Abstract
Synthesis of the largest cytochrome c oxidase (CcO) subunit, Cox1, on yeast mitochondrial ribosomes is coupled to assembly of CcO. The translational activator Mss51 is sequestered in early assembly intermediate complexes by an interaction with Cox14 that depends on the presence of newly synthesized Cox1. If CcO assembly is prevented, the level of Mss51 available for translational activation is reduced. We deleted the C-terminal 11 or 15 residues of Cox1 by site-directed mutagenesis of mtDNA. Although these deletions did not prevent respiratory growth of yeast, they eliminated the assembly-feedback control of Cox1 synthesis. Furthermore, these deletions reduced the strength of the Mss51-Cox14 interaction as detected by co-immunoprecipitation, confirming the importance of the Cox1 C-terminal residues for Mss51 sequestration. We surveyed a panel of mutations that block CcO assembly for the strength of their effect on Cox1 synthesis, both by pulse labeling and expression of the ARG8m reporter fused to COX1. Deletion of the nuclear gene encoding Cox6, one of the first subunits to be added to assembling CcO, caused the most severe reduction in Cox1 synthesis. Deletion of the C-terminal 15 amino acids of Cox1 increased Cox1 synthesis in the presence of each of these mutations, except pet54. Our data suggest a novel activity of Pet54 required for normal synthesis of Cox1 that is independent of the Cox1 C-terminal end.
Keywords: Cytochrome Oxidase, Mitochondria, Protein Assembly, Translation Control, Yeast, Cox1, Cox14, Feedback Assembly, Mss51, Pet54
Introduction
Cytochrome c oxidase (CcO)2 is the terminal electron acceptor of the mitochondrial respiratory chain. It transfers electrons from cytochrome c to oxygen, with a coupled translocation of protons from the matrix to the intermembrane space. In the yeast Saccharomyces cerevisiae, this enzyme is composed of 11 subunits, three of which, Cox1, Cox2, and Cox3, are encoded by the mitochondrial genome, synthesized by organellar ribosomes, and integrated into the inner membrane from the matrix side. Assembly of this enzyme is very complex. It involves not only coordinated assembly of nuclear and mitochondrial encoded subunits, but the addition of metallic prosthetic groups like heme a and copper centers. For this process, more than 30 factors are necessary, although the functions of these proteins are just starting to be elucidated (1, 2).
The yeast model is widely used to study the mechanisms of CcO biogenesis, as several pathogenic mutations affecting CcO assembly have been described in human genes having yeast homologues. The majority of these encephalomyopathies are associated with mutations in nuclear genes encoding CcO assembly factors (3). In recent years some mutations associated with Leigh syndrome have been found in genes affecting expression of mitochondrial genes. This is the case for LRPPRC, a human protein distantly related to the yeast translational activator Pet309 (4–6), and TACO1, a gene specifically required for Cox1 synthesis (7).
Cox1 is the largest subunit of the CcO and spans the mitochondrial inner membrane 12 times (8). Cox1 contains the heme a and heme a3-CuB centers for oxygen reduction. Cox1 is present from the first assembly intermediate, and the rest of the subunits and cofactors are thought to be added in a sequential order (9, 10). Partial assembly of Cox1 is associated with peroxide sensitivity due to pro-oxidant intermediates containing unassembled heme a3 (11). Hence, stoichiometry of Cox1 in the inner membrane has to be highly regulated. For this, many factors have been identified that control Cox1 biogenesis. Pet309 and Mss51 are specific translational activators that function through the COX1 mRNA 5′-UTR (untranslated region) (4, 12). In addition, Mss51 physically interacts with Cox1, suggesting that it has a central role in coordinating the synthesis and assembly of this subunit (13, 14). Cox1 and Mss51 form a high molecular complex with Cox14. Next, Coa1 could insert into this complex (15). Although Mss51 and Coa1 are proposed to be liberated from this complex at early steps (16, 17), Cox14 might remain associated to the assembling CcO until the formation of supercomplexes (16).
The current model proposes that Mss51 limits translational activation of the COX1 mRNA, and is sequestered from this function by its incorporation into assembly-intermediate complexes containing newly synthesized Cox1 and Cox14. In CcO assembly mutants, Mss51 is trapped in these complexes and thus unavailable for efficient COX1 mRNA translational activation (12, 14).
Pulse labeling of Cox1 in vivo with [35S]methionine is specifically reduced in several mutations affecting CcO assembly (for examples, see Refs. 14, 18, and 19). Lower levels of Cox1 labeling have even been documented for mutations affecting the ATP synthase (20, 21) and the CcO substrate cytochrome c (22). This reduction in Cox1 labeling is presumed to be due to decreased COX1 mRNA translation. Here, through mutagenesis of the mitochondrial COX1 gene we have found that the C-terminal domain of Cox1 is necessary for assembly-coupled translational down-regulation. Absence of Cox6, one of the first subunits to be added to the CcO, showed one of the most dramatic C-terminal end-dependent reductions of Cox1 synthesis. In addition, we report that Pet54 is a new component required for normal COX1 mRNA translation. A mutation in Pet54 seems to reduce Cox1 synthesis by a mechanism that is independent of the Cox1 C-terminal end.
