Abstract
Heat shock factor 1 (HSF1) is an important transcription factor in cellular stress responses, cancer, aging, and developmental processes including gametogenesis. Disruption of Hsf1, together with another HSF family member, Hsf2, causes male sterility and complete lack of mature sperm in mice, but the specific role of HSF1 in spermatogenesis has remained unclear. Here, we show that HSF1 is transiently expressed in meiotic spermatocytes and haploid round spermatids in mouse testis. The Hsf1−/− male mice displayed regions of seminiferous tubules containing only spermatogonia and increased morphological abnormalities in sperm heads. In search for HSF1 target genes, we identified 742 putative promoters in mouse testis. Among them, the sex chromosomal multicopy genes that are expressed in postmeiotic cells were occupied by HSF1. Given that the sex chromatin mostly is repressed during and after meiosis, it is remarkable that HSF1 directly regulates the transcription of sex-linked multicopy genes during postmeiotic repression. In addition, our results show that HSF1 localizes to the sex body prior to the meiotic divisions and to the sex chromocenter after completed meiosis. To the best of our knowledge, HSF1 is the first known transcription factor found at the repressed sex chromatin during meiosis.
Keywords: Chromatin Regulation, Gene Regulation, Sperm, Spermatogenesis, Transcription, ChIP-Chip, Heat Shock Factor, Meiosis, Multicopy Genes, Sex Chromatin
Introduction
The mammalian sex chromosomes are highly heteromorphic; the X chromosome is large and gene-rich in contrast to the small, heterochromatic, and degenerate Y chromosome (1). Thus, the sex chromosomes constitute a challenge to the mechanisms that ensure accurate segregation of autosomes during meiosis (2–4). To deal with incomplete synapsis of the sex chromosomes, the sex chromosomes are secluded into a subnuclear compartment called the sex body (5). This subnuclear domain separates the sex chromatin from the rest of the chromatin during meiosis, and it is thought that sex body formation could mask the incompletely synapsed X and Y chromosomes from meiotic surveillance mechanisms (2–4). The sex body is devoid of RNA synthesis, and it contains a unique repertoire of proteins and modified histone variants, which retain the meiotic sex chromosome inactivation (MSCI)4 (2–4). Proteins that have been shown to localize to the sex body belong predominantly to various categories of chromatin proteins, including modified histone variants and proteins associated with DNA damage repair (2–4). For example, the phosphorylated form of the histone variant H2AX (γH2AX) is strongly associated with the sex body as it initiates heterochromatinization of the sex chromosomes (6). Due to MSCI, the X and Y chromosomes are transcriptionally silenced during meiosis at the pachytene stage of spermatogenesis (4). Most sex-linked genes remain repressed during meiosis and, throughout round spermatid development, as several repressive chromatin marks still are present (7–9). The postmeiotic sex chromosome repression might be a direct effect of the MSCI (4).
As a result of no recombination, the Y chromosome ensures its own survival by gene additions from other chromosomes and series of massive inverted repeats, termed palindromes (1, 10). The palindromes give the Y chromosome a possibility of repairing mutations via intrapalindrome, arm-to-arm recombination (1, 10, 11). Repetitive gene families have been found in multiple copies in the mouse Y chromosome: Sly and Ssty1/2 (12–14). Gene duplications give rise to a massive expansion in the so far unknown copy number of genes residing in the repetitive male-specific long arm of the mouse Y chromosome (MSYq) (15). The X chromosome gene content has been primarily based on the analysis of single-copy genes, but large ampliconic regions also reside in the X chromosome (16). These regions contain multicopy gene families, which have not been fully characterized. Nevertheless, 33 X-chromosomal multicopy gene families have been identified in the mouse (16). Some of these genes, such as Slx, have a Y-chromosomal paralogue, Sly (13, 17). Interestingly, the sex chromosomal multicopy genes are expressed at high levels, predominantly in postmeiotic round spermatids (13, 16). The mechanism by which these genes escape the postmeiotic sex chromosome repression has, however, remained obscure.
HSF1 belongs to a family of heat shock transcription factors (HSFs) and is the principal stress-responsive regulator in mammals. HSF1 protects cells from proteotoxic stress through induction of heat shock genes encoding heat shock proteins (Hsps) (18). In addition to heat shock response, HSF1 is important in cancer, aging, and developmental processes like gametogenesis (19–30). Mouse embryos whose mothers lack Hsf1 do not develop beyond the zygotic stage, causing female infertility, and HSF1 is thereby a maternal factor (28). In males, a constitutively active form of HSF1 causes a severe disruption of spermatogenesis and death of pachytene spermatocytes (24), whereas Hsf1−/− males are fertile but produce less sperm and exhibit an increase in disorganized or missing layers of germ cells in the seminiferous tubules (27). Interestingly, disrupting Hsf1 together with another family member Hsf2 causes a clearly potentiated phenotype associated with male infertility and a complete lack of mature spermatozoa, implying that both factors are required for normal spermatogenesis (26). Together, these findings suggest that the activity of HSF1 is tightly intertwined with HSF2 during spermatogenesis, but the specific function of HSF1 in testis is unknown. Intriguingly, there is no correlation between HSF1 and induction of Hsps in male germ cells, highlighting the need to elucidate the HSF1 target genes during sperm maturation.
In this study, we show that the expression of HSF1 was restricted to spermatocytes and round spermatids and that the Hsf1−/− mice displayed dispersed clusters of seminiferous tubules lacking most cell types. Utilizing a chromatin immunoprecipitation on promoter microarray analysis (ChIP-chip), we discovered 742 putative target genes for HSF1 in mouse testis. HSF1 was found to occupy sex chromosomal multicopy genes and regulate their transcription in round spermatids, where the sex chromatin mostly is repressed. Interestingly, HSF1 was localized to the sex chromatin both prior to and after the meiotic divisions in a repressed chromatin environment.
EXPERIMENTAL PROCEDURES
Mice
Hsf1 knock-out mice were maintained in a mixed genetic background bred from a congenic stock (C57BL/6J; Hsf1) intercrossed with BALB/c, and they have been described previously (31). Hsf2 knock-out mice were obtained by the mating of heterozygous mice that has been described earlier (32) and were maintained in a C57BL/6N background. Male hybrid mice of the B6129SF2/J strain were used in the ChIP-chip screen. The animals were kept in a pathogen-free facility under controlled environmental conditions with a 12-h light: 12-h dark cycle and were provided with food and tap water. Protocols for animal experiments were approved by the Departmental Veterinary Office (Haute-Garonne, France) according to French legislation, and by the institutional animal care policy of the Åbo Akademi University (Turku, Finland). Adult (60–80 days old) mice were used for isolation of testes.
