Abstract
The YjgF/YER057c/UK114 family of proteins is highly conserved across all three domains of life and currently lacks a consensus biochemical function. Analysis of Salmonella enterica strains lacking yjgF has led to a working model in which YjgF functions to remove potentially toxic secondary products of cellular enzymes. Strains lacking yjgF synthesize the thiamine precursor phosphoribosylamine (PRA) by a TrpD-dependent mechanism that is not present in wild-type strains. Here, PRA synthesis was reconstituted in vitro with anthranilate phosphoribosyltransferase (TrpD), threonine dehydratase (IlvA), threonine, and phosphoribosyl pyrophosphate. TrpD-dependent PRA formation in vitro was inhibited by S. enterica YjgF and the human homolog UK114. Thus, the work herein describes the first biochemical assay for diverse members of the highly conserved YjgF/YER057c/UK114 family of proteins and provides a means to dissect the cellular functions of these proteins.
Keywords: Bacterial Genetics, Bacterial Metabolism, Purine, Pyridoxal Phosphate, Thiamine, TrpD, YjgF/YER057c/UK114, Anthranilate Phosphoribosyl Transferase, Phosphoribosylamine
Introduction
The accumulation of genomic sequences continues to identify conserved protein families for which no function has been experimentally defined. Analysis of thiamine biosynthesis in Salmonella enterica has proven to be a useful approach to provide insights into some of these genes of unknown function (1–3). In its cofactor form, thiamine pyrophosphate is essential for cell growth. In wild-type Salmonella, the first step of both thiamine and purine synthesis is catalyzed by the enzyme glutamine phosphoribosyl pyrophosphate amidotransferase (PurF; EC 2.4.2.14), which forms phosphoribosyl amine (PRA)3 (4, 5). A strain lacking purF, in combination with a null mutation in gnd, encoding gluconate-6-phosphate dehydrogenase, is unable to generate PRA and thus cannot grow in the absence of thiamine (6, 7). However, if a third gene, yjgF, is eliminated, PRA synthesis and thus thiamine-independent growth are restored (2, 8).
YjgF is a member of the YjgF/YER057c/UK114 family of proteins present in all three domains of life and defines the Cluster of Orthologous Groups 0251 (9). A general biochemical function for the YjgF family has not been demonstrated nor is one suggested by the sequence or structure of family members. Thus, why deleting yjgF restores PRA formation is unknown. Based on previous work by our laboratory and by others, it has been proposed that YjgF interacts with and possibly sequesters small, potentially toxic molecules, thus altering the metabolic environment of the cell (10, 11).
Past work implicated the tryptophan biosynthetic complex anthranilate synthase-phophoribosyltransferase (EC 4.1.3.27, 2.4.2.18) in PurF-independent PRA formation in a yjgF background (8). During tryptophan biosynthesis, the TrpDE complex catalyzes the conversion of chorismate to anthranilate while deaminating glutamine, and subsequently TrpD combines anthranilate and phosphoribosyl pyrophosphate (PRPP) to form phosphoribosyl anthranilate and inorganic phosphate (12, 13). Additional studies suggested that the activity of the first isoleucine biosynthetic enzyme threonine dehydratase (IlvA; EC 4.3.1.19) was involved in PurF-independent PRA formation in a yjgF mutant (2). IlvA is a pyridoxal-5′-phosphate (PLP)-dependent enzyme that dehydrates threonine and releases an enamine intermediate, which tautomerizes to a primary imine. This imine is then hydrolyzed nonenzymatically, forming free ammonia and 2-ketobutyrate, an intermediate in isoleucine synthesis (14, 15).
In this study a combination of in vivo results led to a testable in vitro model that described the mechanism of PRA formation in a yjgF mutant background. The data herein are consistent with the model that IlvA generates a threonine-derived metabolite that TrpD condenses with PRPP to form PRA. The in vitro formation of PRA was reduced by the addition of the S. enterica YjgF protein and the human homolog, UK114. Together, these results identified not only a new physiologically relevant mechanism of PRA formation but defined an in vitro assay for YjgF that can be utilized to probe the biochemical function of the YjgF/YER057c/UK114 family of proteins.
EXPERIMENTAL PROCEDURES
Media and Chemicals
No-carbon E (NCE) medium of Vogel and Bonner (16, 17) with MgSO4 (1 mm) and glucose as a carbon source (11 mm) was used as a minimal medium. When necessary, the following compounds were provided at the indicated final concentrations: adenine, 0.4 mm; thiamine, 100 nm; tryptophan, 0.1 mm; threonine, 600 μm; and anthranilate, 7.5 μm. Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) and Luria-Bertani (LB) broth were used as rich media. Difco BiTek agar was added (15 g/liter) for solid medium. Superbroth (32 g of tryptone, 20 g of yeast extract, 5 g of sodium chloride, and 5 ml of 1 n sodium hydroxide/liter) was used for protein purification. Antibiotics were added when needed at the following concentrations to rich and minimal media, respectively: ampicillin, 30 and 15 mg/ml; chloramphenicol, 20 and 4 mg/ml; kanamycin, 50 and 125 mg/ml; and tetracycline, 20 and 10 mg/ml. Antibiotics and chemicals were purchased from Sigma-Aldrich.
