Abstract
Mitoquinone (MitoQ) is a synthetically modified, redox-active ubiquinone compound that accumulates predominantly in mitochondria. We found that MitoQ is 30-fold more cytotoxic to breast cancer cells than to healthy mammary cells. MitoQ treatment led to irreversible inhibition of clonogenic growth of breast cancer cells through a combination of autophagy and apoptotic cell death mechanisms. Relatively limited cytotoxicity was seen with the parent ubiquinone coenzyme Q10. Inhibition of cancer cell growth by MitoQ was associated with G1/S cell cycle arrest and phosphorylation of the checkpoint kinases Chk1 and Chk2. The possible role of oxidative stress in MitoQ activity was investigated by measuring the products of hydroethidine oxidation. Increases in ethidium and dihydroethidium levels, markers of one-electron oxidation of hydroethidine, were observed at cytotoxic concentrations of MitoQ. Keap1, an oxidative stress sensor protein that regulates the antioxidant transcription factor Nrf2, underwent oxidation, degradation, and dissociation from Nrf2 in MitoQ-treated cells. Nrf2 protein levels, nuclear localization, and transcriptional activity also increased following MitoQ treatment. Knockdown of Nrf2 caused a 2-fold increase in autophagy and an increase in G1 cell cycle arrest in response to MitoQ but had no apparent effect on apoptosis. The Nrf2-regulated enzyme NQO1 is partly responsible for controlling the level of autophagy. Keap1 and Nrf2 act as redox sensors for oxidative perturbations that lead to autophagy. MitoQ and similar compounds should be further evaluated for novel anticancer activity.
Keywords: Anticancer Drug, Apoptosis, Cancer Therapy, Mitochondria, Oxidative Stress
Introduction
Mitochondria are central to many cellular processes, including energy production and apoptosis. A by-product of electron transfer in the mitochondria is the formation of superoxide. When superoxide is not rapidly detoxified, there is higher likelihood for generation of additional oxidants in mitochondria, leading to oxidative damage. Hence, targeting the mitochondria with agents capable of scavenging oxidants is an attractive cytoprotective strategy (1). Conjugation with a cationic triphenylphosphonium moiety is a method for targeting therapeutic molecules to the mitochondria (2). Mitoquinone (MitoQ)3 is one such conjugate. It is derived from the mitochondrial respiratory chain component ubiquinone (3–5). The ubiquinone moiety is covalently linked by an aliphatic 10-carbon chain to a triphenylphosphonium cation. As a lipophilic cation, it can pass easily through lipid bilayers. Driven by the large membrane potential in the mitochondria, it selectively accumulates in the mitochondria. MitoQ is orally active and is being pursued as a potential therapy in degenerative conditions, such as Parkinson disease (2). Although initially designed as an antioxidant, mitochondrial redox cycling of mitoquinone has been shown to lead to superoxide production and apoptotic cell death (6, 7).
Nrf2 (NF-E2-related factor 2) is a major transcription factor for antioxidant response element (ARE)-dependent gene transcription in response to agents that cause oxidative stress (including by the quinone oxidant tBHQ) (8). Nrf2 is critical in the regulation of antioxidant and cytoprotective genes. In the absence of oxidative stress, Nrf2 is sequestered within the cytosol by its inhibitory partner and principle regulator, Keap1 (Kelch-like ECH-associating protein 1). Nrf2 activity is normally limited by Keap1, which constitutively facilitates ubiquitination of Nrf2 and causes its ongoing proteasomal degradation (9). Keap1 can act as a sensor for oxidative stress due to its highly reactive cysteine residues. In vitro, in the presence of free radicals, Nrf2 is released from Keap1 and translocates to the nucleus, where it binds to the ARE and activates gene expression. One such Nrf2-regulated gene is NQO1 (NAD(P)H:quinine oxidoreductase 1). NQO1 is a cytoprotective flavoprotein that catalyzes the two-electron reduction of various quinones and redox dyes (10).
The role of mitochondrial oxidative phosphorylation and signaling in cancer cells is complex and controversial (11, 12). Cancer cells have a greater mitochondrial membrane potential than healthy cells (13). Rhodamine 123, a lipophilic cationic probe, selectively accumulates in the mitochondria and consequently induces greater toxicity in cancer cells (14, 15). Likewise, we found that mitoquinone has significant and selective anticancer activity, unlike untargeted ubiquinone (CoQ10). Here, we demonstrate that the Nrf2-Keap1 proteins act as redox sensors that relay the signal for a failed cytoprotective response to MitoQ. Data are provided to elucidate the cellular response to MitoQ and to determine its novel underlying mechanism of action.
EXPERIMENTAL PROCEDURES
Synthesis of MitoQ
A new method was developed to synthesize MitoQ starting from the commercially available idebenone (see scheme in supplemental Fig. 1).
Idebenone (3.38 g, 0.01 mol) was stirred in 50 ml of dichloromethane containing triethylamine (2 g, 0.02 mol) in an ice bath. To this mixture, methanesulfonyl chloride (2.24 g, 0.02 mol) in 20 ml of dichloromethane was added dropwise. The mixture was stirred on ice for 1 h, and the solution was shaken with 1 m HCl until washings became acidic. The solution was dried, and solvent was removed using a rotary evaporator. The orange semisolid was chromatographed on a silica gel column, and the orange band was eluted with ether-hexane (1:1) solvent. Homogeneous fractions were combined, and solvent was subsequently removed to obtain an orange solid. The yield was almost quantitative. Thin layer chromatography (TLC) on silica gel plates showed a single spot. The methane sulfonyl ester was dissolved in 50 ml of dry acetone, and lithium bromide (1.75 g, 0.02 mol) was added and stirred at room temperature for 24 h. The solvent acetone was removed by evaporation, and the product was extracted with 50 ml of ether. The ether extract was washed with water (2 × 25 ml), the orange solution was dried, and solvent was removed. The compound was purified by silica gel column chromatography, and the bromoderivative of idebenone was eluted with ether/hexane (1:1). Homogeneous fractions were combined, and solvent was removed to get an orange solid. The yield was 3.56 g (89%). TLC in ether/hexane solvent showed a single spot that moved ahead of the starting methanesulfonyl ester.
The bromoderivative was reduced with sodium borohydride (NaBH4) to the corresponding quinol. The bromoderivative was dissolved in 50 ml methanol, and sodium borohydride (1.5 g, 0.04 mol) was slowly added in small aliquots with stirring. After the addition, stirring was continued for an additional 30 min, and the colorless solution was acidified with dilute hydrochloric acid and extracted with 50 ml of chloroform. The organic layer was dried, solvent was removed by blowing nitrogen gas, and the volatiles were removed by vacuum. The resulting colorless oil (quinol) was immediately used in the next step to prevent further oxidation in air to the corresponding quinone.
In 25 ml of dioxane, triphenylphosphene (3.9 g, 0.015 mol) was added to the quinol and kept refluxed under a nitrogen atmosphere for 48 h. The solvent was partially removed by rotary evaporation, and 50 ml of ether was added with stirring. The precipitated product was collected by decantation, and the residual ether was removed by blowing nitrogen gas. The semisolid product was dissolved in 5 ml of dichloromethane and added to the solution in drops to 50 ml of ether with stirring. The precipitated product was collected as before by decantation. The dried product was dissolved in dichloromethane and precipitated from ether, and this process was repeated four times. The purified product consisted of mitoquinol with trace amounts of the oxidized compound mitoquinone. The product was dissolved in 50 ml of ethanol and was gently bubbled with air for 24 h to allow complete oxidation of mitoquinol to mitoquinone. The final quantity of mitoquinone was 3.1 g with an overall yield of 47%. The purity was confirmed by HPLC and LC/MS (mass = 583).
Other Chemicals
Doxorubicin was provided by NCI-Frederick, National Institutes of Health. Co-enzyme Q10, acridine orange, bafilomycin A, pepstatin, E64d, tBHQ, rapamycin, and propidium iodide (PI) were purchased from Sigma-Aldrich. Recombinant human TRAIL was generously provided by Dr. Baolin Zhang (Food and Drug Administration, Bethesda, MD).
Cell Culture
MDA-MB-231 and MCF-7 breast cancer cells were obtained from ATCC (Manassas, VA) and maintained in DMEM/F-12 medium. MCF-12A healthy human mammary epithelial cells were obtained from BioWhittaker Clonetics (Walkersville, MD) and maintained in mammary epithelial cell growth medium (16). Isogenic MDA-MB-231-NQO1-negative and -positive were generously provided by Dr. Erik Bey (University of Texas Southwestern Medical Center, Dallas, TX) (17). MDA-MB-231 cells were stably transfected with a CMV-driven NQO1 cDNA (MDA-MB-231-NQO1-positive) or the pcDNA3 vector alone (MDA-MB-231-NQO1-negative). Control experiments were performed to confirm the NQO1 levels (see Fig. 7A, inset).
