Abstract
A phosphoramidate prodrug of 2′-deoxy-2′-α-fluoro-β-C-methyluridine-5′-monophosphate, PSI-7851, demonstrates potent anti-hepatitis C virus (HCV) activity both in vitro and in vivo. PSI-7851 is a mixture of two diastereoisomers, PSI-7976 and PSI-7977, with PSI-7977 being the more active inhibitor of HCV RNA replication in the HCV replicon assay. To inhibit the HCV NS5B RNA-dependent RNA polymerase, PSI-7851 must be metabolized to the active triphosphate form. The first step, hydrolysis of the carboxyl ester by human cathepsin A (CatA) and/or carboxylesterase 1 (CES1), is a stereospecific reaction. Western blot analysis showed that CatA and CES1 are both expressed in primary human hepatocytes. However, expression of CES1 is undetectable in clone A replicon cells. Studies with inhibitors of CatA and/or CES1 indicated that CatA is primarily responsible for hydrolysis of the carboxyl ester in clone A cells, although in primary human hepatocytes, both CatA and CES1 contribute to the hydrolysis. Hydrolysis of the ester is followed by a putative nucleophilic attack on the phosphorus by the carboxyl group resulting in the spontaneous elimination of phenol and the production of an alaninyl phosphate metabolite, PSI-352707, which is common to both isomers. The removal of the amino acid moiety of PSI-352707 is catalyzed by histidine triad nucleotide-binding protein 1 (Hint1) to give the 5′-monophosphate form, PSI-7411. siRNA-mediated Hint1 knockdown studies further indicate that Hint1 is, at least in part, responsible for converting PSI-352707 to PSI-7411. PSI-7411 is then consecutively phosphorylated to the diphosphate, PSI-7410, and to the active triphosphate metabolite, PSI-7409, by UMP-CMP kinase and nucleoside diphosphate kinase, respectively.
Keywords: Enzymes, Hepatitis Virus, Hepatocyte, Nucleoside Nucleotide Analogs, Nucleoside Nucleotide Metabolism, Phosphoramidate, Prodrug, Stereospecificity
Introduction
Nucleoside analogs have long been the backbone therapy for the treatment of viral diseases such as HIV, HBV, and HSV infections (1–5). Recent studies have suggested that nucleoside analogs may be useful for treating hepatitis C virus (HCV)3 infection (4, 6–8). The most advanced anti-HCV nucleoside, RG7128, is a diisobutyrate nucleoside prodrug of β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylcytidine (PSI-6130) and is currently in phase IIb clinical studies. PSI-6130 demonstrated potent activity in the subgenomic HCV replicon assay (9); the incubation of radiolabeled PSI-6130 with either replicon cells or primary human hepatocytes resulted in the formation of the 5′-mono-, di-, and triphosphate metabolites of PSI-6130 (10–12). The triphosphate metabolite (PSI-6130-TP) was shown to be a potent inhibitor of HCV NS5B RNA-directed RNA polymerase (RdRp) (11). However, incubation of replicon cells with the uridine analog, PSI-6206, resulted in no inhibition of HCV RNA production due to the inability of PSI-6206 to be phosphorylated by cellular nucleoside kinases to its monophosphate, PSI-7411 (10, 12). Biochemical studies showed that PSI-7411 was consecutively phosphorylated to its diphosphate, PSI-7410, by UMP-CMP kinase and its triphosphate, PSI-7409, by nucleoside diphosphate kinase (12). Inhibition studies using the replicase assay and purified recombinant HCV NS5B RdRp showed that PSI-7409 was a potent inhibitor of HCV RNA synthesis (10, 12). Therefore, the phosphoramidate prodrug approach was applied to bypass the nonproductive phosphorylation step.
PSI-7851 is a phosphoramidate prodrug of PSI-7411 and is a mixture of two diastereoisomers, PSI-7976 and PSI-7977. Here, we describe the metabolic pathway for PSI-7851 and its two diastereoisomers, PSI-7976 and PSI-7977.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human cathepsin A (CatA), carboxylesterase 1 (CES1), and carboxylesterase 2 (CES2) were purchased from R & D Systems (Minneapolis, MN); human trypsin, chymase, and neutrophil elastase were from Calbiochem; chymotrypsin was from MP Biomedicals (Solon, OH); cathepsin B, cathepsin D, cathepsin L, and lipase were from Sigma; cathepsin H, calpain 1, and caspase 1–10 were from BioVision (Mountain View, CA). Human liver cytosol from 10 different single donors, pooled human liver cytosol, and primary human hepatocytes (HHPC) were purchased from CellzDirect, Inc. (Durham, NC). Clone A HCV replicon cells were obtained from Apath, LLC (Brooklyn, NY). Radiolabeled [3H]PSI-7851, [14C]PSI-7976, and [14C]PSI-7977 were synthesized by Moravek Biochemicals (Brea, CA). Anti-CES1 antibody was purchased from R & D Systems (Minneapolis, MN); anti-CatA antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-Hint1 antibody was from ProteinTech Group (Chicago). PSI-7851, PSI-7976, PSI-7977, PSI-352707, PSI-7411, PSI-7410, and PSI-7411 were synthesized at Pharmasset. Telaprevir (VX-950) was synthesized by ACME Bioscience, Inc. (Palo Alto, CA). Bis(4-nitrophenyl)phosphate (BNPP), p-nitrophenylacetate (4-NPA), and trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E-64) were purchased from Sigma.
Cellular Metabolism Study
Clone A cells were seeded into T75 flasks at about 5 × 106 cells/flask in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 100 IU/ml penicillin/100 μg/ml streptomycin (Invitrogen) and 10% fetal bovine serum (Cellgro, Manassas, VA). Similarly, human primary hepatocytes (CellzDirect) were seeded in cell plating medium (CellzDirect) into T75 flasks at about 5 × 106 cells/flask. After overnight incubation to allow the cells to attach, cells were incubated with 50 μm PSI-7851, PSI-7976, or PSI-7977 in fresh medium for clone A cells or in cell maintenance medium (CellzDirect) for primary hepatocytes for up to 24 h at 37 °C in a 5% CO2 atmosphere. The same procedures were applied when radiolabeled PSI-7851 was used in the study except that 1 × 106 cells per well were seeded into a 6-well plate, and the cells were incubated with 5 μm [3H]PSI-7851. At selected times, the medium was removed, and the cell layer was washed with cold phosphate-buffered saline (PBS). After trypsinization, cells were counted and centrifuged at 1,200 rpm for 5 min. The cell pellets were suspended in 1 ml of cold 60% methanol and incubated overnight at −20 °C. The samples were centrifuged at 14,000 rpm for 5 min, and the supernatants were collected and dried using a SpeedVac concentrator (Thermo Electron Corp.) and stored at −20 °C until they were analyzed by high performance liquid chromatography (HPLC). Residues were suspended in 100 μl of water, and 50-μl aliquots were injected into HPLC.
The metabolites were separated by ion exchange HPLC with a Whatman 10-μm SAX column (Whatman) using a Series 200 HPLC system (PerkinElmer Life Sciences). The mobile phase consisted of buffer A (0.02 m KH2PO4, pH 3.5) and buffer B (1 m KH2PO4, pH 3.5). Elution was performed using a linear gradient of buffer B from 0 to 100% for 90 min. When [3H]PSI-7851 was used, radioactive metabolites were analyzed using a 610TR radiometric flow scintillation analyzer (PerkinElmer Life Sciences). When unlabeled PSI-7851 was used, UV absorption at 254 nm was followed to detect the products. A standard curve for PSI-7409 was generated by running a series of dilutions of PSI-7409 with the same injection volume as samples. The intracellular concentration (pmol/106 cells) of PSI-7409 was calculated using the PSI-7409 standard curve and converted to micromolars based on a 3-μl volume/1,000,000 cells for normal human liver parenchymal cells (13). PSI-7851 and its respective metabolites were identified by running known chemically synthesized standards.