Bacterial Strains and Plasmids
All strains used in this study are derivatives of S. enterica serovar Typhimurium strain LT2 and are listed along with their respective genotypes in Table 1. The strain carrying trpE8 was provided by J. R. Roth. Deletion of the trp operon was generated by linear recombination (18) and has been described (8). Plasmid pTrpD (pIR-DW) contained wild-type trpD in pSU19 under the control of the plasmid encoded lac promoter and has been described (19). UK114/hp14.5 cDNA was provided by G. Schmitz (20).
TABLE 1.
Bacterial strains
All strains are derivatives of S. enterica serovar Typhimurium LT2 unless otherwise indicated.
| Strain | Genotypea |
|---|---|
| DM7436 | purF2085 gnd-181 yjgF3::MudJ |
| DM8813 | purF2085 gnd-181 yjgF3::MudJ ΔtrpEDCBA3618 |
| DM8822 | purF2085 gnd-181 yjgF3::MudJ ΔtrpEDCBA3618 (pSU19) |
| DM8823 | purF2085 gnd-181 yjgF3::MudJ ΔtrpEDCBA3618 (pSU19-trpD) |
| DM8988 | purF2085 gnd-181 ΔtrpEDCBA3618 |
| DM9593 | E. coli BL21AI pTYB2-purD |
| DM9813 | purF2085 gnd-181 yjgF3::MudJ zxx4099::Tn10d (Tc) trpD16 |
| DM9814 | purF2085 gnd-181 yjgF3::MudJ zxx4099::Tn10d (Tc) trpD12 |
| DM9815 | purF2085 gnd-181 yjgF3::MudJ zxx4099::Tn10d (Tc) trpD122 |
| DM10000 | LT2 wild-type |
| DM10339 | purF2085 gnd-181 yjgF3::MudJ trpE8 |
| DM11108 | E. coli BL21AI pET28a-trpD |
| DM12740 | E. coli BL21AI pET20b-yjgF |
| DM12816 | E. coli BL21AI pET20b-hp14.5/UK114 |
| DM12944 | E. coli BL21AI pET20b-ilvA |
Growth Quantification
Nutritional requirements were assessed in liquid medium. Strains were grown to full density in rich medium at 37 °C, and cells were pelleted by centrifugation and resuspended in an equal volume of saline (85 mm). Five microliters of the cell suspension was used to inoculate 195 μl of the medium in each well of a 96-well microtiter plate. Growth at 37 °C was monitored using a microplate spectrophotometer (Spectro-Max Plus; Molecular Devices, Sunnyvale, CA). Alternatively, 200 μl of the cell suspension was used to inoculate 5-ml cultures in 18 × 150-mm tubes. Cultures were grown with shaking at 37 °C, and growth was monitored with a Bausch & Lomb Spectronic 20. Specific growth rate was determined as μ = ln (X/Xo)/T, where X = A650 during the linear portion of the growth curve and T = time.
Genetic Techniques
The high frequency general transducing mutant of bacteriophage P22 (HT105/1 int-201) (21, 22) was used to perform all transductions required for mutant strain constructions. Standard genetic techniques were used.
Expression and Purification of Proteins
TrpD
The Escherichia coli BL21AI (Invitrogen) strain containing the wild-type trpD gene cloned into the pET-28a expression vector (23) (DM11108) was used for TrpD His6 tag protein purification. A 10-ml culture of DM11108 in LB medium containing kanamycin was used to inoculate 1 liter of superbroth. The culture was grown at 37°C until the A650 of 0.9 at which point the cells were induced with 1.0 mm isopropyl-β-d-thiogalactopyranoside and grown at 28 °C overnight. The cells were harvested at 4 °C by centrifugation and resuspended in buffer (20 mm Tris-HCl, pH 8.0, and 300 mm sodium chloride). The cells were disrupted on ice by sonication using a Sonic Dismembrator 550 (Fisher Scientific). Cell debris was separated by centrifugation. The supernatant was added to prepared Ni-NTA superflow resin (Qiagen), and imidazole was added to 5 mm before stirring the solution on ice for 1 h. A batch elution was performed by using 5-ml preparations of column buffer with each of the following concentrations of imidazole: 20, 40, 60, 100, and 250 mm. Protein concentration was determined with a BCA protein assay kit (Thermo Scientific). Protein activity was verified using a phosphoribosyltransferase assay, as described previously (19, 24, 25).
IlvA
The E. coli BL21AI strain containing the wild-type ilvA gene cloned into the pET20b+ vector (Novagen, Madison, WI) (DM12944) was used for IlvA His6 tag protein purification. Cells were inoculated into 2 ml of LB medium containing ampicillin and allowed to reach full density before being inoculated into 50 ml of superbroth for 2–3 h and then inoculated into 2 liters of superbroth. Cultures were incubated at 37 °C to A650 of 0.3–0.7, and expression was induced by the addition of 0.2% arabinose for 16 h at 30 °C. Cells were harvested at 4 °C by centrifugation and resuspended in buffer (50 mm KPO4, pH 8.0, 500 mm NaCl, 10 mm imidazole). Cells were disrupted in the presence of 1 mg/ml lysozyme and 5 mg/ml DNase on ice by sonication using a Sonic Dismembrator 550. Lysate was clarified by centrifugation and filtration through a 0.45-mm membrane. The lysates were bound to a Ni-NTA superflow resin and purified using the manufacturer's protocol (Qiagen, Valencia, CA). Purified protein was eluted in 50 mm Tris-HCl buffer, pH 8.0, with 10 μm pyridoxal-5′-phosphate. Protein activity was verified by assaying the conversion of threonine to 2-ketobutyrate, following formation of a 2,4-dinitrophenylhydrazine derivative as described previously (26, 27).