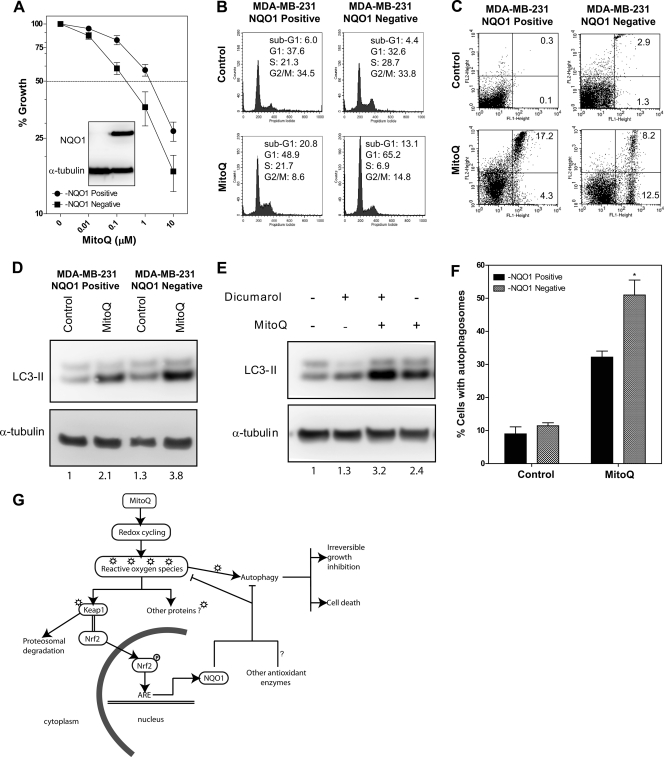
FIGURE 7.
NQO1 plays an essential role in regulating autophagy and cell cycle arrest but not apoptosis after MitoQ treatment. Isogenic MDA-MB-231 cells that were deficient for NQO1 (due to 609C>T polymorphism in NQO1) or with restored expression for NQO1 (using a CMV-driven NQO1 cDNA) were used. A, MDA-MB-231-NQO1-negative cells (with pCDNA3 vector alone, left lane) showed no detectable NQO1 protein in contrast to the MDA-MB-231-NQO1-positive cells (inset). The MDA-MB-231-NQO1-negative cells showed greater reduction in growth than the MDA-MB-231-NQO1-positive cells when exposed to increasing concentrations of MitoQ at 72 h. B, the cell cycle profiles of MDA-MB-231-NQO1-negative cells or MDA-MB-231-NQO1-positive cells exposed to 1 μm MitoQ for 48 h were analyzed by flow cytometry following staining with PI. The percentage of cells in each cell cycle phase was quantified and is provided in each panel. C, apoptosis was measured in live cells after staining with annexin V and propidium iodide following MitoQ treatment at 48 h. The percentage of cells that were annexin V-positive was used as a measure of apoptosis. D, autophagosome-associated LC3-II protein was measured as an indicator of autophagy after 24 h exposure to MitoQ in the MDA-MB-231-NQO1-negative and MDA-MB-231-NQO1-positive cells. E, the NQO1 inhibitor, dicumarol, was used in MDA-MB-231 cells to confirm results from the isogenic cell lines (50 μm, 1-h pretreatment). F, percentage of cells with autophagosomes in the isogenic cell lines followed by MitoQ treatment were counted as in Fig. 3D. *, statistical significance compared with control siRNA cells. G, schematic model for the proposed mechanism for the anticancer cytotoxicity induced by MitoQ (see “Discussion”). Error bars, S.D.
Colony Formation Assay
Cell survival was determined using a colony formation assay after the indicated treatments for 6 h and replating for 7 days followed by washing, fixation, and staining with Wright's Giemsa stain (Sigma) (18). Colonies were counted using a light microscope and triplicate samples from each experiment. Colonies with greater than 30 cells each were counted.
Assessment of Antiproliferative Activity
Growth inhibition was assessed by the sulforhodamine B (SRB; Sigma-Aldrich) assay as described previously (18). GI50 values were calculated using Prism 4 software (GraphPad Software, Inc.).
Flow Cytometric Analysis
For cell cycle analyses, cells were fixed in 70% ethanol and stained with PI after treatment with 5 μg/ml RNase A. Cells were analyzed by a FACSCaliber flow cytometer (BD Biosciences), and cell cycle phases were quantitated using the CellQuest program. For apoptosis analyses, cells were stained with annexin V and PI (16). Both floating and attached cells were collected for FACS analyses.
Ultrastructural Analysis of Autophagy by Transmission Electron Microscopy
Transmission electron microscopy analysis was carried out using either high pressure freezing/freeze substitution or chemical fixation/resin flat embedding as described previously (19).
Quantitation of Percentage of Autophagic Cells
Autophagosomes were identified and counted with a fluorescence microscope using a 40× objective. Autophagosomes were identified from the phase-contrast image using the intracellular structure images obtained by electron microscopy for comparison. A representative example is provided below the graph in Fig. 3D. The percentage of cells with autophagosomes was quantitated by counting cells that stained orange after treatment with acridine orange to stain the acidic vesicular organelles (see supplemental Fig. 2) (20).
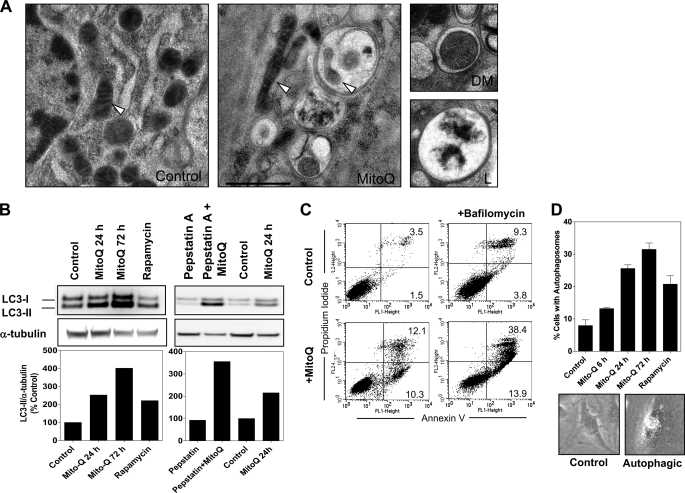
FIGURE 3.
Induction of autophagy by MitoQ in MDA-MB-231 cells. A, transmission electron microscopy was used to study the formation of autophagic vacuoles after 48 h of treatment with 1 μm MitoQ. Representative images are shown from untreated and MitoQ-treated cells with autophagosomes, including one with an engulfed mitochondrion (mitophagy). The mitochondria are marked by white arrowheads. Scale bar, 1 μm. The two smaller images from MitoQ-treated cells show a double-membrane autophagic vacuole (labeled DM) and a fused late stage autophagosome with degraded cellular contents (labeled L). B, the increased levels of the autophagosome-associated LC3-II subunit were used as confirmation of autophagy. MitoQ (1 μm) was tested at 24 and 72 h. Rapamycin was used as a positive control. Increase in LC3-II was confirmed to be from autophagic flux using lysosomal protease inhibitors pepstatin A (10 μg/ml) and E64d (10 μg/ml) in the presence or absence of MitoQ (right). The graphs below show the densitometric quantitation of LC3-II protein levels normalized to α-tubulin levels from a representative analysis. C, measurement of apoptosis following inhibition of autophagy by bafilomycin A using annexin V-PI staining and flow cytometry. MDA-MB-231 cells were treated with MitoQ in the presence or absence of 5 nm bafilomycin A (2-h pretreatment). D, the percentage of cells with autophagosomes (Autophagic) was counted in MitoQ- or rapamycin-treated cells. Error bars, S.D.
Mitochondrial Membrane Potential (ΔΨm)
Cells were incubated for 15 min at 37 °C with the lipophilic cationic probe 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) (Molecular Probes) (5 μg/ml) and assayed using a fluorescence plate reader (Molecular Devices) with the following settings: excitation at 485 nm, emission at 540 and 590 nm. ΔΨm was calculated using the ratio of 590 nm (aggregates)/540 nm (monomeric form).
Immunoprecipitation and Western Blot Analysis
The procedures for lysate preparation, immunoprecipitation, and Western blotting were performed as reported previously (18, 21).
