Abstract
Reversible proline-directed phosphorylation at Ser/Thr-Pro motifs has an essential role in myogenesis, a multistep process strictly regulated by several signaling pathways that impinge on two families of myogenic effectors, the basic helix-loop-helix myogenic transcription factors and the MEF2 (myocyte enhancer factor 2) proteins. The question of how these signals are deciphered by the myogenic effectors remains largely unaddressed. In this study, we show that the peptidyl-prolyl isomerase Pin1, which catalyzes the isomerization of phosphorylated Ser/Thr-Pro peptide bonds to induce conformational changes of its target proteins, acts as an inhibitor of muscle differentiation because its knockdown in myoblasts promotes myotube formation. With the aim of clarifying the mechanism of Pin1 function in skeletal myogenesis, we investigated whether MEF2C, a critical regulator of the myogenic program that is the end point of several signaling pathways, might serve as a/the target for the inhibitory effects of Pin1 on muscle differentiation. We show that Pin1 interacts selectively with phosphorylated MEF2C in skeletal muscle cells, both in vitro and in vivo. The interaction with Pin1 requires two novel critical phospho-Ser/Thr-Pro motifs in MEF2C, Ser98 and Ser110, which are phosphorylated in vivo. Overexpression of Pin1 decreases MEF2C stability and activity and its ability to cooperate with MyoD to activate myogenic conversion. Collectively, these findings reveal a novel role for Pin1 as a regulator of muscle terminal differentiation and suggest that Pin1-mediated repression of MEF2C function could contribute to this function.
Keywords: Differentiation, Mass Spectrometry (MS), Protein Isomerase, Protein Phosphorylation, Protein Stability, Protein Translocation, Transcription Factors, MEF2, Pin1, Muscle
Introduction
Myogenesis is a highly regulated multistep process in which multipotent mesodermal cells give rise to myoblasts that subsequently withdraw from the cell cycle and differentiate into multinucleated myotubes. Vertebrate skeletal muscle differentiation is primarily controlled by the cooperative interactions of members of two families of transcription factors: the muscle-specific basic helix-loop-helix family of myogenic regulatory factors, Myf5, MyoD, myogenin, and MRF4 (myogenic regulatory factor 4), and the ubiquitous MEF2 (myocyte enhancer factor 2) family of MADS (minichromosome maintenance, agamous, deficiens, serum response factor) box transcription factors (MEF2A, -B, -C, and -D) (1). The activities of these two families of myogenic transcription factors are controlled by intracellular signaling pathways in response to extracellular cues. Several studies have demonstrated that MyoD and MEF2 proteins are phosphorylated at Ser or Thr residues that precede Pro (so-called Ser/Thr-Pro motifs) by a number of members of the large superfamily of proline-directed protein kinases, such as cyclin-dependent protein kinases (CDKs),3 mitogen-activated protein kinases (MAPKs), and GSK3 (glycogen synthase kinase-3), that play an essential role in the regulation of the myogenic transcriptional program (2–6).
Ser/Thr-Pro phosphorylation can modulate protein function through the induction of conformational changes that are regulated by the unique parvulin-like peptidyl-prolyl cis/trans-isomerase Pin1, which specifically binds to and isomerizes certain phospho-Ser/Thr-Pro motifs in a defined subset of phosphorylated proteins, thereby affecting their function. Pin1 is a ubiquitous enzyme that regulates a diverse array of cellular processes, often at multiple levels, and aberrant Pin1 function has been implicated in several human diseases (7, 8).
