Abstract
Mitosis must faithfully divide the genome such that each progeny inherits the same genetic material. DNA condensation is crucial in ensuring that chromosomes are correctly attached to the mitotic spindle for segregation, preventing DNA breaks or constrictions from the contractile ring. Histones form an octameric complex of basic proteins important in regulating DNA organization and accessibility. Histone post-translational modifications are altered during mitosis, although the roles of these post-translational modifications remain poorly characterized. Here, we report that N-acetylglucosamine (O-GlcNAc) transferase (OGT), the enzyme catalyzing the addition of O-GlcNAc moieties to nuclear and cytoplasmic proteins at serine and threonine residues, regulates some aspects of mitotic chromatin dynamics. OGT protein amounts decrease during M phase. Modest overexpression of OGT alters mitotic histone post-translational modifications at Lys-9, Ser-10, Arg-17, and Lys-27 of histone H3. Overexpression of OGT also prevents mitotic phosphorylation of coactivator-associated arginine methyltransferase 1 (CARM1) and prevents its correct cellular localization during mitosis. Moreover, OGT overexpression results in an increase in abnormal chromosomal bridge formation. Together, these results show that regulating the amount of OGT during mitosis is important in ensuring correct chromosomal segregation during mitosis.
Keywords: Chromatin, Chromatin Histone Modification, Chromatin Remodeling, Glycosylation, Histone Methylation, Histone Modification, Post-translational Modification, O-GlcNAc, O-GlcNAc Transferase, O-GlcNAcylation
Introduction
Cellular commitment to grow and to divide depends on external cues, as well as a complex regulatory network within the cell. This regulatory network pauses during certain transitions or checkpoints in the cell cycle to assess whether cellular conditions are favorable for growth and division (1). Cell division can occur either symmetrically, important for increasing cell number during development, or asymmetrically, which occurs in adults to maintain tissue homeostasis (2). Clearly, delineating cell cycle control mechanisms is important to understanding not only the molecular basis of maintaining normal multicellularity but also to understanding how its deregulation can lead to cell cycle-related diseases, such as cancer and Alzheimer disease.
One major aspect of cell division is for each daughter cell to inherit the correct genetic material, achieved through DNA compaction and segregation during mitosis (3). The histone complex, composed of two molecules each of histone H2A, H2B, H3, and H4, are the DNA packaging proteins, regulating access to DNA during transcription and replication. This accessibility is signaled partly through the histone tail post-translational modifications. During mitosis, DNA must achieve a level of ∼500-fold compaction to efficiently segregate chromosomes between daughter cells (4). Some post-translational modifications of histone tails are cell cycle-dependent (5), suggesting that the mitotic histone code could impact chromosome condensation and segregation (4). Abnormalities associated with this process result in chromosomal instability and aneuploidy, where chromosomes are partitioned unevenly between daughter cells or lagging chromosomes are trapped and broken at the midbody during cytokinesis (6).
Post-translational modification of nuclear and cytoplasmic proteins on Ser or Thr residues by β-N-acetylglucosamine moieties (O-GlcNAc) is catalyzed by a highly conserved enzyme O-GlcNAc transferase (OGT).2 This sugar modification is abundant, dynamic, and reversible, with the highly conserved enzyme O-GlcNAcase hydrolyzing the sugar. A wide variety of substrates are O-GlcNAcylated, affecting protein turnover, protein-protein interactions, and other diverse functions (7). Importantly, as these same Ser and Thr residues are often phosphorylation sites, it is becoming clear that there is another level of regulation between these two abundant post-translational modifications (8) and that phosphorylation is not simply a binary switch.
Recent studies have shown that O-GlcNAc and OGT are important in cell cycle regulation. Complete ablation of the OGT gene in mice (9) and Drosophila (10) shows that OGT is essential for life. Globally, O-GlcNAc levels decrease during mitosis, increasing once cells enter G1 (11). Concomitantly, OGT mRNA levels decrease during M phase (12). Pharmacologically or genetically altering O-GlcNAc levels and/or OGT levels cause changes in cell cycle rate and alter the expression of cyclins and of immediate early genes (11, 13, 14). Moreover, O-GlcNAcylation of oncogenes, such as c-Myc (15), or tumor suppressors, such as p53 (16), regulates their stability. Finally, mass spectrometric analyses of the midbody phosphoproteome and O-GlcNAc proteome demonstrate that alterations in OGT levels dramatically affect key regulatory points during mitosis, furthering the importance of O-GlcNAc signaling during the cell cycle (17).
Recent reports demonstrate that O-GlcNAc and OGT affect chromatin structure by modulating chromatin-remodeling enzyme activity. For example, O-GlcNAcylation of the histone methyltransferase MLL5 activates its enzymatic activity, stimulating granulopoiesis (18). Building upon these data, as well as the global dependence of cell cycle on O-GlcNAc and on OGT levels, here, we show that OGT overexpression affects M phase dynamics of some histone post-translational modifications.
EXPERIMENTAL PROCEDURES
Antibodies
The O-GlcNAc-specific antibody and OGT antibody have been previously described (19, 20). Antibody to coactivator-associated arginine methyltransferase 1 (CARM1) was from Bethyl Laboratories. Antibodies to H3K4me2, H3K9Ac, and H3S10P were from Cell Signaling. Antibodies to MPM2, H3R17me2, and total H3 were from Millipore. Antibody to H3K27me3 was from Active Motif. Antibody to GFP was from Santa Cruz Biotechnology. Antibodies to α-tubulin, actin, and anti-mouse IgM-HRP were from Sigma. Anti-rabbit-HRP and anti-mouse-HRP were from GE Healthcare.