Detection of Modified Keap1
Oxidative stress-induced Keap1 dimer formation was detected using an electrophoretic mobility shift method (9). After the indicated treatments, cells were lysed in lysis buffer (10 mm sodium phosphate (pH 8.0), 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.2% SDS) containing 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture (Sigma-Aldrich). Whole cell protein samples were electrophoresed using a 4–12% BisTris gel in the presence or absence of reducing agent (β-mercaptoethanol). Following transfer, the Immobilon-P membrane was probed with anti-Keap1 antibody (generously provided by Dr. Mark Hannink, University of Missouri (Columbia, MO)) (9). The slower migrating form of Keap1 (68 kDa) was detected at >120 kDa only under non-reducing conditions.
Confocal Microscopy of Nuclear Protein Localization
Slides were prepared as described previously (22). Antibodies for phospho-Chk1-Ser317, phospho-Chk1-Ser345, and phospho-Chk2-Thr68 were obtained from Cell Signaling Technology (Danvers, MA). Antibody for Nrf2 was obtained from Abcam. Nuclei were stained with DAPI. Immunofluorescence microscopy was performed with a Ziess LSM510 confocal system (Carl Ziess Microimaging, Thornwood, NY).
Hydroethidine Assay with HPLC Analysis
For the detection of superoxide and/or other reactive oxygen species (ROS), cells were incubated in DMEM with 2% FBS and treated with 10 μm HE during the last 30 min of incubation with MitoQ. After the incubation, the cells were processed and analyzed by HPLC with electrochemical detection, as described previously (23).
Transcriptional Activity of Nrf2
Nuclear extracts were prepared from MDA-MB-231 cells using the nuclear extract kit by Active Motif (Carlsbad, CA). The presence of nucleophosmin and absence of GAPDH protein were used as a measure of the purity of nuclear extracts. An ELISA-based assay consisting of an immobilized oligonucleotide containing the ARE consensus binding site (5′-GTCACAGTGACTCAGCAGAATCTG-3′) was used to measure Nrf2 DNA binding activity (TransAM kits, Active Motif, catalog no. 50296). Nrf2 from 2.5 μg of nuclear extract/sample was allowed to bind to the ARE on the 96-well plates. A primary antibody against Nrf2 was then used to detect bound Nrf2. A secondary antibody conjugated to horseradish peroxidase (HRP) provided a colorimetric readout at 450 nm. Nuclear extracts from COS-7 cells transfected with Nrf2 were included as the positive control.
Knockdown of Nrf2
For siRNA experiments, MDA-MB-231 cells were plated in medium lacking antibiotic. After 24 h, the medium was replaced, and 100 nm Nrf2 ON-TARGETplus SMARTPOOL (catalog no. L-003755-00) or ON-TARGETplus non-coding siRNA (catalog no. D-001810-0105) (Dharmacon, Chicago, IL) was transfected using DharmaFECT 1 transfection reagent as described by the manufacturer.
Statistical Analysis
Data shown are representative of at least three independent experiments, unless noted otherwise. Error bars on graphs in all figures denote S.D. values. Student's t test was used to ascertain statistically significant differences (p < 0.05).
RESULTS
Cytotoxicity of MitoQ in Healthy Cells and Cancer Cell Lines
The growth-inhibitory activity of MitoQ was tested in the breast cancer cell lines MDA-MB-231 and MCF-7 and in MCF12A primary mammary epithelial cells using the SRB dye assay (Fig. 1A, left). The effects of MitoQ were compared with untargeted CoQ10 in the same cell lines (Fig. 1A, right). Breast cancer cell lines were more sensitive to MitoQ than healthy mammary cells. The GI50 values for MitoQ in MDA-MB-231, MCF-7, and MCF12A cell lines were 296 nm, 113 nm, and >10 μm, respectively. In contrast, the GI50 values for CoQ10 in these cells were 8.2 μm (MDA-MB-231) and >10 μm (for MCF-7 and MCF-12A). Thus, MitoQ exhibits selective antiproliferative effects in breast cancer cells as compared with healthy mammary cells.
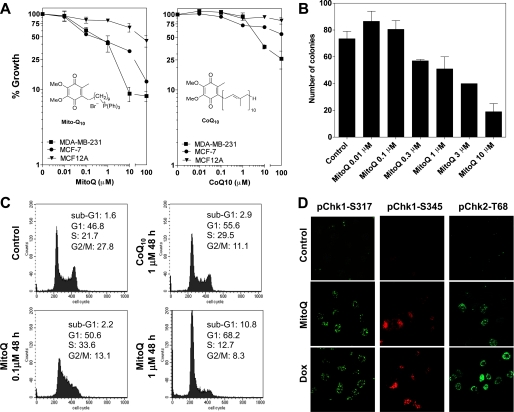
FIGURE 1.
Growth inhibition and cell cycle arrest by MitoQ or CoQ10. A and B, an SRB dye-based assay was used for measuring cell viability following increasing concentrations of either MitoQ (left) or CoQ10 (right) after 72 h. Breast cancer cell lines (MDA-MB-231 or MCF-7) or healthy breast epithelial cells (MCF-12A) were treated as indicated. B, a colony formation assay was performed on MDA-MB-231 cells incubated with MitoQ. Cells were incubated with MitoQ for 6 h, followed by replacement of medium and replating for a colony formation period of 7 days. C, MDA-MB-231 cells were treated with either MitoQ or CoQ10, as indicated, for 48 h. The cells were fixed and stained with PI followed by flow cytometric analysis. The percentage of cells in each cell cycle phase was quantitated and is provided in each panel. D, the phosphorylation of Chk1 and Chk2 kinases was assayed using confocal microscopy. Representative images of increased nuclear foci corresponding to phospho-Chk1-Ser317 (pChk1-S317), phospho-Chk2-Thr68 (pChk1-T68), and cytoplasmic phospho-Chk1-Ser345 (pChk1-S345) after 0.1 μm MitoQ or 0.1 μm doxorubicin (Dox) treatment (24 h) are shown. Error bars, S.D.
One of the hallmark traits of cancer cells is their ability to undergo isolated clonal growth. Results obtained from the SRB growth inhibition assay were confirmed using a colony formation assay for cancer cell killing (Fig. 1B). MDA-MB-231 cells were exposed to MitoQ for 6 h, followed by removal of media and replating. MDA-MB-231 cells were evaluated for their colony formation ability for up to 7 days in drug-free media. MitoQ elicited a dose-dependent decrease in clonogenic growth.
Cell Cycle Arrest by MitoQ
To better understand the mechanism of cytotoxicity by MitoQ, we analyzed the cell cycle and apoptosis profile of MDA-MB-231 cells following drug exposure at 48 h (Fig. 1C). MitoQ arrested MDA-MB-231 cells in the G1/S phase of the cell cycle. Compared with control cells, the percentage of G1 cells increased from 46.8% to 50.6 and 68.2% after exposure to 0.1 and 1 μm MitoQ, respectively. A concentration-dependent increase in sub-G1 DNA-containing cells, indicative of apoptosis, was also detected. With 1 μm MitoQ, 10.8% of cells were apoptotic compared with 1.6% in control cells. The cell cycle arrest by MitoQ was compared with that by the untargeted ubiquinone, CoQ10. In contrast to MitoQ, 1 μm CoQ10 produced a smaller G1/S phase arrest and produced significantly fewer sub-G1 DNA-containing cells (2.9%) (Fig. 1C). To determine the mechanism by which MDA-MB-231 cells undergo G1/S cell cycle arrest, we assayed the phosphorylation of cell cycle checkpoint kinases Chk1 and Chk2. These proteins help maintain the cell cycle checkpoint and may also promote apoptosis (24) via a complex network of phosphorylation-dephosphorylation events (25). MDA-MB-231 cells exposed to 0.1 μm MitoQ showed significant phosphorylation of phospho-Chk1-Ser317, phospho-Chk1-Ser345, and phospho-Chk2-Thr68 (Fig. 1D). Doxorubicin served as a positive control. The data show that MitoQ causes cell cycle arrest and checkpoint kinase activation in cancer cells.
Although 40–60% of the cells underwent growth inhibition (Fig. 1A) with 1 μm MitoQ, this did not appear to translate into apoptotic events (∼11%, as indicated by the number of sub-G1 DNA-containing cells). We performed FACS analysis of annexin V-PI staining of live cells treated with 1 μm MitoQ (Fig. 2A) to further quantify apoptosis. A time course study with MitoQ showed no appreciable apoptosis prior to 24 h. Following treatment with MitoQ for 48 and 72 h, 21.8 and 28.8% of cells were annexin V-positive compared with 3% in control cells. Comparable results were obtained using Hoechst-PI staining to quantify apoptosis (not shown). We conclude that apoptosis only accounts for part of the mechanism by which MitoQ elicits its anticancer growth inhibition.
