Abstract
The PhoP and PhoR proteins from Mycobacterium tuberculosis form a highly specific two-component system that controls expression of genes involved in complex lipid biosynthesis and regulation of unknown virulence determinants. The several functions of PhoP are apportioned between a C-terminal effector domain (PhoPC) and an N-terminal receiver domain (PhoPN), phosphorylation of which regulates activation of the effector domain. Here we show that PhoPN, on its own, demonstrates PhoR-dependent phosphorylation. PhoPC, the truncated variant bearing the DNA binding domain, binds in vitro to the target site with affinity similar to that of the full-length protein. To complement the finding that residues spanning Met1 to Arg138 of PhoP constitute the minimal functional PhoPN, we identified Arg150 as the first residue of the distal PhoPC domain capable of DNA binding on its own, thereby identifying an interdomain linker. However, coupling of two functional domains together in a single polypeptide chain is essential for phosphorylation-coupled DNA binding by PhoP. We discuss consequences of tethering of two domains on DNA binding and demonstrate that linker length and not individual residues of the newly identified linker plays a critical role in regulating interdomain interactions. Together, these results have implications for the molecular mechanism of transmission of conformation change associated with phosphorylation of PhoP that results in the altered DNA recognition by the C-terminal domain.
Keywords: Bacterial Signal Transduction, Gene Regulation, Helix-Loop-Helix Transcription Factors, Protein-DNA Interaction, Protein Domains, Protein Phosphorylation, Linker, M. tuberculosis PhoP, Response Regulator
Introduction
Bacterial adaptation usually takes the form of transcriptional regulation of genes, products of which specifically help the bacterium cope with the given microenvironment. One of the systems bacteria most frequently use to sense chemical or physical changes in the environment is the two-component regulatory system. Such a system typically consists of a histidine protein kinase that functions as an environmental sensor and a response regulator that mediates the transcriptional processes (1, 2). The sensor kinase is usually autophosphorylated in response to environmental signals and subsequently transfers the phosphate to the regulator. In general, the response regulator functions as phosphorylation-activated switches that often mediate a cellular response through transcriptional regulation.
A number of investigations show that inactivation of phoP of Mycobacterium tuberculosis phoP–phoR system leads to significant growth attenuation (3–5). Also, biochemical results reveal that PhoP regulates biosynthesis of sulfolipids (SL),5 diacyltrehaloses (DAT) and polyacyltrehaloses (PAT) and absence of these complex lipid molecules in the phoP mutant is a major reason for its attenuated growth in a mouse model (4–5; for a review, see Ref. 6). While two independent studies show that a point mutation in PhoP contributes to avirulence of M. tuberculosis H37Ra (7) and also accounts for the absence of polyketide-derived acyltrehaloses in M. tuberculosis H37Ra (8), more recently PhoP has been implicated in the ESAT-6 secretion and specific T-cell recognition during virulence regulation of the bacilli (9). Thus, growing evidences strongly suggest that PhoP is a key regulator in M. tuberculosis. However, molecular mechanism of how the regulator functions remains largely unknown.
M. tuberculosis PhoP belongs to the OmpR/PhoB subfamily, the largest subfamily of response regulators comprising two domains, an N-terminal regulatory domain (also called a receiver domain) and a C-terminal transactivation domain (also called an effector domain) that binds specific DNA sequence within the target promoters and interacts with the cellular transcription machinery. The N-terminal domain shares a conserved, doubly wound (α/β)5 fold which include a highly conserved aspartate residue that is the site of phosphorylation (Asp71 in PhoP) (10). The C-terminal domain has been characterized and its crystal structure has been determined (Protein Data Bank code 2PMU) (11). The structural analysis reveals overall folds similar to those of other OmpR family proteins with a winged helix-turn-helix DNA binding motif involved in DNA binding (12–16). Despite strong structural homology within the family members, there exist significant differences in the mechanism of DNA binding functions. For example, although C-terminal structures of OmpR and PhoB are readily superimposed (17), phosphorylation of OmpR enhances DNA binding affinity for specific sites (18), whereas phosphorylation of PhoB results in a relief of inhibition of DNA binding (19). Moreover, while the C terminus of OmpR binds to DNA only weakly (20), the C terminus of PhoB has a higher affinity for specific DNA than the unphosphorylated full-length protein (21). Interestingly, biochemical studies reveal that one of the major differences between OmpR and PhoB reside in the inter-domain linker region that tethers together the N-terminal domain with the C-terminal domain (22). Consistent with this view, a previous study had shown that C-terminal DNA binding by OmpR could influence phosphorylation of the N terminus in which the linker region underwent a conformational change (23), thus suggesting a key role of the linker region in regulation of inter-domain interaction(s).
We are interested in the molecular mechanism responsible for transmission of the conformational change associated with phosphorylation of PhoP that is expected to influence DNA binding by the C-terminal domain. To better understand the functions of the N-terminal domain and inter-domain interaction(s) in effector domain regulation, we sought to investigate domain structure of PhoP. Results reported here identify an 11-residue long inter-domain linker that tethers two functionally-independent domains of PhoP together and regulates inter-domain interactions. While the newly-identified linker region is not required for either domain functions of PhoP, most strikingly, it plays an essential role for phosphorylation-dependent DNA binding to msl3 promoter, previously suggested to be regulated by PhoP (4). These observations invite speculation on additional level(s) of complexity in the regulation of PhoP-controlled transcription processes. Together, our results suggest that although the DNA binding energy and specificity of regulator-promoter interactions is contributed primarily (but not entirely) by the C-domain, linker region of the protein likely allows the regulator to adopt a different phosphorylation-dependent conformation enabling it to discriminate target promoters while it regulates a vast array of genes to either activate or repress transcription.
EXPERIMENTAL PROCEDURES
Recombinant DNA Techniques
All enzymatic manipulations of DNA were performed using standard procedures (24) with reagents purchased from New England Biolabs (restriction endonucleases, T4 DNA ligase, alkaline phosphatase, and Deep Vent DNA polymerase). Plasmid DNA isolation, recovery, and purification of DNA fragments or PCR products from agarose gels were carried out using Qiagen spin columns and procedures (Qiagen). PAGE-purified oligonucleotides were synthesized by Sigma.
Preparation, Amplification, and Cloning of DNA and Site-directed Mutagenesis
Escherichia coli DH5α was used for all cloning procedures. The gene encoding M. tuberculosis H37Rv PhoP was amplified by PCR and cloned into expression vector pET15b (Novagen) as described previously (25). For overproduction and purification of PhoPN and PhoPC domains in E. coli, parts of the phoP gene extending from Met1 to Lys141 of the N-terminal end and from Lys141 to Arg247 of the C-terminal end of PhoP were PCR-amplified using oligonucleotide pairs phoPstart and RPphoPN141 or FPphoPC141 and phoPstop as primers (supplemental Table S1) and the wild-type phoP gene (pET-phoP) as template and introduced between an NdeI and a BamHI site of pET15b (Novagen). The cloning strategy resulted in truncated constructs of PhoP with natural C termini and an N-terminal His tag.