YjgF and UK114
The E. coli BL21AI strain containing the wild-type yjgF gene cloned into the pET20b+ vector (DM12740) was used for YjgF His6 tag protein purification. A 5-ml culture of DM12740 in LB with ampicillin was used to inoculate 3 liters of superbroth with ampicillin, 150 μg/ml. The culture was grown at 37 °C to A650 of 0.8, and cells were induced with 0.2% arabinose and grown at 37 °C for 8 more h. The cells were harvested at 4 °C by centrifugation and resuspended in buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5 mm imidazole, and 12.5% glycerol). The cells were disrupted using a French pressure cell. Cell extract was clarified by centrifugation for 45 min at 20,000 × g at 4 °C. The supernatant was added to a column with Ni-NTA superflow resin and purified according to manufacturer's protocol. The purified protein suspensions were concentrated and dialyzed first into 10 mm HEPES, pH 8.0, with 10 mm EGTA, then into 10 mm HEPES, pH 8.0, and last into 10 mm HEPES, pH 8.0, with 20% glycerol. Human homolog hp14.5/UK114 was purified in the same manner described above, from strain DM12816. Some of the YjgF from the Ni-NTA purification was subsequently subjected to a Superdex size exclusion column (GE Healthcare), and no change in activity was observed after this additional step.
PurD
PurD was expressed and purified using a C-terminal intein tag and chitin affinity chromatography as described previously (28), from strain DM9593.
Phosphoribosylamine Formation Assay
Synthesis of PRA was determined as a function of [14C]glycinamide ribonuclotide ([14C]GAR) produced in a coupled reaction catalyzed by GAR synthetase (PurD). A molecule of [14C]glycine is condensed with a molecule of PRA by PurD to yield a molecule of [14C]GAR. Unless indicated, the standard assay included 100 mm Tris-HCl, pH 8.0, 10 mm magnesium acetate, 2.6 mm adenosine triphosphate, 333 μm dithioerythritol, 27 μm PLP, 1 μm PurD, 2 mm (26 nCi) [14C]glycine, 1 mm PRPP, 1 μm IlvA, and 2 μm TrpD in 75 μl (final volume). The reaction was initiated with the addition of 50 mm threonine, and the mixture was incubated for 4 h at 37 °C. [14C]GAR and [14C]glycine were separated by thin layer chromatography on polyethyleneimine (PEI)-cellulose using an n-propyl alcohol:water system (1:1). The position of radioactive spots was detected using a Cyclone Storage Phosphor System (Packard Instrument Company), and the identities were confirmed using known standards. Radioactivity was measured by scintillation counting and PhosphorImage signal quantification, and [14C]GAR was quantified by correlation based on input counts of glycine standards. Nonenzymatic formation of PRA from 10 mm ribose 5-phosphate and 13 mm ammonia served as a control (29, 30). The assay was performed at pH levels ranging from 5.3 to 10.0, with maximum [14C]GAR produced at pH values between 8.0 and 10.0. Additionally, the assay was performed at temperatures ranging from 15 °C to 45 °C, with maximum [14C]GAR produced at 37 °C.
RESULTS
TrpD Is Necessary for PRA Formation in a yjgF Mutant
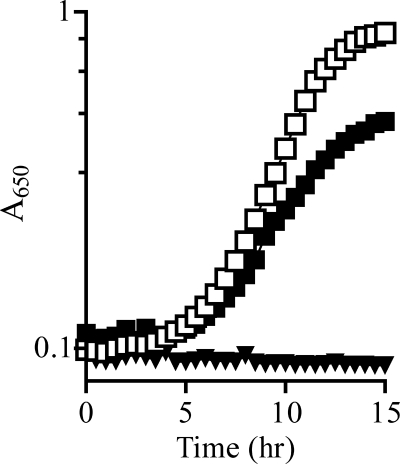
Results from a number of in vivo growth analyses suggested a requirement for TrpD, but not TrpE, in the formation of PRA in a yjgF mutant strain. First, in minimal glucose medium supplemented with adenine and tryptophan, the purF gnd mutant strain was unable to grow (μ ≤ 0.02). In contrast, a purF gnd yjgF (DM7436) mutant strain had a growth rate of μ = 0.32. The introduction of alleles that resulted in catalytically inactive TrpD proteins (trpD16, 12, 122) to a purF gnd yjgF strain prevented detectable growth (μ ≤ 0.02). Thiamine restored full growth (μ ≈ 0.4) to all strains. Second, the introduction of the trpE8 allele, which encodes a protein variant with no anthranilate synthase activity (31), did not affect thiamine-independent growth of the purF gnd yjgF strain (data not shown). Further, although a purF gnd yjgF strain carrying a deletion of the entire tryptophan biosynthetic operon was unable to synthesize thiamine (8), a wild-type copy of trpD provided in trans was sufficient to restore growth to this strain in the absence of thiamine (Fig. 1). Taken together, these results demonstrated that TrpD was the only component of the tryptophan biosynthetic pathway required for PRA formation in a yjgF mutant.