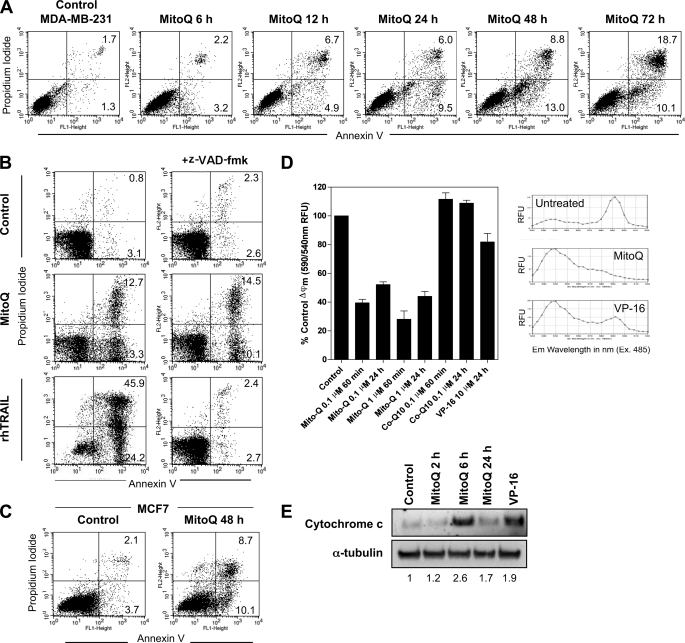
FIGURE 2.
Induction of apoptosis by MitoQ. A, apoptosis was measured by staining with annexin V-PI after a time course of exposure to 1 μm MitoQ from 6 to 72 h. B, cells were also tested after pretreatment for 30 min with the pan-caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone (z-VAD-fmk) prior to MitoQ (48 h, middle). Treatment with recombinant human TRAIL (rhTRAIL; 10 ng/ml) was used as a positive control for caspase-dependent apoptosis (bottom). C, apoptosis was measured in caspase-3-deficient MCF7 cells after 48 h of MitoQ. D, loss of mitochondrial membrane potential was measured by conversion of the monomeric JC-1 dye (fluorescence emission peak at 590 nm) to its aggregated form (fluorescence emission peak at 540 nm). Co-Q10 and VP-16 (10 μm) were included as controls. The JC-1 emission spectra were taken on cells untreated or treated with MitoQ or VP-16 after 1 h. Representative examples of emission spectra are shown. E, release of cytochrome c into the cytosol was measured by Western blot analysis of cytosolic cell fractions following 1 μm MitoQ treatment and using VP-16 treatment for 24 h as a positive control. The numbers below the bands shows the densitometric quantitation of cytochrome c protein levels normalized to α-tubulin levels from a representative analysis. RFU, relative fluorescence units; Error bars, S.D.
To test whether apoptosis by MitoQ was dependent on caspases, we pretreated cells with 10 μm benzyloxycarbonyl-VAD-fluoromethyl ketone for 30 min followed by MitoQ for 48 h (Fig. 2B). Pretreatment with benzyloxycarbonyl-VAD-fluoromethyl ketone did not appreciably lower the total percentage of annexin V-positive cells (18.6% compared with 22.4%). Cells treated with recombinant human TRAIL (10 ng/ml for 16 h) were used as positive control for caspase-dependent apoptosis and showed abrogation of apoptosis in the presence of benzyloxycarbonyl-VAD-fluoromethyl ketone. We also found no significant difference in the percentage of MitoQ-induced annexin V-positive cells in MCF7 cells, which lack caspase-3 (Fig. 2C). Hence, we conclude that MitoQ-induced apoptosis is not caspase-dependent.
Mitochondrial Membrane Depolarization by MitoQ
To determine whether MitoQ causes disruption of the mitochondrial membrane, we studied the mitochondrial membrane potential in MDA-MB-231 breast cancer cells after drug exposure. Conversion of red fluorescent aggregates to green fluorescent monomers of the JC-1 dye upon membrane depolarization marks changes in membrane potential. Exposure of cells to MitoQ (0.1 and 1 μm) led to a rapid membrane depolarization within 60 min of exposure (Fig. 2D). Membrane depolarization appeared to be sustained for up to 24 h after drug exposure. In contrast to MitoQ, CoQ10 did not exhibit any appreciable effect at comparable concentrations. VP-16 was included as a positive control (26). Next, the release of cytochrome c from the mitochondria to the cytosol was measured as an indicator of mitochondrial damage after MitoQ treatment (1 μm). A distinct increase in cytosolic cytochrome c protein levels was observed at 6 h (Fig. 2E). Thus, MitoQ exposure is accompanied by a rapid membrane depolarization and cytochrome c release that may be partly responsible for the apoptosis induced in breast cancer cells.
Induction of Autophagy by MitoQ
Our findings indicate that apoptosis may not be the major mechanism responsible for the antiproliferative activity of MitoQ. We therefore hypothesized that MitoQ may induce autophagy that leads to inhibition of proliferation. Autophagy is one of the consequences of stressing cells, and it is emerging as an important mechanism to explain drug responses in cancer cells (27–29). Autophagy is characterized by formation of autophagic vacuoles (autophagosomes) (30, 31), which can be seen by transmission electron microscopy (31). Autophagosomes were observed after 24 h of incubation with 1 μm MitoQ (Fig. 3A). The presence of mitochondria within the autophagic vacuole (“mitophagy”) was also noted (labeled by arrowheads) (32). Mitophagy is a potential mechanism for turnover of dysfunctional mitochondria, in agreement with the rapid membrane depolarization and cytochrome c release (Fig. 2D) (32). Also shown are representative images of an early autophagic vacuole with a double membrane (labeled DM) and the fusion of two late stage autophagic vacuoles with degraded cellular contents (labeled L) after MitoQ treatment. Additionally, the formation of the proteolytic and lipidated maturation product of the LC3 protein LC3-I (microtubule-associated light chain 3 protein, termed LC3-II) correlates with the extent of autophagic activity (31–34). We found an increase of LC3-II protein at 24 and 72 h after MitoQ treatment of MDA-MB-231 cells (Fig. 3B). Rapamycin (at 10 μm, 24 h) served as the positive control. To confirm that the increase in LC3-II was not transient but indicative of autophagic increase in LC3-II that is subsequently degraded by lysosomal hydrolases after formation of autolysosomes (autophagic flux), we tested the LC3-II in the presence of E64d and pepstatin A, which inhibit lysosomal proteases (31, 35). The presence of lysosomal inhibitors substantially stabilized the LC3-II induction by MitoQ (Fig. 3B, right). It should be noted that due to the massive induction of LC3-II in the presence of lysosomal inhibitors, the Western blot membranes had to be exposed for different periods of time to show clear and quantifiable bands for all samples. Finally, we found that pretreatment with the autophagy inhibitor bafilomycin A1 triggers increased apoptosis in response to 1 μm MitoQ (Fig. 3C). Bafilomycin acts as a vacuolar H+-ATPase inhibitor and prevents acidification of lysosomes and rematuration/fusion of autophagic vacuoles (36). Bafilomycin A1 (a 2-h pretreatment with 5 nm) synergistically increased the percentage of annexin V-positive cells by MitoQ. This result is in agreement with other recent reports showing increased apoptosis by autophagy-inducing agents in the presence of bafilomycin A1 (37, 38). A time course of autophagosome induction in response to MitoQ is shown in Fig. 3D. The percentages of cells with detectable autophagosomes were counted manually. After 72 h, about 31% of cells were autophagic in response to MitoQ (representative cells are shown below the graph). Collectively, this demonstrates that MitoQ induces autophagy at growth-inhibitory concentrations in MDA-MB-231 cells.
MitoQ-induced Modification of Oxidative Stress Sensor Keap1 and Dissociation from the Antioxidant Transcription Factor Nrf2
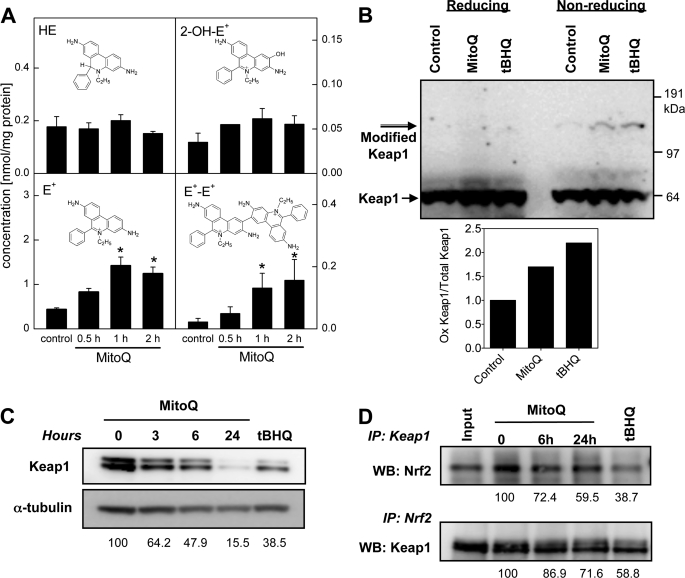
To elucidate the nature of the cellular stress from MitoQ, experiments were performed to investigate if MitoQ generates ROS in breast cancer cells. The effect of MitoQ (1 μm) on ROS production in MDA-MB-231 cells was tested using a hydroethidine (HE) assay with HPLC-based detection of HE-derived oxidation products (39, 40) (Fig. 4A). In the presence of 1 μm MitoQ, there was a significant increase in the HE oxidation products ethidium (E+) and diethidium (E+-E+). Formation of E+ and E+-E+ is attributed to increased levels of one-electron oxidizing species. The increase in 2-hydroxyethidium (2-OH-E+) was not statistically significant. Note that the intracellular level of HE did not change significantly with different treatments. Thus, the observed changes are not due to variations in the availability of the intracellular probe for oxidants (HE) (6, 23). These data are in agreement with a previous report on detection of superoxide generated by MitoQ (6). The formation of E+ and E+-E+ levels indicates that MitoQ stimulates intracellular generation of one-electron oxidizing species.