Cell Culture
HeLa cells were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin at 37 °C in a humidified atmosphere. Early mitotic cells were prepared by incubating HeLa cells in 80 ng/ml nocodazole (Sigma) overnight. Mitotic cells were harvested by shake-off and washed twice with PBS before replating into complete medium. Cells were harvested 1 (late M phase), 3 (early G1), or 5 h (G1) after replating. Alternatively, HeLa cells were arrested in G1/S by double thymidine block as described (11). Mitotic cells were harvested by shake-off ∼11–12 h after release. The adherent cells were incubated for a further 4–5 h (∼16–18 h after release from thymidine) to obtain a population of cells in G1. Adenovirus for GFP was purchased from the Baylor College of Medicine Vector Development Laboratory. Adenovirus for OGT was a generous gift from Dr. Wolfgang H. Dillmann (21). For adenoviral infections, HeLa cells were infected at a multiplicity of infection of 100. Cells were harvested ∼40 h after infection. For O-GlcNAcase inhibitor treatment, Thiamet G (1 mm) or NAG-thiazoline (10 μm) was added at the same time as nocodazole (80 ng/ml) and incubated overnight before harvesting by shake-off.
Western Blotting and Immunoprecipitation
Cells were lysed in 10 mm Tris, pH 8.0, 150 mm NaCl, and 1% Nonidet P-40 supplemented with inhibitors (1 mm NaF, 1 mm β-glycerophosphate, 1 mm sodium orthovanadate, 2 μm O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenyl carbamate, 1 mm sodium butyrate, and PIC1). Insoluble material was pelleted by centrifugation. Immunoprecipitations were performed with the indicated antibodies by incubating at 4 °C for 1–2 h with constant rotation. Protein G beads (GE Healthcare) were added, and immunoprecipitations continued for another 1–2 h. Beads were washed three times with lysis buffer before eluting with Laemmli buffer. For histone extraction, the insoluble pellet was washed with hypotonic lysis buffer (10 mm HEPES, pH 7.9, 10 mm KCl, and 1.5 mm MgCl2 supplemented with inhibitors). The pellet was then resuspended in hypotonic lysis buffer, and an equal volume of 0.4 m HCl was added and incubated on ice for 30 min. The insoluble material was pelleted, and the supernatant was neutralized with 1 m Tris, pH 8.0.
λ-Phosphatase Treatment
CARM1 was immunoprecipitated as described above. On the last wash, the beads were divided equally. The beads were then washed two times with 50 mm HEPES, pH 7.5. Immunoprecipitates were then either mock-treated or treated with λ-phosphatase (New England Biolabs) according to manufacturer's instructions. Reactions were terminated with the addition of Laemmli buffer.
Immunofluorescence
To prepare mitotic cells, HeLa cells were treated with a double thymidine block. After the second release into complete medium for 10–12 h, cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were permeabilized by incubating in PBS with 0.5% Triton X-100 for 10 min. Cells were then washed twice with PBS with 0.1% Triton X-100 (wash buffer) before blocking with 2% BSA prepared in PBS with 0.1% Triton X-100. Cells were incubated in primary antibody prepared in block solution overnight at 4 °C. Cells were then washed twice before incubating with appropriate secondary antibodies for 1 h at room temperature. After incubation, cells were washed five times with wash buffer. Nucleic acid was stained with LOLO-1 (Invitrogen) before mounting coverslips. Images were obtained on an 3i spinning disk confocal microscope using Olympus Slidebook software at the Johns Hopkins University School of Medicine Core Microscopy Facility.
RESULTS
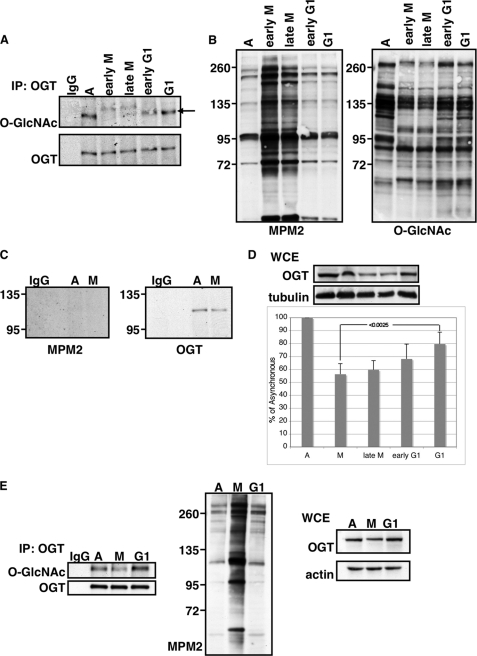
OGT is O-GlcNAcylated, although neither the sites nor the cellular context have been described (20). Additionally, OGT is phosphorylated, and its phosphorylation has been linked primarily with OGT activation. Tyrosine phosphorylation of OGT by the insulin receptor activates OGT (22), as does phosphorylation by calcium/calmodulin-dependent protein kinase IV (23). Recent proteomic analysis of the phosphoproteome and the O-GlcNAcome during mitosis as well as studies showing global decreases in O-GlcNAcylation during mitosis (11, 17) demonstrate the reciprocity that these two modifications, phosphorylation and O-GlcNAcylation, have on one another during mitosis. We therefore wanted to determine whether O-GlcNAcylation and phosphorylation of OGT itself was cell cycle-dependent. HeLa cells were synchronized into early M phase by overnight treatment with 80 ng/ml nocodazole. The mitotic cells were collected by shake-off and were designated early M phase. Alternatively, these mitotic cells were washed, released into complete medium, and harvested 1, 3, or 5 h later, corresponding to late mitosis, early G1, or G1. Equal amounts of OGT were pulled down by performing a non-saturating immunoprecipitation from these cells, and upon Western blotting with an O-GlcNAc-specific antibody (19), we saw that O-GlcNAcylation of OGT decreased during M phase and reappeared when cells entered G1 (Fig. 1A). We verified synchronization of the cells by immunoblotting with an antibody that recognizes Pro-directed phosphorylated Ser and Thr residues (MPM2), which increase during mitosis (24) and are commonly used markers for mitosis (11). We also immunoblotted for total O-GlcNAc levels, which decrease in mitotic cells (Fig. 1B).