We observed that Pin1 is expressed in C2 muscle cells and undergoes differential subcellular relocalization from the nucleus to the cytoplasm during the early phases of myogenic differentiation. Together, these data argue for a direct link between the induction of skeletal muscle differentiation and down-regulation of Pin1 activity in the nucleus. We demonstrate that Pin1 negatively modulates skeletal muscle differentiation because both knockdown of endogenous Pin1 and inhibition of Pin1 activity by overexpression of a dominant negative mutant (Pin1C113A) result in increased differentiation capability of C2 muscle cells. Therefore, we sought to investigate whether the effect of Pin1 on skeletal muscle differentiation could be partially due to a direct modulation of the activity of myogenic transcription factors. MEF2C, a member of the MEF2 family of proteins that plays a prominent role in vertebrate myogenesis, is particularly sensitive to the signaling pathways that regulate muscle formation (9, 10), so we investigated whether it might serve as a target for the inhibitory effects of Pin1 on muscle differentiation. In the present study, we provide several pieces of evidence that Pin1 interacts with MEF2C in vitro and in vivo and that this interaction requires a 77-amino acid region of MEF2C immediately adjacent to the DNA binding and dimerization domain. According to tandem mass spectrometry analysis of the MEF2C protein purified from muscle cells, two phosphoserine residues, Ser98 and Ser110, are present within this region. Mutation of these residues abolishes Pin1/MEF2C interaction. Importantly, Pin1 overexpression negatively modulates MEF2C protein stability and activity as well as the ability of MEF2C to cooperate with MyoD to activate myogenic conversion of 10T1/2 fibroblasts. Taken together, these findings imply that Pin1 is a novel negative regulator of skeletal muscle terminal differentiation, a function that can be explained partly by the inhibition of stability and activity of MEF2C.
EXPERIMENTAL PROCEDURES
Plasmids
pGL3(desMEF2)3, pRSVβ-gal pFLAG-MEF2C, pcDNAI/Amp/MEF2C, and pGEX-Pin1 have been described previously (11, 12). The pcDNA-HA-Pin1 expression vector was generated by subcloning a PCR product of Pin1 cDNA into the pcDNA-HA-HDAC4 vector (13) after removal of the cDNA encoding HDAC4 (BamHI/EcoRI restriction). The pFLAG-MEF2C expression vectors bearing deletions and point mutations on Ser98, Ser110, Ser240, and Ser388, the pcDNAI/Amp/MEF2C 4SA, the pCDNA-HA-Pin1-C113A, and the pGEX-Pin1-W34A mutant plasmids were obtained by mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene). pFLAG-MEF2C-YN and pHA-Pin1-YC were obtained by cloning the PCR products of Pin1 and MEF2C cDNAs, respectively, in the pBiFC-YN and pBiFC-YC vectors. Viral vectors pLKO-puro encoding shRNAs against mouse Pin1 or a control sequence were purchased from Sigma-Aldrich. Viral vectors encoding HA-Pin1 and HA-Pin1 C113A were generated by cloning the respective cDNAs in the pRRL-PGK-GFP transfer vector. The primers used for the PCR and mutagenesis reactions are available in the supplemental material.
Cell Culture and Transfection
The C2C7 murine muscle cells, a subclone of the C2 muscle cell line (14), have been previously described (15). C2C7 cells were grown in advanced Dulbecco's modified Eagle's medium (A-DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; Invitrogen) (growth medium) at low density and, when approaching confluence, induced to differentiate with DMEM (Euroclone), 2% horse serum (Hyclone) (differentiation medium). COS1 simian kidney cells, C3H 10T1/2 mouse fibroblasts, and human embryonic kidney (HEK) 293T cells were maintained in DMEM containing 10% FBS. Cells were transfected using the lipid-based Lipofectamine Plus reagent (Invitrogen). HEK 293T cells were transfected using the standard calcium phosphate precipitation method (16). A myogenic conversion assay of C3H 10T1/2 cells was performed as reported previously (11).
Immunofluorescence and Bimolecular Fluorescence Complementation (BiFC) Assay
Immunostaining of C2C7 cells cultured in 40-mm Petri dishes was performed as described previously (17). The following primary antibodies were used: mouse M2 monoclonal anti-FLAG (F3165; Sigma-Aldrich); rabbit polyclonal anti-HA (H6908; Sigma-Aldrich), and mouse monoclonal anti-myosin heavy chain (MyHC) (MF20 Developmental Studies Hybridoma Bank). Secondary antibodies used were goat anti-mouse IgG rhodamine-conjugated (Pierce), goat anti-rabbit IgG amino methylcoumarin acetate-conjugated (Dako). Nuclei were stained with Hoechst 33342 (Sigma-Aldrich). For the BiFC assay, C2C7 cells were transfected with the indicated plasmids, and 36 h after transfection, they were incubated for 30 min at 30 °C to enhance the fluorophore maturation of the yellow fluorescent protein (YFP).