FIGURE 1.
PRA synthesis in a yjgF mutant requires TrpD. Growth of a purF gnd yjgF Δtrp mutant carrying pSU19 (triangles) or pSU19-trpD (squares) was monitored over time as A650 nm. Strains were grown in glucose media with adenine and tryptophan (filled symbols) or adenine, tryptophan, and thiamine (open symbols), with shaking at 37 °C. All strains grew to full density when thiamine was present in the medium.
Anthranilate Inhibits Thiamine-independent Growth in a yjgF Mutant, and Threonine Overcomes This Inhibition
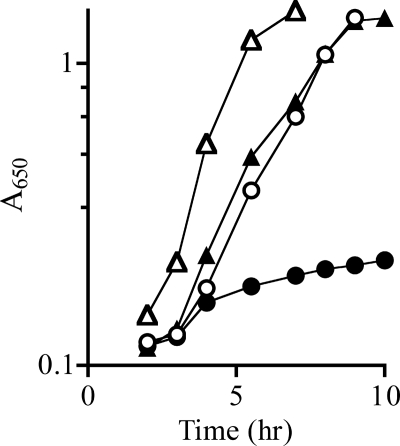
Exogenous anthranilate, a substrate of TrpD, eliminated thiamine-independent growth of a purF gnd yjgF strain (Fig. 2). Titration determined that 7.5 μm anthranilate resulted in full inhibition of growth. This concentration of anthranilate was sufficient to allow the trpE8 mutant to grow to half-density in minimal medium, but was not enough to satisfy the tryptophan requirement of the strain completely (data not shown). The 19 amino acids, excluding tryptophan, were screened for the ability to restore thiamine-independent growth in the presence of anthranilate. Only threonine restored growth (Fig. 2). Titration determined that 500 μm threonine (66-fold excess over anthranilate) was the minimal concentration required to restore full thiamine-independent growth of a purF gnd yjgF strain in the presence of 7.5 μm anthranilate. This effect of threonine was consistent with previous data, suggesting that the isoleucine biosynthetic enzyme IlvA, which uses threonine as a substrate, has a role in PRA formation in a purF gnd yjgF background (2).
FIGURE 2.
Anthranilate inhibits thiamine-independent growth of purF gnd yjgF. Growth of purF gnd yjgF (DM7436) was monitored over time as the absorbance at 650 nm. The strain was grown at 37 °C with shaking in minimal glucose media with adenine (filled triangles), adenine and anthranilate (filled circles), adenine, anthranilate, and threonine (open circles), or adenine, anthranilate, and thiamine (open triangles).
Working Model for PRA Formation in a yjgF Mutant
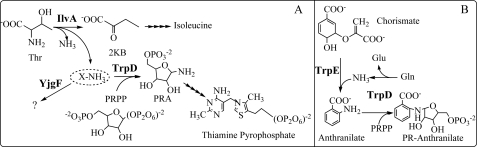
A biochemical model for TrpD-dependent PRA formation was generated based on accumulated in vivo results and is depicted in Fig. 3. This model includes the following predictions: (i) IlvA generates a nitrogen-containing intermediate or side product from threonine; (ii) this IlvA-generated metabolite provides the amino group for PRA; (iii) TrpD forms PRA from the amino donor metabolite and PRPP; and (iv) the presence of YjgF prevents the IlvA-generated amino donor from interacting with TrpD and forming PRA.
FIGURE 3.
Model for PRA formation in yjgF background. A, working model suggesting that IlvA generates an amino-containing product from threonine. This metabolite (X-NH3) then serves as the amino donor for PRA when condensed with PRPP by TrpD. The model suggests that X-NH3 is acted on by YjgF, thus preventing PRA formation by this mechanism. B, schematic of the reactions mediated by TrpD during tryptophan synthesis. The TrpDE complex deaminates glutamine for anthranilate synthesis, and TrpD alone combines anthranilate and PRPP to make PR-anthranilate. 2KB, 2-ketobutyrate.
Reconstitution of TrpD-dependent PRA Formation in Vitro
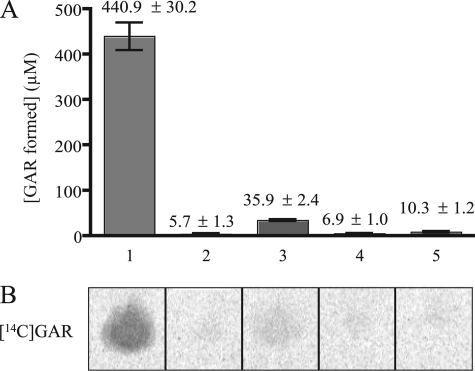
In initial experiments, PRPP, IlvA, and TrpD were combined with an enzyme system that condenses [14C]glycine with PRA (which is unstable) to generate the stable product [14C]glycinamide ribonucleotide ([14C]GAR) (30). Threonine (50 mm) was added to initiate the reaction, and the mixture was incubated for 4 h at 37 °C. The complete reaction yielded [14C]GAR, indicating PRA had been formed (Fig. 4B, lane 1). The concentration of [14C]GAR formed in this reaction was determined to be 440 μm (33 nmol) (Fig. 4A). A completed reaction mixture with unlabeled glycine and proteins removed was submitted for positive time-of-flight mass spectral (+ TOF MS) analysis to verify the production of GAR. A compound with the mass of GAR was detected (expected, m/z 287.1843; observed, m/z 287.0549), supporting the conclusion that PRA had been formed in the assay.
