Abstract
Mycobacterial AdnAB is a heterodimeric DNA helicase-nuclease and 3′ to 5′ DNA translocase implicated in the repair of double strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N-terminal motor domain and a C-terminal nuclease domain. Inclusion of mycobacterial single strand DNA-binding protein (SSB) in reactions containing linear plasmid dsDNA allowed us to study the AdnAB helicase under conditions in which the unwound single strands are coated by SSB and thereby prevented from reannealing or promoting ongoing ATP hydrolysis. We found that the AdnAB motor catalyzed processive unwinding of 2.7–11.2-kbp linear duplex DNAs at a rate of ∼250 bp s−1, while hydrolyzing ∼5 ATPs per bp unwound. Crippling the AdnA phosphohydrolase active site did not affect the rate of unwinding but lowered energy consumption slightly, to ∼4.2 ATPs bp−1. Mutation of the AdnB phosphohydrolase abolished duplex unwinding, consistent with a model in which the “leading” AdnB motor propagates a Y-fork by translocation along the 3′ DNA strand, ahead of the “lagging” AdnA motor domain. By tracking the resection of the 5′ and 3′ strands at the DSB ends, we illuminated a division of labor among the AdnA and AdnB nuclease modules during dsDNA unwinding, whereby the AdnA nuclease processes the unwound 5′ strand to liberate a short oligonucleotide product, and the AdnB nuclease incises the 3′ strand on which the motor translocates. These results extend our understanding of presynaptic DSB processing by AdnAB and engender instructive comparisons with the RecBCD and AddAB clades of bacterial helicase-nuclease machines.
Keywords: DNA-binding Protein, DNA Helicase, DNA Recombination, DNA Repair, Molecular Motors
Introduction
Nucleolytic resection of DNA double strand breaks (DSBs)2 is an essential early step in bacterial homologous recombination that eventuates in a RecA-coated 3′ single-stranded tail that invades a homologous sister chromatid. Bacterial DSB resection is performed by multisubunit helicase-nuclease machines encoded in operon-like gene clusters (1). Three different clades of DSB-resecting machines are typified by Escherichia coli RecBCD (2, 3), Bacillus subtilis AddAB (4–7), and Mycobacterium smegmatis AdnAB (8, 9), respectively. These enzymes differ in subunit content and the number of motor and nuclease domains contained therein.
Mycobacterial AdnAB is a heterodimeric helicase-nuclease. The AdnA and AdnB subunits are each composed of an N-terminal UvrD-like motor domain and a C-terminal RecB-like nuclease module (8). Genetic ablation of the adnAB locus in M. smegmatis results in hypersensitivity to ionizing radiation, comparable with the sensitization conferred by deletion of recA,3 signifying an important DNA repair function of AdnAB in vivo.
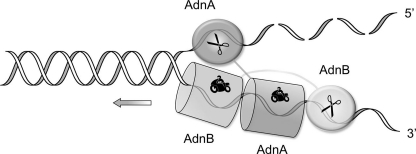
Initial biochemical studies of the DNA-dependent ATPase, dsDNA and ssDNA nuclease, and DSB resection activities of purified AdnAB, together with analyses of the effects of nuclease-inactivating and ATPase-inactivating mutations, prompted a model of AdnAB end-processing that has both shared and unique features vis à vis the E. coli RecBCD motor-nuclease (3, 8, 9). Like RecBCD, AdnAB is a vigorous ssDNA-dependent ATPase (kcat 415 s−1), and it binds stably to DSB ends. In the presence of ATP, the AdnAB motor initiates duplex unwinding from the DSB end without requiring a ssDNA loading strand. AdnAB is a unidirectional ATP-powered translocase on ssDNA (9). During translocation, the “leading” AdnB and “lagging” AdnA motor domains track in tandem, 3′ to 5′, along the same DNA single strand (Fig. 1). This contrasts with RecBCD, in which the RecB and RecD motors track in parallel along the two separated DNA single strands (11–13).
FIGURE 1.
Model for division of labor during AdnAB unwinding and processing DSB ends. Binding of AdnAB to the DSB end initiates ATP-dependent translocation of the tandem motor in the 3′ to 5′ direction along one of the DNA strands, with the dominant AdnB motor domain in the vanguard. Unwinding of the DNA duplex by the advancing motor results in pumping of the 3′ ssDNA strand into the AdnB nuclease domain and threading of the 5′ ssDNA strand through the AdnA nuclease domain. The model is based on experiments in Ref. 9 and is interrogated further by this study.
RecBCD has one nuclease module appended to the RecB motor domain; the RecB nuclease can digest either of the strands displaced by the RecB and RecD helicase motors (2, 3). By contrast, AdnAB has two nuclease domains that behave like an ATP-regulated “molecular ruler” when acting on ssDNA substrates. Absent ATP, AdnAB incises ssDNA by measuring the distance from the free 5′ end to dictate the sites of cleavage, which are predominantly 5 or 6 nucleotides from the 5′ end. ATP hydrolysis elicits a distal displacement of the cleavage sites 16–17 nucleotides from the 5′ terminus. We demonstrated a strict division of labor between the AdnAB subunits, whereby mutations in the nuclease active site of the AdnB subunit ablated the ATP-inducible cleavages, whereas synonymous changes in the AdnA nuclease active site abolished ATP-independent cleavage. By studying the effects of mutations in the AdnA and AdnB phosphohydrolase active sites on the nuclease activities, we showed that ATP hydrolysis by the AdnB motor triggers the AdnB nuclease in cis (8). Analysis of the effects of 5′- and 3′-terminal obstacles on ssDNA cleavage by AdnAB (9) suggested that the AdnA nuclease module receives and processes the displaced 5′ strand, whereas the AdnB nuclease module most likely cleaves the displaced 3′ strand (Fig. 1).
Here, we extend our studies of the dsDNA helicase and end-resection activities of AdnAB to long dsDNA substrates that more accurately report on its putative recombination functions, the end point of which is to yield a 3′-tailed DNA for RecA-mediated strand invasion. To focus on the helicase function of the AdnAB motor, we exploit a “nuclease-dead” AdnAB heterodimer with synonymous mutations in the AdnA and AdnB nuclease domains that abolish ssDNA incision (8). We are especially concerned with the fate of the unwound DNA strands and their interactions with the mycobacterial single strand DNA-binding protein (SSB), which has an imputed role in the generation of a RecA-bound recombination intermediate (14). By studying the unwinding of linear plasmids in the absence and presence of M. smegmatis SSB, we gained new insights to the rate and processivity of the AdnAB motor as it unwinds duplex DNA, the coupling of DNA unwinding to ATP hydrolysis, and the apparent sufficiency of the AdnB motor to power unwinding by the AdnAB heterodimer. Analysis of the resection of end-labeled plasmid DNAs by wild-type AdnAB and single nuclease-crippled mutants supports the proposed division of labor whereby the AdnA subunit acts on the displaced 5′ ssDNA strand, and the AdnB subunit incises the 3′ ssDNA strand.