Immunohistochemistry
Whole WT and Hsf1−/− testes were fixed in 4% paraformaldehyde overnight at room temperature, embedded in paraffin, and cut into 4-μm sections. Sections were incubated overnight at 4 °C with primary antibodies HSF1 (33) and γH2AX (Millipore) and with secondary antibodies mouse Alexa 488 and rabbit Alexa 568 (Invitrogen) for 1 h. Slides were counterstained with Hoechst 33342 (H-1399, Molecular Probes). Images for all channels were sequentially captured from a single confocal section using a Zeiss Meta510 confocal microscope. After background correction, the colocalizations were analyzed using ImageJ MacBiophotonics colocalization finder tool.
Western Blot Analysis
Testes or cauda epididymes from WT and Hsf1−/− males were lysed and boiled in 3× Laemmli buffer and subjected to SDS-PAGE followed by transfer to nitrocellulose membrane (Protran nitrocellulose, Schleicher & Schuell). Proteins were analyzed from three biological replicates. The primary antibodies were as follows: HSF1 (33), transition protein 1, protamine 1, protamine 2 (M-88, M-51, and M-107 respectively, Santa Cruz Biotechnology), β-tubulin (ab6046, Abcam), α-actin (clone AC-40, Sigma). Secondary antibodies were horseradish peroxidase-conjugated (GE Healthcare). The blots were developed with an enhanced chemiluminescence method (ECL kit, GE Healthcare).
Histology
Whole WT and Hsf1−/− testes were fixed in 4% paraformaldehyde overnight, embedded in paraffin, and cut into 4-μm sections. Testis sections were stained with SYBR Green (catalog no. S7563, Invitrogen).
Seminiferous Tubule Characterization
Whole WT and Hsf1−/− testes were isolated. After decapsulation, the seminiferous tubules were dissected free from the interstitial tissue in a Petri dish containing DMEM. The trans-illumination pattern of stages IX–XI (pale zone), XII–I (weak spot), II–VI (strong spot), and VII–VIII (dark zone) were recognized as described previously (34). Images were captured using a Nikon 800i stereo microscope.
Analysis of Sperm Head Morphology
Adult WT and Hsf1−/− male cauda epididymes were isolated, and sperm head morphology was classified into groups of normal, slightly abnormal, and grossly abnormal, as described earlier (35).
ChIP
Testes were isolated from adult WT mice and lysed in 4 ml of buffer, and the ChIP assays for the ChIP-chip screen were performed as described earlier (36). For ChIP verifications, the ChIP-IT Express kit (Active Motif) was used. The following antibodies were used: HSF1 Ab-4 (Thermo Scientific), HSF1 SPA-901, (Stressgen), normal rabbit IgG, sc-2027 (Santa Cruz Biotechnology), and acetylated histone H4 (Upstate Biotechnologies). The following PCR primers were used: Sly (forward (F), 5′-agc aca gaa gga tgc ggt tt-3′ and reverse (R), 5′-gtg ttt cta agg gat cct gaa tat-3′); Ssty1 (F, 5′-gct cct gaa ctc caa ctt gtt c-3′ and R, 5′-cta aac tgg atc aac cat gcc-3′); Ssty2 (F, 5′-aag cag aac gaa act tct-3′ and R, 5′-tgc ttt aca acc ctg g-3′); Srsy (F, 5′-cag gac att tgt tat ctg ttc aag aa-3′ and R, 5′-cct ggg aag aat cag aaa gtc c-3′); Slx (F, 5′-atg cat caa agc tct ct-3′ and R, 5′-ccg gct aac cct aat-3′); Ott, (F, 5′-agg agc aga gta gca ggc at-3′ and R, 5′-atc ctt ttc tga cca gtg gc-3′); Ssxb2 (F, 5′-tgc acg ttt caa cag tca aa-3′ and R, 5′-ctg gag aca caa gaa agg ca-3′); and Hsp70 (F, 5-cac cag cac gtt ccc ca-3′ and R, 5′-ccc gcc tcc ctt gag taa tc-3′).
ChIP-chip
The DNA amplification, hybridization, and data analysis were performed as described previously (35). In short, DNA amplification of material obtained from three biological replicates was performed using ligation-mediated PCR according to NimbleGen Systems' protocol. The experimental HSF1 amplicons were labeled with Cy5 dye, and the total input amplicons were labeled with Cy3 dye (including one dye-swap) and then cohybridized to high density oligonucleotide tiling arrays. The HSF1 ChIP signal was compared with a control input signal. The two-channel raw data were normalized between channels with the Lowess normalization method and ChIP-to-input log2 ratios were produced separately from all three replicates. The target promoters were ranked separately in the three replicates according to the average log2 ratios of all probes covering each promoter using RankProd (37). The data were filtered with p < 0.005, which resulted in a list of 742 HSF1-bound promoters (supplemental Table S1).
Quantitative Real-time RT-PCR
Whole WT and Hsf1−/− testes were isolated. Real-time RT-PCR reactions were prepared and run as described earlier (38). Relative quantities of the target gene mRNAs were normalized against the round spermatid-specific Pfn3, and the fold induction from WT samples was calculated. All reactions were made in triplicate, using samples derived from at least three biological repeats. These primers and probes were used: Sly (F, 5′-att caa tga aga aaa aga aaa atc agt-3′; R, 5′-cca tgg act tct atg cat tt-3′; probe (P), 5′-Fam gga agc ag Q-3′); Ssty2 (F, 5′-ctc cac atc att cca gag acc-3′; R, 5′-aag aag tca ttg tca tca cct gaa-3′; P, 5′-Fam ctg gct gg Q-3′); Slx (F, 5′-gtt gga taa act tgg aga aaa cg-3′; R, 5′-tct cta cag aac gtg caa aac g-3′; P, 5′-Fam cag agg aa Q-3′); Ott (F, 5′-cac acc tca gca agt gga tct-3′; R, 5′-aga tat ctc agc tgt cta att tcg tct-3′; P, 5′-Fam agt ccc ag Q-3′); and Pfn3 (F, 5′-ctg ctg cgt tat ccg tga-3′; R, 5′-aga ttg cac gcc cgt cta-3′; P, 5′-Fam acc tgc tg Q-3′). Q stands for dark quencher dye.
