enzymol., 276, 344–358. [PubMed] [Google Scholar]
- 11.Furey W. and Swaminathan,S. (1997) PHASES-95: a program package for processing and analyzing diffraction data from macromolecules. Methods Enzymol., 277, 590–620. [DOI] [PubMed] [Google Scholar]
- 12.Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr., D52, 30–42. [DOI] [PubMed] [Google Scholar]
- 13.Jones T.A. and Kjeldgard,M. (1997) Electron-density map interpretation. Methods Enzymol., 277, 173–208. [DOI] [PubMed] [Google Scholar]
- 14.Brünger A.T. (1992) X-PLOR. A System for X-ray Crystallography and NMR, v.3.1. Yale University, New Haven, CT.
- 15.Navaza J. and Saludjian,P. (1997) AmoRe: an automated molecular replacement program package. Methods Enzymol., 276, 581–594. [DOI] [PubMed] [Google Scholar]
- 16.Yoder J.A., Soman,N.S., Verdine,G.L. and Bestor,T.H. (1997) DNA (cytosine-5) methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol., 270, 385–395. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson C.R., Bartlett,R., Nurse,P. and Bird,A.P. (1995) The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res., 23, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauster R., Trautner,T.A. and Noyer-Weidner,M. (1989) Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domians. J. Mol. Biol., 206, 305–312. [DOI] [PubMed] [Google Scholar]
- 19.Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilkaitis G., Dong,A., Weinhold,E., Cheng,X. and Klimasauskas,S. (2000) Functional roles of the conserved threonine-250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem., 275, 38722–38730. [DOI] [PubMed] [Google Scholar]
- 21.Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of Adomet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, Singapore, pp. 1–38.
- 22.O’Gara M., Zhang,X., Roberts,R.J. and Cheng,X. (1999) Structure of a binary complex of HhaI methyltransferase with S-adenosyl-l-methionine formed in the presence of a short non-specific DNA oligonucleotide. J. Mol. Biol., 287, 201–209. [DOI] [PubMed] [Google Scholar]
- 23.Pinarbasi E., Elliott,J. and Hornby,D.P. (1996) Activation of a yeast pseudo DNA methyltransferase by deletion of a single amino acid. J. Mol. Biol., 257, 804–813. [DOI] [PubMed] [Google Scholar]
- 24.Tweedie S., Ng,H.-H., Barlow,A.L., Turner,B.M., Hendrich,B. and Bird,A. (1999) Vestiges of a DNA methylation system in Drosophila melanogaster? Nature Genet., 23, 389–399. [DOI] [PubMed] [Google Scholar]
- 25.Pinarbasi E., Kan,M.S., Duran,C., Ford,G.C. and Hornby,D.P. (1998) Substitution of the conserved phenylalanine in the S-adenosyl-l-methionine binding site of M.MspI with tyrosine modifies the kinetic properties of the enzyme. Biol. Chem., 379, 591–594. [PubMed] [Google Scholar]
- 26.Lange C., Wild,C. and Trautner,T.A. (1996) Identification of a subdomain within DNA-(cytosine-C5)-methyltransferase responsible for the recognition of the 5′ part of their DNA target. EMBO J., 15, 1443–1450. [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng X. and Blumenthal,R.M. (1996) Finding a basis for flipping bases. Structure, 4, 639–645. [DOI] [PubMed] [Google Scholar]
- 28.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 29.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Protein Struct. Funct. Genet., 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 31.O’Gara M., Klimasauskas,S., Roberts,R.J. and Cheng,X. (1996) Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase-DNA-AdoHcy complexes. J. Mol. Biol., 261, 634–645. [DOI] [PubMed] [Google Scholar]
- 32.Santi D.V., Garrett,C.E. and Barr,P.J. (1983) On the mechanism of inhibition of DNA-cytosine methyltransferase by cytosine analogs. Cell, 33, 9–10. [DOI] [PubMed] [Google Scholar]
- 33.Chen L., MacMillan,A.M., Chang,W., Ezaz-Nikpay,K., Lane,W.S. and Verdine,G.L. (1991) Direct identification of the active site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry, 30, 11018–11025. [DOI] [PubMed] [Google Scholar]
- 34.Renbaum P., Abrahamove,D., Fainsod,A., Wilson,G.G., Rottem,S. and Razin,A. (1990) Cloning, characterization and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1 (M.SssI). Nucleic Acids Res., 18, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J.C. (1991) DNA topoisomerases: why so many? J. Biol. Chem., 266, 6659–6662. [PubMed] [Google Scholar]
- 36.Malagnac F., Wendel,B., Goyon,C., Faugeron,G., Zickler,D., Rossignol,J.L., Noyer-Weidner,M., Vollmayr,P., Trautner,T.A. and Walter,J. (1997) A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell, 91, 281–290. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y. and Santi,D.V. (2000) m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl Acad. Sci. USA, 97, 8263–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moshous D., Li,L., Chasseval,R., Philippe,N., Jabado,N., Cowan,M.J., Fischer,A. and de Villartay,J.P. (2000) A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum. Mol. Genet., 9, 583–588. [DOI] [PubMed] [Google Scholar]
- 39.Schupbach G. and Wieschaus,E. (1991) Female sterile mutations on the second chromosome of Drosophila melanogaster. Genetics, 129, 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warburton P.E. (1999) Making CENs of mammalian artificial chromosomes. Mol. Genet. Metab., 68, 152–160. [DOI] [PubMed] [Google Scholar]
- 41.Steiner N.C. and Clarke,L. (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell, 79, 865–874. [DOI] [PubMed] [Google Scholar]
- 42.Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]