FIGURE 1.
OGT O-GlcNAcylation and protein levels are regulated by cell cycle. A, OGT was immunoprecipitated from asynchronous HeLa cells (A) or HeLa cells synchronized into early M, late M, early G1, or G1 by arresting cells with nocodazole (for early M phase) or by releasing M phase arrested cells into complete medium. OGT immunoprecipitates (IP) were probed for O-GlcNAc. Blots were stripped and reprobed for OGT. IgG indicates nonspecific antibody control immunoprecipitation. Arrow indicates the band corresponding to OGT. O-GlcNAcylated OGT was normalized relative to the amount of OGT immunoprecipitated. B, total cell extract was probed using the mitosis-specific marker MPM2, which recognizes Pro-directed phosphorylation of Ser and Thr. These extracts were also probed for O-GlcNAc, which decreases during M phase. OGT levels in total cell extract were normalized to tubulin. C, OGT was immunoprecipitated from asynchronous (A) or mitotic (M) HeLa cells and probed with MPM2. Mitotic samples were prepared by arresting cells with nocodazole. These blots were then stripped and reprobed for OGT. IgG indicates nonspecific antibody control immunoprecipitation. D, whole cell extract (WCE) was probed for either OGT or tubulin from asynchronous, early M phase, late M phase, early G1, and G1 cells. Densitometric analysis shows relative amounts of OGT to asynchronous extracts after normalization to tubulin (n = 5). Error bars indicate S.E. The p values were calculated using the paired two-tailed Student's t test. E, OGT was immunoprecipitated from asynchronous HeLa cells (A) or HeLa cells synchronized into M phase (M) or G1. Cells were synchronized at the G1/S border by double thymidine block. Cells were then released into complete medium, and mitotic cells were collected by shake-off ∼11–12 h after release. G1 samples were harvested ∼16–18 h after release. Immunoprecipitates were probed for O-GlcNAc and OGT. Whole cell extract was probed for MPM2, OGT, and actin.
The cyclin B-Cdk1 complex, whose minimal consensus motif is (S/T)P, phosphorylates numerous substrates (25) and is responsible for driving mitosis (26). We wanted to investigate whether OGT becomes phosphorylated by proline-directed kinases during mitosis, perhaps resulting in the observed decrease in O-GlcNAc levels. We took asynchronous or mitotic extracts from HeLa cells, immunoprecipitated OGT, and probed with MPM2. We could not detect proline-directed phosphorylation of OGT using this antibody (Fig. 1C). The lack of reactivity with the MPM2 antibody, however, does not preclude the possibility that OGT may be phosphorylated by other mitotic kinases. In fact, recent proteomic analysis of the mitotic phosphoproteome mapped phosphorylation sites on OGT, although none increased during mitosis (27).
When probing for total OGT levels, we also found that there were lower OGT protein levels in the mitotic samples (Fig. 1D), corroborating experimental evidence showing that OGT mRNA levels decrease during M phase (12). Using another synchronization method, we were able to obtain these same results (Fig. 1E). HeLa cells were synchronized at the G1/S border using a double thymidine block. Cells were then released into complete medium. This method produces a population of cells that enter mitosis between 10 and 12 h after release. We collected the non-adherent cells as the mitotic fraction and allowed the remaining adherent cells to progress to G1. Using this method, we saw the same results as with the nocodazole method, demonstrating that the decrease in OGT protein levels and in OGT O-GlcNAcylation at mitosis was not an artifact of inhibitor treatment with nocodazole.
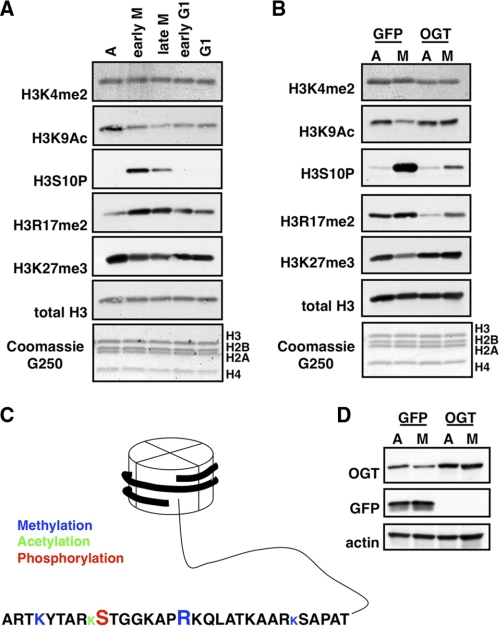
Some post-translational modifications on histones are cell cycle-dependent (5). O-GlcNAc can directly affect chromatin remodelers. As mentioned previously, O-GlcNAcylation of the histone methyltransferase MLL5 is necessary for activity (18). More recent studies have established that Drosophila OGT is encoded by the Polycomb group (PcG) gene super sex combs (sxc), thereby linking O-GlcNAcylation with PcG-mediated gene silencing (10, 28). Because OGT protein levels decreased during mitosis, we asked whether overexpression of OGT during this phase of cell cycle affected mitotic histone post-translational modifications. First, we determined how several histone post-translational modifications normally change during mitosis (Fig. 2A). The nomenclature we use in describing these modifications is first the histone modified, then the amino acid residue followed by the type of modification (P for phosphorylation, Ac for acetylation, me2 for dimethylation, and me3 for trimethylation).
FIGURE 2.