EXPERIMENTAL PROCEDURES
Strains and Genetic Methods
S. cerevisiae strains used in this study, all congenic or isogenic to D273-10B (ATCC 24657), are listed under supplemental Table S1. Genetic methods and media were as previously described (23, 24). Complete fermentable media were YPD or YPGal (containing 2% glucose or 2% galactose). Non-fermentable medium was YPEG (3% glycerol, 3% ethanol). Minimal medium was synthetic complete (0.67% yeast nitrogen base, 2% glucose) lacking the indicated amino acids. The nuclear deletion constructs with KanMX4, LEU2, or URA3 cassettes were obtained by PCR. Plasmids carrying the cox1 mutations were transformed into rho0 strain NAB69 by high-velocity microprojectile bombardment (25). Transformants were selected by their ability to rescue respiratory growth when mated with a strain carrying a Cox1 D369N mutation, L45 (26). Transformants were mated with XPM10b (containing the cox1Δ::ARG8m construct) or XPM13a (containing the cox2-62 and the cox1Δ::ARG8m construct) (13). Cytoductants were selected for their ability to grow on YPEG as haploids or after mating to a strain with the mutation G253D (27). In all cases, correct integration of the different constructs into the mtDNA was confirmed by PCR and DNA sequencing.
Construction of the cox1 Mutant Genes
Plasmid pXPM57, containing the full-length, intronless COX1 gene was used as template for PCR amplifications. This plasmid contains 395 and 990 nucleotides of the COX1 5′-UTR and 3′-UTR, respectively, and was cloned in the XbaI-XhoI sites from pBluescript (Stratagene). All cox1 mutant plasmids were generated by the fusion PCR technique (28) using Accuzyme (Bioline). The 3′ half of the COX1 coding region was amplified with primers that incorporated the mutations. These products were digested with NdeI and AflII and ligated into pXPM57 equally digested. Plasmids were sequenced to confirm the presence of the desired mutations in COX1.
Analysis of Mitochondrial Proteins
Yeast cells were grown in complete or minimal galactose media until late log phase. Crude mitochondria were obtained by disruption of cells with glass beads or by zymoliase 20T treatment as described (29). Proteins were separated by SDS-PAGE on a 16% gel (30), and Western blots were probed with antibodies to HA (Roche Applied Science), c-Myc (Roche), or citrate synthase. Secondary goat anti-mouse or anti-rabbit (Sigma) conjugated to horseradish peroxidase was detected with the ECL kit (GE Healthcare).
In vivo pulse labeling of cells with [35S]methionine was performed as previously described (31). After pulse labeling, cells were chilled on ice and disrupted by vortexing with glass beads to obtain mitochondria (29). The radiolabeled proteins were separated on a 16% polyacrylamide gel and transferred to a PVDF membrane before they were analyzed with a Typhoon 8600 Phosphorimager (GE Healthcare).
Translation in isolated mitochondria (3 mg of protein/ml) in the presence of [35S]methionine was performed as previously described (32). After translation, mitochondria were washed with 0.6 m sorbitol, 20 mm HEPES, pH 7.4, and lysed with a buffer containing 100 mm NaCl, 20 mm Tris, pH 7.4, and either 1% digitonin (w/v) or 1% dodecyl maltoside (w/v). Immunoprecipitation of labeled mitochondrial products with an HA-specific antibody coupled to protein A-agarose (Invitrogen) or Myc-specific antibody coupled to protein A-agarose (Santa Cruz) was performed according to the provider instructions. Proteins were analyzed as described for in vivo labeling experiments.
RESULTS
The Carboxyl-terminal End of Cox1 Is Required for Assembly-mediated Reduction of Cox1 Synthesis
Synthesis of Cox1 is reduced in several mutants affecting CcO assembly. Mss51 has a central role in this process by interacting with the Cox1 protein (13, 14).
We explored which regions of Cox1 might be involved in regulating synthesis. Based on the bovine CcO crystallographic structure (8), a model for the yeast Cox1 structure was generated using the program SWISS MODEL (33). The largest hydrophilic portion of Cox1 was the C-terminal region, comprising 59 amino acids, exposed on the matrix side of the inner membrane. This domain has an extended secondary structure that turns and covers the bottom of Cox1 (Fig. 1A). We reasoned that this portion of Cox1 could contain sites for interaction of peripheral membrane proteins like Mss51.
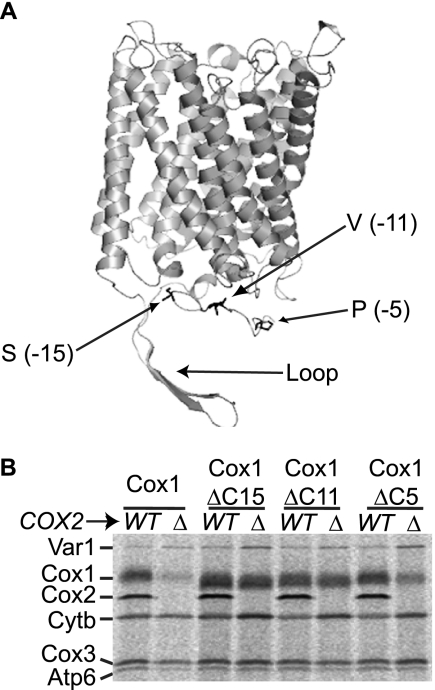
FIGURE 1.