All samples were examinated in a Zeiss Axioskop 40 fluorescence microscope equipped with an Axiocam HRC camera for image acquisition. Quantitative estimates of nuclei present in MyHC-positive cells were performed using the Cell Counter plugin of Image J (available on the National Institutes of Health Web site) by analyzing at least three fields for each sample (3 × 103 nuclei). This experiment was repeated twice.
Lentivirus Production and Transduction
Lentiviral particles were produced by transient transfection of the transfer vectors in association with the packaging vectors (pREV, pΔ8.74, and pVSV-G) in HEK 293T cells as described (18). After 48 h, culture medium was filtered (0.45 μm) and used for infection. Transduction of C2C7 myoblasts was performed by adding the viral preparation to cells in the presence of Polybrene (8 μg/ml; Sigma-Aldrich).
Nucleus-Cytoplasm Fractionation
Subcellular fractions of C2C7 cells at different stages of differentiation were obtained using a digitonin-based lysis buffer. Cells were harvested by trypsinization, washed once with ice-cold 1× PBS and then with TB buffer (20 mm HEPES, pH 7.3, 110 mm potassium acetate, 5 mm sodium acetate, 2 mm magnesium acetate, 1 mm EGTA, 2 mm dithiothreitol, and protease inhibitors). Subsequently, cells were collected and homogenized by incubation in 1 volume of ice-cold TB buffer supplemented with digitonin (40 μg/ml) (D141; Sigma-Aldrich) and protease inhibitors for 4 min. Cytoplasmic protein extracts were harvested, and nuclei were collected by centrifugation. Nuclei were then washed twice with Buffer A (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 1.5 mm dithiothreitol, and protease inhibitors) and lysed with radioimmune precipitation (RIPA) buffer.
Western Blot and Antibodies
Western blot assays were performed as described previously (11). The following antibodies were used: mouse M2 monoclonal anti-FLAG (F3165; Sigma-Aldrich), rabbit polyclonal anti-Pin1 (PC270; Calbiochem), rabbit polyclonal anti-MEF2 (sc-313X; Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-MEF2 (sc-17785; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-MEF2C (a gift from J. McDermott, York University), goat polyclonal anti-enolase antibody (sc-7455; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-NF-YB (a gift from R. Mantovani, University of Milan), mouse monoclonal anti-GST (G1160; Sigma-Aldrich), mouse monoclonal anti-actin (MAB 1501; Chemicon), rabbit polyclonal anti-HA (H6908; Sigma-Aldrich), and mouse monoclonal anti-MyHC (MF20 Developmental Studies Hybridoma Bank).
Glutathione S-Transferase (GST) Pull-down Assay, Far Western, and Co-immunoprecipitation
For the GST pull-down assay, cells were lysed with single detergent lysis buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Triton X-100 supplemented with the Complete protease inhibitor mixture (Roche Applied Science), and phosphatase inhibitors (sodium orthovanadate and sodium fluoride). The same amount for each protein extract was incubated for 2 h at 4 °C with the bacterially purified GST fusion proteins bound to agarose-glutathione beads (Sigma-Aldrich). After three washings, 2× Laemmli loading buffer was added to the beads, and proteins were analyzed by Western blot. GST proteins were obtained as described (12). The far Western experiment was performed as follows. COS1 cells were transfected with pFLAG tag or pFLAG MEF2C expression vectors; after 36 h, cells were lysed in single detergent lysis buffer, and FLAG-tagged proteins were purified with anti-FLAG M2-agarose (Sigma). λ-Phosphatase (100 units) (New England Biolabs) was added to protein extracts for 30 min at 30 °C. Immunoprecipitated proteins were separated by SDS-PAGE and blotted on PVDF membrane. After blocking, membranes were incubated for 1 h at 4 °C with 2 μg/ml of the bacterially purified GST fusion proteins and then analyzed by Western blot with anti-GST antibody. For the co-immunoprecipitation assays, cells were cross-linked with dithiobis(succinimidylproprionate) (DSP; Pierce) for 30 min at 4 °C. The cross-linking was stopped by the addition of glycine at a 0.2 m final concentration for 30 min at 4 °C. After two washings with ice-cold PBS, cells were scraped and then lysed with RIPA precipitation buffer, and proteins were incubated with the anti-FLAG M2-agarose (Sigma-Aldrich) or the polyclonal anti-MEF2 antibody. Protein-antibody complexes were recovered with protein G-Sepharose.