FIGURE 4.
Oxidation of Keap1 and its dissociation from Nrf2 in response to MitoQ. A, the generation of ROS by MitoQ was assayed by measuring HE and its oxidation products by HPLC with electrochemical detection in cells treated with HE (10 μm) for the final 30 min of the MitoQ exposure. HPLC with electrochemical detection peak areas of HE, 2-OH-E+, E+, and E+-E+ extracted from MDA-MB-231 cells are shown after normalization to the protein concentration in the cell lysates. Each panel includes the structure of the analyzed compound. B, Keap1 protein oxidation was confirmed by the appearance of a higher molecular weight Keap1 band that only appears under non-reducing conditions (without β-mercaptoethanol). tBHQ (50 μm) was used as a positive control. Keap1 (68 kDa) and modified Keap1 (>120 kDa) are indicated with arrows. Quantitation of the bands representative of the oxidized and total Keap1 proteins is shown after normalization to α-tubulin levels. C, degradation of Keap1 in response to MitoQ was analyzed by Western blotting after exposure to 3, 6, or 24 h with 1 μm MitoQ. tBHQ (at 50 μm) was used as a positive control. D, interaction between Keap1 and Nrf2 is disrupted after MitoQ treatment. Top, Keap1 was immunoprecipitated (IP) from MitoQ- or tBHQ-treated lysates, followed by Western blotting (WB) with anti-Nrf2 antibody. Bottom, Nrf2 was immunoprecipitated from Mito-Q or tBHQ-treated lysates, followed by Western blotting with anti-Keap1 antibody. Input control samples were precleared with agarose beads and were not immunoprecipitated prior to Western blotting. Error bars, S.D.
To define the molecular mechanism of MitoQ-induced growth arrest, we investigated the ability of oxidants generated by MitoQ to modify Keap1 and engage Nrf2 antioxidant transcriptional activity. The cysteine residues (Cys273, Cys288, and Cys151) on Keap1 act as redox sensors that undergo reversible disulfide bond formation that can be detected as Keap1 dimers with altered electrophoretic mobility under non-reducing conditions (9). MDA-MB-231 cells were exposed to MitoQ, and whole cell lysates were used for Western blot analysis in the presence or absence of a reducing agent, β-mercaptoethanol. The oxidant tBHQ was used as a positive control (9). We detected a higher molecular weight band corresponding to oxidized Keap1 protein in the presence of MitoQ (10 μm, 2 h). This modified Keap1 band was not detected under reducing conditions (left lanes), indicating that it represents disulfide-bonded (oxidized) Keap1 (Fig. 4B). Upon modification, Keap1 is known to undergo proteasome-independent degradation (41). Consistent with this, significant Keap1 degradation was observed after exposure to MitoQ (Fig. 4C). Thus, MitoQ induces oxidative modification and degradation of the oxidant-sensitive regulatory protein Keap1.
We next evaluated whether MitoQ-induced Keap1 modification leads to disruption of Keap1 binding to Nrf2. Keap1 was immunoprecipitated from lysates of MDA-MB-231 cells exposed to MitoQ (1 μm, 6 or 24 h). The anti-Keap1 immunoprecipitates were subjected to immunoblot analysis with anti-Nrf2 antibody. A significant decrease in Keap1-Nrf2 interaction was detected after MitoQ treatment (Fig. 4D, top). tBHQ (50 μm, 16 h) was used as a positive control. To confirm that the association between Keap1 and Nrf2 was disrupted by treatment with MitoQ, Nrf2 was also immunoprecipitated from the whole cell lysates, and immunoblots were probed with anti-Keap1 antibody (Fig. 4D, bottom). The results confirm a reduced amount of Keap1 protein bound to Nrf2 after MitoQ treatment.
Increased Nuclear Protein Levels and Transcriptional Activity of Nrf2
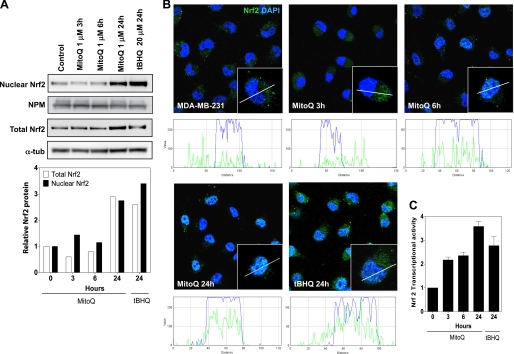
Disruption of Keap1 binding stabilizes Nrf2 protein levels (9). Release from Keap1 under conditions of oxidative stress also triggers the nuclear translocation of Nrf2, where it functions as an antioxidant transcription factor (8). We used whole cell lysates and nuclear fractions from MitoQ-treated MDA-MB-231 cells to investigate Nrf2 protein levels (Fig. 5A). After a 24-h exposure to MitoQ, a 3-fold increase in nuclear protein levels was observed (Fig. 5A). tBHQ was used as a positive control and also showed a greater than 3-fold increase in nuclear Nrf2. Next, to confirm nuclear translocation of Nrf2, we performed confocal microscopy analysis in cells exposed to 1 μm MitoQ (Fig. 5B). A clear increase in nuclear Nrf2 protein was observed after 6 h with MitoQ. Finally, the transcriptional activity of Nrf2 was measured with an assay using an immobilized oligonucleotide containing the ARE consensus binding site (Fig. 5C). tBHQ was used as a positive control. Treatment of MDA-MD-231 cells with MitoQ for 24 h led to a 3.5-fold increase in transcriptional activity of Nrf2.
FIGURE 5.
Increased nuclear protein levels and transcriptional activity of the antioxidant Nrf2. A, nuclear and whole cell fractions of MitoQ-treated MDA-MB-231 cells were probed by Western blotting with anti-Nrf2 antibody. Nucleophosmin (NPM) and α-tubulin (α-tub) levels were used as loading controls for nuclear and whole cell extracts, respectively. B, nuclear translocation of Nrf2 following exposure to MitoQ in MDA-MB-231 cells was examined by confocal microscopy. Nrf2 (green) was probed using anti-Nrf2 antibody, and DAPI staining (blue) was used to localize the nucleus. Inset, single representative cell. A line scan analysis of the confocal images was performed to determine localization of Nrf2 with respect to the nucleus. White color indicates overlapping staining by DAPI and anti-Nrf2 antibody. C, the transcriptional activity of Nrf2 was measured with an assay with immobilized oligonucleotide containing the ARE consensus binding site. tBHQ was used as a positive control. Error bars, S.D.