DNA fragment comprising regions upstream of msl3 was PCR-amplified from the M. tuberculosis genomic DNA using ProofStart DNA polymerase (Qiagen) and oligonucleotide pair FPmsl3up/RPmsl3up (supplemental Table S1). Site-directed mutagenesis of individual PhoP residues (described under “Results”) was carried out in the phoP gene (pET-phoP) by a PCR-based two-stage overlap extension method (26) using complementary oligonucleotides with the mutated codon (supplemental Table S2) and Deep Vent DNA polymerase. All mutants were verified by DNA sequence analysis to confirm the presence of the desired mutation and absence of any unintentional mutations.
Protein Purification
The mutant proteins or the truncated variants of PhoP were overexpressed in E. coli BL21(DE3) cells and purified by immobilized metal affinity chromatography (nickel-nitrilotriacetic acid, Qiagen) as described previously (10, 12, 25). The molecular masses determined by mass spectrometry were in agreement with the values predicted from primary sequence analysis. When appropriate, His tag was cleaved using the Thrombin Clean Cleavage kit (Sigma). The purity of all of the proteins used here was greater than 95% as judged by Coomassie Blue staining of overloaded SDS-polyacrylamide gels. Protein concentrations were estimated by Bradford reagent using bovine serum albumin as a calibration standard.
Trypsin Digestion of PhoP and Identification of Tryptic Fragments
Reaction mixtures (20 μl) containing 4 μg of PhoP were incubated in the digestion mixture containing 50 mm Tris-HCl, pH 7.90, 100 mm NaCl, 3 mm magnesium acetate, and 400 ng of trypsin at 37 °C for 15 min, keeping the ratio of PhoP to trypsin at 100:1 (w/w). Proteolysis was terminated by the addition of SDS, and the aliquots from the digestion mixtures were resolved by 12% SDS-polyacrylamide gel electrophoresis and visualized by Coomassie Blue staining. To determine the N-terminal sequence of the tryptic fragments, resolved polypeptides from an SDS-polyacrylamide gel were transferred electrophoretically onto a polyvinylidene difluoride membrane (Millipore) and visualized by Amido Black (Sigma) staining. Membrane slices containing individual proteolytic products were excised and subjected to automated Edman sequencing using an Applied Biosystems protein sequencer.
PhoP Phosphorylation
Phosphorylation of PhoRC (C-terminal end of M. tuberculosis PhoR comprising residues Thr193–Pro485; see Ref. 10) was carried out in phosphorylation buffer (50 mm HEPES, pH 7.5, 50 mm KCl, 10 mm MnCl2) containing 25 μm of [γ-32P]ATP (BRIT, Hyderabad, India) at 25 °C for 1 h. For phosphotransfer assays, PhoP or its truncated variants were added in phosphorylation buffer at 18 °C for the indicated times. The reactions were terminated by the addition of 10 mm EDTA, and the products were resolved by 12% SDS-polyacrylamide gels. After electrophoresis, the gels were dried and visualized by autoradiography. When required, acetyl phosphate (AcP of ≥85% purity; Sigma) was used to phosphorylate recombinant PhoP in vitro. Previously, we provided a direct demonstration of phosphorylation of PhoP using AcP as the phospho-donor (27). Briefly, purified PhoP was added to phosphorylation buffer supplemented with 50 mm AcP and 10 mm MgCl2, and the mixtures were incubated at 37 °C for 60 min.
CD Spectroscopy
Circular dichroism measurements were carried out with a JASCO spectropolarimeter, model J-810 (Jasco, Tokyo, Japan). All measurements were performed using a 0.1-cm cell. Residue molar ellipticity (θ) was defined as 100 θobs (lc)−1, where θobs was observed ellipticity; l was length of the light path in cm, and c was residue molar concentration of each protein. Scan speed was 100 nm/min, and 10 scans were signal-averaged to increase signal/noise ratio.
Oligodeoxyribonucleotide Probes
Concentrations of PAGE-purified oligonucleotides (Sigma) were determined from absorbance at 260 nm, using calculated extinction coefficients. The 60-bp DR1,2 DNA fragment comprising a 9-bp direct repeat motif was used to assess sequence-specific DNA binding by PhoP (or its variants) as described previously (12, 25). This sequence of the phoP upstream region is within the PhoP-protected DNase I footprint (10, 28). A nonspecific probe was derived from a distal region of phoP promoter and lacked any PhoP binding site (sequence as described in Ref. 25). The target binding site(s) in each set of oligonucleotides were flanked by matching nonspecific sequences, chosen to avoid fortuitous binding. 5′-32P labeling of oligonucleotide probes was carried out using T4 polynucleotide kinase and [γ-32P]ATP. Unincorporated nucleotides and labeled oligonucleotide were separated using a Sephadex-G50 quick spin column (GE Healthcare). DNA probes for EMSA were generated by annealing the labeled strand to its unlabeled complement in 10 mm Tris, pH 8.0, containing 50 mm NaCl following slow cooling to room temperature after heating it to 95 °C for 5 min. Annealing efficiency was verified by native PAGE.
Gel Mobility Shift Assay
Binding reactions (10 μl) contained DNA (10 nm) and PhoP in 20 mm HEPES-Na+ (pH 7.5), 50 mm NaCl, 0.2 mg/ml bovine serum albumin, 10% glycerol, 1 mm dithiothreitol, and 200 ng of sheared herring sperm DNA. Incubation was for 20 min at 20 °C. The reactions were analyzed by electrophoresis through non-denaturing 6% (for msl3up1 DNA substrate) or 8% (for oligonucleotide-based substrate) polyacrylamide gels (acrylamide/bisacrylamide 40:1.1 in 1× Tris-EDTA buffer). The gels were run at 4 °C for 6–8 h at 70 V or 12–14 h at 40 V, dried, and visualized by autoradiography. Following electrophoresis and gel drying, radioactive bands were quantified using a PhosphorImager.