FIGURE 4.
PRA formation assay is dependent on TrpD, IlvA, PRPP and threonine. A standard assay for PRA formation included threonine, IlvA, PRPP, TrpD, cofactors, [14C]glycine, and PurD, as described under “Experimental Procedures.” PRA formation was determined as a function of [14C]GAR production. Lane 1, all reaction components present; lane 2, IlvA omitted; lane 3, TrpD omitted; lane 4, threonine omitted; lane 5, PRPP omitted. A, quantification of [14C]GAR produced under each condition, using [14C]glycine standards of known concentrations. Error bars represent the S.D. of two replicates. B, representative phosphorimage of [14C]GAR spots after separation from [14C]glycine by thin-layer chromatography.
Control reactions determined that each of the four assay components IlvA, TrpD, PRPP, and threonine was necessary for PRA formation. First, no significant amount of [14C]GAR was detected in the absence of IlvA (Fig. 4, lane 2), indicating that IlvA was required to catalyze a reaction with threonine and threonine itself was not a direct substrate for TrpD. Second, in the absence of TrpD, minimal [14C]GAR was detected (Fig. 4, lane 3). A small amount of PRA synthesis in this case was expected, as PRA can be generated from the nonenzymatic condensation of ribose 5-phosphate and ammonium (29). PRPP spontaneously breaks down into ribose 5-phosphate and would combine with ammonia generated from threonine by IlvA to form PRA. Finally, no significant amount of [14C]GAR was detected in the absence of either threonine or PRPP (Fig. 4, lanes 4 and 5). To address the possibility that TrpD utilized ammonia (from deaminated threonine) and PRPP to make PRA, threonine was replaced by equimolar ammonium chloride in the assay. These conditions failed to generate PRA (data not shown), indicating that TrpD did not use free ammonia as a substrate.
IlvA is a PLP-requiring enzyme, and it was formally possible that threonine was reacting nonenzymatically with PLP to form the substrate for TrpD. However, when IlvA was absent and PLP was present, no significant [14C]GAR was formed (Fig. 4, lane 2), showing that the substrate for TrpD was not generated by a nonenzymatic reaction of threonine with PLP or any other reaction component. It was also possible that IlvA generated an inactive form of PLP, pyridoxamine 5′-phosphate, during the dehydration of threonine and pyridoxamine 5′-phosphate, then served as a substrate for TrpD. No [14C]GAR was formed when either pyridoxamine or pyridoxamine 5′-phosphate was added to PRA formation assays in the absence of threonine, IlvA, and PLP (data not shown). Additional attempts to identify the product of IlvA that served as the substrate of TrpD were unsuccessful because this metabolite was unstable and disappeared quickly from the reaction mixture (data not shown).
S. enterica has an additional threonine dehydratase, TdcB, that catalyzes the same reaction as IlvA but is regulated differently and only expressed under anaerobic conditions (32, 33). Not surprisingly, [14C]GAR was formed when IlvA was replaced with equimolar TdcB in the coupled assay. However, branched chain amino acid aminotransferase, (IlvE; EC 2.6.1.42) another PLP-dependent enzyme, was unable to substitute for IlvA when provided with either threonine or its natural substrate, isoleucine. These results indicated that TrpD did not utilize a metabolite produced by all PLP enzymes to generate PRA.
A Minority of the Threonine Is the Amino Donor for PRA Formation
Mass spectrometry analysis confirmed that threonine was serving as an amino source for PRA formation. An equimolar mixture of natural and [15N]threonine (50 mm) was used in the complete reaction. After incubation, the proteins were removed, and the sample was submitted for + TOF MS analysis. Two peaks were identified in the spectrum (m/z 287.0559 and m/z 288.0667), corresponding to the masses of natural GAR and 15N-labeled GAR, respectively.
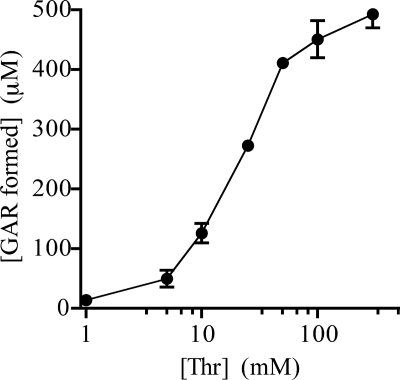
The amount of [14C]GAR formed in vitro was dependent on threonine concentration. When the concentration of threonine was titrated under standard assay conditions (Fig. 5), half-maximal [14C]GAR was formed with 50 mm threonine. At this concentration of threonine, 440 μm [14C]GAR was formed, indicating that ≤1% of the threonine participated in the formation of PRA. When the threonine concentration was held constant at 50 mm and IlvA, TrpD, and PRPP were individually increased in the assay, no increase in GAR formation was observed. However, [14C]GAR formation was reduced if the concentration of PRPP or TrpD was decreased (10-fold, 2-fold, respectively). In the standard assay conditions, the concentration of IlvA could be reduced 10-fold without an effect on GAR formation. Together, these data indicated that under the assay conditions used, IlvA, TrpD, and PRPP were saturating.