EXPERIMENTAL PROCEDURES
M. smegmatis SSB
The ORF encoding M. smegmatis SSB (MSMEG6896) was PCR-amplified from genomic DNA using a sense strand primer that introduced an NdeI site at the start codon and an antisense strand primer designed to introduce an XhoI site at the translation stop codon while converting it to a serine codon. The PCR product was digested using NdeI and XhoI and inserted between the NdeI and XhoI sites of pET21b (Novagen). The resulting pET-SSB plasmid encodes full-length SSB fused to a 9-amino acid C-terminal His6 tag (-SLEHHHHHH). pET-SSB was electroporated into E. coli BL21(DE3) cells. A 1-liter culture derived from a single ampicillin-resistant transformant was grown at 37 °C in Luria-Bertani medium containing 0.1 mg/ml ampicillin until the A600 reached 0.6. The culture was chilled on ice for 1 h and then adjusted to 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and 2% (v/v) ethanol, followed by incubation for 17 h at 17 °C with constant shaking. Cells were harvest by centrifugation, and pellets were stored at −80 °C. All subsequent steps were performed at 4 °C. The thawed bacterial cell pellet was resuspended in 25 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 10% sucrose, 250 mm NaCl, 0.05% Triton X-100). The suspension was sonicated on ice for 5 min to achieve lysis. The lysate was centrifuged at 16,000 rpm for 1 h at 4 °C to remove insoluble material. The supernatant was then applied to a 4-ml nickel-nitrilotriacetic acid-agarose column (Qiagen) that had been equilibrated with buffer A (50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 10% glycerol, 0.05% Triton X-100). The column was washed with buffer A and then eluted stepwise with 100, 200, and 1000 mm imidazole in buffer A. The polypeptide compositions of the fractions were monitored by SDS-PAGE. The recombinant SSB protein was recovered predominantly in the 1000 mm imidazole fraction, which was then applied to a 2.5-ml DEAE-Sephacel column that had been equilibrated with buffer A. SSB protein was recovered in the DEAE flow-through fraction. The SSB was then applied to a 2-ml heparin-agarose column equilibrated with buffer A. The SSB protein was again recovered in the flow-through fraction. After adjusting the buffer to 500 mm NaCl, the SSB was salted out by adding an equal volume of 4 m ammonium sulfate. The SSB precipitate was recovered and was resuspended in 5 ml of buffer B (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm DTT, 10% glycerol). The suspension was clarified by centrifugation, and the supernatant was applied to a Superdex 200 gel filtration column equilibrated with buffer B. The peak SSB-containing fractions were pooled and stored at −80 °C. The yield of SSB was 20 mg/liter bacterial culture.
M. smegmatis AdnAB
Wild-type AdnAB heterodimer and versions containing inactivating mutations in the nuclease or ATPase modules of one or both subunits were produced in E. coli and purified from a soluble extract by nickel affinity and anion exchange chromatography and glycerol gradient sedimentation as described previously (8, 9). The protein concentrations were determined by SDS-PAGE analysis of the AdnAB preparations and densitometry of the stained gel, with interpolation of the polypeptide staining intensities to an internal BSA standard curve (8). The figure legends specify the amounts of added enzyme with respect to the AdnB subunit.
Preparation of 3′ 32P-Labeled dsDNA
Reaction mixtures (25 μl) containing 10 mm Tris-HCl, pH 7.9, 50 mm NaCl, 1 mm DTT, 10 mm MgCl2, 3 μm [α-32P]dATP, 3 μm dTTP, 2.7 μg of EcoRI-digested pUC19 DNA, and 10 units of DNA polymerase I Klenow fragment (New England Biolabs) were incubated for 15 min at 25 °C. The fill-in reaction was terminated by adding 10 μl of a solution containing 100 mm EDTA, 60% glycerol, and 0.25% Orange G, pH 8.0. The pUC19 DNA was then purified by electrophoresis through a 0.8% native agarose gel in 50 mm Tris acetate, 2.5 mm EDTA. The DNA was recovered from an excised gel slice by using a QIAquick gel extraction kit (Qiagen) according to the manufacturer's protocol.
Preparation of 5′ 32P-Labeled dsDNAs
BamHI-digested pUC19 or SmaI-digested pUC-H plasmid DNAs were treated with alkaline phosphatase and then recovered by phenol/chloroform extraction and precipitation with ethanol. 5′ end-labeling reaction mixtures (25 μl) containing 70 mm Tris-HCl, pH 7.6, 5 mm DTT, 10 mm MgCl2, 40 μm [γ-32P]ATP, 2.5 μg of 5′-OH plasmid DNA, and 30 units of T4 polynucleotide kinase (New England Biolabs) were incubated for 1 h at 37 °C. The labeled DNA was gel-purified as described above.
RESULTS
SSB Captures the Strands Unwound by the AdnAB Motor
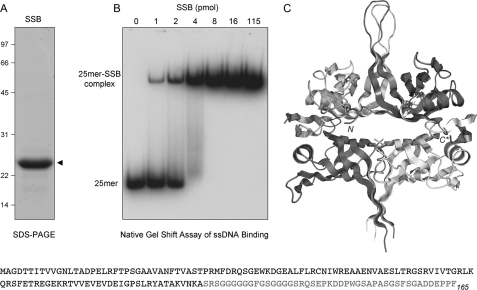
M. smegmatis SSB is a 165-amino acid polypeptide composed of a 120-amino acid N-terminal DNA binding domain and a disordered 45-amino acid C-terminal glycine-rich module (Fig. 2). The OB fold of the DNA binding domain of M. smegmatis SSB and its homotetrameric quaternary structure (Fig. 2C) are both similar to those of M. tuberculosis and E. coli SSBs (15–17). Here, we sought to analyze the impact of M. smegmatis SSB on the activities of M. smegmatis AdnAB. Thus, we produced M. smegmatis SSB in E. coli as a C-terminal His-tagged fusion and then purified the protein from a soluble bacterial extract. SDS-PAGE analysis of the SSB preparation revealed a single polypeptide migrating at ∼24 kDa (Fig. 2A). When incubated with 1 pmol of a 5′ 32P-labeled 25-mer ssDNA oligonucleotide, the recombinant SSB formed a single discrete SSB·DNA complex that was readily separated from free DNA by native PAGE (Fig. 2B). The yield of the SSB·DNA complex depended on SSB concentration up to 4 pmol of SSB monomer, at which point most of the DNA was protein-bound (Fig. 2B); this result is consistent with the SSB homotetramer being the functional unit of DNA binding.
FIGURE 2.
Recombinant M. smegmatis SSB. A, purification. An aliquot (10 μg) of the Superdex-200 fraction of recombinant SSB was analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown, with the positions and sizes (kDa) of marker polypeptide indicated on the left. B, DNA binding. Reaction mixtures (10 μl) containing 20 mm Tris-HCl, pH 8.0, 0.1 μm (1 pmol) 5′ 32P-labeled 25-mer ssDNA (5′-CCAGAACAAACTCATCGTCGTCTAC), 5% glycerol, and increasing amounts of SSB as specified (expressed as pmol of SSB monomers) were incubated for 30 min at 4 °C. The mixtures were adjusted to 15% glycerol and then analyzed by electrophoresis through a 15-cm native 6% polyacrylamide gel containing 22.5 mm Tris borate, 0.625 mm EDTA. The gel was run at 110 V in the cold room for 3 h and then dried under vacuum on DE81 paper. The free 32P-labeled DNA and slower migrating protein·[32P]DNA complexes were visualized by autoradiography of the dried gel. C, crystal structure of the M. smegmatis SSB-(1–120) tetramer is shown (Protein Data Bank code 1X3E); the terminal residues of the SSB-(1–120) protomer at top right are indicated by N and C*, respectively. The primary structure of the M. smegmatis SSB polypeptide is shown at the bottom of the figure.