PSI-7851 Hydrolysis by Various Enzymes
Hydrolysis of PSI-7851 was assayed using various human recombinant enzymes in a 100-μl reaction volume containing 100 μm PSI-7851. The amount of protein in the reaction was 1 μg except for caspases 1–10 where 1 unit of enzyme (enzyme activity that cleaves 1 nmol of the individual caspase substrate per h at 37 °C) was used in the reaction. Different buffer systems were used depending on the enzyme as follows: 200 mm Tris/HCl, pH 7.5, 20 mm CaCl2 for trypsin, chymotrypsin, and chymase; 100 mm Tris/HCl, pH 7.5, 500 mm NaCl for elastase; 50 mm Hepes, pH 7.0, 100 mm NaCl, 0.5% Nonidet P-40 for cathepsin A; 100 mm Tris/HCl, pH 7.5, 100 mm NaCl, 5 mm DTT for cathepsin B and L; 400 mm sodium acetate, pH 5.2, 10 mm DTT, 5 mm EDTA for cathepsin D; 50 mm Tris/HCl, pH 7.5, for carboxylesterase 1 and 2, and lipase; 100 mm Tris/HCl, pH 7.5, 5 mm CaCl2, 5 mm DTT for calpain 1; and 50 mm Hepes, pH 7.2, 50 mm NaCl, 0.1% CHAPS, 10 mm DTT, 10 mm EDTA, 5% glycerol for cathepsin H and caspase 1–10. After incubation at 37 °C for 1 h, the reaction mixture was applied to a YM-10 Microcon filter (Millipore, Billerica, MA) to remove the protein. The flow-through from the filter was collected and analyzed by HPLC using a PARTISIL 10 SAX column (Whatman). The mobile phase consisted of buffer A (0.02 m KH2PO4, pH 3.5) and buffer B (1 m KH2PO4, pH 3.5). Elution was performed using a linear gradient of buffer B from 0 to 100% for 90 min.
Standard Assays for Cathepsin A and CES1
For the CatA assay, the enzyme was activated by following the manufacturer's instructions (R & D Systems, Minneapolis, MN). Briefly, 10 μg/ml CatA was incubated with 1 μg/ml CatL in 25 mm MES buffer, pH 6.0, and 5 mm DTT for 30 min at 37 °C. CatL was then inactivated by adding 10 μm E-64, a CatL inhibitor. CatA activity assays were performed according to manufacturer's instructions using a fluorogenic peptide substrate, (7-methoxycoumarin-4-yl)acetyl-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys-(2,4-dinitrophenyl)-OH (R & D Systems, Minneapolis, MN). CES1 activity assays were performed according to the manufacturer's instructions (R & D Systems, Minneapolis, MN) using 4-NPA as a substrate and following the absorbance change at 405 nm using a Victor3 plate reader (PerkinElmer Life Sciences).
Hydrolysis of PSI-7851/7976/7977
The CatA assay was performed in a 100-μl reaction mixture containing activated CatA (0.1 μg), 100 μm PSI-7851 in 25 mm MES, pH 6.0, 100 mm NaCl, 1 mm DTT, and 0.1% Nonidet P-40, and the CES1 assay was performed in 50 mm Tris/HCl buffer, pH 7.5, containing 100 μm PSI-7851 and 0.4 μg of CES1. After incubation at 37 °C for 1 h, the reaction mixture was applied to a YM-10 Microcon filter (Millipore, Billerica, MA) to remove the protein. The flow-through from the filter was collected and analyzed by HPLC using a PARTISIL 10 SAX column (Whatman). Elution was performed as described above.
Steady-state kinetic parameters for PSI-7976 and PSI-7977 with CatA and CES1 were determined using 14C-labeled compounds. For CatA reactions, a buffer containing 50 mm Hepes, pH 7.0, 100 mm NaCl, and 0.1% Nonidet P-40 was used. For CES1 reactions, a 50 mm Tris/HCl, pH 7.5, buffer was used. Reactions were performed at 37 °C in a 100-μl reaction volume containing varied concentrations of PSI-7976 or PSI-7977, and the final protein concentration for activated CatA and CES1 was 0.1 and 10 μg/ml, respectively. A 10-μl aliquot of the reaction mixture was spotted on a sheet of DE81 paper at every time point. The paper was washed three times with 1 mm ammonium formate for 5 min, followed by an ethanol wash. After the paper was dried, the spots were cut out and counted in a liquid scintillation counter.
HCV Replicon Assay
For each assay, 50 μl of 2× serial drug dilutions were added (in duplicate) per well to a 96-well plate. Clone A cells were added to the plate at 1,500 cells/well in 50 μl of medium to give a total volume of 100 μl per well. The plate was incubated at 37 °C in a humidified 5% CO2 atmosphere for 4 days. After incubation, the supernatant was discarded, and total RNA was extracted from cells using RNeasy 96 kit from Qiagen (Valencia, CA). The extracted RNA was amplified as described by Stuyver et al. (14). The ΔCt values for HCV RNA were determined, and the EC90 was calculated for each sample. A “no drug” (medium only) control was used to determine maximum amount of HCV RNA.
Quantitative RT-PCR for CES1, CatA, and Hint1
The mRNA levels for CES1, CatA, and Hint1 in primary human hepatocytes from two donors (HHPC-1 and HHPC-2) and clone A cells were determined using RT-PCR. Total RNA was extracted from the cells using the Qiagen RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) was used to synthesize cDNA from 1 μg of total RNA. Quantitative RT-PCR was performed using TaqMan® Universal PCR MasterMix and quantified with an ABI 7500 real time PCR system. The TaqMan® gene expression assays used for CES1, CatA, and Hint1 were Hs00275607_ml, Hs00264902_m1, and Hs00602163_m1, respectively (Applied Biosystems, Foster City, CA). 18 S rRNA was used as an endogenous control. All experiments were performed in triplicate. Relative mRNA levels were calculated according to the ΔΔCt method and expressed as fold change relative to the HHPC-1 sample for the CatA/CES1 experiment and relative to the clone A sample for the Hint1 experiment based on the manufacturer's protocols (Applied Biosystems, Foster City, CA).
Western Blot Analysis
Protein extracts were prepared by incubating cells with preheated 1× NuPAGE® LDS sample buffer (Invitrogen) at 65 °C for 10 min. Equivalent amounts of protein extracts were loaded into each well of a 10% BisTris NuPAGE polyacrylamide gel and resolved by electrophoresis using a NuPAGE MOPS running buffer (Invitrogen). Proteins were transferred to a nitrocellulose membrane using an iBlotTM gel transfer device (Invitrogen). Blotting and antibody incubation were performed using the Snap ID protein detection system (Millipore, Billerica, MA) according to the manufacturer's protocol. Nitrocellulose membranes were blotted with SuperBlock T20 PBS blocking buffer (Thermo Scientific, Rockford, IL). Mouse monoclonal CatA, goat polyclonal CES1, and rabbit polyclonal Hint1 antibodies were diluted in blocking buffer, and the corresponding HRP-conjugated secondary antibodies were used. Blots were developed with SuperSignal West Dura extended duration substrate (Thermo Scientific), and the signal was detected using Gel Logic 2200 imaging system (Eastman Kodak).