RESULTS
Phosphorylation-coupled DNA Binding by PhoP
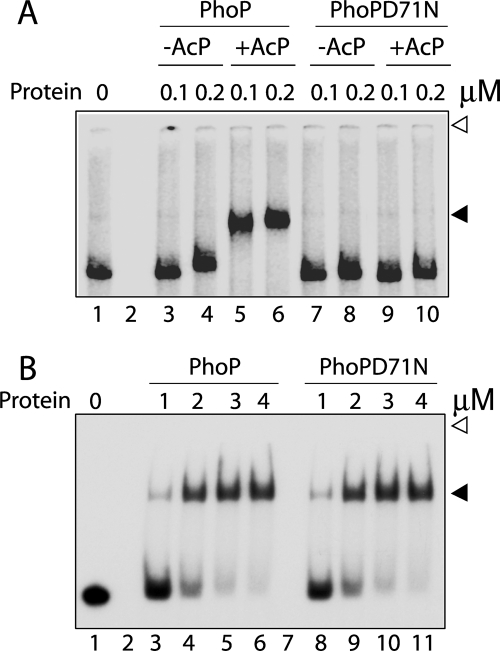
Although PhoP regulates 114 genes, until now it has only been reported to bind to its own promoter (10, 25, 28). Of the several PhoP-regulated genes associated with lipid metabolism, one cluster of genes includes msl3, a polyketide β-ketoacyl synthase that is involved in the synthesis of polyacyltrehaloses (4). The finding that a PhoP knock-out mutant of M. tuberculosis H37Rv lacks diacyltrehaloses and polyacyltrehaloses in the cell envelope also provides direct evidence in support of the role of PhoP in regulating expression of msl3 (4, 5). To determine if PhoP plays a regulatory role by direct binding to the target promoter region, the ability of PhoP to bind to DNA sequence upstream of msl3 (msl3up1 comprising −350 to +60 with respect to the GTG start site) was investigated by EMSA. Purified PhoP was unable to form a slower moving complex stable to gel electrophoresis (Fig. 1A). To investigate if phosphorylation of PhoP was required for in vitro binding to the msl3 upstream region, binding experiments were carried out with PhoP preincubated in phosphorylation mix with AcP as a phospho-donor. Strikingly, PhoP preincubated in phosphorylation mix containing AcP showed efficient DNA binding leading to formation of a stable complex with reduced electrophoretic mobility (compare lanes 3 and 4 with lanes 5 and 6). A quantitative analysis suggested that under identical binding conditions, at least 10-fold stimulation in DNA binding by phosphorylated PhoP was observed with the msl3 promoter DNA compared with the unphosphorylated protein (based on the limits of detection in this assay and based on other gels (data not shown)). As a control experiment, the PhoPD71N mutant (with impaired phosphorylation) preincubated in phosphorylation mix with or without AcP failed to generate a detectable PhoP-DNA complex formation with msl3 promoter DNA (lanes 7–10). From these results, we conclude that a specific interaction(s) between PhoP and the msl3 upstream region is dependent on PhoP phosphorylation. It is also noteworthy that phosphorylated PhoP showed effective msl3 promoter DNA binding at ∼5-fold lower protein concentration. However, these results are in striking contrast with phosphorylation-independent DNA binding of PhoP to the phoP promoter region, as reported previously (10, 12). In agreement with these reports, Fig. 1B shows that wild-type PhoP and PhoPD71N are comparably effective in their relative abilities to bind radiolabeled DR1,2 DNA (phoP promoter-derived oligonucleotide-based DNA substrate) (compare lanes 3–6 and 8–11). Thus, our results suggest for the first time that PhoP retains the ability to utilize its phosphorylation domain in a promoter-specific manner.
FIGURE 1.
Phosphorylation-coupled DNA binding by M. tuberculosis PhoP. A, EMSA of radiolabeled promoter region of msl3 for binding of PhoP (lanes 3–6) or PhoPD71N (lanes 7–10) preincubated in phosphorylation mix with or without AcP. B, EMSA of radiolabeled DR1,2 probe with PhoP (lanes 3–6) and PhoPD71N (lanes 8–11). DNA-protein complexes were analyzed by native PAGE and detected by autoradiography as described under “Experimental Procedures.” Note that the bound complexes resolved from the unbound probes differently in A and B because the DNA probes used were of very different size, a 410-bp full-length promoter and a 60-bp oligonucleotide-based duplex DNA in A and B, respectively. Open and filled arrows, origins of the polyacrylamide gel and retarded complexes (with band shifts in the presence of PhoP proteins), respectively. The gels are representative of at least three independent experiments.
Domain Functions of PhoP
To determine the role of N-domain and/or its phosphorylation in regulating PhoP function, we next investigated the domain structure of the protein. We previously studied histidine-aspartate phosphorelay between M. tuberculosis PhoR and PhoP proteins and showed that Asp-71 is the phospho-acceptor group of PhoP (10). A BLAST search of PhoP within the Protein Data Bank shows the highest sequence identity of 45% with M. tuberculosis PrrA (16) (Protein Data Bank code 1YS6) and the second highest identity of 33% with OmpR/PhoB homologue (14) (Protein Data Bank code 1KGS) from Thermotoga maritima. Comparison of sequence analysis of PhoP with its family members along with the available structural data of PhoPC (11) allowed us to design truncated versions of two domains based on the likely domain boundary (Fig. 2A). To this end, PhoPN141 and PhoPC141 spanning residues Met1–Lys141 and Lys141–Arg247, respectively, were cloned, expressed, and purified to homogeneity as described under “Experimental Procedures” (Fig. 2B). To assess the secondary structural content, we have measured far-UV CD spectra of the purified domains. Both proteins displayed a double trough shape with negative maxima at 209 and 221 nm, characteristic of α-helical structure (supplemental Fig. S1). Thus, our results indicate that the recombinant proteins are folded and retain stability even when expressed as individual domains.
FIGURE 2.
Domain structure of PhoP. A, schematic presentation of bipartite domain structure of M. tuberculosis PhoP showing an N-terminal receiver domain (empty), which comprises the site of phosphorylation (Asp-71 for PhoP) (10) and a relatively shorter C-terminal transactivation domain (gray), which includes the structurally characterized helix-turn-helix DNA binding motif (HTH; dark gray; Protein Data Bank entry 2PMU) (11). Truncated PhoP domains were generated by fragmenting PhoP at Lys141, as indicated by an arrow. B, PhoP, PhoPN141, and PhoPC141 (2 μg/lane), expressed and purified as described under “Experimental Procedures,” were analyzed by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie Blue staining. As a reference, molecular mass markers are resolved in lane 1, and the sizes in kDa are indicated to the left. C, purified PhoP (lanes 2–4) and PhoPN141 (lanes 5–7) at ∼3 μm concentration were compared in transphosphorylation assays for their abilities to accept labeled phosphate from ∼3 μm radiolabeled PhoRC in phosphorylation mix at 18 °C for 30, 60, and 120 s, respectively. As a reference, labeled PhoRC was resolved in lane 1. Products were analyzed by denaturing PAGE and detected by autoradiography. D, EMSA of radiolabeled DR1,2 DNA for binding of ∼3.0 μm PhoPC141 in the absence of any competitor (lane 2), in the presence of increasing concentrations of 6.25, 12.5, 25, and 50-fold excess of specific (lanes 3–6) or nonspecific (lanes 7–10) competitor DNA. E, EMSA of radiolabeled promoter region of msl3 for binding of PhoPC141 (lanes 2 and 3). In all cases, protein-DNA complexes were visualized by autoradiography and quantified by scanning the gels on a PhosphorImager. Open and filled arrowheads indicate origins of the polyacrylamide gel and retarded complexes, respectively.
To determine the functionality of PhoPN141, we next compared its phosphorylation efficiency with full-length PhoP using radiolabeled PhoRC as the phospho-donor (10). Transphosphorylation experiments under the conditions examined and at comparable protein concentration clearly show that PhoPN141 on its own is just as efficient as the full-length protein in its ability to accept radiolabeled phosphate from labeled PhoRC (Fig. 2C, compare lanes 2–4 and 5–7).
For certain OmpR homologues, the purified winged helix domain is sufficient for binding to the target DNA (13, 29). To compare the functionality of PhoPC141 and PhoP, we tested the ability of the homogeneous protein samples to bind radiolabeled DR1,2 DNA (phoP promoter-derived oligonucleotide-based DNA substrate) consisting of a direct repeat motif (10, 12, 25, 28). Purified PhoPC141 was found to form a single slower moving complex stable to gel electrophoresis (Fig. 2D, lane 2). To test the specificity of interaction, we next examined PhoPC141 binding to DR1,2 in the presence of different concentrations of unlabeled DR1,2 as specific competitor (lanes 3–6) or unlabeled nonspecific competitor (lanes 7–10; sequence as described previously (25)), which lacks a consensus PhoP binding site but has comparable base composition. Although more than 64 ± 1.8% of reduction in PhoPC141 binding was observed with an ∼50-fold excess of specific competitor (compare lane 2 and lane 6), an insignificant variation of DNA binding (5 ± 1% in the presence of a 50-fold molar excess of nonspecific competitor; compare lane 2 with lane 10) suggests that PhoPC141 binding to DR1,2 site is sequence-specific. However, PhoPC141, as expected, was unable to form a complex of reduced electrophoretic mobility with msl3 promoter DNA (Fig. 2E). Thus, PhoPC141 is a reliable model to study interactions governing phosphorylation-independent DNA binding of PhoP.