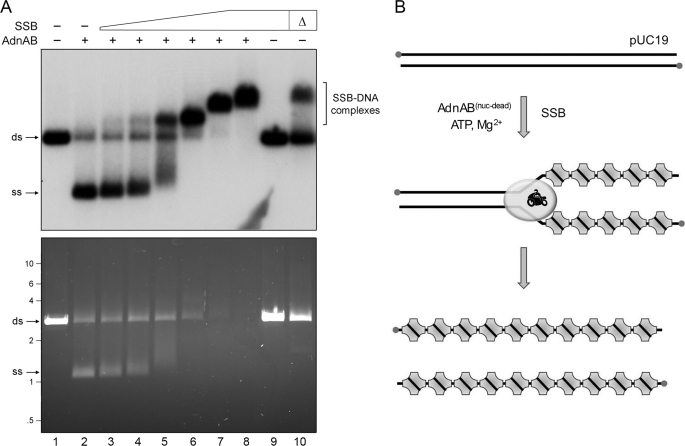
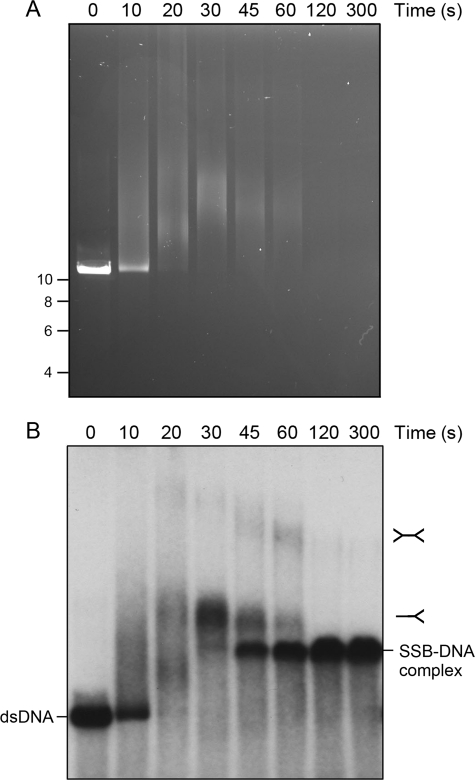
We gauged the effects of SSB on the ability of the AdnAB motor to fully unwind a 2.7-kb linear plasmid DNA substrate, prepared by digestion of pUC19 with BamHI and 5′ 32P-labeling of the DSB ends. The purified AdnA(D934A)-AdnB(D1014A) heterodimer, bearing nuclease-inactivating mutations in both subunits, was incubated with linear pUC19 DNA for 10 min in the presence of 1 mm ATP and 2 mm magnesium. The reactions were quenched with EDTA, and the products were resolved by native agarose gel electrophoresis. Staining the DNA in the gel with ethidium bromide revealed conversion of the input dsDNA to a discrete, more rapidly migrating ssDNA product (Fig. 3A, bottom panel, lane 2) that comigrated with the ethidium-stained ssDNA generated by heating and quick cooling the linear pUC19 plasmid (data not shown). Unwinding of the linear plasmid by nuclease-dead AdnAB was confirmed by autoradiography of the dried gel (Fig. 3A, top panel, lane 2). Note that autoradiography provides a clearer indicator of the extent of the helicase reaction than ethidium staining, insofar as the displaced single strands will bind less ethidium than the intact duplex substrate. AdnAB per se unwound ∼71% of the input 32P-labeled plasmid substrate (Fig. 3A, lane 2).
FIGURE 3.
SSB captures the strands unwound by the AdnAB motor. A, reaction mixtures (10 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 200 ng of 5′ 32P-labeled pUC19 DNA (BamHI-digested; 230 fmol of DSB ends), 1.06 pmol of nuclease-dead AdnAB (where indicated by +), and increasing amounts of SSB, either 3.6 (lane 3), 7.2 (lane 4), 14.4 (lane 5), 28.8 (lane 6), 57.5 (lane 7), or 115 (lanes 8 and 9) pmol of SSB monomer, were incubated for 10 min at 37 °C. The reactions were quenched by adjusting the mixtures to 50 mm EDTA, 15% glycerol, 0.125% Orange-G dye. The mixture in lane 10, lacking AdnAB, was heated for 5 min at 95 °C (Δ) and then chilled on ice prior to adding SSB (115 pmol). The reaction products were analyzed by electrophoresis through a 0.8% native agarose gel in 50 mm Tris acetate, 2.5 mm EDTA. After visualizing the DNA by staining with ethidium bromide (bottom panel), the gel was dried under vacuum on DE81 paper, and radiolabeled DNA was visualized by autoradiography of the dried gel (top panel). B, reaction scheme is illustrated, whereby SSB tetramers bind to the single-stranded DNA formed in the wake of the advancing AdnAB motor to yield SSB·ssDNA complexes as end products. ds, double strand; ss, single strand.
Supplementation of the reaction mixtures with SSB increased in the extent of dsDNA unwinding, concomitant with the concentration-dependent formation of radiolabeled SSB·ssDNA complexes that migrated more slowly than either free ssDNA or dsDNA (Fig. 3A, top panel, lanes 3–8). Indeed, nearly all of the input DNA was unwound by AdnAB and captured by SSB at 5.8 μm SSB (Fig. 3A, lane 7), which corresponded to ∼1 SSB protomer per 11 nucleotides of available plasmid ssDNA. Note the key control showing that the highest level of SSB included in this experiment (11.5 μm) had no effect on the electrophoretic mobility of linear pUC19 dsDNA that had not been exposed to AdnAB (Fig. 3A, lane 9). However, when the pUC19 DNA substrate (absent AdnAB) was heated for 5 min at 95 °C and then quick cooled prior to adding 11.5 μm SSB, about half of the labeled DNA was converted to SSB·ssDNA complexes that comigrated with the SSB·ssDNA complexes generated in the presence of AdnAB (Fig. 3A, lane 10 versus lane 8). We surmise that the effect of SSB is to bind to the ssDNA strands formed behind the advancing AdnAB motor and thereby prevent reannealing of the duplex in the wake of the motor (Fig. 3B). We presume that the progressive effacement of ethidium-staining material with increasing SSB concentrations, although the radiolabel is preserved (Fig. 3A), reflects poor binding of ethidium to the SSB-bound ssDNA versus protein-free ssDNA.
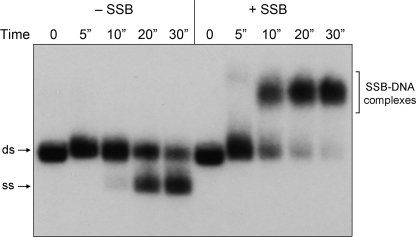
Estimation of the Rate of dsDNA Unwinding by the AdnAB Motor
A kinetic analysis of the reaction of AdnAB with pUC19 DNA is shown in Fig. 4. Absent SSB, we could detect a low level of unwound 32P-labeled ssDNA within 10 s, which increased sharply at 20 and 30 s. In the presence of sufficient SSB to coat the ssDNA product, at least half of the dsDNA was converted to mature SSB·ssDNA complexes within 10 s, and nearly all of the 2.7-bp linear plasmid was unwound by 30 s (Fig. 4). Thus, we estimate that the AdnAB motor can unwind duplex pUC19 DNA at a rate of ∼270 bp s−1.
FIGURE 4.
Estimation of the rate of pUC19 unwinding by the AdnAB motor. Reaction mixtures (50 μl) contained 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 1 μg of 5′ 32P-labeled pUC19 DNA (BamHI-digested; 1.14 pmol of DSB ends), either no SSB (−SSB) or 11.5 μm SSB monomers (+SSB), and 3.8 pmol (76 nm) of nuclease-dead AdnAB. The reactions were initiated by adding AdnAB to reaction mixtures prewarmed to 37 °C. Aliquots (10 μl) were then withdrawn after incubation at 37 °C for the times specified, and the reactions were quenched immediately with EDTA. The time 0 samples were taken prior to adding AdnAB. The products were analyzed by native agarose gel electrophoresis. Radiolabeled DNA was visualized by autoradiography of the dried gel. ds, double strand; ss, single strand.