Cloning, Expression, and Purification of Hint1 and Hint3
Human histidine triad nucleotide-binding proteins 1 and 3 (Hint1 and Hint3) were amplified from human fetal poly(A)+ RNA (Clontech) and ligated into pET17b containing a pre-inserted N-terminal FLAG (DYKDDDK) affinity-tagged Escherichia coli dihydrofolate reductase gene followed by a PreScission protease cleavable linker site (PS) (GE Healthcare) (15, 16). Human fetal poly(A)+ RNA was reverse-transcribed using a Superscript III first strand synthesis kit (Invitrogen) with random hexamers. Hint1 was amplified with forward primer 5′-GGG GCC AAG CTT TGC AGA TGA GAT TGC CAA GGC TCA GG and reverse primer 5′-CTA GTG GAT CCT TAA CCA GGA GGC CAA TGC ATT TGC CGA C. Hint3 was amplified with forward primer 5′-GGG GCC AAG CTT TGC GGA GGA ACA GGT GAA CCG CAG and reverse primer 5′-CTA GTG GAT CCT CAT GTT CTT AGT TTT TCA ATC AAG TGA TCA GCT GTG. Forward primers contained HindIII sites, and reverse primers contained BamHI sites for insertion into the pET17b-FLAG-ecDHFR-PS vector. The constructs were transformed into BL21-gold (DE3) E. coli (Stratagene, La Jolla, CA), and the cells were grown at 37 °C in LB medium containing 0.4% glucose and 100 μg/ml ampicillin until the absorbance at 600 nm reached 0.4, at which point IPTG (0.5 mm) was added. Cells were harvested after 2.5 h growth at 37 °C. The cell pellets were suspended in lysis buffer A (50 mm Tris/HCl, pH 8.0, 5 mm EDTA, 1 mg/ml lysozyme, and 50 μg/ml NaN3) at a ratio of 25 ml/g of cells. After 5 min of incubation at room temperature, lysis buffer B (1.5 m NaCl, 0.1 m CaCl2, 0.1 m MgCl2, 20 μg/ml DNase I, and 1 mm PMSF) was added at 2.5 ml/g cells (1 ml of lysis buffer B/10 ml of cell suspension). The lysate was incubated at room temperature for an additional 5 min followed by the addition of DTT (5 mm). The cell-free extract was collected by centrifugation at 15,000 rpm for 30 min at 4 °C. The supernatant was loaded onto a 1-ml methotrexate-agarose column (Sigma), and the column was washed with 100 ml of buffer C (20 mm Tris/HCl, pH 7.0, 1 mm EDTA, and 1 mm DTT), followed by 600 ml of buffer D (1 m NaCl in buffer C). The fusion protein was eluted with buffer E (buffer D containing 150 μm trimethoprim (Sigma)). All fractions were analyzed by 10% SDS-PAGE (Invitrogen), and fractions containing the fusion protein were combined. The salt concentration was reduced by diluting the protein solution with buffer C (∼7-fold). The dihydrofolate reductase tag was removed by treating with PreScission protease (10 units/mg, GE Healthcare). To run anion exchange column chromatography, the salt concentration of the protein solution was further reduced to <10 mm NaCl by concentrating the protein using an Amicon Ultra-4 centrifugal filter unit with an Ultracel-10 membrane from Millipore (Billerica, MA) and diluting the solution with buffer C. The protein was applied to a Mono Q 5/50GL column (GE Healthcare). The target protein was eluted in the flow-through with buffer C, although dihydrofolate reductase and proteases were retained on the column. The purity of the protein was determined by SDS-PAGE, and fractions containing the target protein were pooled. Glycerol was added to a final concentration of 10%, and the purified protein was aliquoted and stored at −80 °C.
Hint1 and Hint3 Assays
HPLC analysis was used to measure the enzymatic activity of human Hint1 and Hint3. Various concentrations of PSI-352707 were incubated with purified Hint1 or Hint3 in 50 mm Hepes, pH 7.0, and 1 mm MgCl2 buffer at 37 °C. At defined time points the reaction was stopped by shock-freezing in dry ice. All reaction mixtures were filtered through Microcon YM-10 concentrator (Millipore, Billerica, MA) to remove the protein. The products were separated using a Gemini 2 C-18 column. Elution was performed with a linear gradient of 100 to 50% acetonitrile containing 0.1% formic acid for 40 min. As a control, activity of the enzyme was tested using 100 μm AMP-NH2 (Sigma) under the same conditions as described above.
siRNA-mediated Gene Silencing
Huh7 cells were plated in 96-well plates (5 × 103 cells per well) and in 48-well plates (1.25 × 104 cells per well) and incubated overnight at 37 °C in a humidified 5% CO2 atmosphere in 100 μl of growth medium (DMEM containing 100 IU/ml penicillin/100 μg/ml streptomycin, and 10% fetal bovine serum). The next day, the growth medium was removed, and cells were washed in Accell delivery media, and Accell siRNA was delivered to the cells according to the manufacturer's protocol (Dharmacon, Thermo Scientific, Rockford, IL). Accell SMART pool human CTSA siRNA and human HINT1 siRNA were used for CatA or Hint1 silencing, respectively. Accell NonTargeting Pool siRNA was used as a negative control, and Accell GAPD siRNA was used as a positive control. An untreated sample was also included where the growth medium was changed to the Accell delivery medium (Dharmacon, Thermo Scientific, Rockford, IL), but no siRNA was added. Plates were incubated at 37 °C with 5% CO2. The siRNA-mediated knockdown experiments were performed in triplicate. Analysis of mRNA levels was preformed 96 h post siRNA delivery. Knockdown was calculated using untreated cells as reference.
After a 96-h incubation of the cells with the siRNA, the Accell delivery medium in 48-well plates was changed to growth medium containing 5 μm [3H]PSI-7851, and the Accell delivery medium in the 96-well plates was changed to growth medium. All plates were incubated for an additional 24 h. Following the incubation, the cells from 96-well plates were collected for Western blot and RT-PCR analyses, and the cells from the 48-well plates treated with radiolabeled compound were harvested and extracted as described above using methanol. Intracellular metabolites of PSI-7851 were identified and measured using HPLC as described above.
RESULTS
Metabolism of PSI-7851 in Clone A and Primary Human Hepatocytes
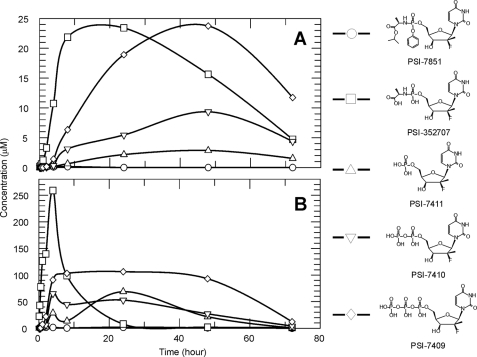
Intracellular concentrations of radiolabeled PSI-7851 and its metabolites in clone A HCV replicon cells or primary human hepatocytes were measured over 72 h by HPLC (Fig. 1). High concentrations of the intermediate metabolite, PSI-352707, formed rapidly in both cell types, followed by the formation of other metabolites suggesting that the conversion of PSI-352707 to PSI-7411 was the overall rate-limiting step in the metabolism of PSI-7851 to the active triphosphate metabolite, PSI-7409. In clone A cells, the levels of PSI-7409 gradually increased to a maximum concentration of about 25 μm over a period of 48 h. PSI-7409 formed at a much faster rate in primary human hepatocytes, achieving a maximum intracellular concentration of ∼100 μm at 4 h and remained at that concentration for 48 h. Concentrations of PSI-7411 and PSI-7410 gradually increased for 48 h up to ∼3 and 9 μm, respectively, in clone A cells. In primary human hepatocytes, the highest levels of PSI-7411 and PSI-7410 were observed at 24 h (69 μm) and 4 h (65 μm), respectively.
FIGURE 1.
Metabolism of PSI-7851 in clone A and primary human hepatocytes. Clone A cells (A) or primary human hepatocytes (B) were treated with 5 μm 3H-labeled PSI-7851, and formation of the metabolites was followed up to 72 h using HPLC methodology. Cellular concentrations of PSI-7851 (○), PSI-352707 (□), PSI-7411 (△), PSI-7410 (▽), and PSI-7409 (♢) are shown.
Hydrolysis of the Carboxyl Ester of PSI-7851
The first step in the activation of PSI-7851 is the hydrolysis of the carboxyl ester linkage between the carboxylic acid from the alaninyl moiety and the isopropyl alcohol of PSI-7851. To identify the enzyme(s) responsible for this reaction, the hydrolysis of PSI-7851 was tested with various human proteases, carboxylesterases, and lipase (Table 1). Of the enzymes tested, only neutrophil elastase, CatA, and CES1 were able to hydrolyze PSI-7851. The target cell type of PSI-7851 is HCV-infected cells in the liver. Because neutrophil elastase is not a liver enzyme, it is unlikely that this enzyme contributes to the hydrolysis of PSI-7851 in liver cells. Therefore, we confined our study to CatA and CES1, which appear to be primarily involved in the initial step of activation of PSI-7851.
TABLE 1.