Knockdown of Nrf2 Increases Autophagy and G1 Cell Cycle Arrest but Not Apoptosis from MitoQ
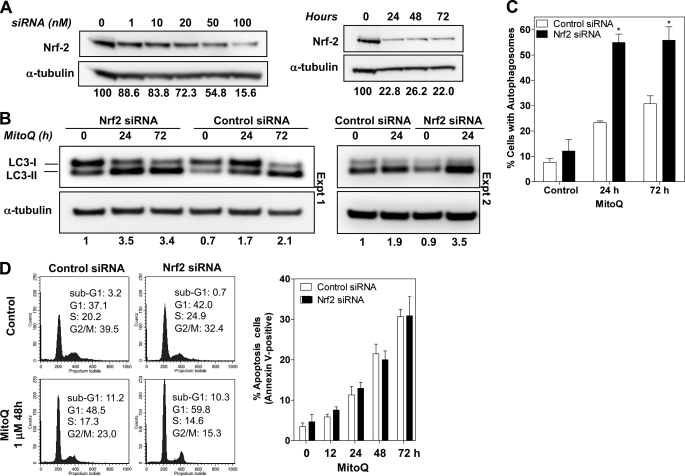
The critical role of Nrf2 as an antioxidant defense mechanism against the MitoQ-induced cellular effects of cell cycle arrest, apoptosis, and autophagy was investigated using siRNA against Nrf2. We identified the optimal conditions for knockdown of Nrf2 protein (100 nm siRNA oligonucleotides for 24 h) (Fig. 6A). We observed ∼84% knockdown of Nrf2 under these conditions. Non-coding siRNA oligonucleotides were used as controls. The relative levels of LC3-II in Nrf2 siRNA-transfected cells were higher than in control siRNA-transfected cells at 24 h. The 1.4-fold increase in LC3-II in Nrf2-siRNA cells remained relatively steady for 72 h after transfection. After exposure of Nrf2 and control siRNA cells to MitoQ for 24 or 72h, we tested for autophagy. A significantly greater level of LC3-II protein was detected in Nrf2 siRNA cells compared with control siRNA, confirming greater levels of autophagy in Nrf2 knockdown cells exposed to MitoQ (Fig. 6B). The difference was most significant at 24 h. A detailed time course is shown in supplemental Fig. 3. We confirmed the role of Nrf2 in down-regulating autophagy by observing greater percentages of cells with autophagosomes in Nrf2 siRNA-transfected cells (Fig. 6C). We also observed a greater percentage of cells in G1 with MitoQ in the Nrf2 siRNA cells compared with cells with control siRNA (Fig. 6D). We conclude that this increase corresponds to a greater percentage of cells in autophagic growth arrest. Finally, we measured apoptosis under knockdown conditions at various time points after adding MitoQ using annexin V-PI staining and FACS analysis. We did not observe a significant difference in the percentage of annexin V-positive cells at any tested time point (Fig. 6D, right). We also did not find significant differences in the sub-G1 fraction of cells (indicative of subdiploid apoptotic cells) following treatment of Nrf2 siRNA cells with MitoQ. Thus, MitoQ-induced Nrf2 signaling is a critical antioxidant defense mechanism that negatively controls autophagy in response to ROS generated by MitoQ.
FIGURE 6.
Knockdown of Nrf2 increases autophagy and cell cycle arrest but not apoptosis by MitoQ. A, conditions for Nrf2 knockdown with siRNA were optimized in MDA-MB-231 cells. Increasing concentrations (at 72 h) and incubation times (with 100 nm) siRNA oligonucleotides were tested. The bands were quantitated using densitometric scanning and normalized to α-tubulin. The relative levels of Nrf2 are indicated below the images. Data shown is representative of three independent experiments. B, increased levels of the autophagosome-associated LC3-II subunit were used as a measure of autophagy 24 h after siRNA. MitoQ (1 μm) was tested at 24 and 72 h. Non-coding siRNA oligonucleotides were used as control siRNA. Data from two individual experiments (Expt.) are shown. C, percentage of cells with autophagosomes after Nrf2 or control siRNA transfection followed by MitoQ treatment were counted as in Fig. 3D. *, statistical significance compared with control siRNA cells. D, cell cycle profiles of Nrf2 or control siRNA cells exposed to MitoQ were analyzed by flow cytometry following staining with PI. The percentage of cells in each cell cycle phase was quantified and is provided in each panel. Right, apoptosis was measured in live Nrf2 siRNA MDA-MB-231 cells treated with 1 μm MitoQ for 72 h. The percentage of cells that were annexin V-positive was used as a measure of apoptosis. Error bars, S.D.
NQO1-deficient MDA-MB-231 Cells Undergo Higher Levels of Autophagy and G1 Cell Cycle Arrest but Not Apoptosis from MitoQ
NQO1 is a downstream target of Nrf2. We hypothesized that NQO1 might be one of the antioxidant enzymes regulated by Nrf2 that could negatively control the level of autophagy after MitoQ treatment. MDA-MB-231 cells have no detectable NQO1 protein under regular growth conditions (42). Previously generated isogenic MDA-MB-231 cells stably transfected with NQO1 were used for studying the role of NQO1 (17). We first confirmed the NQO1 status in these cells using Western blot analysis (Fig. 7A, inset). Next we probed the growth inhibition profile by using two approaches. We tested cellular proliferation using the SRB dye assay and cell cycle arrest using propidium iodide staining/FACS analyses. MDA-MB-231 NQO1-negative cells showed greater levels of growth inhibition in response to increasing concentrations of MitoQ at 72 h (Fig. 7A). MDA-MB-231-NQO1-negative cells also showed higher percentage of cells in the G1 cell cycle phase by FACS analysis at 48 h with 0.1 μm MitoQ (Fig. 7B). In contrast, NQO1-positive and NQO1-negative cells showed no significant difference in the total percentage of apoptotic cells (Fig. 7C). It was noted that the NQO1-negative cells had a greater percentage of cells that were in early apoptosis following MitoQ, as indicated by the annexin V-positive and propidium iodide-negative fraction of cells. However, the total percentage of annexin V-positive cells was not significantly different between the two cell lines. These results were confirmed using Hoechst-PI staining of live cells (not shown). We then assayed for autophagy using LC3 Western blotting and light fluorescence microscopy. MDA-MB-231-NQO1-negative cells showed higher levels of LC3-II than MDA-MB-231-NQO1-positive cells in response to MitoQ (Fig. 7D). The higher levels of LC3-II in the absence of NQO1 were also confirmed using the NQO1 inhibitor dicumarol in MDA-MB-231-NQO1-positive cells (43). Cells pretreated with dicumarol showed greater levels of LC3-II in response to MitoQ than with MitoQ alone (Fig. 7E). The NQO1-negative cells also showed a greater percentage of cells with autophagosomes as seen by microscopy (Fig. 7F). These results suggest that NQO1 is, at least in part, responsible for the downstream signaling from Nrf2 and controlling the level of autophagy in response to MitoQ.
DISCUSSION
MitoQ was first designed as an antioxidant intended to block mitochondrial oxidative damage by preventing lipid peroxidation (4). Within the mitochondria, MitoQ undergoes redox cycling by complex II and is reduced to mitoquinol. In contrast to the toxic effects of MitoQ on tumor cells, this compound is protective to normal cells (4, 44, 45). Consistent with these previous findings, our preliminary data suggest that MitoQ does not cause cytotoxicity in healthy mammary epithelial cells. Sensitivity of breast cancer cells (MDA-MB-231 and MCF-7) to MitoQ was 30-fold higher than for healthy mammary epithelial cells (MCF-12A). MitoQ also enhanced the anticancer cytotoxicity of doxorubicin when tested in breast cancer cells (not shown). We therefore hypothesized that MitoQ might possess unique anticancer activity as a single agent.
Our studies show that MitoQ is cytotoxic and induces both autophagy and apoptosis in human breast cancer cells. MitoQ caused irreversible inhibition of clonogenic cell growth, a G1/S cell cycle arrest, and checkpoint kinase activation. Rapid mitochondrial membrane depolarization was accompanied by a release of cytochrome c, a response that was not observed with the untargeted ubiquinone (CoQ10). MitoQ treatment led to a greater percentage of cells undergoing autophagy than apoptosis under the same conditions. Irreversible growth arrest, observed as a loss of clonogenicity, suggests that autophagy might ultimately lead to cell death in response to MitoQ. Agents such as rapamycin and tunicamycin that preferentially cause a growth arrest in the G1/S phase of the cell cycle are also known to induce autophagy (34). Inhibitors of mitochondrial electron transport chain complexes also induce a caspase-independent, autophagic cell death (46, 47). Autophagy under such circumstances occurs as a consequence of mitochondrially generated oxidants, including superoxide, hydrogen peroxide, and hydroxyl radicals (48). We therefore hypothesized that autophagy is a plausible mechanism for MitoQ-induced growth inhibition. Cell death following autophagy is characterized by morphological changes that are distinct from apoptosis (49). It involves the caspase-independent degradation of organelles, as observed with MitoQ in caspase-deficient MCF-7 cells or in MDA-MB-231 cells pretreated with a pan-caspase inhibitor. The role of autophagy in cancer chemotherapy is not fully understood (49, 50). Although it could be a mechanism for drug resistance (i.e. through the degradation of unnecessary cellular components), it could also serve as a mechanism for drug-induced cell death (i.e. due to the degradation of critical cellular components). The observation that a short term exposure to MitoQ leads to an irreversible loss of clonogenic growth suggests that autophagy was unable to mitigate the cytotoxic and irreversible damage by MitoQ and is not a successful means of drug resistance. Cell death following autophagy occurs if the cells are unable to repair the damage. We found that cells exposed to an inhibitor of autophagy (bafilomycin A1) together with MitoQ underwent increased apoptosis compared with cells treated with MitoQ alone. This switch to apoptosis is a common observation for autophagic cells (37, 38).