We extended this analysis by tracking the unwinding of a 5′ 32P-labeled 11.2-kb linear dsDNA (pUC-H) composed of the pUC vector and an 8.5-kb segment of vaccinia virus genomic DNA. Our presumption was that the time required for the AdnAB motor to unwind the linear DNA in the presence of excess SSB would be proportional to DNA length and that the extent of unwinding ought to provide a crude indicator of AdnAB processivity. As shown in Fig. 5, we detected unwinding intermediates of retarded electrophoretic mobility at 10, 20, and 30 s that coalesced into discrete, “mature,” unwound SSB·ssDNA product complexes as early as 45 s. The conversion of dsDNA substrate to unwinding intermediates was complete within 20 s, and the subsequent conversion of the intermediates to unwound product complexes was effectively completed between 60 and 120 s (Fig. 5). By simple division (11,200 bp unwound within 45 s), we surmised that AdnAB unwound the longer pUC-H plasmid substrate at ∼250 bp s−1. Thus, the lower bound estimates of the rate of DNA unwinding by the AdnAB motor were concordant for the two linear DNAs over a 4-fold length range. We infer also that the AdnAB motor is fairly processive once its has initiated unwinding, being capable of displacing up to 11 kb of dsDNA in the presence of saturating SSB levels that, as we show below, sequester ssDNA and impede it from serving as a substrate for AdnAB (thereby making it unlikely that AdnAB could reinitiate on a partially unwound “Y” duplex with SSB-coated tails).
FIGURE 5.
Unwinding of an 11.2-kb dsDNA by the AdnAB motor. Reaction mixtures (80 μl) contained 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 0.8 μg of 5′ 32P-labeled pUC-H DNA (SmaI-digested; 910 fmol of DSB ends), either no SSB (−SSB) or 23 μm SSB monomers (+SSB), and 9.7 pmol (120 nm) of nuclease-dead AdnAB. Aliquots (10 μl) were withdrawn after incubation at 37 °C for the times specified and then quenched immediately with EDTA. The reaction products were analyzed by electrophoresis through a 0.6% native agarose gel. DNA was visualized by staining with ethidium bromide (A) before the gel was dried, and radiolabeled DNA was visualized by autoradiography (B). The positions and sizes (kbp) of linear dsDNA markers are indicated on the left in A.
SSB Inhibits ssDNA-dependent ATP Hydrolysis by AdnAB
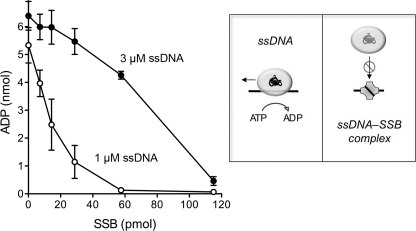
The ATP phosphohydrolase activity of AdnAB is strictly dependent on ssDNA. The nuclease-dead AdnAB motor is readily triggered by a 24-mer ssDNA oligonucleotide (9). Here, we gauged the effects of SSB on the hydrolysis of 1 mm [α-32P]ATP in the presence of 1 or 3 μm 24-mer ssDNA cofactor. We found that SSB inhibited ATPase activity in a concentration-dependent manner, with an inverse relationship between SSB potency and ssDNA concentration (Fig. 6). At 1 μm ssDNA, an 80% decrement in ATP hydrolysis was seen at 29 pmol (2.9 μm) of input SSB monomer, and ATPase activity was virtually abolished at 58 pmol (5.8 μm) of SSB (Fig. 6), which corresponds to an ∼1.5:1 ratio of SSB tetramer to 24-mer ssDNA. At 3 μm ssDNA, ATP hydrolysis was inhibited 93% by 115 pmol (11.5 μm) of SSB monomer (Fig. 6). SSB inhibition was not a mere consequence of increased protein concentration, insofar as control experiments showed that addition of 29, 58, or 115 pmol of bovine serum albumin to reaction mixtures containing 1 μm ssDNA had no effect on the extent of ATP hydrolysis by AdnAB (data not shown). We surmise that an ssDNA segment bound to SSB is unavailable to serve as a platform for directional translocation by the AdnAB ATPase motor (Fig. 6).
FIGURE 6.
SSB inhibits ssDNA-triggered ATP hydrolysis by the AdnAB motor. Reaction mixtures (10 μl) containing 20 mm Tris-HCl, pH 8.0, 0.5 mm DTT, 1 mm MgCl2, 1 mm [α-32P]ATP, 1 or 3 μm 24-mer ssDNA (5′-GCCCTGCTGCCGACCAACGAAGGT) as specified, 8.5 nm nuclease-dead AdnAB, and increasing amounts of SSB polypeptide as specified (pmol of SSB monomer) were incubated for 10 min at 37 °C. The reactions were quenched with 2 μl of 5 m formic acid. Aliquots (2 μl) were analyzed by ascending PEI-cellulose TLC with 0.45 m ammonium sulfate as the mobile phase. [α-32P]ATP and [α-32P]ADP were quantified by scanning the TLC plate with a Fujix BAS2500 imager. The extents of [α-32P]ADP formation are plotted as a function of input SSB. Each datum is the average of three experiments (±S.E.). The schematics at right illustrate how ssDNA-triggered ATP hydrolysis is coupled to translocation of the AdnAB motor and how sequestration of the ssDNA cofactor by SSB progressively masks its ability to trigger the motor ATPase.
Estimating the Coupling of ATP Hydrolysis and Duplex Unwinding
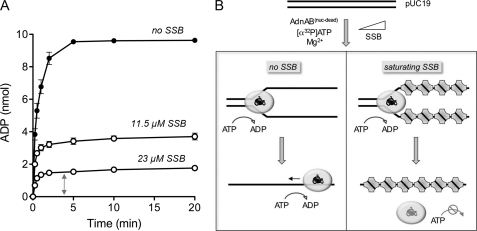
The coupling between ATP hydrolysis and duplex unwinding is a longstanding concern in the helicase field, the issue being how many base pairs are unwound for each ATP consumed. Biochemical and crystallographic studies of the prototypal SF1 helicases UvrD and PcrA favor a tight coupling whereby each catalytic cycle of ATP hydrolysis leads to a 1 nucleotide translocation of the helicase, in the 3′ to 5′ direction, along the ssDNA to which it is bound, which in turn results in unwinding of 1 bp of duplex DNA ahead of the advancing helicase (18–20). The motor domains of AdnAB resemble those of UvrD and PcrA, so one might predict a similarly tight coupling. However, the prospect of tandem motors translocating along the same DNA strand lends added complexity to the AdnAB system. Moreover, it is not trivial to gauge the macroscopic efficiency of the AdnAB motor during unwinding of a long DNA duplex when the DNA can anneal behind the helicase (thereby replenishing the substrate for unwinding) and the motor can just as well exploit the ssDNA product to trigger ongoing ATP hydrolysis and translocation uncoupled from duplex unwinding (Fig. 7B, left panel). This problem is evident when we analyzed the kinetic profile of ATP hydrolysis during the reaction of AdnAB with linear pUC19 dsDNA in the absence of SSB. Although the plasmid is unwound by AdnAB in less than 1 min (Fig. 3), we see that ATP hydrolysis continued for 2 min until all of the input ATP was converted to ADP (Fig. 7A). Thus, ATP hydrolysis and duplex unwinding are uncoupled under these conditions. (The rate of ATP hydrolysis by AdnAB in the absence of linear pUC19 was slowed by >1000-fold; data are not shown.)
FIGURE 7.