The Cox1 carboxyl-terminal end is required to down-regulate Cox1 synthesis in a cox2Δ mutant. A, model of the S. cerevisiae Cox1 protein, based on the crystal structure of the bovine CcO. The model was constructed using SWISS MODEL, and visualized with MacPymol. Alignment of the yeast and bovine Cox1 sequences revealed that S. cerevisiae has ∼23 additional residues in the C-terminal region, from Lys483 to Asn505, which are located at −52 to −29 with respect to the C-terminal end of Cox1 (Loop). Arrows indicate residues on the Cox1 C-terminal end where deletions start. The number in parentheses indicates the position of these residues with respect to the last amino acid of Cox1. B, mitochondrial translation products were labeled with [35S]methionine in the presence of cycloheximide, and proteins were analyzed as described under “Experimental Procedures.” Cells carried either the wild-type Cox1 protein (Cox1), or the Cox1 protein lacking 15 (Cox1ΔC15), 11 (Cox1ΔC11), or 5 (Cox1ΔC5) amino acids of the carboxyl-terminal end. The cox2Δ mutation (Δ) was introduced as indicated. Abbreviations are as follows: cytochrome c oxidase subunit 1, Cox1; subunit 2, Cox2; subunit 3, Cox3; cytochrome b, Cytb; subunit 6 of ATPase, Atp6; and the ribosomal protein, Var1.
By site-directed mutagenesis and mitochondrial transformation we deleted codons encoding the last 5 (Cox1ΔC5; PAVQS), 11 (Cox1ΔC11; VHSFNTPAVQS), or 15 (Cox1ΔC15; SPPAVHSFNTPAVQS) residues from the C-terminal portion of Cox1. The three mutant strains retained the ability to grow on non-fermentable YPEG medium at levels comparable with wild-type (data not shown), demonstrating that these Cox1 variants are assembled into active CcO.
To disrupt assembly, we first introduced a mitochondrial cox2 deletion (cox2-62) (34) into mtDNA encoding the truncated forms of Cox1. Mitochondrial translation products were pulse-labeled in cells incubated with cycloheximide and [35S]methionine. As expected, in cells synthesizing wild-type Cox1, the cox2Δ mutation induced a dramatic reduction of Cox1 labeling (Fig. 1B). Similarly, synthesis of the Cox1ΔC5 protein was reduced by the cox2Δ mutation. In contrast, the synthesis of Cox1ΔC15 and Cox1ΔC11 was not reduced by the cox2Δ mutation. These data indicate that residues between −5 and −11 from the Cox1 C terminus, VHSFNT, are required to down-regulate Cox1 synthesis in a cox2 mutant.
The Last 15 Residues of Cox1 Facilitate the Interaction between Mss51 and Cox14
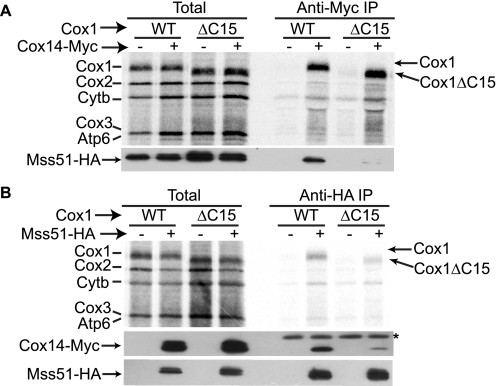
Assembly-mediated control of Cox1 synthesis involves sequestration of Mss51 in complexes containing Cox14 and newly synthesized Cox1. Interaction of Mss51 and Cox14 from wild-type mitochondrial extracts has been previously observed (14), and is known to be dependent upon synthesis of Cox1 (12). We therefore tested whether interactions among these components were affected by deletion of the last 15 residues of Cox1. We attached a triple Myc epitope to the C terminus of Cox14, and a triple hemagglutinin (HA) epitope to the C terminus of Mss51. The respiratory competence of the Mss51-HA, Cox14-Myc strains were comparable with wild-type levels, indicating that the tagged proteins were functional (data not shown). We first asked whether immunoprecipitation of Cox14-Myc would co-precipitate newly synthesized Cox1ΔC15. Mitochondria isolated from strains containing Cox14-Myc, Mss51-HA, and either wild-type Cox1 or Cox1ΔC15, were allowed to synthesize proteins in the presence of [35S]methionine. After solubilization in 1% digitonin, the mitochondrial extracts were immunoprecipitated with a Myc-specific antibody. The immunoprecipitated proteins were separated by SDS-PAGE and transferred to a PVDF membrane. Co-precipitation of newly synthesized Cox1 and Cox1ΔC15 with Cox14-Myc was equally efficient (Fig. 2A). However, probing the PVDF membrane with HA-specific antibody to detect Mss51-HA revealed that almost no Mss51-HA was co-precipitated with Cox14-Myc in the presence of the truncated Cox1ΔC15, in contrast to wild-type Cox1. We were unable to analyze the interaction of unlabeled Cox1ΔC15 with Cox14-Myc by Western blotting because the truncation apparently removed the epitope recognized by the Cox1-specific antibodies we tested (data not shown).
FIGURE 2.
The C-terminal end of Cox1 is necessary for stable interaction of Mss51 and Cox14. A, translation in isolated mitochondria from wild-type Cox1 or Cox1ΔC15 strains was performed in the presence of [35S]methionine. Mitochondria were washed and solubilized with 1% digitonin. Mitochondrial extracts were immunoprecipitated with a Myc-specific antibody to precipitate Cox14-Myc or (B) HA-specific antibody to precipitate Mss51-HA. As control, strains lacking the Myc or HA epitopes in Cox14 and Mss51, respectively, were included as indicated. Translation products were separated by SDS-PAGE, and transferred to PVDF membrane before autoradiography. The membranes were incubated with HA-specific antibody to detect Mss51-HA or Myc-specific antibody to detect Cox14-Myc as indicated. The immunoprecipitated fractions in the Western blot with anti-Myc antibody showed an additional, unspecific band, which is probably due to the IgG light chain of the antibody used for co-immunoprecipitation (IP) (*). Total samples represent 5% of the aliquots used for immunoprecipitation.