Statistical Analysis
The chromosomal distribution analysis was performed with the GeneMerge tool, which uses hypergeometric distribution for calculation of the p value (39). The investigation of the biological processes associated with target genes was performed with the DAVID analysis tool, which uses Fisher's exact test for calculation of the p value (40). Unpaired Student's t test was used for calculation of the p value in comparisons between WT and Hsf1−/−.
RESULTS
HSF1 Is Transiently Expressed in Spermatocytes and Round Spermatids
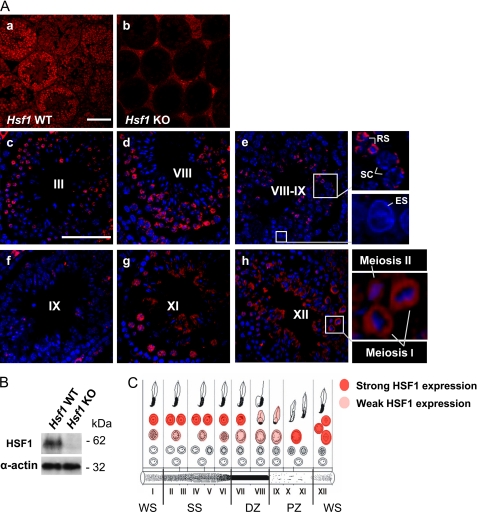
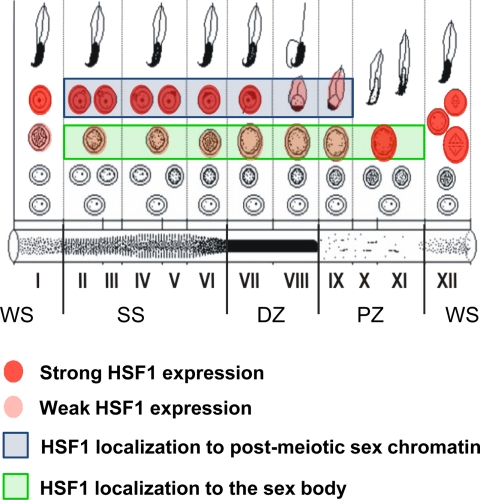
It has been reported previously that in mice kept under normal physiological conditions, HSF1 is expressed in all testicular cells (24). However, the localization of HSF1 in specific germ cell types has remained unclear. Therefore, we investigated HSF1 expression in mouse testis, where the cycle of the seminiferous epithelium is composed of 12 stages (I–XII), each comprising a distinct repertoire of developing germ cell types (34). The specificity of the HSF1 antibody was confirmed by a total lack of HSF1 expression in Hsf1−/− seminiferous tubules when compared with the WT (Fig. 1A, panels a and b, and B). A detailed analysis of the WT testis revealed that early stages of the cycle of the seminiferous epithelium (I–III) displayed high expression of HSF1 in the nuclei of round spermatids, whereas only discrete spots were present in the spermatocytes (Fig. 1A, panel c). Similar HSF1 expression patterns were detected until stage VIII (Fig. 1A, panel d). In the transition between stages VIII–IX, HSF1 was still present in round spermatids and vaguely in spermatocytes, but in early elongating spermatids, HSF1 disappeared (Fig. 1A, panel e, insets). In accordance, stage IX almost completely lacked expression of HSF1 (Fig. 1A, panel f). A striking reappearance of HSF1 occurred at stage XI, displaying strong expression in spermatocytes (Fig. 1A, panel g). At the last stage of the epithelial cycle (XII), where the meiotic divisions take place, HSF1 was found in germ cells undergoing meiosis I and II (Fig. 1A, panel h, inset). Specifically during meiosis I, HSF1 was localized in a ring-like fashion around the DNA. The complete HSF1 expression profile in distinct cell types, throughout the cycle of the seminiferous epithelium, is schematically presented in Fig. 1C, showing that HSF1 is transiently expressed in late meiosis and round spermatid development.
FIGURE 1.
HSF1 is transiently expressed during mouse spermatogenesis. A, the localization and expression of HSF1 in specific cell types were examined by immunohistochemistry. Panels a and b, HSF1 immunostaining of wild-type (Hsf1 WT) and Hsf1 knock-out (Hsf1 KO) testis. No signal in the seminiferous epithelium of Hsf1 KO testis indicates specificity of the antibody. Panels c–h, HSF1 immunostaining (red) of WT testis, counterstained with Hoechst (blue). Representative stages are denoted by Roman numerals. Insets show blowups of the selected regions. SC, spermatocyte; RS, round spermatid; ES, elongating spermatid. Scale bars, 100 μm. B, Western blot analysis of HSF1 levels in Hsf1 WT and KO testis. Equal protein loading was assessed by β-tubulin and α-actin. C, a schematic drawing displaying the cell type-specific expression of HSF1 throughout the seminiferous epithelial cycle. Each vertical column, designated by a Roman numeral, depicts a stage of the seminiferous epithelial cycle and is comprised of a defined set of developing germ cells. In the schematic (C), the developmental progression of a cell is followed from the bottom row, left to right. The cycle ends with the release of morphologically mature sperm to the lumen of the tubule. The light absorption patterns of corresponding stages of the seminiferous tubule are shown at the bottom. Stages IX–XI, pale zone (PZ); stages XII–I, weak spot (WS); stages II–VI, strong spot (SS); and stages VII–VIII, dark zone (DZ).