Hydrolysis of PSI-7851 by various enzymes
| Enzyme | Type | Source | Activity |
|---|---|---|---|
| nmol/min/μg | |||
| Trypsin | Serine protease | Pancreas | NDa |
| Chymotrypsin | Serine protease | Pancreas | ND |
| Chymase | Serine protease | Skin | ND |
| Elastase | Serine protease | Neutrophil | 6.5 |
| Cathepsin A | Serine protease | Recombinant | 27.0 |
| Cathepsin B | Cysteine protease | Liver | ND |
| Cathepsin D | Aspartic protease | Liver | ND |
| Cathepsin H | Cysteine protease | Liver | ND |
| Cathepsin L | Cysteine protease | Liver | ND |
| Carboxylesterase 1 | Serine esterase | Recombinant | 1.8 |
| Carboxylesterase 2 | Serine esterase | Recombinant | ND |
| Calpain 1 | Cysteine protease | Plasma | ND |
| Caspase 1–10 | Cysteine protease | Recombinant | ND |
| Lipase | Serine esterase | Pancreas | ND |
a ND means not detected after a 2-h incubation with 100 μm of PSI-7851.
Expression Levels of CatA and CES1 in Clone A and Primary Human Hepatocytes
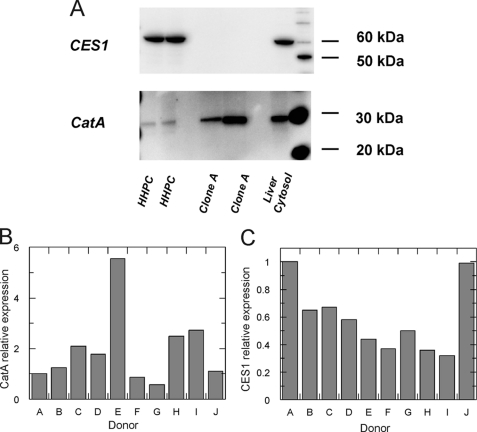
To assess the involvement of CatA and/or CES1 in the metabolism of PSI-7851, the mRNA and protein expression levels of CES1 and CatA in both subgenomic HCV replicon cells (clone A) and primary human hepatocytes (HHPC) were examined by quantitative RT-PCR and Western blot, respectively. Results indicated that the mRNA encoding CatA was expressed in both clone A and HHPC, whereas CES1 was expressed in HHPC but was undetectable in clone A cells (Table 2). The level of CatA expression in clone A cells was ∼5 times higher than in HHPC. Western blot analysis was also performed to examine the levels of CatA and CES1 protein expression in both clone A cells and HHPC. Pooled human liver cytosol was used as a positive control. Protein extracts from HHPC or clone A cells were separated by SDS-PAGE followed by immunoblotting with CES1- and CatA-specific antibodies. The results confirmed that only CatA was expressed in clone A, although both CatA and CES1 were expressed in HHPC (Fig. 2A). Donor dependence of CatA and CES1 was assessed using liver cytosol from 10 different single donors (donors A–J). As shown in Fig. 2B, CatA expression was significantly different from donor to donor, and ∼10-fold difference was observed between the lowest (donor G) and the highest (donor E). CES1 expression was somewhat donor-dependent with ∼3-fold difference between the lowest (donor I) and the highest (donor A) (Fig. 2C).
TABLE 2.
Gene expression as fold changes relative to HHPC-1 cells
The results are presented as an average of triplicate experiments with the maximum and the minimum values in parentheses.
| Target gene | HHPC-1 | HHPC-2 | Clone A |
|---|---|---|---|
| CES1 | 1 | 1.49 (1.39–1.61) | <0.001 |
| CatA | 1 | 0.87 (0.79–0.96) | 4.91 (4.64–5.19) |
FIGURE 2.
Expression of CES1 and CatA in HHPC and clone A cells. Western blot analysis was performed in extracts of clone A replicon cells, primary human hepatocytes, and pooled human liver cytosol using CES1-specific (A, upper panel) and CatA-specific (A, lower panel) antibodies. The same amount of total protein was loaded on the gel for the Western blot analysis. CatA (B) and CES1 (C) expression levels in human liver cytosol from 10 different single donors were examined by Western blot and compared. The expression levels were normalized against the amount of total protein loaded on the gel. Results are shown as fold-change relative to donor A.
Effects of CatA and CES1 Inhibitors on the Metabolism of PSI-7851
We have demonstrated that CatA and CES1 were capable of hydrolyzing PSI-7851 in the enzyme assays, and both enzymes were expressed in primary hepatocytes, but only CatA was detected in clone A cells. These results suggested that the contribution of CatA and CES1 to the first step in PSI-7851 metabolism was different in clone A cells compared with primary hepatocytes. One way to assess the contribution of each enzyme to the metabolism of PSI-7851 was to monitor the intracellular metabolites in the presence of a specific inhibitor of CatA or CES1. Telaprevir (VX-950), a potent HCV NS3/4A protease inhibitor, was recently shown to inhibit chymotrypsin, chymase, elastase, and cathepsins F, K, L, S, and V in vitro (17). The effect on the enzymatic activity of CatA or CES1 was evaluated. Results showed that telaprevir inhibited CatA-mediated cleavage of the fluorogenic peptide substrate, (7-methoxycoumarin-4-yl)acetyl-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys-(2,4-dinitrophenyl)-OH, with an IC50 value of 0.21 μm, whereas telaprevir did not affect the ability of CES1 to hydrolyze 4-NPA (Table 3). BNPP, a known inhibitor of carboxylesterases (18, 19), inhibited the hydrolysis of 4-NPA by CES1 (IC50 = 0.11 μm) but did not inhibit the activity of CatA (Table 3).
TABLE 3.
Inhibition of CatA and CES1 by telaprevir or BNPP
| Enzyme | Substratea | IC50 |
|
|---|---|---|---|
| Telaprevir | BNPP | ||
| μm | |||
| CatA | Fluorogenic peptide | 0.21 ± 0.04 | >100 |
| CES1 | 4-NPA | >100 | 0.11 ± 0.06 |
a See “Experimental Procedures” for details.
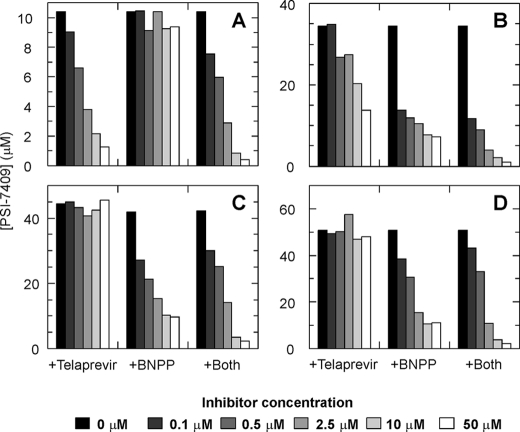
The effect of telaprevir and BNPP on the metabolism of PSI-7851 was studied by following the intracellular concentrations of the active triphosphate PSI-7409 in clone A cells and HHPC from three different donors (donors 1- 3) incubated with 5 μm [3H]PSI-7851. In the absence of inhibitors, ∼10 μm PSI-7409 was formed in clone A cells, whereas 35–50 μm PSI-7409 was observed in HHPC depending on the donor (Fig. 3). In clone A cells, telaprevir inhibited the formation of PSI-7409 in a dose-dependent manner, although BNPP did not affect the metabolism of PSI-7851 in these cells (Fig. 3A). Inhibition of PSI-7409 formation in the presence of both telaprevir and BNPP was similar to that in the presence of only telaprevir (Fig. 3A). These results indicated that CatA was the major enzyme hydrolyzing PSI-7851 in clone A cells. In HHPC, inhibition of PSI-7409 formation by telaprevir was different among the donors. Dose-dependent inhibition of PSI-7409 formation was seen in HHPC from donor 1 (Fig. 3B), and no inhibition by telaprevir was observed in HHPC from donors 2 and 3 (Fig. 3, C and D). BNPP inhibited the formation of PSI-7409 in a dose-dependent manner in HHPC from all three donors (Fig. 3, B–D). When the two inhibitors were combined, a similar inhibition pattern was observed compared with BNPP alone at lower concentrations of inhibitors, although a greater inhibition was seen at higher concentrations (Fig. 3, B–D).
FIGURE 3.
Effect of telaprevir and BNPP on formation of PSI-7409. Clone A (A) or primary human hepatocytes from three different donors 1–3 (B–D) were incubated with a CatA inhibitor, telaprevir, a CES1 inhibitor, BNPP, or both telaprevir and BNPP, and the formation of PSI-7409 was followed. The cellular concentrations of PSI-7409 at various concentrations of inhibitors are shown in the graphs.