Our findings are also in concordance with previous reports relating the production of ROS to the induction of autophagic cell death in cancer cells (27, 47, 50). The pro-oxidant arsenic trioxide and celecoxib-derived OSU-03012 induce ROS and autophagy in cancer cells (27, 51). We speculate that the production of ROS might be involved in the induction of autophagic cell death by MitoQ. In contrast, other to-be-determined mechanisms might be responsible for the apoptosis induced by MitoQ. A combination of autophagy and apoptosis has also been observed after camptothecin, tamoxifen, and oridonin treatment in breast cancer cells (52, 53).
Given the nature of its redox cycling, it is not surprising that MitoQ was also found to generate ROS in isolated mitochondria (6). Oxidative perturbations within cells can trigger numerous defense signals that protect cells and tissues from irreversible damage. Based on our findings, we propose a model for the mechanism for MitoQ-induced cell death via autophagy and signaling by the Nrf2-Keap1 pathway (Fig. 7G). Nrf2 is a critical transcription factor that regulates antioxidant responses by increasing the expression of antioxidant enzymes. The cysteine-rich, redox-sensitive protein Keap1 acts as a negative regulator of Nrf2. We propose that the MitoQ-induced oxidation of Keap1 is a redox sensor for the cellular response and results in subsequent degradation of Keap1. This modification of Keap1 leads to the release, stabilization, and nuclear translocation of Nrf2 protein, ultimately resulting in increased Nrf2 transcriptional activity. Although we observed degradation of Keap1, this could not directly account for the disruption of Keap1-Nrf2 complex formation. We draw this conclusion based on the observation that Nrf2 protein stabilization and translocation (in Fig. 5) are dependent on the oxidation-induced degradation of Keap1 and disruption of Keap1-Nrf2 complex formation (9, 21). Our reverse immunoprecipitation experiments also confirm the reduced protein-protein interaction. Earlier studies with oxidant-producing agents, such as tBHQ, sulforoaphane, and quercetin, showed a similar engagement of the Keap1-Nrf2 pathway (54, 55). Our data demonstrate the critical role of Nrf2 in controlling the level of autophagy and growth inhibition in response to oxidative stress. We speculate that the induction of an Nrf2-mediated antioxidant response limits but does not abolish the induction of autophagy and growth arrest by MitoQ. Hence, Nrf2 is a cytoprotective signal that limits the extent of growth arrest and autophagy under basal conditions. In the absence of Nrf2 (e.g. following knockdown by siRNA), the antioxidant defense to MitoQ is decreased, resulting in increased unquenched ROS and increased autophagy and G1 cell cycle arrest. We note that Nrf2 transfection alone caused a slight increase in the LC3-II protein level, which may indicate that Nrf2 is also responsible for regulating basal levels of autophagy in these cancer cells. This finding is unique and requires further investigation because, to our knowledge, there are no known Nrf2-responsive genes that inhibit or stimulate autophagy. With MitoQ, we observed an increase in the levels of LC3-I, in addition to LC3-II (Fig. 3B). We speculate that this previously unreported increase in LC3-I is unique to oxidative stress and is related to Nrf2 because it was not observed with rapamycin or in the absence of Nrf2 (Fig. 6B). We investigated NQO1, an Nrf2-regulated enzyme, for its role in regulating autophagy and apoptosis. Our data suggest that NQO1, in agreement with the role for Nrf2, limits the extent of autophagy induced by MitoQ. Isogenic cells deficient in NQO1 had elevated levels of autophagy and G1 cell cycle arrest (Fig. 7). There was no observed effect on apoptosis by MitoQ. We speculate that NQO1 limits the oxidant-induced autophagy that is triggered by MitoQ. The role for NQO1 in controlling autophagy is also evident in the higher levels of LC3-II observed under basal conditions in the NQO1-deficient MDA-MB-231 cells, compared with other cancer cell lines (35). This is in agreement with the protective role for NQO1 observed in response to menadione in NQO1-null mice (56).
It is likely that the control of autophagy downstream of Nrf2 occurs with multiple antioxidant enzymes playing a role in clearing the reactive oxygen species. It is also plausible that other proteins indirectly regulated by Nrf2 might directly and negatively regulate autophagic processes.
The combination of MitoQ and the knockdown of Nrf2 may also point to a novel therapeutic opportunity to enhance the antitumor effects of MitoQ. Recent reports propose a new model for Nrf2-Keap1 signaling where the dissociation of Nrf2 from Keap1 is proposed to be an in vitro phenomenon resulting from the experimental use of excess drug concentrations (57). This model suggests that there are two separate pools of Nrf2 (free and bound to Keap1) that translocate to the nucleus to mediate antioxidant signaling. Our data cannot rule out this mechanism, but the combined effects of MitoQ on Keap1 and Nrf2 are consistent with the view that Keap1 down-regulates Nrf2 in the absence of oxidative stress.
Our observation that oxidative stress mediates autophagic signaling by an antiproliferative agent underscores the need to understand the multifaceted responses to oxidative stress induced by cancer drugs. Autophagic mechanisms can trigger a reversible growth arrest that allows cells to escape death after elimination of damaged cellular components. This defense mechanism could be responsible for the failure of many promising investigational drugs to be successfully translated into the clinic. In the case of MitoQ, the autophagic growth arrest and apoptosis appear to be irreversible, as manifested by reduced clonogenic growth even after removal of the drug. It is likely that MitoQ operates in a manner that is similar mechanistically to tBHQ and that it acts as a weak electrophile. It is conceivable that MitoQ is more cytotoxic to cancer cells than CoQ10 because of the enhanced intracellular accumulation of MitoQ. We recently showed that MitoQ can reverse the cardiotoxic effects of doxorubicin, a widely used cancer chemotherapy drug, and that MitoQ enhances doxorubicin-mediated cytotoxicity in breast cancer cells (58). These attributes of MitoQ provide us with a unique opportunity to synergize with doxorubicin to better destroy breast tumor cells while providing protection from the cardiotoxic side effects of doxorubicin. These investigations are currently under way in our laboratory.
Supplementary Material
Acknowledgments
We thank members of the Laboratory of Biochemistry (FDA), Dr. Yves Pommier (NCI, National Institutes of Health), Dr. Melanie Simpson (NCI, National Institutes of Health), and Dr. V. Speransky (NIBIB, National Institutes of Health) for helpful discussions and Ksenia Guoliaeva (FDA) for technical assistance. We thank Dr. Mark Hannink (University of Missouri, Columbia, MO) for generously providing anti-Keap1 antibody. We thank Dr. Alasdair Steven (NIAMS, National Institutes of Health) for facilitating use of an electron microscope.
This work was supported in part by National Institutes of Health Intramural Research Programs of NIAMS and NCI and by National Institutes of Health Grants RO1CA125112 and RO1CA136799.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- MitoQ
- mitoquinone
- ARE
- antioxidant response element
- CoQ10
- coenzyme Q-10 or ubiquinone
- HE
- hydroethidine
- ROS
- reactive oxygen species
- tBHQ
- tert-butylhydroquinone
- VP-16
- etoposide
- PI
- propidium iodide
- SRB
- sulforhodamine B
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- JC-1
- 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide.