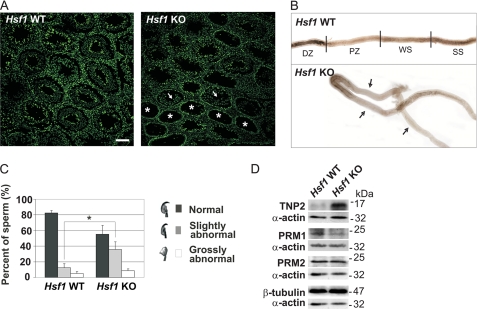
The cell type-specific HSF1 expression prompted us to assess the Hsf1−/− testis phenotype. It has been reported previously that 5–30% of the Hsf1−/− seminiferous tubules show disorganized or missing layers of germ cells (27). We investigated which cell types were missing in the HSF1-deficient tubules. Employing histological analyses, we detected that most of the affected tubules contained only spermatogonia, displaying a total lack of other germ cell types (Fig. 2A). Nevertheless, some of the disrupted tubules lacked spermatids and only contained cell layers composed of spermatogonia and spermatocytes. The disrupted tubules of the Hsf1 knock-out were often found in clusters (Fig. 2A). The seminiferous tubules are organized as highly convoluted loops (41). The clustered patterns reflect the long stretches of pale tubules frequently seen in the knock-out (Fig. 2B), and the faint appearance of the Hsf1−/− tubule is most likely due to the loss of cells, whereas the darker areas between the pale regions could constitute of apoptotic cells.
FIGURE 2.
Characterization of the phenotype of Hsf1−/− testis and mature sperm. A, SYBR green staining of testis cross sections of wild-type (Hsf1 WT) and Hsf1 knock-out (Hsf1 KO) mice, was used to determine the morphology of the seminiferous tubules, in addition to characterization of missing cell types. Asterisks indicate tubules containing only spermatogonia, and arrows indicate tubules containing spermatogonia and spermatocytes. Scale bar, 100 μm. B, isolated seminiferous tubules from Hsf1 WT and KO males. Arrows indicate abnormally long pale zones. To visualize long stretches of tubules, several images were merged into one using Adobe Photoshop. Stages IX–XI, pale zone (PZ); stages XII–I, weak spot (WS); stages II–VI, strong spot (SS); and stages VII–VIII, dark zone (DZ). C, analysis of hematoxylin-stained sperm smears from WT and KO males. The sperm heads were classified into categories of normal, slightly abnormal and grossly abnormal. 400–500 sperm in each category were counted in a blind experiment. Mean ± S.D. (n = 3); *, p < 0.05. D, Western blot analysis of transition protein 2 (TNP2), protamine 1 (PRM1), and protamine 2 (PRM2) levels in cauda epididymis isolated from WT and KO mice. The blots are representative of three biological repeats. Equal protein loading was assessed by β-tubulin and α-actin.
Next, the phenotype of Hsf1−/− sperm was examined. In addition to lowered sperm count (27), we noticed that epididymis lacking HSF1 contained 1.8-fold more sperm with slightly abnormal head structures than the WT (Fig. 2C). Moreover, aberrant levels of chromatin packing proteins, such as transition protein 2 and protamine 1, were detected in HSF1-deficient epididymes (Fig. 2D). Our results, which displayed abnormal sperm head morphology and disturbed replacement of transition proteins with protamines, imply defects in the chromatin packing in Hsf1−/− sperm.
HSF1 Has Target Genes in Common with HSF2 in Testis
To unravel the functions of HSF1 in spermatogenesis, our strategy was to identify its target genes in testis. Although earlier studies have reported genome-wide HSF targets in response to stress in different model organisms (42–44), no screen for mammalian HSF1 targets under physiologically normal conditions has been performed. Using a ChIP-chip approach (see “Experimental Procedures”), we discovered 742 novel targets for HSF1 in mouse testis (supplemental Table S1). The complete data set is available at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo under accession no. GSE22492). The highest ranked target genes (p < 0.001) were predominantly associated to biological and molecular functions regarding transcription and nucleic acid binding (Table 1). This result implies that the majority of HSF1 targets in testis are involved in transcriptional regulation and that HSF1, through its target genes, also can indirectly modulate transcription.
TABLE 1.
Functional annotation classes of HSF1 target genes analyzed by the David analysis tool (40)
| Enriched Gene Ontology terms | Proportion of enriched genes | p valuea |
|---|---|---|
| % | ||
| Biological process | ||
| Nucleotide and nucleic acid metabolic process | 31.8 | 0.001 |
| Regulation of transcription | 22.7 | 0.007 |
| RNA biosynthetic process | 20.5 | 0.02 |
| Molecular function | ||
| Nucleic acid binding | 31.8 | 0.002 |
| Zinc ion binding | 20.5 | 0.02 |
| Transcription regulator activity | 13.6 | 0.1 |
a Values indicate the probability that the category has been identified by random chance. The HSF1 target genes with a p value <0.001 were included in this analysis in a single copy. The default thresholds in the DAVID functional annotation tool were used (minimum of two genes in the class and maximum p value of 0.1).
Analysis of the chromosomal distribution of HSF1 target genes revealed that they were dispersed over the whole genome, but interestingly, a prominent accumulation was detected on the Y chromosome (supplemental Table S2). As we observed previously that a similar enrichment of HSF2 targets in mouse testis (35), we wanted to determine how many targets are specific for HSF1 and how many are shared with HSF2. Both ChIP-chip screens were analyzed utilizing RankProd (37), and the same cut-off criteria (p < 0.005) were applied when defining the number of HSF1 and HSF2 targets, enabling comparison of the two target gene lists (supplemental Table S1) (35). The results showed that ∼15% of the targets were shared by both HSFs. Based on the obtained results, we concluded that HSF1 binds to some target genes independently of HSF2, whereas other targets may require the presence of both factors at their promoters.
HSF1 Regulates Transcription of Multicopy Genes in Post-meiotic Cells
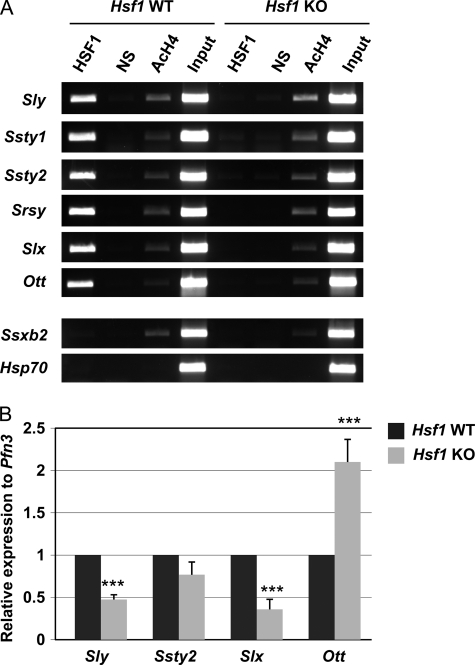
Of all the putative HSF1 targets, we focused on the X- and Y-chromosomal genes. Interestingly, all of the sex chromosomal target genes were multicopy genes (supplemental Table S1). A substantial proportion of these multicopy genes is transcribed and maintains an open reading frame (17, 45, 46). Y-chromosomal multicopy genes Sly and Ssty1/2, which we characterized previously as HSF2 targets (35), were also validated as target genes for HSF1 (Fig. 3A). The most recently discovered and still uncharacterized Y-chromosomal multicopy gene family, Srsy (Entrez Gene accession no. 385550), also was occupied by HSF1. Furthermore, the X-linked multicopy genes Slx and Ott (which was not included in the microarray of the ChIP-chip screen) were both bound by HSF1 (Fig. 3A), whereas another X-chromosomal multicopy gene, Ssxb2, was not targeted by HSF1 (Fig. 3A). Thus, our results suggest that HSF1 is present on certain but not all X-linked multicopy gene promoters.