Stereoselectivity of CatA and CES1
PSI-7851 contains a chiral phosphorous atom and therefore is a mixture of two diastereomers, PSI-7976 and PSI-7977. Both isomers form a common nonisomeric intermediate (PSI-352707) once the carboxyl ester is hydrolyzed. Therefore, the difference in the anti-HCV activity between the two isomers could be due to differences in the kinetics or the substrate specificity of the enzymes responsible for the first hydrolytic step. The ability of PSI-7976, PSI-7977, or PSI-7851 to inhibit HCV RNA replication in clone A replicon cells was measured. Results of this in vitro assay showed that PSI-7977 was a more potent inhibitor of HCV RNA replication than PSI-7976; the isomeric mixture, PSI-7851, demonstrated intermediate activity (Table 4).
TABLE 4.
Anti-HCV activity of PSI-7976, PSI-7977, and PSI-7851 in clone A replicon
| Compound | EC50 | EC90 |
|---|---|---|
| μm | μm | |
| PSI-7976 | 1.07 ± 0.04 | 2.99 ± 0.82 |
| PSI-7977 | 0.092 ± 0.005 | 0.29 ± 0.08 |
| PSI-7851 | 0.149 ± 0.001 | 0.51 ± 0.16 |
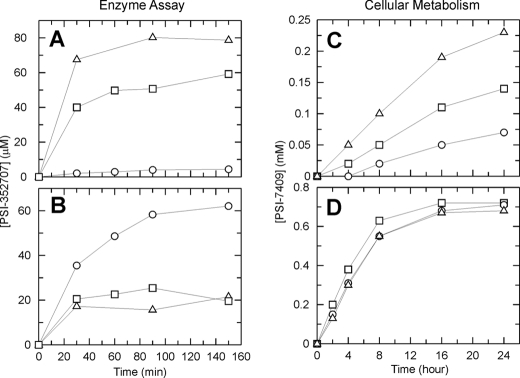
Because of the difference in potency between PSI-7976 and PSI-7977, we studied the hydrolysis of PSI-7976 and PSI-7977 by CatA and CES1 with respect to time (Fig. 4, A and B). Results showed that when CatA was incubated with PSI-7977 or PSI-7976 for 150 min, ∼18-fold more PSI-352707 was formed when PSI-7977 was the substrate compared with PSI-7976. Moreover, the catalytic efficiency for PSI-7977 with CatA was ∼30-fold higher than that for PSI-7976 (Table 5). The isomeric mixture, PSI-7851, demonstrated intermediate activity (Fig. 4A). These results indicate that CatA preferred PSI-7977 as a substrate over PSI-7976.
FIGURE 4.
Stereospecificity. Stereoselectivity was studied by enzyme assays (A and B) and cellular metabolism assays (C and D). PSI-352707 product formation was followed by incubating PSI-7851 (□), PSI-7976 (○), or PSI-7977 (△) with human recombinant CatA (A) or CES1 (B). Time-dependent formation of PSI-7409 (active triphosphate form) in clone A cells (C) or primary human hepatocytes (HHPC) (D) was by incubating with PSI-7851 (□), PSI-7976 (○), or PSI-7977 (△). See “Experimental Procedures” for the experimental conditions.
TABLE 5.
Kinetic parameters for PSI-7976 and PSI-7977 with CatA and CES1
| Enzyme | Compound | Km | kcat | kcat/Km |
|---|---|---|---|---|
| (μm) | s−1 | s−1μm−1 | ||
| CatA | PSI-7976 | 880 ± 430 | 0.30 ± 0.07 | 0.0003 |
| PSI-7977 | 700 ± 90 | 7.4 ± 0.4 | 0.0106 | |
| CES1 | PSI-7976 | 51 ± 11 | 0.27 ± 0.02 | 0.0053 |
| PSI-7977 | Not determineda | |||
a The kinetic parameters were not determined due to complex kinetics.
CES1 hydrolyzed both isomers and the isomeric mixture (Fig. 4B). However, the CES1-mediated hydrolysis of PSI-7977 and PSI-7851 did not progress in a time-dependent manner. When the enzyme was incubated with 100 μm PSI-7977 or PSI-7851, ∼20 μm PSI-352707 was formed within 30 min, and the formation of product no longer increased in a time-dependent manner (Fig. 4B). Because of this complex kinetic behavior, we were not able to determine the steady-state kinetic parameters for PSI-7977 (Table 5). On the other hand, PSI-7976 showed a time-dependent increase in product formation (Fig. 4B), and the kinetic parameters were determined (Table 5). These results indicate that CES1 preferentially hydrolyzes PSI-7976 over PSI-7977. The kinetic data also indicated that PSI-7976 is a better substrate for CES1 than for CatA (Table 5).
The intracellular concentration of PSI-7409 was also compared when clone A or HHPC were incubated with 50 μm PSI-7976, PSI-7977, or PSI-7851. As described above, CES1 expression was undetectable in clone A cells. Therefore, the concentration of PSI-7409 should be higher in clone A cells incubated with PSI-7977 because only CatA was expressed in clone A cells and PSI-7977 was the preferred substrate for CatA. As shown in Fig. 4C, incubating clone A cells with PSI-7977 resulted in a higher concentration of PSI-7409 than clone A cells incubated with PSI-7976. As expected, PSI-7851 gave an intermediate concentration of PSI-7409 in clone A cells. This result correlates well with the enzymatic studies (Fig. 4, A and C) and explains why PSI-7977 was the more potent inhibitor in the clone A HCV replicon assay (Table 4). The intracellular concentration of PSI-7409 was similar when HHPC were incubated with PSI-7976, PSI-7977, or PSI-7851 (Fig. 4D). Because both CES1 and CatA are expressed in HHPC, both enzymes should contribute to the activation of PSI-7976 and PSI-7977. Therefore all three compounds would be expected to generate similar concentrations of PSI-7409.
Conversion of PSI-352707 to PSI-7411 by Hint Proteins
Hint proteins belong to the HIT (histidine triad) protein superfamily and have been shown to be capable of catalyzing the deamination of adenosine 5′-monophosphoramide (AMP-NH2) to AMP (20). Because Hint proteins have been shown to possess phosphoramidase activity (15, 21, 22), recombinant human Hint1 and Hint3 were cloned, purified, and tested for the ability to convert PSI-352707 to the corresponding monophosphate, PSI-7411. The activity of purified Hint1 and Hint3 was first tested with the control compound AMP-NH2, and both enzymes were able to deaminate AMP-NH2 to AMP (data not shown). When PSI-352707 was used as the substrate, Hint1 was able to catalyze the conversion of PSI-352707 to PSI-7411, but the kinetic parameters (kcat and Km) could not be determined because the Km value for PSI-352707 appeared to be higher than the highest concentration of PSI-352707 tested (1.5 mm). Conversion of PSI-352707 to PSI-7411 by Hint3 was not detected when the enzyme was incubated with 1 mm PSI-352707 at 37 °C for more than 12 h.
Expression of Hint1
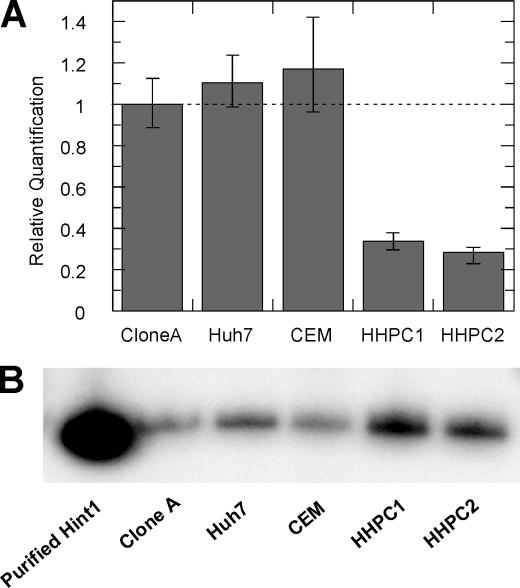
Because Hint1 appeared to be involved in the metabolic pathway of PSI-7851 and its isomers, the gene and protein expression of Hint1 was examined in various cell types. We compared Hint1 gene expression levels in clone A replicon cells (Huh7-derived), a hepatoma cell line (Huh7), a human T-lymphoblast leukemia cell line (CEM), and HHPC from two different donors. Results showed that Hint1 mRNA was expressed in the different culture cell lines and in HHPC (Fig. 5A). The mRNA expression level in HHPC was ∼30% of that in clone A. Hint1 protein expression was also examined by Western blot (Fig. 5B). The Hint1 protein was detected in all four cell types with slightly higher amounts in HHPC.