Defining the Minimal Functional N-terminal Domain of PhoP (PhoPN)
Having established independent domain functions, it became important to more closely investigate domain boundaries of the functional domains. To define the minimal domain capable of phosphorylation on its own, two shorter N-terminal constructs, terminating at residues Arg137 (PhoPN137) and Arg138 (PhoPN138), respectively, were cloned, expressed, and purified (supplemental Fig. S2A). The homogeneous protein samples were tested in transphosphorylation assays with labeled PhoRC. When compared for phosphorylation efficiency, PhoPN138 was found to be as efficient as PhoPN141 (Fig. 3A, compare lanes 5–7 and 8–10). However, PhoPN137 at a comparable protein concentration showed ∼3-fold lower transphosphorylation efficiency (compare lanes 2–4 and 5–7). Note that under the conditions examined, PhoPN139 (residues 1–139) was as effective as PhoPN141 in accepting radiolabeled phosphate from labeled PhoRC (data not shown). Thus, residues 1–138 are necessary and sufficient for phosphorylation of PhoP. In agreement with these results, structural alignment of the N-terminal domain of PhoP orthologs shows that the minimal region corresponding to the (α/β)5 topology of PhoPN comprises Met1–Arg138 (Fig. 3B).
FIGURE 3.
Defining the minimal N-terminal domain of PhoP. A, the indicated N-terminal fragments of PhoP (∼3 μm), expressed and purified as described under “Experimental Procedures,” were compared in phosphotransfer assays with radiolabeled PhoRC for 30, 60, and 120 s, respectively. Reaction conditions, sample analysis, and detection of phosphorylated proteins were as described in the legend to Fig. 2C; labeled PhoRC was resolved in lane 1. B, ClustalW alignment of the amino acid sequences of N-terminal domain of PhoP and its orthologs from E. coli (EcOmpR) and Streptomyces coelicolor (ScPhoP). Conserved and similar amino acids are indicated by black and gray shadings, and α-helices and β-sheets are indicated by filled arrows and empty rectangles, respectively. Six highly conserved residues of the protein subfamily important for phosphorylation and signal propagation are indicated by asterisks.
Identification of an Interdomain Linker
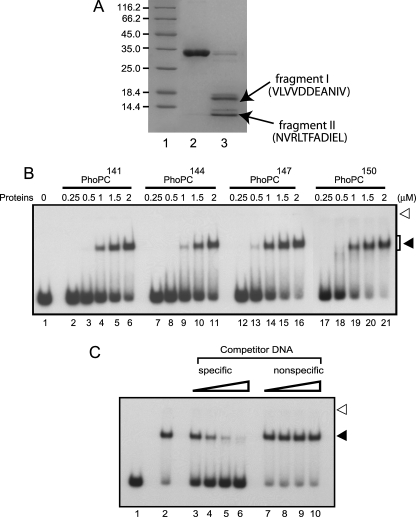
Having defined the minimal phosphorylation domain within the PhoP protein, we sought to investigate the proximal boundary of the C-terminal domain. Individual protein domains can often be identified by limited proteolysis under native conditions because the flexible unstructured interdomain linkers are considerably more susceptible to cleavage (30, 31). To determine likely domain boundaries within PhoP, purified protein was digested with trypsin and analyzed by SDS-polyacrylamide gel electrophoresis. The results showed that trypsin yielded two specific fragments (I and II) of ∼15.5 and 11.2 kDa, respectively (Fig. 4A, arrows). To probe the cleavage sites, both peptide fragments were transferred from the gel by electroblotting the samples onto polyvinylidene difluoride (Millipore) membrane and subjected to N-terminal sequencing. The results showed unambiguously that the fragments I and II were generated by trypsin cleavage of the peptide bonds between Arg22 and Val23 and between Arg147 and Asn148 of the PhoP primary sequence, respectively. Although the former cleavage site maps within PhoPN, interestingly, the latter site lies within the region of the PhoPC domain, which has been suggested as a part of the PhoP linker region (11).
FIGURE 4.
Defining the minimal C-terminal DNA binding domain of PhoP. A, proteolysis of PhoP was carried out with trypsin, and samples were resolved by SDS-PAGE and visualized by Coomassie Blue staining. Trypsin cleavage sites were identified by N-terminal sequencing of peptides from lane 3 (see “Results” for a description of proteolytic fragments) as described under “Experimental Procedures.” Untreated PhoP was resolved in lane 2; the sizes of marker proteins (lane 1) in kDa are indicated to the left. B, EMSA of radiolabeled DR1,2 DNA for binding of increasing concentrations of indicated PhoPC constructs, expressed and purified as described under “Experimental Procedures.” C, EMSA of radiolabeled DR1,2 DNA for binding of 3.0 μm PhoPC150 (residues 150–247) in the absence of any competitor (lane 2), in the presence of a 6.25-, 12.5-, 25-, and 50-fold molar excess of specific (lanes 3–6) or nonspecific (lanes 7–10) competitor DNA as described in the legends to Fig. 2D. Open and filled arrows indicate origins of the polyacrylamide gel and retarded complexes with band shifts produced in presence of PhoP proteins, respectively.
To precisely define the linker, we constructed a collection of plasmids encoding a nested series of C-terminal PhoP fragments starting at residues Lys144, Arg147, and Arg150 extending up to the C-terminal end (Arg247) of full-length PhoP. These proteins, which are referred to as PhoPC144–PhoPC150, were overexpressed and purified (supplemental Fig. S2B). Interestingly, in EMSA experiments with 32P-labeled DR1,2 DNA, all of the purified PhoPC fragments (PhoPC144, PhoPC147, and PhoPC150) were found to be equally effective in forming slower moving complexes stable to gel electrophoresis (Fig. 4B). Over a range of protein concentrations examined, all of the three recombinant proteins (i) formed a single retarded complex in native polyacrylamide gel like that of PhoPC141 and (ii) displayed comparable DNA binding affinity based on the limits of detection in this assay. However, two additional C-domain constructs, PhoPC152 (residues 152–247) and PhoPC154 (residues 154–247), of PhoP failed to show any detectable expression, presumably due to the lack of in vivo stability (data not shown). Thus, the deletion analyses described above have established that the domain spanning residues Arg150–Arg247 of PhoP is directly involved in and essential for DNA binding. In order to determine whether this domain is sufficient (on its own) for specific binding to the target DNA sites, the specificity of binding was demonstrated by its resistance to competition by an unlabeled heterologous competitor relative to the unlabeled homologous competitor (Fig. 4C). Quantification of the competition experiments shows that although there was a greater than 60 ± 2% decrease in radioactivity of bound DNA by a 50-fold excess of specific competitor (compare lanes 2 and 6), there was a less than 4 ± 1% variation of radioactivity of bound DNA in the presence of an identical -fold excess of nonspecific competitor (compare lanes 2 and 10). Thus, this domain is both necessary and sufficient for specific DNA binding activity of PhoP.