Dynamics of some histone post-translational modifications. A, acid-extracted histones were probed using the indicated antibodies from asynchronous (A), early mitotic, late mitotic, early G1, and G1 cells. Total histones were stained with Coomassie G250. B, acid-extracted histones were probed using the indicated antibodies from asynchronous or mitotic cells overexpressing either GFP or OGT. C, the diagram depicts the histone tail of H3 and the modifications investigated in this study. The type of post-translational modification is indicated by color: methylation in blue, acetylation in green, and phosphorylation in red. The relative size of the amino acid code represents how that particular modification is affected by mitosis; a larger letter indicates that the modification increases, a smaller letter indicates that the modification decreases, and a letter of the same size indicates that the modification is not cell cycle-dependent. D, control blots demonstrating that GFP and OGT were effectively overexpressed.
We chose to examine one marker that is known to increase dramatically during mitosis, H3S10P (29). We also chose to analyze several other histone post-translational modifications, H3K4me2, H3K9Ac, H3R17me2, and H3K27me3. H3K4me2 and H3K9Ac marks are usually associated with active loci (30, 31). Although there seems to be no cell cycle dependence of H3K4me2, acetylation at H3K9 decreases during mitosis (5). H3R17me2, catalyzed by CARM1, is generally associated with gene activation, and as yet, no cell cycle dependence has been reported (32, 33). Trimethylation of Lys-27 is found in heterochromatic regions and is thought to be important in maintaining X chromosome inactivation (30). This modification has also been found to decrease in M phase (5) (all modifications summarized in Fig. 2C). Histones were acid-extracted from asynchronously growing HeLa cells or HeLa cells synchronized into early M phase, late M phase, early G1, and G1. As expected, we observed an increase in H3S10P during mitosis, and it decreased once cells entered G1. We did not observe any changes in H3K4me2 as has been previously reported (5). Additionally, H3K9Ac and H3K27me3 decreased in M phase, returning once cells had exited mitosis. Unexpectedly, we saw dimethylation of Arg-17 of H3 increase during mitosis, returning to basal conditions upon entry into G1.
We then determined whether OGT overexpression altered histone post-translational modifications. We overexpressed either OGT or GFP as a control in asynchronous or mitotic cells (Fig. 2B). Overexpression of OGT had no effect on H3K4me2; however, although acetylation of H3K9 and trimethylation of H3K27 decreased during mitosis under control conditions, with OGT overexpression, we did not observe this diminishment. Furthermore, the increase in mitotic phosphorylation of Ser-10 and mitotic dimethylation of Arg-17 was not as dramatic when OGT was overexpressed. Again, we observed a decrease in OGT protein during M phase with GFP overexpression (Fig. 2D).
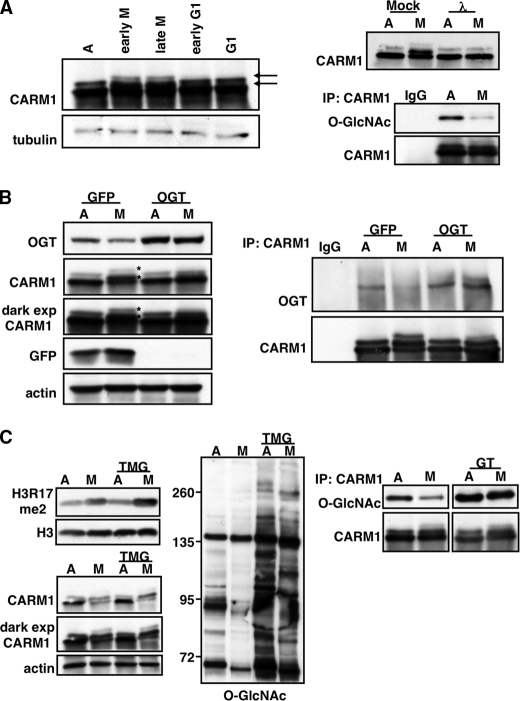
Because our laboratory has previously characterized the association of OGT with CARM1 (34), we decided to focus on H3R17me2 as this modification is a known CARM1-specific target (35). As reported previously, we were able to detect M phase-specific phosphorylation of CARM1 by immunoblotting with a CARM1-specific antibody (36, 37) (Fig. 3A, left panels). Under asynchronous conditions, CARM1 migrated as a doublet; however, in mitotic lysates, both bands exhibited a shift due to phosphorylation that we confirmed by treating CARM1 immunoprecipitates with λ-phosphatase (Fig. 3A, right panels). Our previous studies demonstrate that CARM1 is also O-GlcNAcylated. We therefore investigated whether CARM1 O-GlcNAcylation is cell cycle-dependent by immunoprecipitating CARM1 from asynchronous and mitotic extracts and probing with an O-GlcNAc antibody. Indeed, CARM1 O-GlcNAcylation decreased during mitosis (Fig. 3A, lower right panels).
FIGURE 3.
CARM1 is affected by OGT overexpression. A, left panels, extracts from synchronized cells were probed for CARM1 to demonstrate that it becomes mitotically phosphorylated (indicated by arrows). Upper right panel, phosphorylation was confirmed by immunoprecipitating CARM1 from asynchronous (A) and mitotic (M) extracts and treating with phosphatase. CARM1 was immunoprecipitated from asynchronous and mitotic extracts and probed for total O-GlcNAc levels. IgG was included as a specificity control. Lower right panels, blots were stripped and reprobed for total CARM1 in the immunoprecipitates (IP). λ, λ-phosphatase. B, total cell extract from asynchronous or mitotic cells overexpressing either GFP or OGT were probed using antibodies directed against OGT, CARM1, GFP, or tubulin. Overexpression of OGT prevents CARM1 mitotic phosphorylation as indicated with asterisks. CARM1 was immunoprecipitated from asynchronous and mitotic cells overexpressing either GFP or OGT and probed for CARM1 and OGT. OGT does not associate with CARM1 during mitosis in control cells, whereas in OGT-overexpressing cells, OGT associates with CARM1 regardless of cell cycle state. dark exp, dark exposure. C, treatment of cells with the O-GlcNAcase inhibitor Thiamet G (TMG) does not exhibit the same effects as OGT overexpression. Left panel, acid-extracted histones were probed for the CARM1-specific target H3R17me2. Middle and lower left panels, total cell extract was probed using antibodies directed against O-GlcNAc (middle panel) and CARM1 and actin (lower left panels). Right panels, inhibition of O-GlcNAcase using NAG-thiazoline dramatically increases CARM1 O-GlcNAcylation without affecting mitotic phosphorylation. CARM1 was immunoprecipitated from asynchronous and mitotic extracts treated with or without NAG-thiazoline and probed for O-GlcNAc. Blots were stripped and reprobed for total CARM1 in the immunoprecipitates. GT, NAG-thiazoline.