FIGURE 5.
Enzyme activity and gene and protein expression of Hint1. A, relative Hint1 mRNA expression levels in clone A, Huh7, CEM, and primary human hepatocytes from two different donors determined by RT-PCR. B, Western blot showing expression of Hint1 protein in extracts from clone A, Huh7, CEM, and primary human hepatocytes from two different donors. The same amount of protein was loaded on the gel for the Western blot analysis.
siRNA-mediated Gene Silencing
As shown above, Hint1 was able to catalyze deamination of PSI-352707 to give PSI-7411 and was expressed in various cell types, including Huh7 and HHPC. It is likely that Hint1 is involved in PSI-7851 metabolism in the cells. To confirm that Hint1 is involved in the metabolism of PSI-7851 and its isomers, knockdown experiments were performed with Huh7 cells using a Hint1-specific siRNA. A similar experiment was also performed using CatA-specific siRNA to examine the role of CatA in the metabolism of PSI-7851. Delivery of CatA or Hint1 siRNA into cells resulted in reduced CatA or Hint1 mRNA expression, respectively (Fig. 6A). The mRNA levels were quantified by RT-PCR, and expression of both genes was reduced by 85–95% (Fig. 6A). CatA siRNA treatment did not affect Hint1 mRNA expression, and Hint1 siRNA treatment had no effect on CatA gene expression (Fig. 6A). CatA and Hint1 gene silencing in the siRNA-treated cells was also confirmed at the protein level by Western blot analysis using specific antibodies against the targeted proteins. As shown in Fig. 6B, the level of CatA and Hint1 protein expression appeared to be significantly decreased in the corresponding siRNA-treated cells although the level of GAPDH remained unchanged.
FIGURE 6.
siRNA gene silencing. A, quantification of CatA or Hint1 mRNA expression in extracts of Huh7 cells treated with CatA siRNA, Hint1 siRNA, or only delivery media (DM). Gene expression levels relative to the DM control are shown in the graph. B, Western blot analyses of expression of CatA or Hint1 in the extract from Huh7 cells treated with CatA- or Hint1-specific siRNAs. The control experiment was done using the extract from the siRNA-untreated cells. The effect of the siRNAs on expression of GAPDH was also examined. C, amounts of PSI-352707 or total metabolites in the CatA or Hint1 knocked down cells treated with 5 μm [3H]PSI-7851 for 24 h.
To determine the effects of CatA and Hint1 depletion on PSI-7851 metabolism, siRNA-treated Huh7 cells were incubated with [3H]PSI-7851 for 24 h. An HPLC analysis of the extracts prepared from the CatA-depleted cells showed that the concentrations of both PSI-352707 and total metabolites were reduced by ∼50% compared with untreated cells (Fig. 6C). In the Hint1-depleted cells, the amount of PSI-352707 was ∼1.5-fold higher than in the untreated cells, whereas the amount of the total metabolites was similar to that in the untreated cells. In other words, the decrease in the amount of CatA slowed the hydrolysis of PSI-7851 and the formation of PSI-352707. Decreasing the amount of Hint1 in cells resulted in a slowing of the deamination of PSI-352707, which led to an increase of this metabolite. This result indicates that Hint1 is, at least in part, involved in converting PSI-352707 to PSI-7411.
DISCUSSION
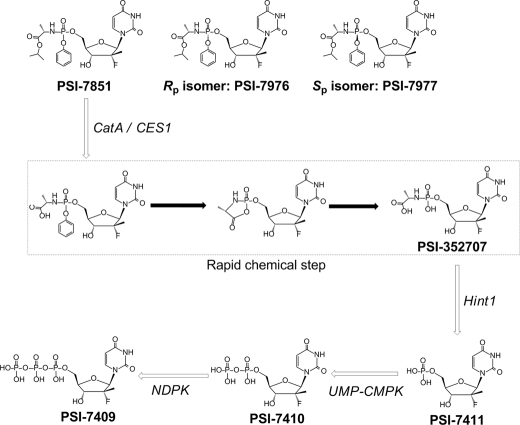
PSI-7851, a mixture of two diastereoisomers (PSI-7977 and PSI-7976), is a potent phosphoramidate prodrug of 2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine-5′-monophosphate. To inhibit the HCV NS5B RNA polymerase, these prodrugs must be metabolized to the active 5′-triphosphate form. In this study, enzymes involved in the metabolism of PSI-7851 and its diastereoisomers were identified. As illustrated in Fig. 7, the proposed metabolic pathway in primary human hepatocytes involves four enzymatic steps and one nonenzymatic chemical step.
FIGURE 7.
Proposed metabolic pathway for PSI-7851 and its diastereoisomers.
The first step in the metabolism of phosphoramidate prodrugs of nucleoside monophosphate analogs involves the hydrolysis of the carboxyl ester moiety (23, 24). Examination of 23 different enzymes, including serine proteases, cysteine proteases, aspartic protease, and serine esterases, resulted in the identification of two serine proteases, cathepsin A and neutrophil elastase, and a serine esterase, carboxylesterase 1 that were capable of hydrolyzing the carboxyl ester of PSI-7851. Because neutrophil elastase is not a liver enzyme and therefore not likely to be involved in PSI-7851 activation in the liver, further studies were performed with CatA and CES1. The hydrolysis of PSI-7851 by CatA was about 15-fold better than by CES1. Indeed, CatA has been reported to be the major enzyme hydrolyzing the nucleotide phosphonoamidate prodrugs, GS-7340 and GS-9131, which are inhibitors of HIV (25). Although the activity for CES1 was lower than that for CatA, the involvement of this enzyme in the metabolism of PSI-7851 and its isomers was considered to be important because this enzyme was present in primary human hepatocytes. RT-PCR and Western blot experiments demonstrated that both CatA and CES1 were expressed in primary human hepatocytes although CES1 expression in clone A cells was undetectable. Clone A cells are derived from Huh7 cells, a hepatoma cell line that is permissive for HCV replicon replication (26). The suppression of CES1 may be common in hepatoma cell lines; Yang et al. (27) have previously reported that HepG2 cells, another hepatoma cell line, expressed much lower levels of CES1 than primary hepatocytes.