We also immunoprecipitated these mitochondrial extracts with HA-specific antibody. As previously reported, newly made Cox1 co-precipitates with Mss51-HA (13). This interaction was similarly efficient in Cox1ΔC15 mitochondria (Fig. 2B). However, co-immunoprecipitation of Cox14-Myc with Mss51-HA was dramatically reduced in the presence of Cox1ΔC15, compared with wild-type Cox1. Taken together, these results indicate that the ability of the truncated Cox1ΔC15 protein to bridge the interaction between Cox14-Myc and Mss51-HA is compromised.
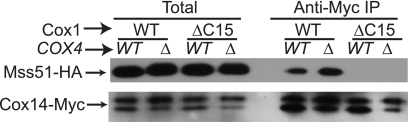
Taken together, these data indicate that the C-terminal 15 residues of Cox1 are required for normal stability of a complex (or complexes) containing Mss51, Cox14, and newly synthesized Cox1. Instability of this complex could account for the robust synthesis of Cox1ΔC15 in a mutant unable to assemble CcO, because Mss51 would not be efficiently sequestered and therefore available to activate COX1 mRNA translation. To test this, we next examined the Mss51-Cox14 interaction in strains unable to assemble CcO due to a cox4Δ mutation that contained either wild-type Cox1 or Cox1ΔC15 (Cox1 synthesis in a cox4Δ mutant was also regulated by the Cox1 C-terminal end, see below). Mitochondria were isolated, allowed to synthesize unlabeled proteins, and solubilized with 1% dodecyl maltoside. Cox14-Myc was precipitated with a Myc-specific antibody, and the precipitates were analyzed by Western blotting. We observed an increased efficiency of Mss51-HA co-immunoprecipitation in cox4Δ as compared with COX4 mitochondria in the presence of Cox1. However, the Cox14-Mss51 interaction was not detected in the presence of Cox1ΔC15, regardless of whether the cox4Δ mutation prevented CcO assembly (Fig. 3).
FIGURE 3.
Interaction of Mss51 with Cox14 is not stable in CcO assembly mutants lacking the C-terminal end of Cox1. Mitochondria from Cox1 or Cox1ΔC15 in the presence of either wild-type COX4 (WT) or cox4Δ (Δ) mutation were solubilized with 1% dodecyl maltoside and immunoprecipitated with a Myc-specific antibody. The immunoprecipitated (IP) fraction was analyzed by Western blot with an antibody to HA, the membrane was stripped and then reprobed with an antibody to Myc. The total fractions represent 5% of mitochondria before solubilization. Western blot with anti-Myc antibody showed a doublet, which is probably due to partial cleavage of the triple Myc epitope.
The Carboxyl-terminal End of Cox1 Regulates Cox1 Synthesis in Several Mutants Affecting CcO Assembly
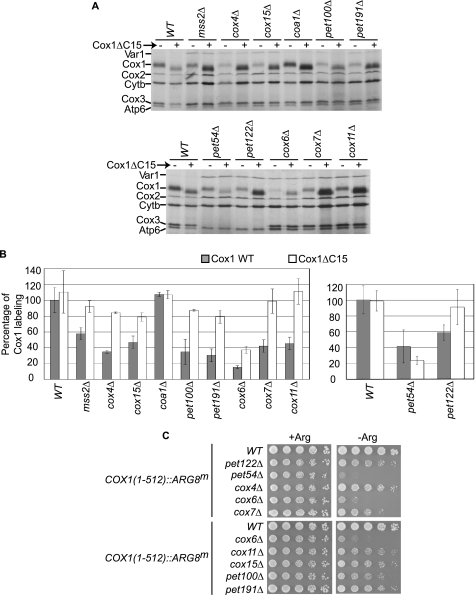
A wide range of mutations that affect CcO assembly show reduced levels of Cox1 labeling in the presence of [35S]methionine (14, 18, 19). We tested whether labeling of the truncated variant Cox1ΔC15 would also remain unaffected when CcO assembly was disrupted by mutations other than cox2Δ. Two groups of CcO mutants were created. In the first group, synthesis of CcO subunits was prevented by cox4Δ, cox6Δ, and cox7Δ mutations, or by COX3 mRNA translational activator mutations pet122Δ and pet54Δ. Cox6 is added to the first assembly intermediate containing Cox1, whereas Cox3, Cox4, and Cox7 are assembled later (35). In the second group, assembly chaperones were eliminated: mss2Δ (necessary for assembly of Cox2 (36)), cox11Δ, cox15Δ (involved in formation of CuB and heme a centers in Cox1, respectively (37, 38)), coa1Δ (participates in Cox1 assembly (16, 17)), pet100Δ (involved in formation of intermediates containing Cox7, Cox7a, and Cox8 (39), and pet191Δ (twin Cx9C protein necessary for full assembly of CcO (40)). Cox11, Cox15, and Coa1 seem to participate in the early stages of CcO assembly (1, 15).