We then determined what effects OGT overexpression would have on CARM1. We overexpressed GFP and OGT in asynchronous and mitotic HeLa cells and probed for CARM1 from these extracts. CARM1 did not become phosphorylated in mitosis with OGT overexpression (Fig. 3B, left panels). Moreover, the cell cycle dependence of the association between CARM1 and OGT was altered when OGT was overexpressed. In control cells, CARM1 associated with OGT in asynchronous cells but not in mitotic cells; however, in OGT-overexpressing cells, OGT and CARM1 co-immunoprecipitated regardless of cell cycle state (Fig. 3B, right panels).
To demonstrate that the effects we observed with OGT overexpression, the attenuation in mitotic H3R17me2 and in CARM1 phosphorylation, were due to increases in OGT protein expression and not to increases in global protein O-GlcNAcylation, we treated asynchronous and mitotic cells with Thiamet G, a potent and specific O-GlcNAcase inhibitor (38). After treatment overnight with Thiamet G (TMG), we saw a dramatic increase in total protein O-GlcNAcylation (Fig. 3C, middle panel). There was no difference in dimethylation of Arg-17 of H3 (Fig. 3C, upper left panels) nor in CARM1 mitotic phosphorylation (Fig. 3C, lower left panels), suggesting that it is the association of CARM1 with OGT and not increases in global O-GlcNAc levels that mediates the effects we observed. Using another O-GlcNAcase inhibitor, NAG-thiazoline (39), we were able to dramatically increase CARM1 O-GlcNAcylation without affecting mitotic phosphorylation, again suggesting that interaction with OGT during mitosis and not increases in global O-GlcNAcylation is the mechanism that prevents CARM1 mitotic phosphorylation.
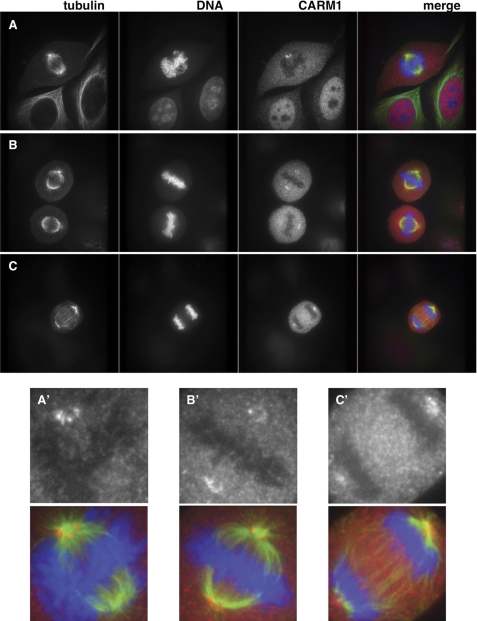
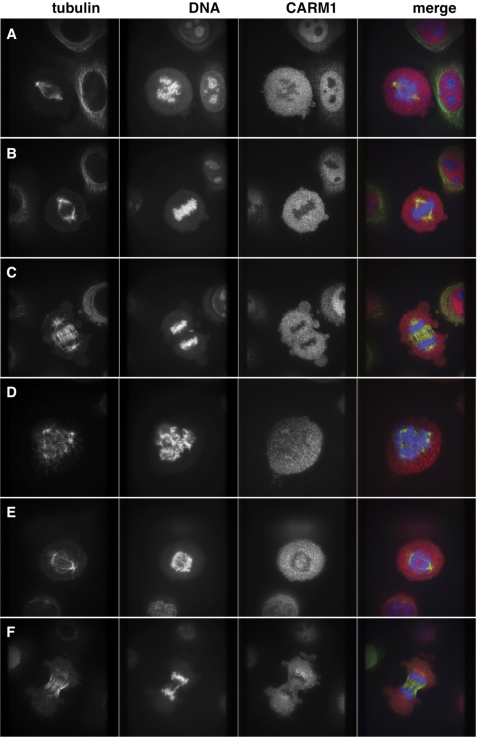
We then used indirect immunofluorescence to detect CARM1 cytolocalization during mitosis. Previous studies have shown that mutation of one of the mitotic phosphorylation sites of CARM1 from Ser to Glu, mimicking its mitotic phosphorylation, caused its exportation from the nucleus (36). As reported previously, CARM1 was enriched in the nucleus in interphase cells (Fig. 4A). As DNA condensed and began to align on the metaphase plate, CARM1 concentrated to the pericentriolar area (Fig. 4A). In fact, CARM1 seemed to form a ring around the centrosomes (Fig. 4, A′–C′), and the localization to this area was stable throughout mitosis (Fig. 4, A–C).
FIGURE 4.
CARM1 localizes to the pericentriolar region during mitosis. HeLa cells were stained for tubulin (green), DNA (blue), and CARM1 (red) from cells in prometaphase (A), metaphase (B), and anaphase (C). CARM1 is found concentrated near the centrosomes. A′, B′, and C′ are enlargements of CARM1 (black and white) and merged images from A, B, and C to show the ring CARM1 forms around the centrosome.