FIGURE 5.
GAR formation as a function of threonine. PRA formation assays were performed as described under “Experimental Procedures.” The quantity of [14C]GAR produced with varying levels of threonine in the assay (1, 5, 10, 25, 50, 100, and 300 mm) was determined. Error bars represent the S.D. of two replicates.
YjgF Homologs from S. enterica and Human Decrease PRA Formation in Vitro
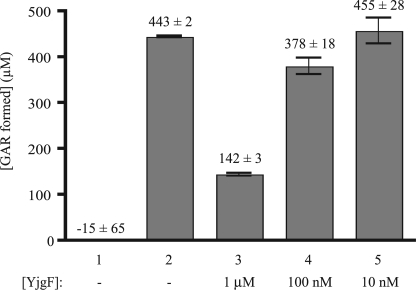
TrpD-dependent PRA formation only occurs in a yjgF mutant in vivo, consistent with the working model in which YjgF inhibits this mechanism of PRA formation (Fig. 3). To test this aspect of the model, purified YjgF was added to the in vitro PRA formation assay. The addition of YjgF (≥1 μm) to the assay decreased the amount of [14C]GAR formed more than 3-fold (Fig. 6), with lower concentrations of protein showing a graded response. Bovine serum albumin added to the same protein concentrations had no significant effect on [14C]GAR formation, and digestion of the YjgF protein by protease eliminated the inhibitory effect (data not shown). Significantly, 1 μm YjgF was able to prevent the formation of ∼300 μm GAR, suggesting that YjgF was catalytic or altered the kinetics of IlvA. Additionally, this result suggested that the TrpD-dependent PRA formation in vitro reconstituted the physiological mechanism occurring in vivo in the absence of yjgF.
FIGURE 6.
Addition of YjgF decreases PRA formation. PRA formation assays were performed as described under “Experimental Procedures,” and the amount of [14C]GAR was quantitated. Assays contained the indicated components and included purified YjgF protein as follows: lane 1, standard assay lacking TrpD; lane 2, reaction components of standard assay; lanes 3–5, purified YjgF present at the indicated final concentrations (1 μm, 100 nm, and 10 nm, respectively) in the standard assay. Error bars represent the S.D. of two replicates.
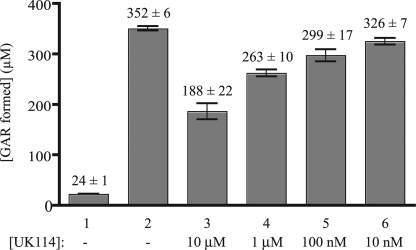
The broad conservation of the YjgF/YER057c/UK114 family of proteins prompted inclusion of the Homo sapiens homolog, UK114 (also called hp14.5), in the in vitro assay. The sequence of UK114 is 44% identical to S. enterica YjgF and is considered to be in the “high identity group” of this protein family (10). Purified UK114 was also able to impact TrpD-dependent PRA formation in vitro (Fig. 7). This result was consistent with the observation that expression of UK114 functionally complemented S. enterica yjgF phenotypes in vivo.4 Taken together, these data emphasized the potential of the in vitro PRA formation assay system to probe the biochemical function inherent to all members of the YjgF family.
FIGURE 7.
Human homolog of YjgF, UK114, decreases PRA formation. PRA formation assays were performed as described under “Experimental Procedures,” and the amount of [14C]GAR was quantitated. Assays contained the indicated components and included purified UK114 protein as follows: lane 1, standard assay lacking TrpD; lane 2, reaction components of standard assay; lanes 3–6, purified UK114 present at the indicated final concentrations (10 μm, 1 μm, 100 nm, and 10 nm, respectively) in the standard assay. Error bars represent the S.D. of two replicates.
DISCUSSION
YjgF is a member of the highly conserved YjgF/YER057c/UK114 family of proteins present in all three domains of life. In S. enterica, lack of this protein generates a number of metabolic phenotypes, including the ability to generate PRA in the absence of glutamine amidotransferase. Nutritional studies identified two enzymes and two metabolites that were considered to be involved in PRA formation in the absence of YjgF in vivo. Here, PRA formation was reconstituted in vitro with these four components: IlvA, TrpD, threonine, and PRPP. Importantly, addition of the YjgF protein to the assay inhibited PRA formation, suggesting that this reconstituted system reflected the in vivo situation from which it was derived.
The model for PRA formation addressed in this work (Fig. 3) was consistent with key in vivo findings: (i) isoleucine inhibited PRA formation (2), (ii) anthranilate inhibited PRA formation, (iii) threonine antagonized anthranilate inhibition, and (iv) PRA was not formed in strains with a functional YjgF. In the context of the model, allosteric inhibition of IlvA by isoleucine would reduce its activity(ies), resulting in less product, generically denoted X-NH3. Anthranilate is the natural substrate for TrpD and would be expected to compete with X-NH3 at the active site. Consistently, the presence of increased threonine would generate more X-NH3 and thus compete out the anthranilate.