We constructed strains carrying each nuclear mutation with either wild-type mtDNA or the mtDNA encoding Cox1ΔC15, and carried out [35S]methionine labeling (Fig. 4, A and B). With the exception of the coa1Δ strain, which was previously demonstrated to have normal levels of Cox1 [35S]methionine labeling (16, 17), and in our hands had no respiratory growth defect in the D273-10B nuclear genetic background used here, the CcO mutants showed reduced labeling of wild-type Cox1 by 42–85%. In contrast, labeling of the truncated Cox1ΔC15 protein was not reduced by most of these mutations, indicating that the C-terminal end of Cox1 regulates Cox1 synthesis independently of the stage where CcO assembly is interrupted. Two mutants consistently showed the most dramatic reduction of both Cox1 and Cox1ΔC15 [35S]methionine labeling: cox6Δ and pet54Δ.
FIGURE 4.
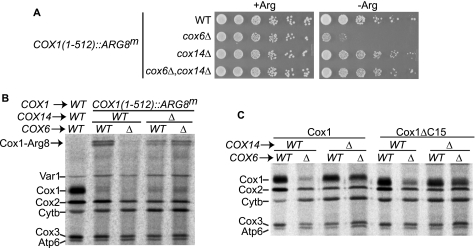
The Cox1 C-terminal end regulates Cox1 synthesis in many CcO mutants. A, Cox1 (−) or Cox1ΔC15 (+) cells with a deletion in the indicated genes were pulse-labeled with [35S]methionine in the presence of cycloheximide, and proteins were analyzed as described under “Experimental Procedures.” B, quantification of the Cox1 signals from A. The level of Cox1 labeling was normalized to the Cox3/Atp6 signal, and was expressed as a percentage of the wild-type, Cox1 signal (except for signals from the pet54Δ and pet122Δ mutants, which were normalized with respect to Cytb). Error bars indicate standard deviations from 3 independent experiments. We also compared the signal of the cytochrome b to the Cox3/Atp6 signal (or the signal from Cox2 to the cytochrome b in the pet54Δ and pet122Δ mutants), and in those cases no significant difference was observed (data not shown). C, translation of the mitochondrial reporter gene COX1(1–512)::ARG8m was analyzed by growth of the indicated mutants on media lacking (−Arg) or containing arginine (+Arg). In this construct the precursor of Arg8 was fused to the C-terminal end of the complete Cox1. Cells were spotted as serial dilutions and grown for 3 days at 30 °C.
The cox6Δ mutation reduced labeling of wild-type Cox1 by 85%, and also reduced labeling of Cox1ΔC15 by 63%. Thus, the cox6Δ mutation strongly reduced Cox1 labeling, and this effect is only slightly ameliorated by the C-terminal truncation of Cox1. The pet54Δ mutation reduced labeling of wild-type Cox1 by 60%. In contrast to other CcO assembly mutants, the pet54Δ mutation similarly reduced labeling of the truncated Cox1ΔC15. This was the only mutant analyzed whose Cox1 labeling was not increased by the C-terminal truncation of Cox1, suggesting that this effect might not be due simply to the lack of CcO assembly. Indeed, deletion of another COX3 mRNA translational activator, Pet122, resulted in a labeling pattern similar to those of the majority of CcO assembly mutants.
The more dramatic reduction of Cox1 labeling in cox6Δ and pet54Δ could be due to decreased synthesis or a more rapid degradation of newly made Cox1. To distinguish these possibilities, we analyzed expression of the mitochondrial reporter gene ARG8m, which codes for a soluble biosynthetic enzyme in the matrix, and whose activity does not depend on the presence of CcO (41). ARG8m was fused in-frame to the end of the COX1 coding region to create the construct COX1(1–512)::ARG8m (13). This ARG8m sequence specifies the cleavage site for the pre-Arg8 mitochondrial targeting signal, such that accumulation of mature Arg8 should not be affected by the stability of Cox1. Thus, expression of ARG8m from this construct provides a readout of COX1 mRNA translation. Furthermore, the Cox1 moiety encoded by COX1(1–512)::ARG8m is assembled into active CcO complexes, supporting normal respiratory growth (13).
We combined some of the nuclear mutations described in Fig. 4A with the COX1(1–512)::ARG8m construct. All the CcO mutants analyzed showed reduced growth in medium lacking arginine as compared with a wild-type strain with the COX1(1–512)::ARG8m construct (Fig. 4C). However, absence of Cox6 and Pet54 consistently showed the most dramatic reduction in Arg+ growth. [35S]Methionine labeling of the Cox1-Arg8 fusion protein in these cells was reduced in all the CcO mutants, but most dramatically reduced in the cox6Δ and pet54Δ mutants (supplemental Fig. S1). These data confirm that the CcO assembly defect caused by the loss of Cox6 or Pet54 reduced synthesis of the reporter fused to full-length Cox1 more than other CcO mutants.
Cox6, together with Cox5a, are the first subunits to assemble with Cox1 (35). It has been suggested that Cox5a and Cox6 confer stability to newly synthesized Cox1 (42). To further test whether deletion of Cox6 confers a strong decrease in Cox1 synthesis we first asked whether conditions that alter Cox1 pulse labeling in CcO assembly-defective mutants similarly alter expression of the COX1(1–512)::ARG8m reporter. In cox14Δ cells, [35S]methionine labeling of Cox1 is restored to wild-type levels even in the presence of mutations affecting CcO assembly (14). Consistent with this finding, double mutant cox6Δ, cox14Δ cells grew on medium lacking arginine as well as wild-type COX6, COX14 cells (Fig. 5A). Mitochondrial [35S]methionine labeling of these strains showed a band corresponding to the Cox1-Arg8 fusion protein. Labeling of the Cox1-Arg8 precursor, as well as the small amount of processed Cox1 was reduced by cox6Δ. However, Cox1 labeling was largely restored in the cox6Δ cox14Δ double mutant (Fig. 5B). Together these data confirm that translation of the Arg8 reporter is decreased in the absence of Cox6.