Cycloheximide Treatment and Transcription Reporter Assays
Protein half-life was assessed in transfected COS1 cells that were treated with 25 μm cycloheximide (Sigma-Aldrich) and collected at the indicated time points, and then proteins were analyzed by Western blot. The intensity of the protein bands was quantitated by densitometry. Transactivation assays were performed by cotransfecting C2C7 muscle cells with pGL3(desMEF2)3, pRSVβ-gal, and the expression vector for HA Pin1. Twenty-four hours after transfection, cells were shifted to differentiation medium and kept in culture for an additional 24 h. Cell lysates were obtained by three freeze/thaw cycles and processed as described previously (11). In another series of experiments, C3H 10T1/2 cells were cotransfected with the indicated combination of plasmids. Luciferase/Renilla assays were performed using the DLR assay system (Promega). Luciferase and Renilla activities were measured in a Chameleon luminometer (Hidex Ltd.).
Mass Spectrometry Analysis
FLAG-tagged MEF2C was purified both from proliferating and differentiating C2C7 cells by affinity chromatography. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis was performed as described previously (11). Phosphorylated peptides were analyzed by tandem MS experiments performed on a Q-Star pulsar (QqTof hybrid system from PE SCIEX Instruments (Toronto, Canada). The phosphopeptides purified on immobilized gallium affinity chromatography columns (Pierce) were concentrated and desalted over a capillary column manually packed with 200 μl of POROS R3 material, conditioned with 5% formic acid. The peptide mixture was eluted using 1 μl of 50% methanol, 5% formic acid directly into the nanoelectrospray needle (Protana).
RESULTS
Pin1 Modulates Skeletal Muscle Differentiation
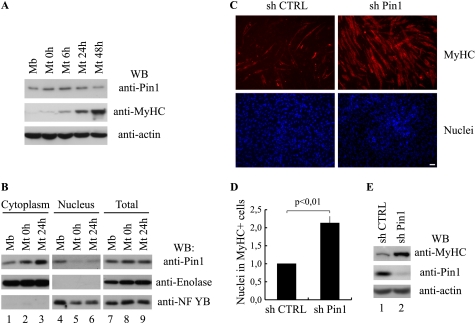
Given the key role played by proline-directed Ser/Thr kinases in the control of skeletal muscle differentiation, coupled with emerging evidence that the enzymatic activity of Pin1 promotes regulatory postphosphorylation events of proteins at sites of proline-dependent phosphorylation, we sought to test whether Pin1 is a regulator of muscle differentiation. We initially looked for the presence of Pin1 in C2C7 muscle cells, a well defined model for ex vivo differentiation. These cells proliferate as myoblasts in high serum concentration. When cells reach confluence, they can be induced to differentiate by reducing the serum concentration from 10% to 2% (see “Experimental Procedures”) (14). In order to examine whether changes in Pin1 expression could be observed during the course of myogenic differentiation, the expression of Pin1 protein was investigated by Western blot analysis of protein extracts taken at different times after serum reduction. To follow the process of differentiation, we monitored for the induction of MyHC, a well known marker of terminal differentiation. As shown in Fig. 1A (top), Pin1 is already present in myoblasts (Mb), where a single band is recognized at a molecular mass of 18 kDa. Pin1 expression does not change when cells reach confluence (Mt 0h) or at any time after switching into differentiation medium examined. Under our culture conditions, we detected MyHC expression within 6 h (Mt 6h in Fig. 1A (middle)). Pin1 expression in skeletal muscle cells was further confirmed by Western blot analysis of murine satellite cells and embryonic and fetal primary muscle cells extracts (data not shown). These analysis confirmed that Pin1 expression level does not change during terminal differentiation. We next tested the subcellular localization of Pin1 during muscle cell