FIGURE 3.
HSF1 is a transcriptional regulator of sex chromosomal multicopy genes in testis. A, ChIP analysis of HSF1 binding to several sex chromosomal multicopy genes in wild-type (Hsf1 WT) and Hsf1 knock-out (Hsf1 KO) testis. In addition, examples of an X-chromosomal multicopy gene, Ssxb2, and a single-copy gene, Hsp70, not bound by HSF1 are shown. NS, nonspecific antibody; AcH4, acetylated histone H4. Input represents 1% of the total material used in the ChIP assay. B, real-time RT-PCR analysis of gene expression in whole wild-type (Hsf1 WT) and knock-out (Hsf1 KO) testes. The mRNA levels were normalized to the round spermatid-specific Pfn3. The relative expression was calculated from the Hsf1 WT sample, which arbitrarily was set to 1. Mean ± S.D. (n = 3); ***, p < 0.005.
Real-time RT-PCR was used to decipher whether HSF1 participates in the transcription of multicopy genes. Because multicopy genes are mostly expressed in post-meiotic cells (13, 14, 16, 17, 47), the mRNA levels were normalized to the round spermatid-specific Pfn3. mRNA levels of the Y-linked multicopy gene Sly were clearly reduced in Hsf1−/− testis, whereas a minor reduction in Ssty2 levels was observed when compared with the WT (Fig. 3B). The mRNA levels of the X-linked multicopy gene Slx also were decreased in the Hsf1−/− testis (Fig. 3B). However, the levels of Ott mRNA were increased in the HSF1-deficient testis, implying that HSF1 can either have an activating or a repressing effect on X-chromosomal multicopy genes. These results demonstrate that HSF1 regulates transcription of X- and Y-chromosomal multicopy genes after meiosis in round spermatids.
HSF1 Is Localized to the Meiotic and Postmeiotic Sex Chromatin
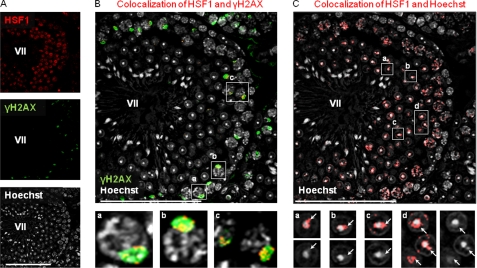
Due to the prominent localization of HSF1 in discrete loci of the pachytene spermatocytes (Fig. 1A), we investigated whether HSF1 occupies the sex chromatin already during meiotic stages of the spermatogenesis. We analyzed HSF1 colocalization with γH2AX, a marker for the sex body (6), in the cycle of the seminiferous epithelium of WT testis (Fig. 4 and supplemental Fig. S1). In stages XII–I, HSF1 did not colocalize with the intense, elongated γH2AX structure that hallmarks X- and Y-chromosomes at the transition between the zygotene and pachytene spermatocytes (supplemental Fig. S1A). Intriguingly, in the following stages II–III, HSF1 localized at the sex chromatin in early pachytene spermatocytes (stage II–III, supplemental Fig. S1B) and was most abundant in the sex body of pachytene spermatocytes in stages VI–VIII (Fig. 4, A and B, and supplemental Fig. S1, C and D). In stage XI diplotene spermatocytes, HSF1 disappeared from the sex chromatin and localized to the surroundings of metaphase I and II chromosomes (supplemental Fig. S1, A and F). After completed meiosis, HSF1 was not detected at the sex chromatin in stage I round spermatids (supplemental Fig. S1A), where the postmeiotic sex chromatin can be visualized as the Hoechst-dense structure at the periphery of chromocenter (8). However, HSF1 relocalized to the sex chromatin in stage II–III round spermatids (supplemental Fig. S1B) and was observed at the sex chromatin throughout round spermatid development (supplemental Fig. S1, B–E). In elongating spermatids, HSF1 levels decreased rapidly, even though HSF1 still was detected at the sex chromatin (supplemental Fig. S1E). After stage IX, HSF1 was not detected in late elongating spermatids (supplemental Fig. S1F). It is worth noticing that the degree of observed colocalization in both spermatocytes and round spermatids varied (Fig. 4, B and C, and supplemental Fig. S1), depending on the section of the confocal three-dimensional stack (supplemental Fig. S2). A schematic illustration of HSF1 localization to the meiotic sex chromatin in spermatogenesis is shown in Fig. 5. Our results suggest that HSF1 localizes to the meiotic and postmeiotic sex chromatin in a repressive environment.
FIGURE 4.
HSF1 localizes to meiotic and postmeiotic sex chromatin. A, detection of HSF1 (upper panel) and γH2AX (middle panel) by immunofluorescence and detection of DNA by Hoechst (lower panel) in a single confocal section of stage VII seminiferous tubule. B, colocalization of HSF1 with γH2AX (red) is superimposed on γH2AX-stained sex bodies (green), designating HSF1 localization to meiotic sex chromatin. Insets a, b, and c show HSF1 localization to sex bodies in stage VII pachytene spermatocytes. C, postmeiotic sex chromatin (white arrows in panels a–d) can be observed as a cloud-like structure peripherally to the Hoechst-dense chromocenter. The HSF1 Hoechst colocalization (red) reveals HSF1 localization to the postmeiotic sex chromatin of round spermatids. Compare HSF1 Hoechst colocalization in insets a–d (upper panels) with Hoechst-only staining (lower panels), where the sex chromocenter is clearly visible. Note that the same confocal section of the stage VII seminiferous tubule is shown in A, B, and C. The specificity of secondary antibodies that were used in the colocalization analyses was confirmed (supplemental Fig. S3). Colocalization was analyzed using ImageJ MacBiophotonics colocalization finder tool. Representative stages are shown in the figure, and the blowup is shown in the insets. Stages are denoted by Roman numerals. Scale bars, 100 μm.