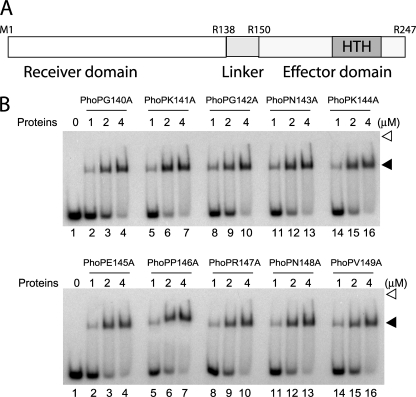
Based on the above experiments and in conjunction with previous results, we propose that residues Met1–Arg138 comprise the minimal, functional N-terminal domain of PhoP, and residues Arg150–Arg247 comprise the shortest C-terminal domain. Residues Ala139–Val149 do not have any significant effect on the function of either domain in vitro, suggesting that this segment simply tethers two independent domains of the PhoP protein together. Fig. 5A shows a schematic presentation of the revised domain structure with residues indicating domain boundaries of PhoP.
FIGURE 5.
Alanine-scanning mutagenesis of PhoP linker region. A, revised domain structure of M. tuberculosis PhoP showing newly identified linker region (residues 139–149) shown in light gray. Note that Arg138 and Arg150 represent the last residue of the proximal N-terminal domain and the first residue of the distal C-terminal domain of PhoP, respectively. B, EMSA of radiolabeled DR1,2 DNA for binding of the indicated single alanine mutants of PhoP, expressed and purified as described under “Experimental Procedures.” Sample analysis and detection of protein-DNA complexes were as described in the legend to Fig. 1B.
To examine the importance of the linker residues in effector domain regulation, we next performed alanine-scanning mutagenesis to obtain single alanine substitutions of 10 non-alanine residues of the linker, and their effects on DNA binding were evaluated. We introduced the mutations into the phoP gene residing in plasmid pET-phoP. In order to expedite purification and analysis of mutant PhoPs, these proteins were expressed like wild-type PhoP containing an N-terminal polyhistidine tag and purified by nickel affinity chromatography (supplemental Fig. S3) as described under “Experimental Procedures.” His tags were cleaved using the Thrombin Clean Cleavage kit from Sigma. To examine the effect of Ala substitution, DNA binding of purified mutant proteins was investigated by EMSA using an end-labeled DR1,2 probe. All of the point mutants, under the conditions examined and at comparable protein concentrations, displayed wild-type PhoP-like DNA binding affinity by efficiently forming a single retarded complex stable to gel electrophoresis (Fig. 5B). Thus, we surmise that amino acid residues of the linker region do not appear to have any significant role in DR1,2 DNA binding affinity of M. tuberculosis PhoP.
Role of the Linker Region in Phosphorylation-coupled DNA Binding by PhoP
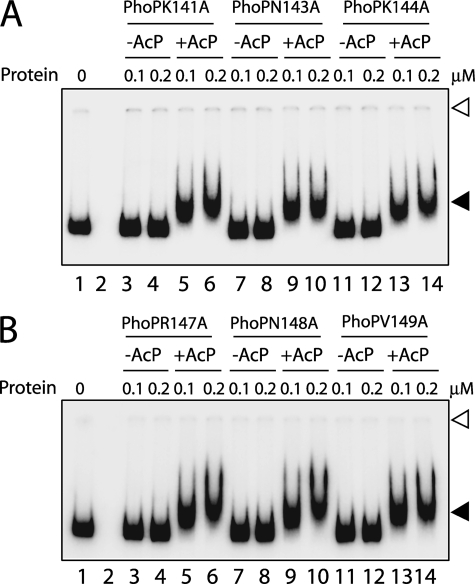
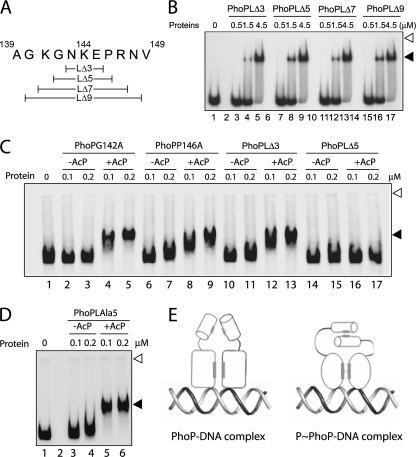
We next examined msl3 promoter DNA binding by linker mutants of PhoP. Strikingly, all of the linker mutants with a single alanine replacing a nonalanine linker residue of PhoP showed efficient phosphorylation-dependent DNA binding similar to the wild-type protein (Fig. 6). These results suggest that a single residue of the linker region does not appear to regulate phosphorylation-coupled DNA binding to the msl3 promoter. To further investigate the importance of linker, we generated a number of linker deletion mutants with three, five, seven, and nine amino acid residues of the PhoP linker deleted, as shown schematically in (Fig. 7A). The deletion mutants were overexpressed, purified to homogeneity (data not shown), and examined for DNA binding activity. Importantly, all of the linker deletion mutants, including PhoPLΔ3–PhoPLΔ9, showed wild-type PhoP-like DNA binding ability when examined using end-labeled DR1,2 DNA (Fig. 7B). Although these observations indicate overall structural stability of linker deletion constructs of PhoP, consistent with our previous observations (Figs. 2 and 4), these results suggest that the affinity and specificity of DNA binding are primarily determined by the residues of the PhoP C-terminal domain.
FIGURE 6.
A and B, EMSA of radiolabeled msl3 promoter region for binding of the indicated PhoP linker mutants carrying single alanine substitutions. Sample analysis and detection of protein-DNA complexes were as described in the legend to Fig. 1. Open and filled arrows indicate origins of the polyacrylamide gel and retarded complexes with band shifts produced in the presence of PhoP proteins, respectively. The gels are representative of at least three independent experiments.
FIGURE 7.
A, scheme showing the linker deletion constructs of M. tuberculosis PhoP. B, EMSA of radiolabeled DR1,2 DNA for binding of PhoP linker deletion mutants. Sample analysis and detection of protein-DNA complexes were as described in the legend to Fig. 1A. C, EMSA of radiolabeled msl3 promoter region for binding of the indicated single alanine mutants as well as linker deletion mutants of PhoP, preincubated in phosphorylation mix with or without AcP. D, EMSA of radiolabeled msl3 promoter region for binding of PhoPLAla5 mutant, preincubated in phosphorylation mix with or without AcP. Each of the gels is representative of at least three independent experiments with at least two different preparations of protein stocks. Sample analysis and detection of protein-DNA complexes were as described in the legends to Fig. 1A. E, dual mode of DNA binding by unphosphorylated and phosphorylated PhoP, respectively, employing mechanisms that are either linker-independent or require the presence of at least eight residues of the PhoP linker (see “Results”). The N-terminal domain is represented as a cylinder, and the C-terminal DNA binding domain is represented as an ellipse for PhoP and a circle for phosphorylated PhoP (to suggest phosphorylation-coupled conformational change), respectively; the protein-protein interaction interface is shaded in gray.