Estimating the coupling of ATP hydrolysis and duplex unwinding. A, reaction mixtures (80 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm [α-32P]ATP, 1.6 μg of pUC19 DNA (BamHI-digested; 1.82 pmol of DSB ends), 9.7 pmol (120 nm) of nuclease-dead AdnAB, and either no SSB, 11.5 μm SSB, or 23 μm SSB as specified were incubated at 37 °C. Aliquots (10 μl, containing 10 nmol input ATP) were withdrawn at the times specified and quenched with formic acid. The products were analyzed by PEI-cellulose TLC. The extents of conversion of [α-32P]ATP to [α-32P]ADP are plotted as a function of time. Each datum is the average of three experiments ±S.E. B, illustration of the ATP reaction outcomes in the absence of SSB, where unwound ssDNA is available to trigger ongoing ATP hydrolysis, versus in the presence of saturating SSB, where the unwound SSB-coated DNA strands are largely unavailable to serve as platforms for further ATP hydrolysis.
We reasoned that inclusion of SSB might confer tighter coupling by preventing reannealing of the ssDNA strands and shielding them from utilization as an ATPase activator by AdnAB (Fig. 7B, right panel). This turned out to be the case, insofar as increasing concentrations of SSB progressively lowered the extent of ATP hydrolysis by AdnAB during unwinding of pUC19 DNA (Fig. 7A). At a concentration of 23 μm SSB, the ATPase reaction was complete in 1 min (concomitant with unwinding of the plasmid), and no further hydrolysis occurred even up to 20 min (Fig. 7A). Taking into account the extent of ATP hydrolysis in this reaction and the amount of input dsDNA, we estimated an efficiency of ∼5 ATP molecules consumed by AdnAB per base pair. This value is in the same ballpark as that determined for E. coli RecBCD by Roman and Kowalczykowski (21, 22), who reported that ∼3 ATPs were consumed per base pair unwound.
Duplex Unwinding by AdnAB with a Crippled AdnA Phosphohydrolase Module
RecBCD and AdnAB each contain two motor domains in separate subunits, but they appear to be organized in fundamentally distinct ways. In RecBCD, the RecB and RecD motors act in parallel. The RecB subunit engages and translocates along the 3′ DNA strand during duplex unwinding, and the RecD subunit translocates along the 5′ DNA strand (11, 13). By contrast, mycobacterial AdnAB seems to behave like a serial motor, in which ATP hydrolysis by the leading (dominant) AdnB subunit precedes ATP hydrolysis by the lagging (dependent) AdnA subunit. This framework predicts correctly that mutating the lead motor abolishes all ATP hydrolysis and eliminates ssDNA translocation and unwinding of short DNA duplexes (9). It is proposed that the AdnB subunit uses ATP hydrolysis to pump single-stranded DNA through its N-terminal domain in a path that feeds the polynucleotide into the N-terminal motor domain of the AdnA subunit, where it can then trigger the AdnA phosphohydrolase activity (9). However, it is not clear whether an active AdnA motor aids translocation and duplex unwinding by the AdnAB complex.
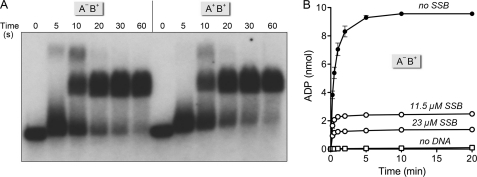
Here, we interrogated the role of the AdnA motor domain in plasmid unwinding by introducing a phosphohydrolase-inactivating mutation D285A (designated AdnA−) into the AdnA subunit of the nuclease-dead AdnAB complex. We assayed the plasmid helicase activity of the AdnA−B+ complex in parallel with wild-type AdnA+B+ motor. The kinetics of single turnover pUC19 unwinding in the presence of SSB were virtually identical for the AdnA+B+ and AdnA−B+ enzymes, both of which yielded mature unwound SSB·ssDNA complexes within 10 s (Fig. 8A). Thus, within the limits of the assay, we see no effect of a crippling mutation in the AdnA phosphohydrolase active site on the rate or extent of pUC19 unwinding. By contrast, an AdnA+B− mutant that has an ATPase-inactivating mutation in the AdnB motor (9) failed to unwind pUC19 DNA in the presence of SSB (data not shown). We conclude that the AdnB subunit motor is necessary and sufficient for the processive helicase activity of the AdnAB heterodimer.
FIGURE 8.
Duplex unwinding by AdnAB with a crippled AdnA phosphohydrolase module. A, reaction mixtures (60 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 1.2 μg of 5′ 32P-labeled pUC19 DNA (BamHI-digested; 1.37 pmol DSB ends), 11.5 μm SSB, and 7.3 pmol (120 nm) of nuclease-dead AdnAB heterodimers with either two wild-type motor domains (A+B+) or a wild-type AdnB motor plus a AdnAD255A mutant motor (A−B+) were incubated at 37 °C. Aliquots (10 μl) were withdrawn at the times specified and quenched with EDTA. The mixtures were analyzed by native agarose gel electrophoresis, and the radiolabeled DNA was visualized by autoradiography of the dried gel. B, reaction mixtures (80 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm [α-32P]ATP, 1.6 μg of pUC19 DNA (BamHI-digested; 1.82 pmol of DSB ends), 9.7 pmol (120 nm) of nuclease-dead AdnA−B+ motor, and either no SSB, 11.5 μm SSB, or 23 μm SSB as specified were incubated at 37 °C. The “no DNA” control reaction mixture contained AdnA−B+ motor, 23 μm SSB, and other components but no pUC19 DNA. Aliquots (10 μl, containing 10 nmol of input ATP) were withdrawn at the times specified and quenched with formic acid. The products were analyzed by PEI-TLC. The extents of conversion of [α-32P]ATP to [α-32P]ADP are plotted as a function of time. Each datum is the average of three experiments ±S.E.
The preceding mutational results raise the issue of whether the AdnA motor even fires during dsDNA unwinding by the AdnAB heterodimer. An analysis of the kinetics of ATP hydrolysis during the reaction of AdnA−B+ with pUC19 (Fig. 8B) suggests that it does, albeit modestly. Although the kinetic profiles ±23 μm SSB are qualitatively similar to what we observed for the wild-type AdnAB motor, the end point values for ADP formation were lower in the case of AdnA−B+. From the data in Fig. 8B, we estimated a macroscopic efficiency of ∼4.2 ATPs consumed by the AdnA−B+ heterodimer per bp of DNA. (Note the control reactions showing that ATP hydrolysis in the presence of AdnA−B+ and 23 μm SSB was contingent on inclusion of linear pUC19 DNA; see Fig. 8B.)
Action of the AdnAB Nucleases on the Displaced ssDNA Strands
A key issue in understanding the mechanism of DSB processing by AdnAB is how the nuclease modules of the AdnA and AdnB subunits are disposed with respect to the displaced DNA strands. Previously, we proposed that the two nucleases are physically and functionally segregated (9). In our model, the AdnB nuclease is situated behind the tandem motor on the displaced 3′ strand, so that access of the 3′ strand to the nuclease active site requires pumping of the DNA through the tandem motor, whereas the AdnA nuclease is poised to receive and degrade the displaced 5′ strand directly at the “Y-fork” of the dsDNA-ssDNA junction (Fig. 1). The experiments supporting this model derived from analyses of AdnAB nuclease action on ssDNA oligonucleotide substrates. Here, our aim was to study the processing of the individual strands displaced by nuclease-competent AdnAB as it unwinds a linear dsDNA substrate and to gauge the impact of SSB on the processing reactions.