FIGURE 5.
Schematic presentation of HSF1 localization to the meiotic sex chromatin. HSF1 localizes to the meiotic sex chromatin in spermatocytes of stages II–XI, where the sex chromosomes are retained silent in a γH2AX-enriched sex body (Fig. 4 and supplemental Fig. S1). After the meiotic divisions, HSF1 localizes to the postmeiotic sex chromatin in spermatids of stages II–IX, where the sex chromatin is observed as a cloud-like structure peripherally to the Hoechst-dense chromocenter (Fig. 4 and supplemental Fig. S1). Roman numerals depict the stage of the seminiferous epithelial cycle, each of which is comprised of a defined set of developing germ cells. The light absorption patterns of corresponding stages of the seminiferous tubule are shown at the bottom. Pale zone (PZ), stages IX–XI; weak spot (WS), stages XII–I; strong spot (SS), stages II–VI, and dark zone (DZ), stages VII–VIII.
DISCUSSION
HSF1 is best known as the principal regulator of the heat shock response. In addition, HSF1 is a developmental factor, but its function in testis is not demonstrated conclusively. To establish the impact of HSF1 on spermatogenesis, we examined its expression and knock-out phenotype and searched for its direct target genes. The expression analyses revealed that HSF1 is localized specifically in spermatocytes and round spermatids. These results are well in line with HSF1 expression studies previously conducted with rat testis (48). Moreover, a novel transient expression pattern of HSF1 was detected (Fig. 1). The HSF1 expression observed in pachytene spermatocytes prior to the meiotic divisions, in cells undergoing meiotic divisions and right after the completion of meiosis in haploid spermatids, strongly indicates that HSF1 plays a role in meiotic cell divisions and early spermatid differentiation.
In spermatogenesis, ∼75% of the germ cells are estimated to undergo apoptotic cell death in the testis (49). In addition to spontaneous cell death, the developing germ cells are highly susceptible to stress (50, 51). Because the process of spermatogenesis is sensitive to high temperatures, quality control mechanisms are important to eliminate injured or abnormal cells in spermatogenesis. Of all cell types in spermatogenesis, the pachytene spermatocytes are most vulnerable to elevated temperatures (52, 53). We found that HSF1 was expressed in discrete loci of these sensitive pachytene spermatocytes under physiological conditions (Fig. 1B, panel g). Interestingly, constitutively active HSF1 causes apoptosis of pachytene spermatocytes, and the heat-inducible apoptosis of pachytene spermatocytes is markedly inhibited in Hsf1−/− testes (24, 25). Although HSF1 is activated, the Hsps are not induced in the spermatocytes in response to heat stress. These results indicate that HSF1 promotes apoptotic cell death of pachytene spermatocytes exposed to thermal stress, thereby protecting the organism from abnormal development in the next generation (25).
Recently, it was demonstrated that HSF1 and HSF2 can form heterotrimers upon stress (54), which could provide an efficient mechanism to integrate the trans-activating capabilities of these factors. Moreover, HSF1 has been found to interact with HSF2 in mouse testis (54). These findings, combined with our new results on their common target genes (supplemental Table S3), suggest that HSF1 and HSF2 can form heterotrimers in testis and thereby facilitate transcriptional fine-tuning of a subset of target genes. The functional relationship between HSF1 and HSF2 in spermatogenesis is intriguing, and several obvious questions, e.g. the stoichiometry in a possible heterocomplex in testis, remain to be answered. Given the slightly different DNA-binding preferences of HSF1 and HSF2 (55, 56), the composition of heat shock elements on the target promoters could direct the formation of a specific heterocomplex.
In contrast to HSF1, the impact of HSF2 on spermatogenesis has earlier been examined. Hsf2−/− males display reduced size of testis, disruption of spermatogenesis at the pachytene stage, lowered number of germ cells, and increased sperm head abnormalities (32, 35, 57). Inactivation of both Hsf1 and Hsf2 results in a more severe phenotype, manifested by arrested spermatogenesis and complete sterility (26). This phenotype proposes that both HSFs are essential for male fertility. The Hsf1−/− testis phenotype was clearly different from that of Hsf2−/− (32, 57) because long pale zones of seminiferous tubules and regions lacking spermatocytes and spermatids were found in the Hsf1−/− testis (Fig. 2). In contrast to the distinct testis phenotype, mature sperm lacking HSF1 displayed close resemblance to Hsf2−/− sperm as abnormal head morphology and defects in the replacement of chromatin packing proteins were detected in sperm from either Hsf1−/− or Hsf2−/− male mice (35). The phenotypical analyses indicate that HSF1 has unique and overlapping functions with HSF2 in spermatogenesis. Our results also revealed that HSF1 and HSF2 have shared target promoters in testis (supplemental Table S3) and that both HSFs are required for the transcriptional regulation of sex chromosomal multicopy genes (Fig. 3) (35). It has been suggested that HSF1 requires cooperation with HSF2 in development (54) and that the synergistic action of both HSFs is crucial for sperm production (26). It is albeit possible that the different HSFs could to certain extent compensate each other.
We discovered that the transcription of sex chromosomal multicopy genes is regulated by both HSF1 and HSF2, although they seem to have opposing actions on certain promoters (Fig. 3) (35), leading to either activation or repression of the target genes. Due to MSCI, the X and Y chromosomes are transcriptionally silenced in meiosis at the pachytene stage of spermatogenesis (4). The repressive state of the sex chromosomes continues throughout the development of round spermatids (7–9). Interestingly, the sex-linked multicopy genes are expressed predominantly in round spermatids (13, 16, 45). Specifically, the multicopy genes residing in the MSYq-region are critical for sperm differentiation and for correct packing of the chromatin in male germ cells (13, 45, 58, 59). The existence of multiple copies of the X- and Y-chromosomal genes has been proposed to counteract the repressive sex chromatin state in postmeiotic cells (16). In light of these reports, we propose that the presence of both HSF1 and HSF2 is required for the transcriptional regulation of certain X- and Y-chromosomal multicopy gene promoters in postmeiotic cells.