In contrast, with OGT overexpression, we saw a marked decrease in the number of cells showing CARM1 localization to the pericentriolar area. CARM1 appeared diffuse throughout the cell during mitosis (Fig. 5, A–C). We counted over 250 mitotic cells, and although greater than 87% of control cells displayed CARM1 localization to the pericentriolar area during mitosis, 46% of OGT-overexpressing cells did not. There was also an increase in the number of cells in which the centrosomes had been amplified (Fig. 5D) with OGT overexpression. Additionally, OGT-overexpressing cells displayed errors in chromosomal separation as we observed less condensed chromatin during anaphase (Fig. 5E) and the formation of chromosomal bridges (Fig. 5F). These phenotypes were apparent even if CARM1 was found near the centrioles (Fig. 5, D–F), suggesting that there are multiple mechanisms whereby OGT is acting to produce DNA segregation errors. We counted over 200 cells that were in anaphase or telophase from either control cells or OGT-overexpressing cells. From these cells, 3% of control cells displayed errors in chromosome segregation, which we define as DNA trapped within the plane of division. On the other hand, in OGT-overexpressing cells, 73% of these cells displayed chromosomal abnormalities. Results are summarized in Table 1.
FIGURE 5.
OGT overexpression causes mislocalization of CARM1 and DNA abnormalities. OGT-overexpressing cells were stained for tubulin (green), DNA (blue), and CARM1 (red) from cells in prometaphase (A), metaphase (B), and anaphase (C). CARM1 does not concentrate near the centrosome in panels A–C. In panels D–F, CARM1 is found in the pericentriolar region; however, there is centrosome amplification in panel D, the degree of DNA condensation in panel E is less than in control cells, and the formation of a chromosomal bridge is shown in panel F; all are characteristics of cells overexpressing OGT.
TABLE 1.
Summary of immunofluorescence data
DNA segregation errors are defined as DNA caught within the plane of division.
| Total | Percentage | ||
|---|---|---|---|
| DNA segregation errors | |||
| Control | 7 | 220 | 3.1 |
| OGT | 177 | 241 | 73.4 |
| CARM1 not at pericentriolar area | |||
| Control | 38 | 295 | 12.8 |
| OGT | 128 | 277 | 46.2 |
DISCUSSION
Here, we demonstrate that OGT expression levels must be balanced during the cell cycle. We found that OGT protein amounts decreased during M phase, which correlates with previously reported data showing that OGT mRNA levels decrease during this time (12). This partial decrease in OGT amounts seems crucial in preventing inappropriate signaling complexes, as was demonstrated in an earlier report showing that OGT overexpression causes changes in mitotic phosphorylation and mitotic O-GlcNAcylation of the cytoskeletal protein vimentin (40). Additional support for this hypothesis comes from a recent proteomic analysis of O-GlcNAc and phosphate in mitotic cells. OGT overexpression alters several of the key signaling pathways controlling the cell cycle, including increasing the inhibitory phosphorylation of Cdk1 due to alterations in upstream pathways leading to Cdk1 activation in mitosis (17).
Additionally, we show that overexpressing OGT caused changes in mitotic post-translational modifications of histones. Mitotic phosphorylation of the N-terminal tail of H3 occurs at Thr-3 by haspin (41, 42), occurs at Ser-10 and -28 by Aurora kinase B (29, 43), and occurs at Thr-11 by death-associated protein (DAP)-like kinase (Dlk), also known as zipper interacting protein (ZIP) kinase (44). Although they display a distinct spatio-temporal pattern, no specific function has been ascribed for each modification. These mitotic histone marks have been suggested to alter protein interaction with chromatin as K9me3 adjacent S10P has been shown to evict heterochromatin protein 1 (HP1) from chromatin during mitosis (45, 46). Alternatively, these mitotic phosphorylation sites may mark certain parts of the chromatid as S10P is found in pericentromeric regions and T11P is found in centromeres (44). These modifications may also play roles in compaction and segregation (47, 48) and, although not crucial for mitotic progression (4), prevent abnormal distribution of DNA between daughter cells. The attenuation of H3S10P and increase in H3K9Ac and H3K27me3 observed with OGT overexpression impacts all these possibilities. OGT overexpression does not prevent mitotic progression per se (11), but in the studies shown here, affected histone tail modifications and DNA segregation. Of note, treatment of cells with histone deacetylase inhibitors has been shown to cause chromosomal instability through faulty mitotic checkpoints (47, 49), consistent with our observation of increased H3K9Ac in OGT-overexpressing cells and the formation of chromosomal bridges.
The mechanism by which OGT overexpression affects these various histone modifications remains an area under active investigation. It is possible that OGT can interact with histone-modifying enzymes, preventing them from recognizing their substrates, as we have observed for CARM1. This possibility could, in fact, be the mechanism that resulted in less H3S10P in OGT-overexpressing cells as the kinase that phosphorylates this site during mitosis, Aurora kinase B, is a known OGT-interacting protein (40). Additionally, OGT overexpression could increase O-GlcNAcylation of histone-modifying proteins, thereby affecting enzymatic activity. In fact, several of these enzymes have been identified as O-GlcNAc proteins in separate studies such as MLL5 (18) and Jumonji domain containing demethylase 1C (17).
Interestingly, OGT localization during mitosis is similar to the mitotic kinases Polo-like kinase 1 (Plk1), Aurora kinase A, and Aurora kinase B (50–53). OGT localizes to the spindle poles at prophase, localizes at the spindles during metaphase and anaphase, and concentrates at the cleavage furrow at cytokinesis (11). One intriguing possibility is that in normal cells, the OGT that is not associated with the mitotic spindle is degraded. OGT co-immunoprecipitates and co-localizes with some of these mitotic kinases, suggesting that the association of OGT with these kinases could protect it from degradation. Overexpression of OGT would then override this normal process, causing abnormal signaling complex formation and sequestration of histone-modifying enzymes from their mitotic substrates.