FIGURE 5.
Synthesis of Cox1 is reduced in a cox6Δ mutant. A, translation of COX1(1–512)::ARG8m in the indicated mutants was analyzed as described in the legend to Fig. 4C. B, mitochondrial translation products of cells from A were obtained in the presence of cycloheximide and [35S]methionine. In addition, a strain with wild type COX1 was included as control. The Cox1-Arg8 precursor protein is indicated with an arrow. For unknown reasons this fusion is detected as a doublet. C, Cox1 or Cox1ΔC15 cells with a deletion in the indicated genes were pulse-labeled with [35S]methionine in the presence of cycloheximide, and proteins were analyzed as described under “Experimental Procedures.”
We next performed [35S]methionine labeling in the presence of cycloheximide of Cox1 or Cox1ΔC15 cells. The cox6Δ mutation reduced labeling of Cox1 and Cox1ΔC15. However, labeling was restored to almost wild-type levels in the cox6Δ cox14Δ double mutant (Fig. 5C). Cox1 pulse labeling in CcO assembly mutants can also be increased by overexpression of Mss51 (14). We found that overexpression of MSS51 partially restored labeling of Cox1 and Cox1ΔC15 in a cox6Δ mutant (supplemental Fig. S2).
Taken together, these data indicate that the assembly defect caused by the cox6Δ reduces Cox1 synthesis more strongly than the other assembly defects tested. It apparently acts by reducing the level of Mss51 available for translational activation, because Cox1 synthesis is partially restored by the cox14Δ and C-terminal truncation of Cox1, both of which weaken the Cox1-Mss51 interaction.
DISCUSSION
Assembly of the CcO is a multistep process that involves the coordinated incorporation of mitochondrially and nuclearly encoded subunits, as well as prosthetic groups. It is now recognized that mitochondrially encoded Cox1 is the foundation upon which further assembly occurs. A common observation is that pulse labeling of Cox1 is reduced in the majority of mutants with defects in CcO assembly, reflecting decreased synthesis of Cox1. The mechanisms by which the CcO assembly state is sensed to regulate Cox1 synthesis are not completely understood. The proteins Mss51 (13, 14), Cox14 (14), and Coa1 (16, 17) form complexes with newly synthesized Cox1, coupling regulation of Cox1 synthesis to CcO assembly.
One question that remains is the mechanism by which the assembly state of Cox1 is sensed. To map the portions of Cox1 involved in this regulation we analyzed the C-terminal domain of Cox1 by site-directed mutation. This 59-amino acid region is exposed on the matrix side of the inner membrane in the assembled enzyme, and could interact with peripheral proteins like Mss51. We found that deletion of the last 11 or 15 residues of Cox1 disrupted the assembly-feedback control of Cox1 synthesis without inactivating CcO. These mutants showed wild-type levels of Cox1 synthesis when the CcO assembly was impaired. These C-terminal deletions of Cox1 also reduced the interaction between Mss51 and Cox14 during otherwise normal assembly. It is unknown whether Mss51 and Cox14 interact directly or via intermediate proteins in early assembly complexes. In any event, Mss51 and Cox14 do not interact normally even in the absence of CcO assembly when Cox1 lacks its last 15 residues. This weakened interaction could reduce sequestration of Mss51 in CcO assembly intermediates, making more Mss51 available for activation of COX1 mRNA translation.
Interestingly, during the analysis of a wide range of CcO-deficient mutants, we found that Pet54 is required for normal levels of Cox1 synthesis. Pet54 is required both for COX3 translation (43) and for splicing of the aI5β intron present in the COX1 gene of many yeast strains (44), and these activities are genetically separable (45). However, the COX1 gene in the strains employed here lacks introns. This novel Pet54 activity is independent of the C-terminal end of Cox1.
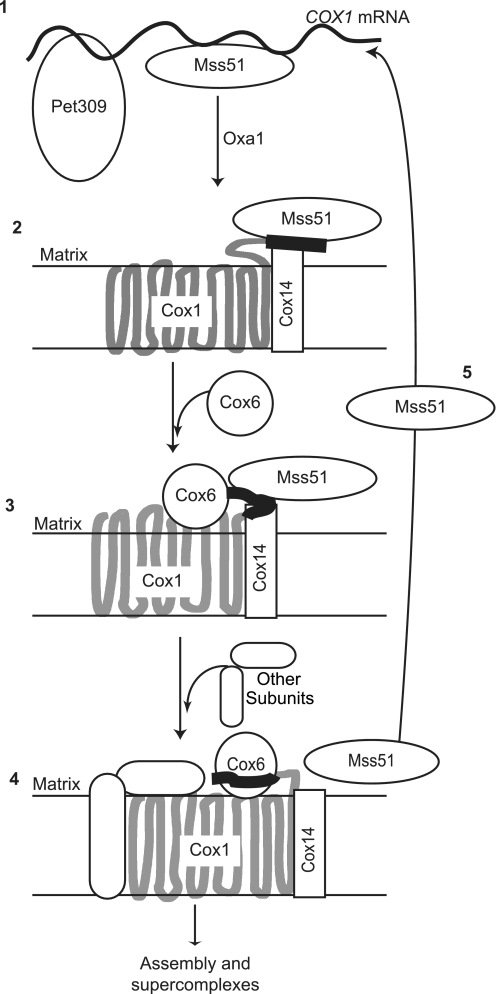
The hydrophilic Cox1 C-terminal end could adopt different conformations during assembly of the newly synthesized protein in response to its association with assembly factors and other subunits. These conformations could differ in their ability to sequester Mss51. We observed that among the CcO mutants analyzed in the present study, absence of Cox6 caused one of the most dramatic reductions in Cox1 synthesis, as analyzed by [35S]methionine labeling experiments or expression of the ARG8m reporter fused to COX1. We propose that early in assembly the C-terminal end of Cox1 has a conformation that strongly stabilizes the association of Mss51 with a high molecular weight complex containing Cox14 and Cox1 (Fig. 6, step 2). When Cox6 and possibly Cox5a are added to the complex, and/or when hemes are inserted (15), the C-terminal end of Cox1 could change. The crystal structure of assembled bovine CcO shows that these subunits are in close proximity to the C-terminal end of Cox1 (8). This conformational change could weaken the Cox14-dependent interaction of Mss51 with the assembly complex. It was previously proposed that Mss51 is liberated from the assembling enzyme early at (16) or before (15) the point where Cox5a and Cox6 are added. However, it is possible that some Mss51 remains weakly bound to the complex until further steps of assembly are completed. This would help explain how mutations blocking assembly downstream in the pathway, such as the cox4Δ mutation, also elicit reduced Cox1 synthesis by sequestration of Mss51, but less strongly than the cox6Δ (Fig. 6, step 3). At the latest, once Cox1 is assembled within the CcO and its C-terminal end acquires the final or close to final conformation, Mss51 must be liberated from the complex (Fig. 6, step 4) and available for further rounds of translational activation of the COX1 mRNA (Fig. 6, steps 1 and 5).