We next investigated phosphorylation-coupled DNA binding of PhoP linker deletion mutants. Interestingly, PhoPLΔ3 showed phosphorylation-dependent msl3 promoter DNA binding similar to single alanine mutants of PhoP (Fig. 7C, lanes 10–13). In striking contrast, with identical labeled probe, PhoPLΔ5 failed to generate a stable DNA-protein complex with reduced electrophoretic mobility (Fig. 7C, lanes 14–17). It should be noted that alanine substitution of two residues, Gly142 and Pro146, which makes up the difference between PhoPLΔ3 and PhoPLΔ5, did not influence phosphorylation-coupled DNA binding by PhoP (Fig. 7C, lanes 2–5 and lanes 6–9, respectively). From these results, we conclude that not a single residue of the linker but rather a constellation of linker residues is important for phosphorylation-coupled DNA binding by PhoP. One additional control experiment examined the effect of alanine substitution for all of the five residues spanning Gly142–Pro146 together in a single PhoP mutant (PhoPLAla5) on DNA binding by PhoP. Strikingly, the mutant PhoPLAla5 with unchanged linker length showed phosphorylation-dependent DNA binding to the msl3 promoter region as efficiently as the wild-type PhoP (Fig. 7D, lanes 3–6). Thus, our results suggest that linker length and not the residues of the PhoP linker region is essential for phosphorylation-dependent DNA binding by the regulator. Together, these results are summarized as a scheme showing two distinctly different modes of DNA binding by PhoP (Fig. 7E). According to this scheme, unphosphorylated PhoP binds to DNA independently of its linker; however, phosphorylated PhoP binding to a specific promoter is dependent on its linker length and not amino acid residues of the linker.
DISCUSSION
M. tuberculosis PhoP regulates more than 110 genes, acting both as a transcriptional activator and repressor of the target promoters (4). However, biochemical studies on its mechanism of action have been lacking. Here, we determine the precise domain structure of the protein and show that interdomain interactions between two functionally independent domains tethered together by an 11-residue-long linker play a key role in regulating functionality of this important regulator. Although the newly identified linker does not appear to be critical for phosphorylation-independent DNA binding, we demonstrate the importance of the linker in phosphorylation-dependent DNA binding of the regulator at a specific promoter.
We have recently shown that PhoP appears to bind symmetrically on tandem direct repeats, suggesting that one of the two repeat units is being recognized backwards (i.e. one recognizes 5′ to 3′ and the other recognizes 3′ to 5′) (25). Efficient recognition of downstream repeat motif by the second PhoP protomer in reverse orientation concomitant with the binding of the first PhoP molecule to the upstream site is likely to involve flexibility sufficient to allow changes in the relative orientation of PhoP domains. In this work, we identify a linker region extending from at least Ala139 to Val149. Because the residues of the linker region do not have any role in either of the domain functions (Fig. 2) and one of the trypsin cleavage sites maps within this region (Fig. 4A), we suppose that the linker region remains largely unstructured. An unstructured (and potentially long) linker between two domains might impart flexibility, allowing changes in the relative orientation of the two domains of PhoP.
The full-length form of DrrD (14) and DrrB (15) from T. maritima, two members of PhoP family, have been crystallized and shown to be composed of two domains: an N-terminal one at which phosphorylation occurs and the DNA binding domain at the C-terminal end. However, the role of N-terminal domain and/or its phosphorylation in effector domain regulation remains unclear. The fact that affinity and specificity of DR1,2 DNA binding by PhoP is determined by the residues of the C-domain alone (Fig. 4; see also Ref. 12) is consistent with the DNA binding domain of PhoB, the E. coli orthologue of M. tuberculosis PhoP, which shows higher affinity for DNA than the unphosphorylated full-length PhoB protein (21). However, this is in striking contrast with the DNA binding domain of Bacillus subtilis PhoP, which requires a more than 5-fold higher protein concentration for DNA binding to the target site compared with the unphosphorylated full-length PhoP (32). Thus, despite sharing largely similar structures, clear differences in their mode of action suggest that structural homology does not always lead to functional homology, even among closely related members of the protein subfamily, and linker length and/or sequence presumably contributes, at least in part, to the functional diversity of the response regulators. In agreement with this view, OmpR and PhoB with largely superimposable C-terminal structure (17) undergo phosphorylation-coupled activation via two distinctly different mechanisms (18, 19). The difference that could account for the two different mechanisms of action resides in the interdomain linker region (15 residues versus 6 residues, respectively) (22). What is particularly interesting and, we think, mechanistically important is that here we show that M. tuberculosis PhoP comprises a linker, which is approximately average in length, to that of OmpR and PhoB, to retain the ability to bind the target promoters both in phosphorylation-independent and phosphorylation-dependent form.
Although phosphorylation is thought to play an important role to bind the target DNA to activate or repress gene expression in vivo, response regulators under in vitro conditions often bind their cognate recognition sequence in the absence of protein phosphorylation. Consistent with this view, PhoP has been shown to recognize DR1,2 independent of phosphorylation (12, 25) (also see Fig. 1B). Also, members like PhoP from Salmonella enterica and B. subtilis form dimers in solution and bind DNA regardless of its phosphorylation status (33, 34). However, phosphorylation of PhoB from E. coli induces dimerization and increases its affinity for the target DNA (35). In sharp contrast, structural data from DrrD (14) and DrrB (15) from T. maritima indicate that in the unphosphorylated state, the recognition helix is freely exposed to the solvent, which would make it available for DNA binding. In agreement with these findings, PhoPC bound to the PhoP boxes with an affinity comparable with that of the full-length protein. This suggests that the N-terminal end of PhoP does not influence binding to the target sequences (DR1,2 DNA) regardless of its phosphorylation state, a result further confirmed by efficient DR1,2 binding by PhoPD71N (Fig. 1B).
Because PhoP was shown to be a positive regulator of the synthesis of three classes of polyketide-derived acyltrehaloses known as SL, DAT, and PAT (4, 5), we set out to study msl3 promoter recognition by PhoP. Interestingly, striking similarities in growth, attenuation, morphological, and cytochemical properties between M. tuberculosis H37Ra and M. tuberculosis phoP mutant have been proposed to be a direct consequence of the absence of these three kinds of complex lipids (5, 36–38). It is noteworthy that within the Mycobacterium genus, SL, DAT, and PAT are relatively restricted to the virulent strains of the M. tuberculosis complex, suggesting that these lipids contribute to pathogenicity of the tubercle bacilli, an assumption that has been supported by a number of in vitro and in vivo studies (for a review, see Refs. 39–46). In addition, recent studies establish a link between PhoP function and the secretion of ESAT-6 and CFP-10 (9), the secreted mycobacterial proteins that are immunodominant antigens in a majority of human tuberculosis cases (47). These reports in conjunction with recent results showing that among the reasons for the attenuation of the M. tuberculosis H37Ra strain is a single nucleotide polymorphism affecting codon 219 (TCG to TTG) in the phoP gene and changing a Ser to a Leu that (i) is incapable of restoring polyketide-derived acyltrehalose synthesis in a phoP-phoR knock-out mutant of H37Rv (8) and (ii) prevents the secretion of proteins that are important for virulence (7) explain why PhoP is considered a virulence-associated response regulator of M. tuberculosis.