A hallmark of the heat shock response is the extinction of bulk transcription, whereas transcription of Hsp is activated rapidly and strongly (60). The transcriptional activation of Hsp promoters is mainly mediated by HSF1, as HSF2 has only a modulating role upon heat stress (38). Nevertheless, both factors can bind to the Hsp promoters in response to heat shock (38) when most other promoters are silenced. Similarly to heat shock, transcription of all single-copy X- and Y-chromosomal genes is silenced during meiosis, and the genes remain repressed throughout spermatid development (4, 8, 9). Intriguingly, our results indicated that HSF1 and HSF2 occupy sex-linked multicopy genes in round spermatids (Fig. 3 and Ref. 35), allowing the multicopy genes to escape postmeiotic sex chromosome repression. It is likely that HSF1 and HSF2, possibly with help from other factors, are capable of binding to target promoters in transcriptionally repressive environments.
Localization studies revealed that HSF1 occupies the sex chromatin of the sex body during meiotic stages of the spermatogenesis (Fig. 4 and supplemental Fig. S1). The sex body is a specialized meiotic chromatin domain distinct from the autosomal domain, and it is characterized by a lack of RNA synthesis and by sequestration of a unique array of proteins to retain MSCI (2–4). Proteins that have been found to localize to the sex body are mainly chromosomal proteins, such as histone variants and proteins associated with DNA damage repair (2–4). To our knowledge, this is the first time a transcription factor such as HSF1 is shown to colocalize with γH2AX and the sex body in meiotic cells and with the sex chromocenter of postmeiotic cells. A challenge for forthcoming studies is to establish the specific function of HSF1 at the repressed sex chromatin in distinct cell types.
Supplementary Material
Acknowledgments
We are grateful to Noora Kotaja for insightful discussions. We thank Karoliina Rautoma and Johanna Manninen for expert technical assistance, Pia Roos-Mattjus and the members of our laboratory for critical comments on the manuscript.
This work was supported by The Academy of Finland, The Sigrid Jusélius Foundation, The Finnish Cancer Organizations, and Åbo Akademi University (to L. S.), The Magnus Ehrnrooth Foundation and Foundation of Åbo Akademi Research Institute (to M. Å., A. V., and E. H.), and by the Turku Graduate School of Biomedical Sciences (TuBS) (to M. Å. and A. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S3.
- MSCI
- meiotic sex chromosome inactivation
- HSF
- heat shock factor
- F
- forward
- R
- reverse
- Q
- dark quencher dye
- KO
- knock-out
- ChIP-chip
- chromatin immunoprecipitation on promoter microarray analysis.
REFERENCES
- 1.Ellis P. J., Affara N. A. (2006) Hum. Fertil. (Camb.) 9, 1–7 [DOI] [PubMed] [Google Scholar]
- 2.Hoyer-Fender S. (2003) Cytogenet. Genome Res. 103, 245–255 [DOI] [PubMed] [Google Scholar]
- 3.Handel M. A. (2004) Exp. Cell. Res. 296, 57–63 [DOI] [PubMed] [Google Scholar]
- 4.Turner J. M. (2007) Development 134, 1823–1831 [DOI] [PubMed] [Google Scholar]
- 5.Solari A. J. (1974) Int. Rev. Cytol. 38, 273–317 [DOI] [PubMed] [Google Scholar]
- 6.Mahadevaiah S. K., Turner J. M., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W. M., Burgoyne P. S. (2001) Nat. Genet. 27, 271–276 [DOI] [PubMed] [Google Scholar]
- 7.Greaves I. K., Rangasamy D., Devoy M., Marshall Graves J. A., Tremethick D. J. (2006) Mol. Cell. Biol. 26, 5394–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Namekawa S. H., Park P. J., Zhang L. F., Shima J. E., McCarrey J. R., Griswold M. D., Lee J. T. (2006) Curr. Biol. 16, 660–667 [DOI] [PubMed] [Google Scholar]
- 9.Turner J. M., Mahadevaiah S. K., Ellis P. J., Mitchell M. J., Burgoyne P. S. (2006) Dev. Cell 10, 521–529 [DOI] [PubMed] [Google Scholar]
- 10.Turner J. M. (2005) Chromosoma 114, 300–306 [DOI] [PubMed] [Google Scholar]
- 11.Lange J., Skaletsky H., van Daalen S. K., Embry S. L., Korver C. M., Brown L. G., Oates R. D., Silber S., Repping S., Page D. C. (2009) Cell 138, 855–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conway S. J., Mahadevaiah S. K., Darling S. M., Capel B., Rattigan A. M., Burgoyne P. S. (1994) Mamm. Genome 5, 203–210 [DOI] [PubMed] [Google Scholar]
- 13.Touré A., Clemente E. J., Ellis P., Mahadevaiah S. K., Ojarikre O. A., Ball P. A., Reynard L., Loveland K. L., Burgoyne P. S., Affara N. A. (2005) Genome Biol. 6, R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touré A., Grigoriev V., Mahadevaiah S. K., Rattigan A., Ojarikre O. A., Burgoyne P. S. (2004) Genomics 83, 140–147 [DOI] [PubMed] [Google Scholar]
- 15.Ellis P. J., Ferguson L., Clemente E. J., Affara N. A. (2007) BMC Evol. Biol. 7, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller J. L., Mahadevaiah S. K., Park P. J., Warburton P. E., Page D. C., Turner J. M. (2008) Nat. Genet. 40, 794–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynard L. N., Turner J. M., Cocquet J., Mahadevaiah S. K., Touré A., Höög C., Burgoyne P. S. (2007) Biol. Reprod. 77, 329–335 [DOI] [PubMed] [Google Scholar]
- 18.Lindquist S., Craig E. A. (1988) Annu. Rev. Genet. 22, 631–677 [DOI] [PubMed] [Google Scholar]
- 19.Khaleque M. A., Bharti A., Sawyer D., Gong J., Benjamin I. J., Stevenson M. A., Calderwood S. K. (2005) Oncogene 24, 6564–6573 [DOI] [PubMed] [Google Scholar]