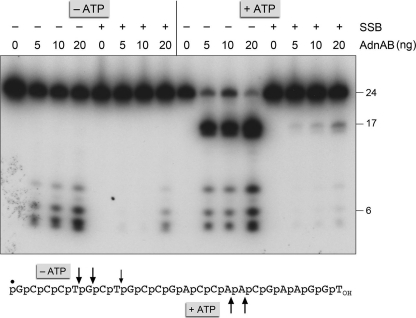
First, we tested the effect of SSB on the ssDNA nuclease activities of the wild-type AdnAB heterodimer, using a 5′ 32P-labeled 24-mer ssDNA substrate. Absent ATP and SSB, the AdnA nuclease module of the AdnAB heterodimer incises the 24-mer close to the 5′ end to yield a cluster of labeled 5-, 6-, and 8-nucleotide products (Fig. 9). Inclusion of ATP in the reaction triggers the AdnB nuclease module to cleave the DNA at two distal sites to yield 5′-labeled 16- and 17-mer products (Fig. 9). The instructive findings were that inclusion of SSB in the reaction mixtures at a 4:1 ratio of SSB monomers to ssDNA suppressed both nuclease activities (Fig. 9), signifying that an ssDNA segment bound to SSB is protected from processing by the AdnA and AdnB nuclease modules.
FIGURE 9.
SSB inhibits the ssDNA nuclease activities of AdnAB. Reaction mixtures (10 μl) containing 20 mm Tris-HCl, pH 8.0, 0.5 mm DTT, 2 mm MgCl2, 1 mm ATP, 100 nm (1 pmol) 5′ 32P-labeled 24-mer ssDNA (shown at bottom with the 5′-label denoted by ●), 4 pmol of SSB (where indicated by +), and AdnAB as specified (5, 10 or 20 ng of AdnB subunit, corresponding to 50, 100, or 200 fmol of AdnAB heterodimer) were incubated for 5 min at 37 °C. The reactions were quenched with formamide/EDTA, heated for 5 min at 95 °C, and then analyzed by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 m urea, 45 mm Tris borate, 1.25 mm EDTA. An autoradiograph of the gel is shown; oligonucleotide sizes are noted on the right. The principal sites of AdnAB incision of the 24-mer DNA in the absence of ATP are indicated by arrows above the oligonucleotide sequence; the cleavage sites induced by ATP are indicated below.
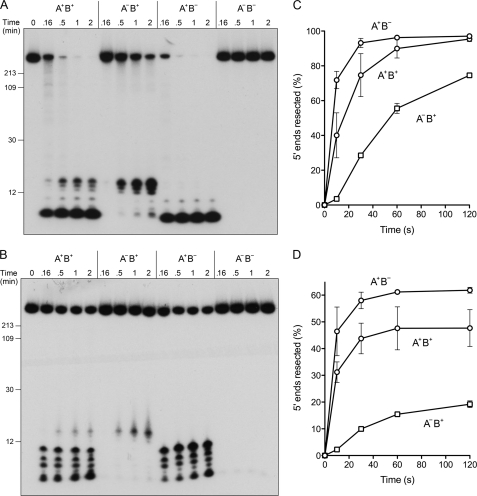
Next, we tracked the fate of the 5′ 32P-labeled DSB ends of linear pUC19 during its unwinding by four different versions of the AdnAB motor with (i) both nucleases active (A+B+); (ii) both nucleases dead (A−B−); (iii) only the AdnA nuclease active (A+B−); and (iv) only the AdnB nuclease active (A−B+). To assay processing of the strands displaced by these AdnAB motors, the reactions were quenched after 10, 30, 60, or 120 s, and the radiolabeled products were analyzed by electrophoresis through a 15% polyacrylamide gel in 7 m urea (Fig. 10), which resolves only the shortest nucleolytic cleavage products. In the absence of SSB, the A+B+ motor cleaved about half of the input 5′ 32P-labeled pUC19 ends within 10 s to yield short oligonucleotides (predominantly <12 nucleotides long) (Fig. 10A). The ends were cleaved completely in 30–60 s by the A+B+ enzyme. As expected, the double nuclease-dead A−B− motor elicited no decay of the labeled DNA ends (Fig. 10A). When the AdnA nuclease alone was active (in A+B−), the 5′ 32P-labeled pUC19 ends were cleaved to yield products <10-nucleotides in length (Fig. 10A). However, when the AdnB nuclease alone was active (in A−B+), the cleavage products were distinctly longer (14–16 nucleotides) (Fig. 10A). Thus, the nuclease ruler phenomenon, first observed with ssDNA substrates (8), also pertains to processing of 5′-labeled DSB ends. The key insight from this experiment concerned the kinetics of processing by the two autonomous nuclease modules, to wit. (i) The AdnA nuclease incised the 5′ ends rapidly, to an extent of 70% within 10 s, at which time the AdnB nuclease had cleaved only 4% of the input pUC19 to release short oligonucleotides (Fig. 10C). (ii) Substantial AdnB cleavage became evident at 30 s (29%) and increased further at 1 min (56%) and 2 min (75%) (Fig. 10C). (iii) The kinetic lag between the two nucleases was apparent even when both nucleases were active, as gauged by inspection of the two size populations of cleavage products (> or <12-nucleotides) generated by the A+B+ enzyme (Fig. 10A). These results suggest that the AdnA nuclease cleaved the displaced 5′ end as soon as it was unwound by the AdnAB motor (i.e. as a “first bite” when the motor starts), whereas the AdnB nuclease cleaved near the 5′ end to generate oligonucleotide products after the AdnAB motor had traversed the length of the duplex (i.e. as a “last bite” when the motor reached the end of the strand along which it translocated 3′ to 5′).
FIGURE 10.
AdnAB nuclease action at 5′-labeled DSB ends. Reaction mixtures (50 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 1 μg of 5′ 32P-labeled pUC19 DNA (BamHI-digested; 1.14 pmol DSB ends), and 6 pmol (120 nm) of AdnAB heterodimers with either two wild-type nuclease domains (A+B+), a wild-type AdnB nuclease plus an inactive AdnAD934A mutant nuclease (A−B+), a wild-type AdnA nuclease plus an inactive AdnBD1014A mutant nuclease (A+B−), or two inactive mutant nucleases (A−B−), and either no SSB (A) or 23 μm SSB (B) were incubated at 37 °C. The reactions were initiated by adding AdnAB to reaction mixtures prewarmed to 37 °C. Aliquots (10 μl) were withdrawn at the times specified and quenched with formamide/EDTA. The mixtures were heated for 5 min at 95 °C and then analyzed by electrophoresis through 15-cm 15% polyacrylamide gels containing 7 m urea, 45 mm Tris borate, 1.25 mm EDTA. Autoradiographs of the gels are shown. The positions and sizes (in nucleotides) of heat-denatured 5′-labeled DNA markers (generated by restriction endonuclease digestion of the 5′ 32P-labeled pUC19 DNA substrate) are indicated on the left. C and D, extents of 5′ strand cleavage to generate short oligonucleotides were quantified by scanning the gels with a phosphorimager and are plotted as a function of time for reactions containing no SSB (C) or 23 μm SSB (D). Each datum is the average of two experiments; error bars indicate the range.
The DSB processing experiment was also performed in the presence of 23 μm SSB, in an effort to limit the observed end-resection to a single round of dsDNA unwinding by the AdnAB motor (because the SSB-coated ssDNA products will be largely protected from new rounds of translocation and incision by AdnAB). This maneuver was apparently effective (Fig. 10B), insofar as the extents of cleavage of the 5′-labeled pUC19 DNA to generate short products at 1–2 min were lower in the presence of SSB (48% for A+B+; 61% for A+B−) than in its absence, and the end-processing reactions of the AdnA nuclease were virtually complete within 10 s, yielding a cluster of short 5′-labeled fragments (≤10 nucleotides) (Fig. 10, B and D). Note that there was again a lag before the first appearance of the longer 5′-labeled oligonucleotide cleavage products generated by the AdnB nuclease (Fig 10, B and D). We infer from the processing pattern in the presence of SSB that not every dsDNA unwinding event results in strand scission to liberate short oligonucleotide products when the nuclease modules are active.