A detailed study on the role of PhoP in complex lipid biosynthesis in M. tuberculosis suggests that PhoP probably regulates the expression of the acyltransferase, polyketide synthase (pks), or pks-associated genes involved in the synthesis or transfer of the methyl branched fatty acyl substituents found in SL, DAT, and PAT (5). However, the presence of PhoR, the cognate sensor kinase of PhoP was not required for the synthesis of these lipids because complementation of 1237ΔphoPR::hyg (a 774-bp fragment of the M. tuberculosis genome encompassing part of the phoP and phoR coding sequences was replaced with a hygromycin resistance cassette from Streptomyces hygroscopicus; see Ref. 5) with phoP alone was sufficient to restore their synthesis (5). Thus, it was suggested that phosphorylation might only serve to increase the affinity of the response regulator for certain promoters and might not be essential for the binding of PhoP to its DNA target (5) as has been shown by PhoQ-independent activation of target genes by S. enterica PhoP (48). In agreement with this view, we observed a striking difference in DNA binding affinity of phosphorylated PhoP to the msl3 promoter compared with the DR1,2 DNA binding (Fig. 1, compare A and B). In fact, this view gets further support from our additional results showing phosphorylation-independent comparably effective msl3 promoter recognition by wild-type PhoP and PhoPD71N at relatively higher protein concentrations (supplemental Fig. S4). Furthermore, recent analyses of lipid composition revealed that M. tuberculosis H37Ra is deficient in the production of phthiocerol dimycocerosates (8), a family of polyketide-derived lipids implicated in the virulence of M. tuberculosis (49–52). Studies with lipid-deficient mutants of M. tuberculosis suggest that loss of phthiocerol dimycocerosate in addition to SL, DAT, and PAT explains the inability of M. tuberculosis H37Ra to fix neutral red (53). It is noteworthy that contrary to the production of SL, DAT, and PAT, the synthesis of phthiocerol dimycocerosate is not under the regulatory control of the phoP–phoR system (5). In fact Chesne-Seck et al. (8) recently showed that synthesis of phthiocerol dimycocerosate of M. tuberculosis H37Ra was not restored upon complementation with phoP from M. tuberculosis H37Rv. Thus, a mutation replacing Asp71 by Asn at the primary phosphorylation site of PhoP is unlikely to influence phenotypic consequences through effects on lipid composition. Nevertheless, our results shown here clearly demonstrate the importance of the linker for transmission of the effect of phosphorylation that influences high affinity DNA binding by the response regulator at the promoter region of the gene involved in lipid biosynthesis.
Taken together, these in vitro data suggest that coupling of two functional domains of PhoP in a single polypeptide chain is essential for interdomain interaction(s), which regulates phosphorylation-dependent DNA binding. We consider different mechanisms to explain these results. First of all, the fact that the truncated PhoPC domain is as efficient as the full-length protein in binding to DR1,2 DNA rules out the possibility of occlusion of DNA binding surfaces by PhoPN. Thus, a model in which interaction of PhoP with DNA can cause the N-domain to move away from the C terminus, a process that might remove a barrier to stable DNA binding, does not fit with our results. However, a potential mechanism involving interdomain interactions between the N-domain and the C-domain facilitate an integrated view of our results. In fact, this is in agreement with our earlier observations suggesting conformational change within C-domain of the protein as a result of phosphorylation at the N terminus of PhoP (27). The fact that a linker deletion mutant with deletion of 5 residues or more fails to recognize msl3 promoter region but is otherwise active for DR1,2 DNA binding is strongly suggestive of the importance of linker length for phosphorylation-dependent DNA binding. Thus, it is tempting to speculate that a long, flexible linker region is essential for the transmission of the conformational change from the N-domain to the C-domain of the protein. Consistent with this view, in the structure of M. tuberculosis PrrA, which shares the highest (45%) sequence identity with PhoP, the recognition helix is involved in interactions with the regulatory domain (16).
Although architectural variations among response regulator family members are not surprising and are even expected because of weak sequence homologies within the diverse target promoters, one wonders how to address promoter-specific recognition by conserved structures to achieve diverse functions. Based on these results and results from other studies (for a review, see Ref. 54) and the considerations discussed above, we suggest that much (but not all) of the binding energy and specificity for interaction with target DNA sites and response regulator proteins come from the C-terminal DNA binding domain and that additional discrimination is provided by the linker region during recognition of the target DNA site. In support of this view, we cite experiments from Kenney and co-workers (22) that attempted to understand the mechanism of effector domain regulation of homologous proteins (E. coli OmpR and PhoB). These experiments, which exploited chimeric constructs of OmpR and PhoB with substituted linkers, measured in vivo transcription activation. Consistent with our suggestion, their transcription assays suggested that the effector domain regulation by either N terminus required its cognate interdomain linker.
In conclusion, our results identify the domain structure of PhoP and provide new insights into the mode of action of the transcription regulator. Although we determine that interdomain interactions play an important role in regulating PhoP function, our results clearly show that coupling of the two functional domains of PhoP together in a single polypeptide chain is essential for phosphorylation-dependent DNA binding. Most strikingly, we also demonstrate that although PhoP recognizes the core DNA site within its own promoter in a linker-independent manner, the linker region of the regulator is essential for phosphorylation-coupled specific recognition of another target promoter (msl3 promoter). Although much work remains to be done in identifying genetic determinants (within the msl3 promoter) regulated by phosphorylated PhoP and mechanism of transcription regulation, the present work clearly shows that the regulator retains the capability of interacting with multiple promoters by at least two distinctly different modes (namely linker-independent and linker-dependent), a novel result of unusual significance with implications for the mechanism of DNA binding and transcription regulation by the key regulator.
Supplementary Material
Acknowledgments
We thank Renu Sharma and Mahendra Yadav for technical assistance, Sharanjeet Kaur and Purnananda Guptasarma for circular dichroism studies, Paramjit Kaur and Girish Sahni for N-terminal sequencing of the peptides, Sankalp Gupta for a few preliminary experiments, Arijit Kumar Das and Dr. Subramanian Karthikeyan for very useful discussions, and Renu Sharma for help with preparation of the manuscript.
This work was supported in part by Council of Scientific and Industrial Research (CSIR) Network Project NWP-05 and Supra Institutional Project SIP-10 and by Government of India Department of Science and Technology Grant SR/SO/BB-16/2007 and Department of Biotechnology Grant BT/PR8545/Med/14/1262/2006 (both to D. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- SL
- sulfolipids
- DAT
- diacyltrehaloses
- PAT
- polyacyltrehaloses
- AcP
- acetyl phosphate.