- 20.Dai C., Whitesell L., Rogers A. B., Lindquist S. (2007) Cell 130, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu A. L., Murphy C. T., Kenyon C. (2003) Science 300, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 22.Morley J. F., Morimoto R. I. (2004) Mol. Biol. Cell 15, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakai A., Suzuki M., Tanabe M. (2000) EMBO J. 19, 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izu H., Inouye S., Fujimoto M., Shiraishi K., Naito K., Nakai A. (2004) Biol. Reprod. 70, 18–24 [DOI] [PubMed] [Google Scholar]
- 26.Wang G., Ying Z., Jin X., Tu N., Zhang Y., Phillips M., Moskophidis D., Mivechi N. F. (2004) Genesis 38, 66–80 [DOI] [PubMed] [Google Scholar]
- 27.Salmand P. A., Jungas T., Fernandez M., Conter A., Christians E. S. (2008) Biol. Reprod. 79, 1092–1101 [DOI] [PubMed] [Google Scholar]
- 28.Christians E., Davis A. A., Thomas S. D., Benjamin I. J. (2000) Nature 407, 693–694 [DOI] [PubMed] [Google Scholar]
- 29.Metchat A., Åkerfelt M., Bierkamp C., Delsinne V., Sistonen L., Alexandre H., Christians E. S. (2009) J. Biol. Chem. 284, 9521–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bierkamp C., Luxey M., Metchat A., Audouard C., Dumollard R., Christians E. (2010) Dev. Biol. 339, 338–353 [DOI] [PubMed] [Google Scholar]
- 31.McMillan D. R., Xiao X., Shao L., Graves K., Benjamin I. J. (1998) J. Biol. Chem. 273, 7523–7528 [DOI] [PubMed] [Google Scholar]
- 32.Kallio M., Chang Y., Manuel M., Alastalo T. P., Rallu M., Gitton Y., Pirkkala L., Loones M. T., Paslaru L., Larney S., Hiard S., Morange M., Sistonen L., Mezger V. (2002) EMBO J. 21, 2591–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarge K. D., Murphy S. P., Morimoto R. I. (1993) Mol. Cell. Biol. 13, 1392–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotaja N., Kimmins S., Brancorsini S., Hentsch D., Vonesch J. L., Davidson I., Parvinen M., Sassone-Corsi P. (2004) Nat. Methods 1, 249–254 [DOI] [PubMed] [Google Scholar]
- 35.Åkerfelt M., Henriksson E., Laiho A., Vihervaara A., Rautoma K., Kotaja N., Sistonen L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang Y., Östling P., Åkerfelt M., Trouillet D., Rallu M., Gitton Y., El Fatimy R., Fardeau V., Le Crom S., Morange M., Sistonen L., Mezger V. (2006) Genes Dev. 20, 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004) FEBS Lett. 573, 83–92 [DOI] [PubMed] [Google Scholar]
- 38.Östling P., Björk J. K., Roos-Mattjus P., Mezger V., Sistonen L. (2007) J. Biol. Chem. 282, 7077–7086 [DOI] [PubMed] [Google Scholar]
- 39.Castillo-Davis C. I., Hartl D. L. (2003) Bioinformatics 19, 891–892 [DOI] [PubMed] [Google Scholar]
- 40.Dennis G., Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 41.Russell L. D., Ettlin R. A., Sinha Hikim A. P., Clegg E. D. (1990) Histological and Histopathological Evaluation of the Testis, 1st Ed., Cache River Press, St. Louis, MO [Google Scholar]
- 42.Hahn J. S., Hu Z., Thiele D. J., Iyer V. R. (2004) Mol. Cell. Biol. 24, 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birch-Machin I., Gao S., Huen D., McGirr R., White R. A., Russell S. (2005) Genome Biol. 6, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinklein N. D., Murray J. I., Hartman S. J., Botstein D., Myers R. M. (2004) Mol. Biol. Cell 15, 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Touré A., Szot M., Mahadevaiah S. K., Rattigan A., Ojarikre O. A., Burgoyne P. S. (2004) Genetics 166, 901–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynard L. N., Cocquet J., Burgoyne P. S. (2009) Biol. Reprod. 81, 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calenda A., Allenet B., Escalier D., Bach J. F., Garchon H. J. (1994) EMBO J. 13, 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alastalo T. P., Lönnström M., Leppä S., Kaarniranta K., Pelto-Huikko M., Sistonen L., Parvinen M. (1998) Exp. Cell. Res. 240, 16–27 [DOI] [PubMed] [Google Scholar]
- 49.Print C. G., Loveland K. L. (2000) Bioessays 22, 423–430 [DOI] [PubMed] [Google Scholar]
- 50.Crew F. A. (1922) J. Anat. 56, 98–106 [PMC free article] [PubMed] [Google Scholar]
- 51.Moore C. R. (1924) Science 59, 41–44 [DOI] [PubMed] [Google Scholar]
- 52.Almon E., Goldfinger N., Kapon A., Schwartz D., Levine A. J., Rotter V. (1993) Dev. Biol. 156, 107–116 [DOI] [PubMed] [Google Scholar]
- 53.Schwartz D., Goldfinger N., Rotter V. (1993) Oncogene 8, 1487–1494 [PubMed] [Google Scholar]
- 54.Sandqvist A., Björk J. K., Åkerfelt M., Chitikova Z., Grichine A., Vourc'h C., Jolly C., Salminen T. A., Nymalm Y., Sistonen L. (2009) Mol. Biol. Cell 20, 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kroeger P. E., Morimoto R. I. (1994) Mol. Cell. Biol. 14, 7592–7603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manuel M., Rallu M., Loones M. T., Zimarino V., Mezger V., Morange M. (2002) Eur. J. Biochem. 269, 2527–2537 [DOI] [PubMed] [Google Scholar]
- 57.Wang G., Zhang J., Moskophidis D., Mivechi N. F. (2003) Genesis 36, 48–61 [DOI] [PubMed] [Google Scholar]
- 58.Ellis P. J., Clemente E. J., Ball P., Touré A., Ferguson L., Turner J. M., Loveland K. L., Affara N. A., Burgoyne P. S. (2005) Hum. Mol. Genet. 14, 2705–2715 [DOI] [PubMed] [Google Scholar]
- 59.Ward M. A., Burgoyne P. S. (2006) Biol. Reprod. 74, 652–658 [DOI] [PubMed] [Google Scholar]
- 60.Kugel J. F., Goodrich J. A. (2006) Mol. Cell 22, 153–154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.