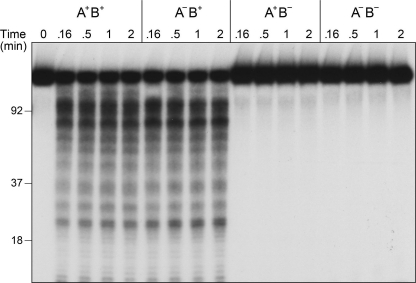
DSB processing reactions in the presence of SSB were also performed using a 3′ 32P end-labeled pUC19 DNA substrate. In this case, we observed that the AdnB nuclease cleaved the DNA within 10 s to generate a heterogeneous array of 3′-labeled cleavage products in the range of 20–200 nucleotides (Fig. 11). The sizes of the AdnB cleavage products did not change appreciably between 10 s and 2 min, implying the following. (i) They arise by incision of the displaced 3′ strand during a single round of duplex unwinding by the AdnAB complex. (ii) AdnB does not cleave at a strictly fixed distance from the initial 3′ end. Ablation of the AdnB nuclease while sparing the AdnA nuclease eliminated nearly all of these 3′-labeled cleavage products (Fig. 11). The differential cleavage patterns and kinetics of short product release from 5′- and 3′-labeled substrates are consistent with separate action of the AdnA and AdnB nucleases on the displaced 5′ and 3′ strands, respectively.
FIGURE 11.
AdnAB nuclease action at 3′-labeled DSB ends. Reaction mixtures (50 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 1 μg of 3′ 32P-labeled pUC19 DNA (EcoRI-digested and 3′-labeled with [32P]dAMP; 1.14 pmol of DSB ends), 23 μm SSB, and 6 pmol (120 nm) of AdnAB heterodimers as indicated were incubated at 37 °C. Aliquots (10 μl) were withdrawn at the times specified and quenched with formamide/EDTA. The mixtures were heated for 5 min at 95 °C and then analyzed by electrophoresis through a 15-cm 15% polyacrylamide gel containing 7 m urea, 45 mm Tris borate, 1.25 mm EDTA. An autoradiograph of the gel is shown. The positions and sizes (in nucleotides) of heat-denatured 3′-labeled DNA markers (generated by restriction endonuclease digestion of the 3′ 32P-labeled pUC19 DNA substrate) are indicated on the left.
To expand the size resolution of the product analysis, we performed a similar series of experiments with wild-type and nuclease-defective AdnAB complexes, with or without SSB, in which the reaction products were analyzed by alkaline agarose gel electrophoresis (Fig. 12). This method does not resolve (or quantitatively recover) the smallest radiolabeled oligonucleotides, but it does recover and separate longer single-stranded DNAs in the range of 200–2000 nucleotides (most of which were not separated from the substrate DNA by the 15% PAGE method). The salient points are illustrated by comparing the patterns of digestion of 5′- and 3′-labeled pUC19 by the AdnAB proteins with single active AdnB (A−B+) and AdnA (A+B−) nuclease modules. To wit, the AdnB nuclease yielded much longer 5′-labeled ssDNA products (500–2000 nucleotides) at 10 s in the presence of SSB than did the AdnA nuclease (<100 nucleotides) (Fig. 12A). The pattern was reversed with 3′-labeled pUC19 (Fig. 12B), such that the AdnB nuclease formed shorter 3′ end-labeled ssDNA products (<500 nucleotides) at 10 s in the presence of SSB than did the AdnA nuclease (900–2100 nucleotides). Here again, inclusion of SSB typically reduced the extent of DNA cleavage, likely by limiting the observed nuclease activity to the initial round of duplex unwinding. These results support the division of nuclease labor as depicted in Fig. 1.
FIGURE 12.
AdnAB product analysis by alkaline-agarose gel electrophoresis. Reaction mixtures (25 μl) containing 20 mm Tris-HCl, pH 8.0, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, either 0.5 μg of 5′ 32P-labeled pUC19 (BamHI-digested; 570 fmol of DSB ends; A) or 0.5 μg of 3′ 32P-labeled pUC19 (EcoRI-digested; 570 fmol of DSB ends; B), 3 pmol (120 nm) of AdnAB heterodimers as indicated, and either no SSB (−) or 23 μm SSB (+) were incubated at 37 °C. After 10 or 30 s, 10-μl aliquots were withdrawn and quenched immediately by adjustment to 72 mm EDTA and 0.85% SDS. The mixtures were supplemented with proteinase K (0.8 units; Sigma) and incubated at 37 °C for 15 min. The samples were then adjusted to 100 mm NaOH, 4% glycerol, and 1% bromphenol blue and analyzed by electrophoresis (10 h at 20 V at room temperature) through a 1.5% alkaline-agarose gel in 50 mm NaOH, 2 mm EDTA. The gel was rinsed twice for 10 min in 5% trichloroacetic acid and then dried under vacuum on DE81 paper. Labeled DNA was visualized by autoradiography of the dried gels. The positions and sizes (in nucleotides) of heat-denatured 5′- or 3′-labeled DNA markers (generated by restriction endonuclease digestion of the 32P-labeled pUC19 DNA substrates) are indicated on the right.
DISCUSSION
This study extends our understanding of DSB resection by mycobacterial AdnAB and provides instructive comparisons to the RecBCD and AddAB clades of bacterial motor-nuclease machines. A nuclease-dead version of the AdnAB motor allowed us to track the unwinding of long linear duplex DNAs in the presence of mycobacterial SSB. SSB had little if any impact on the rate of dsDNA unwinding, but it conveniently sequestered the unwound strands as SSB·ssDNA complexes. Thus, we can regard the reaction with linear plasmid DNA in the presence of saturating SSB as reflecting a single round of DNA unwinding by the AdnAB motor translocating, apparently processively, in the 3′ to 5′ direction on one of the DNA strands (Fig. 1). We infer processivity from the fact that AdnAB can unwind 2.7- or 11.2-kbp linear DNAs with similar facility and similar rates of fork progression. Our estimates of the unwinding rate of the AdnAB motor are predicated on an assumption that many of the unwinding events in the ensemble measurement reflect the action of AdnAB initiating unwinding from one of the DSB ends and then propagating a Y-fork along the entire length of the plasmid substrate (as illustrated in Fig. 3B). If this is the case, then the time required to accumulate a substantial fraction of the SSB·ssDNA end product complexes divided by the DNA length should correspond to a lower bound value of the unwinding rate. (A lower bound value due to this simple formula does not factor in the possibility that end binding and initiation of unwinding might contribute to the time required to detect an end product.) The unwinding time course experiments with pUC19 substrate in Figs. 4 and 8 were performed at a ratio of ∼3–5 AdnABs per DSB end, to promote reaction synchrony. The unwinding intermediates detected by PAGE at 5–10 s migrated slower than the unwound linear SSB-ssDNA end products (Fig. 8); such retarded mobility is expected for Y-fork molecules with two SSB-coated “prongs.” The kinetic analysis of unwinding of the 11.2-kbp substrate was performed at an ∼10:1 ratio of AdnAB to DSB ends, and in this case, we can discern unwinding intermediates of retarded mobility evolving over time along two electrophoretic “arcs” that likely correspond to single-Y and double-Y fork molecules (Fig. 5). In this experiment, the major fraction includes the faster moving single-Y arc, suggesting that single Y molecules are likely to predominate at the lower AdnAB-DSB ratios used in the pUC19 unwinding experiments. (A definitive assessment of the fork distribution will ultimately require EM or atomic force microscopy analysis of the partially unwound SSB-coated molecules.)