REFERENCES
- 1.Fontan P. A., Walters S., Smith I. (2004) Curr. Sci. 86, 122–134 [Google Scholar]
- 2.Av-Gay Y., Deretic V. (2005) in Tuberculosis and the Tubercle Bacillus (Cole S. T. ed) pp. 359–367, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 3.Pérez E., Samper S., Bordas Y., Guilhot C., Gicquel B., Martín C. (2001) Mol. Microbiol. 41, 179–187 [DOI] [PubMed] [Google Scholar]
- 4.Walters S. B., Dubnau E., Kolesnikova I., Laval F., Daffe M., Smith I. (2006) Mol. Microbiol. 60, 312–330 [DOI] [PubMed] [Google Scholar]
- 5.Gonzalo Asensio J., Maia C., Ferrer N. L., Barilone N., Laval F., Soto C. Y., Winter N., Daffé M., Gicquel B., Martín C., Jackson M. (2006) J. Biol. Chem. 281, 1313–1316 [DOI] [PubMed] [Google Scholar]
- 6.Ryndak M., Wang S., Smith I. (2008) Trends Microbiol. 16, 528–534 [DOI] [PubMed] [Google Scholar]
- 7.Lee J. S., Krause R., Schreiber J., Mollenkopf H. J., Kowall J., Stein R., Jeon B. Y., Kwak J. Y., Song M. K., Patron J. P., Jorg S., Roh K., Cho S. N., Kaufmann S. H. (2008) Cell Host Microbe 3, 97–103 [DOI] [PubMed] [Google Scholar]
- 8.Chesne-Seck M. L., Barilone N., Boudou F., Gonzalo Asensio J., Kolattukudy P. E., Martín C., Cole S. T., Gicquel B., Gopaul D. N., Jackson M. (2008) J. Bacteriol. 190, 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frigui W., Bottai D., Majlessi L., Monot M., Josselin E., Brodin P., Garnier T., Gicquel B., Martin C., Leclerc C., Cole S. T., Brosch R. (2008) PLoS Pathog. 4, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S., Sinha A., Sarkar D. (2006) FEBS Lett. 580, 5328–5338 [DOI] [PubMed] [Google Scholar]
- 11.Wang S., Engohang-Ndong J., Smith I. (2007) Biochemistry 46, 14751–14761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A. K., Pathak A., Sinha A., Datt M., Singh B., Karthikeyan S., Sarkar D. (2010) J. Mol. Biol. 398, 647–656 [DOI] [PubMed] [Google Scholar]
- 13.Blanco A. G., Sola M., Gomis-Rüth F. X., Coll M. (2002) Structure 10, 701–713 [DOI] [PubMed] [Google Scholar]
- 14.Buckler D. R., Zhou Y., Stock A. M. (2002) Structure 10, 153–164 [DOI] [PubMed] [Google Scholar]
- 15.Robinson V. L., Wu T., Stock A. M. (2003) J. Bacteriol. 185, 4186–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak E., Panjikar S., Konarev P., Svergun D. I., Tucker P. A. (2006) J. Biol. Chem. 281, 9659–9666 [DOI] [PubMed] [Google Scholar]
- 17.Itou H., Tanaka I. (2001) J. Biochem. 129, 343–350 [DOI] [PubMed] [Google Scholar]
- 18.Head C. G., Tardy A., Kenney L. J. (1998) J. Mol. Biol. 281, 857–870 [DOI] [PubMed] [Google Scholar]
- 19.Allen M. P., Zumbrennen K. B., McCleary W. R. (2001) J. Bacteriol. 183, 2204–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenney L. J. (2002) Curr. Opin. Microbiol. 5, 135–141 [DOI] [PubMed] [Google Scholar]
- 21.Ellison D. W., McCleary W. R. (2000) J. Bacteriol. 182, 6592–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walthers D., Tran V. K., Kenney L. J. (2003) J. Bacteriol. 185, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ames S. K., Frankema N., Kenney L. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11792–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Gupta S., Pathak A., Sinha A., Sarkar D. (2009) J. Bacteriol. 191, 7466–7476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 27.Sinha A., Gupta S., Bhutani S., Pathak A., Sarkar D. (2008) J. Bacteriol. 190, 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalo-Asensio J., Soto C. Y., Arbués A., Sancho J., del Carmen Menéndez M., García M. J., Gicquel B., Martín C. (2008) J. Bacteriol. 190, 7068–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Hackert E., Stock A. M. (1997) Structure 5, 109–124 [DOI] [PubMed] [Google Scholar]
- 30.Schulz G., Schirmer R. (1979) Principles of Protein Structure, Springer, New York [Google Scholar]
- 31.Janin J., Chothia C. (1985) Methods Enzymol. 115, 420–430 [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Abdel-Fattah W. R., Hulett F. M. (2004) J. Bacteriol. 186, 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perron-Savard P., De Crescenzo G., Le Moual H. (2005) Microbiology 151, 3979–3987 [DOI] [PubMed] [Google Scholar]
- 34.Prágai Z., Allenby N. E., O'Connor N., Dubrac S., Rapoport G., Msadek T., Harwood C. R. (2004) J. Bacteriol. 186, 1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCleary W. R. (1996) Mol. Microbiol. 20, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 36.Cason J., Allen C. F., DeAcetis W., Fonken G. J. (1956) J. Biol. Chem. 220, 893–904 [PubMed] [Google Scholar]
- 37.Middlebrook G., Coleman C. M., Schaefer W. B. (1959) Proc. Natl. Acad. Sci. U.S.A. 45, 1801–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daffé M., Lacave C., Lanéelle M. A., Gillois M., Lanéelle G. (1988) Eur. J. Biochem. 172, 579–584 [DOI] [PubMed] [Google Scholar]
- 39.Goren M. B., Brenan P. J. (1979) in Tuberculosis (Youmans G. P. ed) pp. 63–193, W.B. Saunders Co., Philadelphia [Google Scholar]
- 40.Goren M. B. (1990) in Handbook of Lipid Research: Glycolipids, Phosphoglycolipids and Sulfoglycolipids, Vol. 6 (Kates M. ed) pp. 363–461, Plenum Press, New York [Google Scholar]
- 41.Rousseau C., Neyrolles O., Bordat Y., Giroux S., Sirakova T. D., Prevost M. C., Kolattukudy P. E., Gicquel B., Jackson M. (2003) Cell Microbiol. 5, 405–415 [DOI] [PubMed] [Google Scholar]
- 42.Rousseau C., Turner O. C., Rush E., Bordat Y., Sirakova T. D., Kolattukudy P. E., Ritter S., Orme I. M., Gicquel B., Jackson M. (2003) Infect. Immun. 71, 4684–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilleron M., Stenger S., Mazorra Z., Wittke F., Mariotti S., Böhmer G., Prandi J., Mori L., Puzo G., De Libero G., (2004) J. Exp. Med. 199, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Converse S. E., Mougous J. D., Leavell M. D., Leary J. A., Bertozzi C. R., Cox J. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domenech P., Reed M. B., Dowd C. S., Manca C., Kaplan G., Barry C. E., 3rd (2004) J. Biol. Chem. 279, 21257–21265 [DOI] [PubMed] [Google Scholar]
- 46.Graham J. E., Clark-Curtiss J. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11554–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodin P., Rosenkrands I., Andersen P., Cole S. T., Brosch R. (2004) Trends Microbiol. 12, 500–508 [DOI] [PubMed] [Google Scholar]
- 48.Lejona S., Castelli M. E., Cabeza M. L., Kenney L. J., García Véscovi E., Soncini F. C. (2004) J. Bacteriol. 186, 2476–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camacho L. R., Ensergueix D., Perez E., Gicquel B., Guilhot C. (1999) Mol. Microbiol. 34, 257–267 [DOI] [PubMed] [Google Scholar]
- 50.Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999) Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 51.Jackson M., Stadthagen G., Gicquel B. (2007) Tuberculosis 87, 78–86 [DOI] [PubMed] [Google Scholar]
- 52.Rousseau C., Winter N., Pivert E., Bordat Y., Neyrolles O., Avé P., Huerre M., Gicquel B., Jackson M. (2004) Cell Microbiol. 6, 277–287 [DOI] [PubMed] [Google Scholar]
- 53.Cardona P. J., Soto C. Y., Martín C., Giquel B., Agustí G., Andreu N., Guirado E., Sirakova T., Kolattukudy P., Julián E., Luquin M. (2006) Microbes Infect. 8, 183–190 [DOI] [PubMed] [Google Scholar]
- 54.Gao R., Mack T. R., Stock A. M. (2007) Trends Biochem. Sci. 32, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.