CARM1 is phosphorylated mitotically on Ser-217 and Ser-228, rendering CARM1 catalytically inactive (36, 37). Interestingly, Ser-217 is in a Plk1 consensus motif ((D/E)X(S/T)(p)), where p indicates the phosphorylated residue (54). CARM1 cytolocalization to the pericentriolar region during M phase is similar to Plk1 (50). Plk1 could recruit CARM1 to this region and phosphorylate it, thus inactivating it. The possible function for CARM1 in this area is not known. CARM1 could act as a scaffold or protein complex stabilizer, and in fact, previous studies have shown that CARM1 catalytic activity is dispensable for transcription from some NFκB-dependent gene expression (55), suggesting that some functional aspects of CARM1 do not rely on catalysis.
Here, we show that OGT overexpression not only prevented the upstream CARM1 kinase from phosphorylating it, but it also prevented CARM1 from methylating Arg-17 of H3. H3R17me2 is primarily linked with gene activation (32); however, it may also serve as a mitotic histone marker. In fact, H3S10P, another marker for mitosis, is also associated with growth factor stimulation (56). It is possible that a small population of H3R17me2 associates with activated genes during interphase, whereas in mitosis, this modification rises throughout the whole genome, much like the paradoxical phosphorylation of Ser-10 of H3 (57). Interestingly, we also observed a decrease in basal levels of dimethylation on Arg-17 of H3 in asynchronous extracts with OGT overexpression. It is possible that OGT overexpression is affecting events upstream of CARM1 recruitment to promoters. Kinetic chromatin immunoprecipitation (ChIP) studies demonstrate that chromatin remodeling and preinitiation complex formation at an active estrogen receptor promoter follow an ordered assembly (58). Additionally, ChIP-on-chip tiling arrays in Caenorhabditis elegans show that the O-GlcNAc modification is enriched in promoter regions (59). OGT overexpression could be affecting events upstream of CARM1 binding to active gene promoters, thus preventing dimethylation of Arg-17.
O-GlcNAc is a proposed regulator of energy homeostasis because its donor substrate, UDP-GlcNAc, is centered around many metabolic pathways (7). Cell cycle progression is clearly dependent on nutrient availability, all of which are deregulated in diseases such as cancer and Alzheimer disease. Potentially, Alzheimer disease is the result of re-entry of postmitotic neurons into the cell cycle (60). Additionally, the impaired glucose uptake associated with Alzheimer disease could possibly reduce O-GlcNAc levels within neurons (61). One possibility is that in normal postmitotic neurons, O-GlcNAc and OGT levels are sufficiently high enough to prevent entry into cell cycle, perhaps through inhibition of cyclin-dependent kinases. Once glucose metabolism is altered, O-GlcNAc levels decrease, allowing reactivation of these kinases. In fact, recent studies demonstrate that decreases in O-GlcNAc levels correlate with tau hyperphosphorylation, a hallmark of Alzheimer disease (38, 62). With the recent insights in how O-GlcNAc impacts phosphorylation at the site-specific level as well as at the level of kinase regulation (8, 17, 63), our current model of cell cycle progression being controlled solely through the actions of kinases must be adapted.
This work was supported by the National Institutes of Health (R01 DK61671 and R01 CA2486, to G. W. H.).
- OGT
- O-GlcNAc transferase
- NAG-thiazoline
- 1,2-dideoxy-2′-methyl-α-d-glucopyranoso[2,1-d]-Δ2′-thiazoline.
REFERENCES
- 1.Satyanarayana A., Kaldis P. (2009) Oncogene 28, 2925–2939 [DOI] [PubMed] [Google Scholar]
- 2.Yamashita Y. M. (2009) Front. Biosci. 14, 3003–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belmont A. S. (2006) Curr. Opin. Cell Biol. 18, 632–638 [DOI] [PubMed] [Google Scholar]
- 4.Georgatos S. D., Markaki Y., Christogianni A., Politou A. S. (2009) Front. Biosci. 14, 2017–2027 [DOI] [PubMed] [Google Scholar]
- 5.Bonenfant D., Towbin H., Coulot M., Schindler P., Mueller D. R., van Oostrum J. (2007) Mol. Cell. Proteomics 6, 1917–1932 [DOI] [PubMed] [Google Scholar]
- 6.Holland A. J., Cleveland D. W. (2009) Nat. Rev. Mol. Cell Biol. 10, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart G. W., Housley M. P., Slawson C. (2007) Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Gucek M., Hart G. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambetta M. C., Oktaba K., Müller J. (2009) Science 325, 93–96 [DOI] [PubMed] [Google Scholar]
- 11.Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. (2005) J. Biol. Chem. 280, 32944–32956 [DOI] [PubMed] [Google Scholar]
- 12.Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., Alexander K. E., Matese J. C., Perou C. M., Hurt M. M., Brown P. O., Botstein D. (2002) Mol. Biol. Cell 13, 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boehmelt G., Wakeham A., Elia A., Sasaki T., Plyte S., Potter J., Yang Y., Tsang E., Ruland J., Iscove N. N., Dennis J. W., Mak T. W. (2000) EMBO J. 19, 5092–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell N., Zachara N. E., Hart G. W., Marth J. D. (2004) Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamemura K., Hayes B. K., Comer F. I., Hart G. W. (2002) J. Biol. Chem. 277, 19229–19235 [DOI] [PubMed] [Google Scholar]
- 16.Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., Hart G. W. (2010) Sci. Signal. 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiki R., Chikanishi T., Hashiba W., Ito H., Takada I., Roeder R. G., Kitagawa H., Kato S. (2009) Nature 459, 455–459 [DOI] [PubMed] [Google Scholar]