Our estimates of an ensemble AdnAB unwinding rate of 250–270 bp s−1 in the presence of SSB are similar to the value reported for E. coli RecBCD (21, 22). Maximal rates of plasmid unwinding by RecBCD, powered by two motors acting in parallel on the displaced ssDNA strands, are on the order of 1400 bp s−1 (12). Crippling the RecD ATPase active site converts RecBCD into a single-motor machine powered by the 3′ to 5′ translocase of the RecB subunit, with a maximum unwinding rate of 800 bp s−1 (12). Here, we observed little or no difference in the rate of unwinding of pUC19 DNA by AdnAB motor when the AdnA phosphohydrolase active site was crippled. The AdnA−B+ motor can be viewed as analogous to the single motor Bacillus AddAB complex. A recent report characterizing a nuclease-dead version of the Bacillus AddAB motor revealed that it unwound linear DNA at an average rate of 82 bp s−1 in the presence of SSB, with a maximum interval rate of 250 bp s−1 between pauses (23). A similar maximum rate of 250 bp s−1 was reported for Bacteroides fragilis AddAB (24).
Inclusion of SSB in the unwinding reactions also allowed us to gauge the consumption of ATP during a single round of dsDNA unwinding, wherein the unwound SSB·ssDNA product complexes do not serve as platforms for ongoing AdnAB-catalyzed ATP hydrolysis. We derived values of 5 and 4.2 ATPs hydrolyzed by the A+B+ and A−B+ motors, respectively, per bp of available duplex DNA. It was reported recently that the single-motor B. fragilis AddAB motor-nuclease hydrolyzes two ATPs per base pair unwound (24). The observed ATP consumptions provide a lower bound estimate of the energy coupling of the unwinding motor. We can regard the consumption of 1 ATP per bp unwound as the gold standard of tight coupling by superfamily 1 helicases, according to the physicochemical models elaborated for PcrA and UvrD (18–20). The lower efficiency of AdnAB energy coupling could reflect transient uncoupling of ATP hydrolysis from forward translocation of the motor, i.e. if the motor slips out of gear into “neutral” at some point during the unwinding reaction and then re-engages in “drive.”
By tracking the resection of the 5′ and 3′ strand at the DSB ends, we illuminated the division of labor among the AdnA and AdnB nuclease modules during dsDNA unwinding, whereby the AdnA nuclease processes the unwound 5′ strand to liberate a short oligonucleotide product, and the AdnB nuclease incises the 3′ strand on which the motor translocates. Under the reaction conditions employed here in the presence of SSB, the AdnB nuclease makes its first incision at heterogeneous sites within an ∼500-nucleotide interval from the 3′ end of the DSB (Figs. 11 and 12B). Whereas duplex unwinding by the AdnAB motor in the presence of SSB is apparently processive, it is not clear from the present experiments whether the AdnA and AdnB nuclease modules are committed to reiteratively cleaving the displaced strands after the initial scissions have occurred. By analogy to RecBCD, we presume that the nuclease functions of mycobacterial AdnAB are modulated in some fashion to achieve asymmetric processing of the displaced 5′ strand, with preservation of a 3′ single-stranded tail onto which RecA can assemble. Whether AdnAB does this via recognition of χ-like DNA sequences (that regulate RecBCD activity) is not known. Indeed, the mechanism of RecA assembly on the recombinogenic 3′ strand during mycobacterial HR is tabula rasa and is likely to differ from the pathway of RecBCD-dependent RecA loading described for E. coli (10, 25), insofar as mycobacterial RecBCD plays no discernible role in RecA-dependent DNA repair in vivo.3 Although many questions remain concerning the action and interactions of AdnAB, this study consolidates a working model of DSB processing entailing processive unidirectional translocation of the AdnB motor on the 3′ strand and separate nuclease cleavages of the 5′ and 3′ strand by the AdnA and AdnB subunits, respectively (Fig. 1). It sets the stage for efforts to reconstitute subsequent steps of mycobacterial HR, especially RecA recruitment to the AdnAB unwound strands and strand invasion into a homologous DNA.
Acknowledgment
We thank Poulami Samai for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI64693.
R. Gupta, D. Barkan, G. Redelman-Sidi, S. Shuman, and M. S. Glickman, unpublished data.
- DSB
- double strand break
- SSB
- single strand DNA-binding protein.
REFERENCES
- 1.Cromie G. A. (2009) J. Bacteriol. 191, 5076–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B. (2004) Nature 432, 187–193 [DOI] [PubMed] [Google Scholar]
- 3.Dillingham M. S., Kowalczykowski S. C. (2008) Microbiol. Mol. Biol. Rev. 72, 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haijema B. J., Venema G., Kooistra J. (1996) J. Bacteriol. 178, 5086–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haijema B. J., Meima R., Kooistra J., Venema G. (1996) J. Bacteriol. 178, 5130–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeeles J. T., Dillingham M. S. (2007) J. Mol. Biol. 371, 66–78 [DOI] [PubMed] [Google Scholar]
- 7.Yeeles J. T., Cammack R., Dillingham M. S. (2009) J. Biol. Chem. 284, 7746–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha K. M., Unciuleac M. C., Glickman M. S., Shuman S. (2009) Genes Dev. 23, 1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unciuleac M. C., Shuman S. (2010) J. Biol. Chem. 285, 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spies M., Kowalczykowski S. C. (2006) Mol. Cell 21, 573–580 [DOI] [PubMed] [Google Scholar]
- 11.Dillingham M. S., Spies M., Kowalczykowski S. C. (2003) Nature 423, 893–897 [DOI] [PubMed] [Google Scholar]
- 12.Dillingham M. S., Webb M. R., Kowalczykowski S. C. (2005) J. Biol. Chem. 280, 37069–37077 [DOI] [PubMed] [Google Scholar]
- 13.Taylor A. F., Smith G. R. (2003) Nature 423, 889–893 [DOI] [PubMed] [Google Scholar]
- 14.Reddy M. S., Guhan N., Muniyappa K. (2001) J. Biol. Chem. 276, 45959–45968 [DOI] [PubMed] [Google Scholar]
- 15.Raghunathan S., Kozlov A. G., Lohman T. M., Waksman G. (2000) Nat. Struct. Biol. 7, 648–652 [DOI] [PubMed] [Google Scholar]
- 16.Saikrishnan K., Jeyakanthan J., Venkatesh J., Acharya N., Sekar K., Varshney U., Vijayan M. (2003) J. Mol. Biol. 331, 385–393 [DOI] [PubMed] [Google Scholar]
- 17.Saikrishnan K., Manjunath G. P., Singh P., Jeyakanthan J., Dauter Z., Sekar K., Muniyappa K., Vijayan M. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 1140–1148 [DOI] [PubMed] [Google Scholar]
- 18.Velankar S. S., Soultanas P., Dillingham M. S., Subramanya H. S., Wigley D. B. (1999) Cell 97, 75–84 [DOI] [PubMed] [Google Scholar]
- 19.Dillingham M. S., Wigley D. B., Webb M. R. (2000) Biochemistry 39, 205–212 [DOI] [PubMed] [Google Scholar]
- 20.Lee J. Y., Yang W. (2006) Cell 127, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roman L. J., Kowalczykowski S. C. (1989) Biochemistry 28, 2863–2873 [DOI] [PubMed] [Google Scholar]
- 22.Roman L. J., Kowalczykowski S. C. (1989) Biochemistry 28, 2873–2881 [DOI] [PubMed] [Google Scholar]
- 23.Fili N., Mashanov G. I., Toseland C. P., Batters C., Wallace M. I., Yeeles J. T., Dillingham M. S., Webb M. R., Molloy J. E. (2010) Nucleic Acids Res. 38, 4448–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter M., Parry F., Dryden D. T., Blakely G. W. (2010) Nucleic Acids Res. 38, 3721–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson D. G., Kowalczykowski S. C. (1997) Cell 90, 77–86 [DOI] [PubMed] [Google Scholar]