The reconstitution of PRA formation in vitro required an atypical reaction catalyzed by TrpD. The substrate for this reaction, X-NH3, was dependent on IlvA and threonine and could reflect either an intermediate in the formation of 2-ketobutyrate or a byproduct of the reaction. Deamination of threonine by IlvA is mediated by a PLP cofactor and proceeds by a previously defined mechanism (14, 15). Briefly, IlvA dehydrates threonine and releases an enamine intermediate, aminocrotonate, which tautomerizes to the corresponding imine, iminobutyrate, and is then hydrolyzed in the absence of enzyme to form products 2-ketobutyrate and free ammonia. Despite the instability of the enamine and imine intermediates, it is conceivable one of these served as the amino donor in TrpD-mediated PRA formation. In the in vitro assay, 50 mm threonine resulted in formation of 440 μm GAR (representing PRA), with the remaining threonine converted to 2-ketobutyrate within 60 min (data not shown). This low yield of GAR is consistent with the diversion of a minor unknown species to PRA formation or, alternatively, a small percentage of the normal enamine/imine intermediate binding TrpD and forming PRA, rather than spontaneously hydrolyzing to 2-ketobutyrate. Preliminary results suggest the X-NH3 metabolite is unstable, perhaps favoring an explanation involving a reaction intermediate rather than a byproduct.5
The ability of YjgF protein to inhibit the TrpD-dependent PRA formation in vitro was predicted by both in vivo phenotypes and the working model for this class of proteins (8, 10, 11). The inhibition of PRA formation by YjgF displayed several features that provided insights into the function of this protein. First, the inhibition was titratable and declined with decreasing protein concentration. Second, the data suggested that YjgF had a catalytic role in the assay because 1 μm protein reduced PRA formation by ∼300 μm. Significantly, the human homolog (UK114) was active in this assay, consistent with the finding that UK114 complemented the yjgF mutant phenotypes in S. enterica4 and Saccharomyces cerevisiae (34) in vivo. Together, these results emphasized that this protein family is characterized both by high sequence identity and functional conservation.
The data presented here are consistent with a scenario in which a threonine-derived byproduct or intermediate is acted on by YjgF in a way that prevents its availability to TrpD for PRA synthesis. Structural studies identified an affinity of YjgF for analogs of the enamine intermediate formed from the dehydration of threonine, and these authors suggested that YjgF had a role in facilitating the IlvA reaction (10). Based on the broad distribution of this family, including organisms that lack IlvA, we hypothesize a general role of YjgF that involves a class of molecules (11, 35). A broad role for YjgF would account for the wide variety of nutritional and metabolic phenotypes that have been reported to involve YjgF in diverse organisms (11, 20, 27, 34, 36–43). For instance, each phenotype of a yjgF mutant could result from a distinct metabolite impacting a cellular process either positively or negatively. In the assay described here, one such metabolite (X-NH3) and effect (gained PRA synthesis) has been identified and reconstituted in vitro.
In summary, the results described here identified a new enzymatic mechanism of PRA formation that involves nonstandard substrates and/or products of two central metabolic enzymes in S. enterica. Further, this work defined the first assay for diverse members of the YjgF/YER057c/UK114 family of proteins and provides a system to dissect further the biochemical function of these proteins.
Acknowledgments
We thank Melissa Christopherson, Benjamin Bice, George Schmitz and Eugenio Vivas who provided purified proteins (UK114, IlvA, IlvE, and TdcB, respectively) that were used in this study.
This work was supported, in whole or in part, by National Institutes of Health Competitive Grant GM47296. This work was also supported by a 21st Century Scientist Scholars award from the J. S. McDonnell Foundation (to D. M. D.).
M. Christopherson, G. E. Schmitz, and D. M. Downs, unpublished data.
J. A. Lambrecht and D. M. Downs, unpublished data.
- PRA
- phosphoribosyl amine
- GAR
- glycinamide ribonuclotide
- Ni-NTA
- nickel-nitrilotriacetic acid
- PLP
- pyridoxal-5′-phosphate
- PRPP
- phosphoribosyl pyrophosphate.