FIGURE 6.
Model for assembly-feedback translational regulation of the COX1 mRNA. See text for details. The C-terminal 15 residues of Cox1 are indicated with a thick black line. A thick gray line represents the rest of the Cox1 protein.
It remains unclear whether assembly-feedback regulated synthesis of Cox1 in mitochondria occurs in other species. A few examples in mammals suggest that translation of the COX1 mRNA might be reduced by defects associated with CcO assembly. It has been documented that a 15-base pair deletion in the human mitochondrial COX3 gene (46), as well as lack of cytochrome c in mouse fibroblasts (47) leads to a modest reduction of [35S]methionine labeling of Cox1. However, these studies did not clearly distinguish whether decreased Cox1 labeling was due to reduced synthesis or increased turnover.
Regulated synthesis of Cox1 in S. cerevisiae is the first identified example in mitochondria where an organelle-encoded protein has amino acid sequences that couple regulation of its own synthesis to assembly. However, a similar mechanism has been demonstrated in the chloroplast of Chlamydomonas reinhardtii. Synthesis of some organelle-encoded subunits is strongly reduced when other subunits from the photosynthetic complexes are missing (48–50). For the b6f complex, a C-terminal extension of 11 residues within cytochrome f is necessary for this regulation, possibly by stabilizing an interaction with the translational activator Tca1 when the enzyme is not assembled (51). This extension showed no obvious similarity with the Cox1 C terminus.
The C-terminal end of Cox1 contains the consensus motif SPP(P/A)XH, where His503 (as numbered in the bovine sequence) is necessary for the tunneling of protons through the D channel to the heme a3-CuB center (52). Interestingly, this histidine is part of the VHSFNT motif and is removed by our deletion of the last 15 residues of Cox1, demonstrating that it is not essential for oxidative phosphorylation in yeast.
The hydrophilic carboxyl-terminal domain of Cox1 is less conserved overall than the transmembrane domains. However, this region seems to be crucial for CcO activity. In the protist Acanthamoeba castellanii the mitochondrial COX1 gene lacks the region coding for the C-terminal end. Interestingly, a nucleus-encoded protein homologous to this domain is imported into mitochondria, suggesting that the two peptides interact in trans to fulfill the same role as the intact protein (53).
Supplementary Material
Acknowledgments
We thank Dr. A. Tzagoloff and Dr. D. González-Halphen for the kind gift of antisera, Dr. G. del Río-Guerra and Dr. T. Lara-Ortíz for the gift of yeast deletion strains, and Dr. S. Funes for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM029362 (to T. D. F.), research grants from CONACyT (47514) and Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT), and UNAM Grant IN215008 (to X. P.-M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- CcO
- cytochrome c oxidase.