Telaprevir, which was found to be an inhibitor of CatA, inhibited the metabolism of PSI-7851 in clone A. No inhibition of PSI-7409 formation was observed in primary human hepatocytes from two out of three donors tested. Inhibition of PSI-7409 formation by telaprevir was observed in primary human hepatocytes from donor 1, but the degree of inhibition was weaker than that seen in clone A (Fig. 3B). This donor dependence may be due to differential expression of CES1 and CatA. As shown in Fig. 2, CatA and CES1 expression varied in human liver cytosol from different donors. In addition, CES1 expression was previously shown to vary individually (28, 29). Therefore, CatA and/or CES1 expression in primary human hepatocytes from donor 1 may be different from donors 2 and 3, which may result in differences in the relative contribution of these two enzymes in hydrolyzing PSI-7851. Although inhibition of PSI-7409 formation by telaprevir was seen in primary hepatocytes from only one out of three donors, it was important to determine whether telaprevir affects the activity of PSI-7851 or its isomer, PSI-7977, given the clinical potential for combination antiviral therapy. Forestier et al. (30) reported pharmacokinetics of telaprevir at clinically relevant dosing; Cmax values were 3696 ng/ml (5.4 μm) and 3391 ng/ml (5.0 μm) when the patients were treated with telaprevir alone or telaprevir in combination with peginterferon alfa-2a, respectively (30). Our results showed ∼20 and 14 μm PSI-7409 was formed in the presence of 10 μm (∼2-fold the Cmax) and 50 μm (∼10-fold the Cmax) telaprevir in primary human hepatocytes (Fig. 3B). We have reported previously that PSI-7409 is a potent inhibitor of NS5B RdRp with Ki value of 0.42 μm. Therefore, even in the presence of telaprevir at 10-fold the Cmax, the concentration of PSI-7409 in primary human hepatocytes was greater than 30 times the Ki of PSI-7409 for the NS5B RdRp. For that reason, our in vitro results suggest that it is unlikely that co-dosing of telaprevir would affect the antiviral activity of PSI-7851 or its diastereoisomer, PSI-7977, in the liver. Inhibition of PSI-7409 formation in primary human hepatocytes by the combination of telaprevir and BNPP was complex because at lower concentrations, the levels of inhibition were similar to BNPP alone, although at higher concentrations, greater inhibition was observed compared with BNPP alone. Because CES1 is highly expressed in primary hepatocytes, PSI-7851 could be hydrolyzed predominantly by CES1 and thus inhibition of PSI-7409 followed a dose-dependent manner similar to BNPP alone at low concentrations. However, when BNPP concentration was increased to a certain level, CES1 might be highly inhibited so that the contribution of CatA in PSI-7851 hydrolysis might become significant resulting in a greater inhibition of PSI-7409 formation in the presence of high concentrations of the two inhibitors compared with BNPP alone. These results clearly demonstrated that CatA was primarily responsible for the hydrolysis of PSI-7851 in clone A replicon cells, although both CatA and CES1 were involved in hydrolyzing PSI-7851 in primary human hepatocytes.
PSI-7851 is a mixture of two phosphate diastereoisomers, PSI-7976 and PSI-7977. Interestingly, our enzyme studies showed that CatA preferentially hydrolyzes PSI-7977, although CES1 prefers PSI-7976 as a substrate. The stereo-selective hydrolysis of an acyclovir phosphoramidate prodrug was previously demonstrated by using carboxypeptidase Y, a structural homolog of cathepsin A (31, 32). A computer model of carboxypeptidase Y docked with a phosphoramidate prodrug in the active site suggested that positioning of the carbonyl moiety of the carboxyl ester with the R-phosphate diastereoisomer was more preferable for catalysis than with the S-diastereoisomer (31). However, they failed to identify which diastereoisomer was preferentially hydrolyzed by carboxypeptidase Y in their biochemical assay. Our results indicated that the Sp diastereoisomer (PSI-7977) of PSI-7851 is a 35-fold better substrate for CatA than the Rp diastereoisomer (PSI-7976). Several studies have also demonstrated that CES1-mediated hydrolysis of various substrates were also stereo-selective (33–35). In our enzyme assays, the two diastereoisomers (PSI-7976 and PSI-7977) and the isomeric mixture (PSI-7851) were hydrolyzed by CES1, but the kinetics of the reaction with PSI-7977 and PSI-7851 were complex. As shown in Fig. 4B, incubating CES1 with PSI-7977 or PSI-7851 resulted in unusual kinetics. A small fraction of the substrate was converted (∼20%) to the product within 30 min indicating that PSI-7977 was a substrate for the enzyme; however, additional product formation was not observed upon further incubation. Currently, we do not fully understand the mechanism of this kinetic behavior. Our working hypothesis is that PSI-7977 and PSI-7976 bind to the enzyme in two different orientations as follows: a productive or a nonproductive orientation. It is possible that PSI-7977 is preferentially binding in the nonproductive orientation and forms a dead-end complex. Further mechanistic studies are ongoing.
Recently chosen for further clinical development, the Sp diastereoisomer PSI-7977 is the more active isomer when compared with PSI-7976 in the clone A replicon-based assay. Because clone A cells express only CatA and not CES1, the difference in anti-HCV activity between the two isomers is likely due to poor expression of CES1 and the stereo-specificity of CatA for PSI-7977. Indeed, intracellular metabolism studies with clone A cells showed that incubation with PSI-7977 resulted in higher concentrations of PSI-7409. However, in primary hepatocytes, similar amounts of PSI-7409 were formed when these cells were incubated with PSI-7851 or its isomers. This is because both CatA and CES1 were present in primary hepatocytes, and these enzymes were responsible for hydrolyzing PSI-7851 and its isomers. These results suggest that the anti-HCV activities of PSI-7851, PSI-7976, and PSI-7977 could be similar in human liver cells infected with HCV. Screening for anti-HCV agents is typically performed in HCV replicon cells derived from Huh7 cells where CES1 expression is significantly lower than primary hepatocytes. Thus, the anti-HCV activity of phosphoramidate compounds in the replicon-based screening system may not directly correlate with their activity in vivo.
The second step within the metabolic pathway is a nonenzymatic rapid chemical reaction. Once the carboxyl ester is hydrolyzed, the phenol group in the phosphate moiety is released spontaneously by a nucleophilic attack of the free carboxyl group at the phosphate, resulting in the formation of the alaninyl phosphate intermediate (PSI-352707). This chemical reaction has been described previously (36). The third step in the metabolism involves deamination of PSI-352707 to release alanine to form PSI-7411, a monophosphate intermediate. Our enzyme studies demonstrated that Hint1 was able to convert PSI-352707 to PSI-7411 but Hint3 was not. Because binding of PSI-352707 to Hint1 was very weak, we were not able to determine the kinetic parameters for PSI-352707 with this enzyme (Km >1 mm). Chou et al. (21) have studied Hint1 substrate specificity using different phosphoramidates and phosphoramidothiolates, and the reported Km values ranged from 0.13 to 215 μm. The weak binding affinity for PSI-352707 may be due to the 2′-fluoro-2′-methyl disubstitution on the sugar moiety.
Hint1 is expressed in various cell types, including clone A, Huh7, CEM, and primary human hepatocytes. The involvement of Hint1 in the metabolism of PSI-7851 and its diastereoisomers was further confirmed by knockdown studies using siRNA, which resulted in an increased intracellular concentration of PSI-352707, indicating that the Hint1-mediated deamination step was blocked. The total level of metabolites was not affected by Hint1 depletion. This result suggests that Hint1 was, at least in part, involved in the conversion of PSI-352707 to PSI-7411. Gene silencing experiments were also performed using CatA-specific siRNA. When CatA was knocked down, the level of PSI-352707 was reduced by ∼2-fold indicating that CatA was the major enzyme that hydrolyzes PSI-7851 to PSI-352707 in Huh7 cells. The total amount of PSI-7851 and its metabolites also decreased in the CatA-depleted cells, possibly the result of blocking the hydrolysis of the carboxyl ester of PSI-7851 and egress of PSI-7851 and its metabolites out of the cells.
The final two steps leading to the formation of the active 5′-triphosphate form are phosphorylation events catalyzed by cellular kinases. We demonstrated previously that PSI-7411 is phosphorylated to PSI-7410 by UMP-CMP kinase and PSI-7410 is subsequently metabolized to PSI-7409 by nucleoside diphosphate kinase (12). PSI-7409 was shown to be a potent inhibitor of HCV NS5B RNA polymerase with a Ki value of 0.42 μm (12).
In summary, our studies have determined the metabolic pathway and identified enzymes involved in activating PSI-7851 and its diastereoisomers. The first step, hydrolysis of the carboxyl ester, is a stereospecific reaction catalyzed by CatA and CES1 in primary human hepatocytes. However, it is possible that the other enzymes also play a role in this step. The second step involves a rapid chemical reaction of the hydrolyzed product to form PSI-352707, the common phosphonamide intermediate for all three compounds. The third step can be catalyzed by Hint1, which converts PSI-352707 to PSI-7411, the 5′-monophosphate form. The fourth and fifth steps are phosphorylation reactions mediated by UMP-CMP kinase and nucleoside diphosphate kinase, which converts PSI-7411 to PSI-7410 and PSI-7410 to PSI-7409, respectively (12). Stereospecificity of the two isomers occurred at the first hydrolysis step, with PSI-7977 being a better substrate for CatA and PSI-7976 for CES1. Once the carboxyl ester moiety of these compounds is hydrolyzed, the remaining steps leading to the formation of PSI-7409 are identical. In addition, our studies showed that among the enzymes involved in the metabolic pathway (CatA, CES1, and Hint1), levels of mRNA and protein expression could be different between immortalized cell lines and primary human hepatocytes. Nevertheless, the formation of the active triphosphate was almost identical between the two diastereoisomers in primary human hepatocytes suggesting that both isomers will be equally active in the liver cells, where infection of HCV takes place. The in vitro antagonistic drug-drug interactions with telaprevir, an HCV protease inhibitor telaprevir currently in phase 3 studies, are unlikely in clinical studies given the duplicity of enzymatic pathways available for delivery of the triphosphate intracellularly. Overall, the results from this study characterize the intracellular events involved in activating PSI-7851 and its isomers and provide a better understanding of how these nucleotide analogs are metabolized to their active forms that ultimately inhibit HCV replication.