REFERENCES
- 1.Skovran E., Lauhon C. T., Downs D. M. (2004) J. Bacteriol. 186, 7626–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enos-Berlage J. L., Langendorf M. J., Downs D. M. (1998) J. Bacteriol. 180, 6519–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd J. M., Pierik A. J., Netz D. J., Lill R., Downs D. M. (2008) Biochemistry 47, 8195–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tso J. Y., Hermodson M. A., Zalkin H. (1982) J. Biol. Chem. 257, 3532–3536 [PubMed] [Google Scholar]
- 5.Newell P. C., Tucker R. G. (1968) Biochem. J. 106, 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen L., Enos-Berlage J., Downs D. M. (1996) Genetics 143, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enos-Berlage J. L., Downs D. M. (1996) J. Bacteriol. 178, 1476–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne B. A., Ramos A. I., Downs D. M. (2006) J. Bacteriol. 188, 6786–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatusov R. L., Koonin E. V., Lipman D. J. (1997) Science 278, 631–637 [DOI] [PubMed] [Google Scholar]
- 10.Parsons L., Bonander N., Eisenstein E., Gilson M., Kairys V., Orban J. (2003) Biochemistry 42, 80–89 [DOI] [PubMed] [Google Scholar]
- 11.Schmitz G., Downs D. M. (2004) J. Bacteriol. 186, 803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauerle R. H., Margolin P. (1966) Cold Spring Harbor Symp. Quant. Biol. 31, 203–214 [DOI] [PubMed] [Google Scholar]
- 13.Zalkin H. (1973) Adv. Enzymol. Relat. Areas Mol. Biol. 38, 1–39 [DOI] [PubMed] [Google Scholar]
- 14.Chargaff E., Sprinson D. B. (1943) J. Biol. Chem. 151, 273–280 [Google Scholar]
- 15.Phillips A. T., Wood W. A. (1965) J. Biol. Chem. 240, 4703–4709 [PubMed] [Google Scholar]
- 16.Davis R. W., Botstein D., Roth J. R.Cold Spring Harbor Laboratory (1980) Advanced Bacterial Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17.Vogel H. J., Bonner D. M. (1956) J. Biol. Chem. 218, 97–106 [PubMed] [Google Scholar]
- 18.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos I., Downs D. M. (2003) J. Bacteriol. 185, 5125–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmiedeknecht G., Kerkhoff C., Orsó E., Stöhr J., Aslanidis C., Nagy G. M., Knuechel R., Schmitz G. (1996) Eur. J. Biochem. 242, 339–351 [DOI] [PubMed] [Google Scholar]
- 21.Roberts G. P. (1978) Isolation and Characterization of Informational Suppressors in Salmonella typhimurium. Ph.D. thesis, University of California, Berkeley [Google Scholar]
- 22.Schmieger H. (1972) Mol. Gen. Genet. 119, 75–88 [DOI] [PubMed] [Google Scholar]
- 23.Ramos I., Vivas E. I., Downs D. M. (2008) J. Bacteriol. 190, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grieshaber M., Bauerle R. (1974) Biochemistry 13, 373–383 [DOI] [PubMed] [Google Scholar]
- 25.Henderson E. J., Nagano H., Zalkin H., Hwang L. H. (1970) J. Biol. Chem. 245, 1416–1423 [PubMed] [Google Scholar]
- 26.Burns R. O. (1971) Methods Enzymol. 17B, 555–560 [Google Scholar]
- 27.Christopherson M. R., Schmitz G. E., Downs D. M. (2008) J. Bacteriol. 190, 3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koenigsknecht M. J., Ramos I., Downs D. M. (2007) J. Biol. Chem. 282, 28379–28384 [DOI] [PubMed] [Google Scholar]
- 29.Nierlich D. P., Magasanik B. (1965) J. Biol. Chem. 240, 366–374 [PubMed] [Google Scholar]
- 30.Schendel F. J., Cheng Y. S., Otvos J. D., Wehrli S., Stubbe J. (1988) Biochemistry 27, 2614–2623 [DOI] [PubMed] [Google Scholar]
- 31.Blume A. J., Balbinder E. (1966) Genetics 53, 577–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umbarger H. E., Brown B. (1957) J. Bacteriol. 73, 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luginbuhl G. H., Hofler J. G., Decedue C. J., Burns R. O. (1974) J. Bacteriol. 120, 559–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oxelmark E., Marchini A., Malanchi I., Magherini F., Jaquet L., Hajibagheri M. A., Blight K. J., Jauniaux J. C., Tommasino M. (2000) Mol. Cell. Biol. 20, 7784–7797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz G. (2006) A Link between yjgF and Isoleucine Biosynthesis in Salmonella enterica Serovar Typhimurium. Ph.D. thesis, University of Wisconsin-Madison, Madison [Google Scholar]
- 36.Kim J. M., Yoshikawa H., Shirahige K. (2001) Genes Cells 6, 507–517 [DOI] [PubMed] [Google Scholar]
- 37.Goupil-Feuillerat N., Cocaign-Bousquet M., Godon J. J., Ehrlich S. D., Renault P. (1997) J. Bacteriol. 179, 6285–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappu P., Shin B. S., Zalkin H., Mäntsälä P. (1999) J. Bacteriol. 181, 3810–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zelyas N. J., Cai H., Kwong T., Jensen S. E. (2008) J. Bacteriol. 190, 7957–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Z., Spain J. C. (1998) J. Bacteriol. 180, 2502–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melloni E., Michetti M., Salamino F., Pontremoli S. (1998) J. Biol. Chem. 273, 12827–12831 [DOI] [PubMed] [Google Scholar]
- 42.D'Inca R., Marteil G., Bazile F., Pascal A., Guitton N., Lavigne R., Richard-Parpaillon L., Kubiak J. Z. (2010) J. Proteomics 73, 1542–1550 [DOI] [PubMed] [Google Scholar]
- 43.Kim K. S., Pelton J. G., Inwood W. B., Andersen U., Kustu S., Wemmer D. E. (2010) J. Bacteriol. 192, 4089–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castilho B. A., Olfson P., Casadaban M. J. (1984) J. Bacteriol. 158, 488–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. (1984) Gene 32, 369–379 [DOI] [PubMed] [Google Scholar]