REFERENCES
- 1.Fontanesi F., Soto I. C., Barrientos A. (2008) IUBMB Life 60, 557–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrmann J. M., Funes S. (2005) Gene 354, 43–52 [DOI] [PubMed] [Google Scholar]
- 3.Pecina P., Houstková H., Hansíková H., Zeman J., Houstek J. (2004) Physiol. Res. 53, Suppl. 1, S213–223 [PubMed] [Google Scholar]
- 4.Manthey G. M., McEwen J. E. (1995) EMBO J. 14, 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Mitchell G. A., Morin C., Mann M., Hudson T. J., Robinson B., Rioux J. D., Lander E. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E. A., Consortium L. (2010) Mol. Biol. Cell 21, 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J. E., Lochmüller H., Chevrette M., Kaufman B. A., Horvath R., Shoubridge E. A. (2009) Nat. Genet. 41, 833–837 [DOI] [PubMed] [Google Scholar]
- 8.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) Science 272, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 9.Lemaire C., Robineau S., Netter P. (1998) Curr Genet. 34, 138–145 [DOI] [PubMed] [Google Scholar]
- 10.Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., Van den Bogert C. (1998) Eur. J. Biochem. 254, 389–394 [DOI] [PubMed] [Google Scholar]
- 11.Khalimonchuk O., Bird A., Winge D. R. (2007) J. Biol. Chem. 282, 17442–17449 [DOI] [PubMed] [Google Scholar]
- 12.Perez-Martinez X., Butler C. A., Shingu-Vazquez M., Fox T. D. (2009) Mol. Biol. Cell 20, 4371–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Martinez X., Broadley S. A., Fox T. D. (2003) EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrientos A., Zambrano A., Tzagoloff A. (2004) EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalimonchuk O., Bestwick M., Meunier B., Watts T. C., Winge D. R. (2010) Mol. Cell. Biol. 30, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mick D. U., Wagner K., van der Laan M., Frazier A. E., Perschil I., Pawlas M., Meyer H. E., Warscheid B., Rehling P. (2007) EMBO J. 26, 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., Winge D. R. (2007) EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabral F., Solioz M., Rudin Y., Schatz G., Clavilier L., Slonimski P. P. (1978) J. Biol. Chem. 253, 297–304 [PubMed] [Google Scholar]
- 19.Carlson C. G., Barrientos A., Tzagoloff A., Glerum D. M. (2003) J. Biol. Chem. 278, 3770–3775 [DOI] [PubMed] [Google Scholar]
- 20.Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P. P., Rytka J., di Rago J. P. (2007) J. Biol. Chem. 282, 10853–10864 [DOI] [PubMed] [Google Scholar]
- 21.Soto I. C., Fontanesi F., Valledor M., Horn D., Singh R., Barrientos A. (2009) Biochim. Biophys. Acta 1793, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrientos A., Pierre D., Lee J., Tzagoloff A. (2003) J. Biol. Chem. 278, 8881–8887 [DOI] [PubMed] [Google Scholar]
- 23.Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Guthrie C., Fink G. R. eds (2002) Guide to Yeast Genetics and Molecular and Cell Biology, Academic Press, San Diego [Google Scholar]
- 25.Bonnefoy N., Remacle C., Fox T. D. (2007) Methods Cell Biol. 80, 525–548 [DOI] [PubMed] [Google Scholar]
- 26.Meunier B., Lemarre P., Colson A. M. (1993) Eur. J. Biochem. 213, 129–135 [DOI] [PubMed] [Google Scholar]
- 27.Meunier B. (2001) Biochem. J. 354, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 29.Diekert K., de Kroon A. I., Kispal G., Lill R. (2001) Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31.Bonnefoy N., Bsat N., Fox T. D. (2001) Mol. Cell. Biol. 21, 2359–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westermann B., Herrmann J. M., Neupert W. (2001) Methods Cell Biol. 65, 429–438 [DOI] [PubMed] [Google Scholar]
- 33.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnefoy N., Fox T. D. (2000) Mol. Gen. Genet. 262, 1036–1046 [DOI] [PubMed] [Google Scholar]
- 35.Horan S., Bourges I., Taanman J. W., Meunier B. (2005) Biochem. J. 390, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broadley S. A., Demlow C. M., Fox T. D. (2001) Mol. Cell. Biol. 21, 7663–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiser L., Di Valentin M., Hamer A. G., Hosler J. P. (2000) J. Biol. Chem. 275, 619–623 [DOI] [PubMed] [Google Scholar]
- 38.Barros M. H., Carlson C. G., Glerum D. M., Tzagoloff A. (2001) FEBS Lett. 492, 133–138 [DOI] [PubMed] [Google Scholar]
- 39.Church C., Goehring B., Forsha D., Wazny P., Poyton R. O. (2005) J. Biol. Chem. 280, 1854–1863 [DOI] [PubMed] [Google Scholar]
- 40.Khalimonchuk O., Rigby K., Bestwick M., Pierrel F., Cobine P. A., Winge D. R. (2008) Eukaryot. Cell 7, 1427–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steele D. F., Butler C. A., Fox T. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontanesi F., Jin C., Tzagoloff A., Barrientos A. (2008) Hum. Mol. Genet. 17, 775–788 [DOI] [PubMed] [Google Scholar]
- 43.Costanzo M. C., Seaver E. C., Fox T. D. (1986) EMBO J. 5, 3637–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valencik M. L., Kloeckener-Gruissem B., Poyton R. O., McEwen J. E. (1989) EMBO J. 8, 3899–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valencik M. L., McEwen J. E. (1991) Mol. Cell. Biol. 11, 2399–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffbuhr K. C., Davidson E., Filiano B. A., Davidson M., Kennaway N. G., King M. P. (2000) J. Biol. Chem. 275, 13994–14003 [DOI] [PubMed] [Google Scholar]
- 47.Vempati U. D., Han X., Moraes C. T. (2009) J. Biol. Chem. 284, 4383–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drapier D., Rimbault B., Vallon O., Wollman F. A., Choquet Y. (2007) EMBO J. 26, 3581–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minai L., Wostrikoff K., Wollman F. A., Choquet Y. (2006) Plant Cell 18, 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wostrikoff K., Girard-Bascou J., Wollman F. A., Choquet Y. (2004) EMBO J. 23, 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choquet Y., Zito F., Wostrikoff K., Wollman F. A. (2003) Plant Cell 15, 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muramoto K., Hirata K., Shinzawa-Itoh K., Yoko-o S., Yamashita E., Aoyama H., Tsukihara T., Yoshikawa S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7881–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gawryluk R. M., Gray M. W. (2010) Mol. Biol. Evol. 27, 7–10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.