REFERENCES
- 1.Murphy M. P. (1997) Trends Biotechnol. 15, 326–330 [DOI] [PubMed] [Google Scholar]
- 2.Murphy M. P. (2008) Biochim. Biophys. Acta 1777, 1028–1031 [DOI] [PubMed] [Google Scholar]
- 3.Kalivendi S. V., Konorev E. A., Cunningham S., Vanamala S. K., Kaji E. H., Joseph J., Kalyanaraman B. (2005) Biochem. J. 389, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelso G. F., Porteous C. M., Coulter C. V., Hughes G., Porteous W. K., Ledgerwood E. C., Smith R. A., Murphy M. P. (2001) J. Biol. Chem. 276, 4588–4596 [DOI] [PubMed] [Google Scholar]
- 5.Koopman W. J., Verkaart S., Visch H. J., van der Westhuizen F. H., Murphy M. P., van den Heuvel L. W., Smeitink J. A., Willems P. H. (2005) Am. J. Physiol. Cell Physiol. 288, C1440–C1450 [DOI] [PubMed] [Google Scholar]
- 6.Doughan A. K., Dikalov S. I. (2007) Antioxid. Redox Signal. 9, 1825–1836 [DOI] [PubMed] [Google Scholar]
- 7.Cochemé H. M., Kelso G. F., James A. M., Ross M. F., Trnka J., Mahendiran T., Asin-Cayuela J., Blaikie F. H., Manas A. R., Porteous C. M., Adlam V. J., Smith R. A., Murphy M. P. (2007) Mitochondrion 7, (suppl.) S94–S102 [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. (2009) Mol. Cell. Biol. 29, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D. D., Hannink M. (2003) Mol. Cell. Biol. 23, 8137–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal A. K., McBride O. W., Adesnik M., Nebert D. W. (1988) J. Biol. Chem. 263, 13572–13578 [PubMed] [Google Scholar]
- 11.Gogvadze V., Orrenius S., Zhivotovsky B. (2008) Trends Cell Biol. 18, 165–173 [DOI] [PubMed] [Google Scholar]
- 12.Warburg O. (1956) Science 124, 269–270 [PubMed] [Google Scholar]
- 13.Armstrong J. S. (2006) Br. J. Pharmacol. 147, 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernal S. D., Lampidis T. J., Summerhayes I. C., Chen L. B. (1982) Science 218, 1117–1119 [DOI] [PubMed] [Google Scholar]
- 15.Davis S., Weiss M. J., Wong J. R., Lampidis T. J., Chen L. B. (1985) J. Biol. Chem. 260, 13844–13850 [PubMed] [Google Scholar]
- 16.Hoke E. M., Maylock C. A., Shacter E. (2005) Free Radic. Biol. Med. 39, 403–411 [DOI] [PubMed] [Google Scholar]
- 17.Bentle M. S., Reinicke K. E., Bey E. A., Spitz D. R., Boothman D. A. (2006) J. Biol. Chem. 281, 33684–33696 [DOI] [PubMed] [Google Scholar]
- 18.Rao V. A., Agama K., Holbeck S., Pommier Y. (2007) Cancer Res. 67, 9971–9979 [DOI] [PubMed] [Google Scholar]
- 19.McDonald K. L., Morphew M., Verkade P., Müller-Reichert T. (2007) Methods Mol. Biol. 369, 143–173 [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., Azad M. B., Gibson S. B. (2010) Can. J. Physiol. Pharmacol. 88, 285–295 [DOI] [PubMed] [Google Scholar]
- 21.Zhang D. D., Lo S. C., Cross J. V., Templeton D. J., Hannink M. (2004) Mol. Cell. Biol. 24, 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao V. A., Fan A. M., Meng L., Doe C. F., North P. S., Hickson I. D., Pommier Y. (2005) Mol. Cell. Biol. 25, 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zielonka J., Hardy M., Kalyanaraman B. (2009) Free Radic. Biol. Med. 46, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirao A., Cheung A., Duncan G., Girard P. M., Elia A. J., Wakeham A., Okada H., Sarkissian T., Wong J. A., Sakai T., De Stanchina E., Bristow R. G., Suda T., Lowe S. W., Jeggo P. A., Elledge S. J., Mak T. W. (2002) Mol. Cell. Biol. 22, 6521–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niida H., Katsuno Y., Banerjee B., Hande M. P., Nakanishi M. (2007) Mol. Cell. Biol. 27, 2572–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Facompré M., Wattez N., Kluza J., Lansiaux A., Bailly C. (2000) Mol. Cell. Biol. Res. Commun. 4, 37–42 [DOI] [PubMed] [Google Scholar]
- 27.Gao M., Yeh P. Y., Lu Y. S., Hsu C. H., Chen K. F., Lee W. C., Feng W. C., Chen C. S., Kuo M. L., Cheng A. L. (2008) Cancer Res. 68, 9348–9357 [DOI] [PubMed] [Google Scholar]
- 28.Vandenabeele P., Vanden Berghe T., Festjens N. (2006) Sci. STKE 2006, pe44. [DOI] [PubMed] [Google Scholar]
- 29.Wang M., Tan W., Zhou J., Leow J., Go M., Lee H. S., Casey P. J. (2008) J. Biol. Chem. 283, 18678–18684 [DOI] [PubMed] [Google Scholar]
- 30.Tasdemir E., Galluzzi L., Maiuri M. C., Criollo A., Vitale I., Hangen E., Modjtahedi N., Kroemer G. (2008) Methods Mol. Biol. 445, 29–76 [DOI] [PubMed] [Google Scholar]
- 31.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., et al. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007) Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 34.Tasdemir E., Maiuri M. C., Tajeddine N., Vitale I., Criollo A., Vicencio J. M., Hickman J. A., Geneste O., Kroemer G. (2007) Cell Cycle 6, 2263–2267 [DOI] [PubMed] [Google Scholar]
- 35.Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2005) Autophagy 1, 84–91 [DOI] [PubMed] [Google Scholar]
- 36.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. (1991) J. Biol. Chem. 266, 17707–17712 [PubMed] [Google Scholar]
- 37.Kanzawa T., Kondo Y., Ito H., Kondo S., Germano I. (2003) Cancer Res. 63, 2103–2108 [PubMed] [Google Scholar]
- 38.Paglin S., Hollister T., Delohery T., Hackett N., McMahill M., Sphicas E., Domingo D., Yahalom J. (2001) Cancer Res. 61, 439–444 [PubMed] [Google Scholar]
- 39.Zielonka J., Srinivasan S., Hardy M., Ouari O., Lopez M., Vasquez-Vivar J., Avadhani N. G., Kalyanaraman B. (2008) Free Radic. Biol. Med. 44, 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zielonka J., Vasquez-Vivar J., Kalyanaraman B. (2008) Nat. Protoc. 3, 8–21 [DOI] [PubMed] [Google Scholar]
- 41.Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W., Hannink M. (2005) J. Biol. Chem. 280, 30091–30099 [DOI] [PubMed] [Google Scholar]
- 42.Ross D., Siegel D. (2004) Methods Enzymol. 382, 115–144 [DOI] [PubMed] [Google Scholar]
- 43.Du J., Daniels D. H., Asbury C., Venkataraman S., Liu J., Spitz D. R., Oberley L. W., Cullen J. J. (2006) J. Biol. Chem. 281, 37416–37426 [DOI] [PubMed] [Google Scholar]
- 44.Dhanasekaran A., Kotamraju S., Kalivendi S. V., Matsunaga T., Shang T., Keszler A., Joseph J., Kalyanaraman B. (2004) J. Biol. Chem. 279, 37575–37587 [DOI] [PubMed] [Google Scholar]
- 45.Kelso G. F., Porteous C. M., Hughes G., Ledgerwood E. C., Gane A. M., Smith R. A., Murphy M. P. (2002) Ann. N.Y. Acad. Sci. 959, 263–274 [DOI] [PubMed] [Google Scholar]
- 46.Chen Y., McMillan-Ward E., Kong J., Israels S. J., Gibson S. B. (2007) J. Cell Sci. 120, 4155–4166 [DOI] [PubMed] [Google Scholar]
- 47.Kunchithapautham K., Rohrer B. (2007) Autophagy 3, 433–441 [DOI] [PubMed] [Google Scholar]
- 48.Chen Y., Gibson S. B. (2008) Autophagy 4, 246–248 [DOI] [PubMed] [Google Scholar]
- 49.Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Nat. Rev. Mol. Cell. Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 50.Azad M. B., Chen Y., Gibson S. B. (2009) Antioxid. Redox. Signal. 11, 777–790 [DOI] [PubMed] [Google Scholar]
- 51.Kanzawa T., Zhang L., Xiao L., Germano I. M., Kondo Y., Kondo S. (2005) Oncogene 24, 980–991 [DOI] [PubMed] [Google Scholar]
- 52.Abedin M. J., Wang D., McDonnell M. A., Lehmann U., Kelekar A. (2007) Cell Death Differ. 14, 500–510 [DOI] [PubMed] [Google Scholar]
- 53.Cui Q., Tashiro S., Onodera S., Minami M., Ikejima T. (2007) Biol. Pharm. Bull. 30, 859–864 [DOI] [PubMed] [Google Scholar]
- 54.Hong F., Freeman M. L., Liebler D. C. (2005) Chem. Res. Toxicol. 18, 1917–1926 [DOI] [PubMed] [Google Scholar]
- 55.Tanigawa S., Fujii M., Hou D. X. (2007) Free Radic. Biol. Med. 42, 1690–1703 [DOI] [PubMed] [Google Scholar]
- 56.Radjendirane V., Joseph P., Lee Y. H., Kimura S., Klein-Szanto A. J., Gonzalez F. J., Jaiswal A. K. (1998) J. Biol. Chem. 273, 7382–7389 [DOI] [PubMed] [Google Scholar]
- 57.Li W., Kong A. N. (2009) Mol. Carcinog. 48, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandran K., Aggarwal D., Migrino R. Q., Joseph J., McAllister D., Konorev E. A., Antholine W. E., Zielonka J., Srinivasan S., Avadhani N. G., Kalyanaraman B. (2009) Biophys. J. 96, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.