- 19.Comer F. I., Vosseller K., Wells L., Accavitti M. A., Hart G. W. (2001) Anal. Biochem. 293, 169–177 [DOI] [PubMed] [Google Scholar]
- 20.Kreppel L. K., Blomberg M. A., Hart G. W. (1997) J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 21.Clark R. J., McDonough P. M., Swanson E., Trost S. U., Suzuki M., Fukuda M., Dillmann W. H. (2003) J. Biol. Chem. 278, 44230–44237 [DOI] [PubMed] [Google Scholar]
- 22.Whelan S. A., Lane M. D., Hart G. W. (2008) J. Biol. Chem. 283, 21411–21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song M., Kim H. S., Park J. M., Kim S. H., Kim I. H., Ryu S. H., Suh P. G. (2008) Cell Signal 20, 94–104 [DOI] [PubMed] [Google Scholar]
- 24.Westendorf J. M., Rao P. N., Gerace L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindqvist A., Rodríguez-Bravo V., Medema R. H. (2009) J. Cell Biol. 185, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 28.Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., Honda B. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. (1997) Chromosoma 106, 348–360 [DOI] [PubMed] [Google Scholar]
- 30.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 31.Sims R. J., 3rd, Reinberg D. (2006) Genes Dev. 20, 2779–2786 [DOI] [PubMed] [Google Scholar]
- 32.Bauer U. M., Daujat S., Nielsen S. J., Nightingale K., Kouzarides T. (2002) EMBO Rep. 3, 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 34.Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. (2008) J. Biol. Chem. 283, 33935–33941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schurter B. T., Koh S. S., Chen D., Bunick G. J., Harp J. M., Hanson B. L., Henschen-Edman A., Mackay D. R., Stallcup M. R., Aswad D. W. (2001) Biochemistry 40, 5747–5756 [DOI] [PubMed] [Google Scholar]
- 36.Feng Q., He B., Jung S. Y., Song Y., Qin J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2009) J. Biol. Chem. 284, 36167–36174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higashimoto K., Kuhn P., Desai D., Cheng X., Xu W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12318–12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., Vocadlo D. J. (2008) Nat. Chem. Biol. 4, 483–490 [DOI] [PubMed] [Google Scholar]
- 39.Knapp S., Vocadlo D. J., Gao Z., Kirk B., Lou J., Withers S. G. (1996) J. Am. Chem. Soc. 118, 6804–6805 [Google Scholar]
- 40.Slawson C., Lakshmanan T., Knapp S., Hart G. W. (2008) Mol. Biol. Cell 19, 4130–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai J., Sultan S., Taylor S. S., Higgins J. M. (2005) Genes Dev. 19, 472–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polioudaki H., Markaki Y., Kourmouli N., Dialynas G., Theodoropoulos P. A., Singh P. B., Georgatos S. D. (2004) FEBS Lett. 560, 39–44 [DOI] [PubMed] [Google Scholar]
- 43.Goto H., Yasui Y., Nigg E. A., Inagaki M. (2002) Genes Cells 7, 11–17 [DOI] [PubMed] [Google Scholar]
- 44.Preuss U., Landsberg G., Scheidtmann K. H. (2003) Nucleic Acids Res. 31, 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D. (2005) Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 46.Hirota T., Lipp J. J., Toh B. H., Peters J. M. (2005) Nature 438, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 47.Cimini D., Mattiuzzo M., Torosantucci L., Degrassi F. (2003) Mol. Biol. Cell 14, 3821–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mora-Bermúdez F., Gerlich D., Ellenberg J. (2007) Nat. Cell Biol. 9, 822–831 [DOI] [PubMed] [Google Scholar]
- 49.Shin H. J., Baek K. H., Jeon A. H., Kim S. J., Jang K. L., Sung Y. C., Kim C. M., Lee C. W. (2003) Oncogene 22, 3853–3858 [DOI] [PubMed] [Google Scholar]
- 50.Arnaud L., Pines J., Nigg E. A. (1998) Chromosoma 107, 424–429 [DOI] [PubMed] [Google Scholar]
- 51.Barr A. R., Gergely F. (2007) J. Cell Sci. 120, 2987–2996 [DOI] [PubMed] [Google Scholar]
- 52.Fuller B. G., Lampson M. A., Foley E. A., Rosasco-Nitcher S., Le K. V., Tobelmann P., Brautigan D. L., Stukenberg P. T., Kapoor T. M. (2008) Nature 453, 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruchaud S., Carmena M., Earnshaw W. C. (2007) Cell 131, 230–231 [DOI] [PubMed] [Google Scholar]
- 54.Malik R., Lenobel R., Santamaria A., Ries A., Nigg E. A., Körner R. (2009) J. Proteome Res. 8, 4553–4563 [DOI] [PubMed] [Google Scholar]
- 55.Jayne S., Rothgiesser K. M., Hottiger M. O. (2009) J. Mol. Biol. 394, 485–495 [DOI] [PubMed] [Google Scholar]
- 56.Chadee D. N., Hendzel M. J., Tylipski C. P., Allis C. D., Bazett-Jones D. P., Wright J. A., Davie J. R. (1999) J. Biol. Chem. 274, 24914–24920 [DOI] [PubMed] [Google Scholar]
- 57.Prigent C., Dimitrov S. (2003) J. Cell Sci. 116, 3677–3685 [DOI] [PubMed] [Google Scholar]
- 58.Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 59.Love D. C., Ghosh S., Mondoux M. A., Fukushige T., Wang P., Wilson M. A., Iser W. B., Wolkow C. A., Krause M. W., Hanover J. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monaco E. A., 3rd, Vallano M. L. (2005) Front. Biosci. 10, 143–159 [DOI] [PubMed] [Google Scholar]
- 61.Dias W. B., Hart G. W. (2007) Mol. Biosyst 3, 766–772 [DOI] [PubMed] [Google Scholar]
- 62.Liu F., Shi J., Tanimukai H., Gu J., Gu J., Grundke-Iqbal I., Iqbal K., Gong C. X. (2009) Brain 132, 1820–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dias W. B., Cheung W. D., Wang Z., Hart G. W. (2009) J. Biol. Chem. 284, 21327–21337 [DOI] [PMC free article] [PubMed] [Google Scholar]