Footnotes
- HCV
- hepatitis C virus
- CatA
- cathepsin A
- RdRp
- RNA-dependent RNA polymerase
- BNPP
- bis(4-nitrophenyl)phosphate
- 4-NPA
- p-nitrophenylacetate
- HHPC
- primary human hepatocytes
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Vivet-Boudou V., Didierjean J., Isel C., Marquet R. (2006) Cell. Mol. Life Sci. 63, 163–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hewlett G., Hallenberger S., Rübsamen-Waigmann H. (2004) Curr. Opin. Pharmacol. 4, 453–464 [DOI] [PubMed] [Google Scholar]
- 3.Naesens L., De Clercq E. (2001) Herpes 8, 12–16 [PubMed] [Google Scholar]
- 4.De Clercq E. (2007) Nat. Rev. Drug Discov. 6, 1001–1018 [DOI] [PubMed] [Google Scholar]
- 5.Férir G., Kaptein S., Neyts J., De Clercq E. (2008) Rev. Med. Virol. 18, 19–34 [DOI] [PubMed] [Google Scholar]
- 6.Kwong A. D., McNair L., Jacobson I., George S. (2008) Curr. Opin. Pharmacol. 8, 522–531 [DOI] [PubMed] [Google Scholar]
- 7.Furman P. A., Lam A. M., Murakami E. (2009) Fut. Med. Chem. 1, 1429–1452 [DOI] [PubMed] [Google Scholar]
- 8.Carroll S. S., Olsen D. B. (2006) Infect. Disord. Drug Targets 6, 17–29 [DOI] [PubMed] [Google Scholar]
- 9.Stuyver L. J., McBrayer T. R., Tharnish P. M., Clark J., Hollecker L., Lostia S., Nachman T., Grier J., Bennett M. A., Xie M. Y., Schinazi R. F., Morrey J. D., Julander J. L., Furman P. A., Otto M. J. (2006) Antivir. Chem. Chemother. 17, 79–87 [DOI] [PubMed] [Google Scholar]
- 10.Ma H., Jiang W. R., Robledo N., Leveque V., Ali S., Lara-Jaime T., Masjedizadeh M., Smith D. B., Cammack N., Klumpp K., Symons J. (2007) J. Biol. Chem. 282, 29812–29820 [DOI] [PubMed] [Google Scholar]
- 11.Murakami E., Bao H., Ramesh M., McBrayer T. R., Whitaker T., Micolochick Steuer H. M., Schinazi R. F., Stuyver L. J., Obikhod A., Otto M. J., Furman P. A. (2007) Antimicrob. Agents Chemother. 51, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami E., Niu C., Bao H., Micolochick Steuer H. M., Whitaker T., Nachman T., Sofia M. A., Wang P., Otto M. J., Furman P. A. (2008) Antimicrob. Agents Chemother. 52, 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duarte M. I., Andrade H. F., Jr., Mariano O. N., Corbett C. E., Sesso A. (1989) J. Submicrosc. Cytol. Pathol. 21, 275–279 [PubMed] [Google Scholar]
- 14.Stuyver L. J., McBrayer T. R., Tharnish P. M., Hassan A. E., Chu C. K., Pankiewicz K. W., Watanabe K. A., Schinazi R. F., Otto M. J. (2003) J. Virol. 77, 10689–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou T. F., Cheng J., Tikh I. B., Wagner C. R. (2007) J. Mol. Biol. 373, 978–989 [DOI] [PubMed] [Google Scholar]
- 16.Chou T. F., Tikh I. B., Horta B. A., Ghosh B., De Alencastro R. B., Wagner C. R. (2007) J. Biol. Chem. 282, 15137–15147 [DOI] [PubMed] [Google Scholar]
- 17.Liverton N. J., Carroll S. S., Dimuzio J., Fandozzi C., Graham D. J., Hazuda D., Holloway M. K., Ludmerer S. W., McCauley J. A., McIntyre C. J., Olsen D. B., Rudd M. T., Stahlhut M., Vacca J. P. (2010) Antimicrob. Agents Chemother. 54, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Block W., Arndt R. (1978) Biochim. Biophys. Acta 524, 85–93 [DOI] [PubMed] [Google Scholar]
- 19.Imai T. (2006) Drug Metab. Pharmacokinet. 21, 173–185 [DOI] [PubMed] [Google Scholar]
- 20.Brenner C. (2002) Biochemistry 41, 9003–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou T. F., Baraniak J., Kaczmarek R., Zhou X., Cheng J., Ghosh B., Wagner C. R. (2007) Mol. Pharm. 4, 208–217 [DOI] [PubMed] [Google Scholar]
- 22.Chou T. F., Bieganowski P., Shilinski K., Cheng J., Brenner C., Wagner C. R. (2005) J. Biol. Chem. 280, 15356–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuigan C., Sutton P. W., Cahard D., Turner K., O'Leary G., Wang Y., Gumbleton M., De Clercq E., Balzarini J. (1998) Antivir. Chem. Chemother. 9, 473–479 [DOI] [PubMed] [Google Scholar]
- 24.McGuigan C., Tsang H. W., Sutton P. W., De Clercq E., Balzarini J. (1998) Antivir. Chem. Chemother. 9, 109–115 [DOI] [PubMed] [Google Scholar]
- 25.Birkus G., Wang R., Liu X., Kutty N., MacArthur H., Cihlar T., Gibbs C., Swaminathan S., Lee W., McDermott M. (2007) Antimicrob. Agents Chemother. 51, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blight K. J., Kolykhalov A. A., Rice C. M. (2000) Science 290, 1972–1974 [DOI] [PubMed] [Google Scholar]
- 27.Yang J., Shi D., Yang D., Song X., Yan B. (2007) Mol. Pharmacol. 72, 686–694 [DOI] [PubMed] [Google Scholar]
- 28.Crow J. A., Borazjani A., Potter P. M., Ross M. K. (2007) Toxicol. Appl. Pharmacol. 221, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D., Pearce R. E., Wang X., Gaedigk R., Wan Y. J., Yan B. (2009) Biochem. Pharmacol. 77, 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forestier N., Reesink H. W., Weegink C. J., McNair L., Kieffer T. L., Chu H. M., Purdy S., Jansen P. L., Zeuzem S. (2007) Hepatology 46, 640–648 [DOI] [PubMed] [Google Scholar]
- 31.Derudas M., Carta D., Brancale A., Vanpouille C., Lisco A., Margolis L., Balzarini J., McGuigan C. (2009) J. Med. Chem. 52, 5520–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiraiwa M. (1999) Cell. Mol. Life Sci. 56, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H., Fleming C. D., Nishi K., Redinbo M. R., Hammock B. D. (2005) Chem. Res. Toxicol. 18, 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai T., Taketani M., Shii M., Hosokawa M., Chiba K. (2006) Drug Metab. Dispos. 34, 1734–1741 [DOI] [PubMed] [Google Scholar]
- 35.Sun Z., Murry D. J., Sanghani S. P., Davis W. I., Kedishvili N. Y., Zou Q., Hurley T. D., Bosron W. F. (2004) J. Pharmacol. Exp. Ther. 310, 469–476 [DOI] [PubMed] [Google Scholar]
- 36.Valette G., Pompon A., Girardet J. L., Cappellacci L., Franchetti P., Grifantini M., La Colla P., Loi A. G., Périgaud C., Gosselin G., Imbach J. L. (1996) J. Med. Chem. 39, 1981–1990 [DOI] [PubMed] [Google Scholar]