differentiation. To this end, a cellular fractionation of nuclear and cytoplasmic compartments was performed. As shown in Fig. 1B, Pin1 protein was equally distributed in the nuclear and cytoplasmic compartments in undifferentiated cells (Mb, lanes 1 and 4, top), whereas we observed an early differentiation-dependent nuclear-cytoplasmic shuttling of Pin1 (Mt 0h and 24h, lanes 2, 5, 3, and 6, top). These data show that Pin1 undergoes specific alterations in cytolocalization as cells transit through early differentiation. To explore the potential role of Pin1 during skeletal myogenesis, we knocked down Pin1 expression in C2C7 myoblasts by RNA interference using lentiviral vectors encoding short hairpin RNAs against mouse Pin1 (sh Pin1) or a control sequence (sh CTRL). Proliferating C2C7 cells were infected with the lentiviral vectors. After 2–3 days in culture, when the cells had reached about 90% confluence, they were induced to differentiate. Immunofluorescent analysis performed on these cells showed that the proportion of MyHC-positive cells (red stain) is considerably higher in sh Pin1-treated cells than in control cells (sh CTRL) (Fig. 1C). Based on the number of nuclei present in MyHC-positive cells, a 2-fold increase of muscle cell differentiation was estimated in Pin1-deficient myotubes (Fig. 1D). The negative effect of Pin1 on skeletal muscle cell differentiation depends on its catalytic activity. An HA Pin1 C113A catalytically inactive mutant, a dominant negative form of Pin1 (19), when transduced into these cells, caused a strong increase of the percentage of nuclei present in MyHC-positive cells (supplemental Fig. S1, A and B). Western blot analysis of C2C7 cells cultured for 48 h in differentiation medium revealed a strong increase of MyHC (Fig. 1E) in Pin1-knockdown cells or in cells overexpressing the dominant negative mutant (supplemental Fig. S1C, MyHC). Altogether, these results suggest that Pin1 catalytic activity helps prevent C2C7 muscle cell differentiation.
FIGURE 1.
Knockdown of Pin1 promotes skeletal muscle differentiation. A, Western blot (WB) analysis of Pin1, MyHC, and actin protein levels during myogenic differentiation. Total protein extracts from proliferating (Mb) or differentiating (Mt 0h, 6h, 24h, and 48h) C2C7 muscle cells were analyzed by Western blot using the antibodies against Pin1, MyHC, and actin. B, subcellular localization of Pin1 during skeletal muscle differentiation. Cytoplasmic (lanes 1–3), nuclear (lanes 4–6), and total (lanes 7–9) protein extracts from proliferating (Mb) or differentiating (Mt 0h and Mt 24h) C2C7 muscle cells were analyzed by Western blot using the antibody against Pin1; furthermore, we checked the quality of the subcellular protein extracts with antibodies against the glycolytic enzyme enolase and the nuclear transcription factor NF-YB. C, C2C7 myoblasts were infected with lentiviruses encoding short hairpin RNAs (a scramble control sequence (sh CTRL) or a Pin1 silencing sequence (sh Pin1), respectively) and then induced to differentiate. After 48 h from serum withdrawal, cells were fixed and subjected to immunostaining using the anti-MyHC antibody (red stain, upper panels), cell nuclei were stained by Hoechst (lower panels). Bar, 50 μm. D, the proportion of nuclei in differentiated cells is reported as ratio between the nuclei incorporated in MyHC-positive cells and total nuclei. The number obtained in Pin1-depleted cells (sh Pin1) is expressed relative to the number evaluated in cells infected with the scramble control vector (sh CTRL) taken as 1. The data are presented in the histogram and represent the mean ± S.D. (error bars) of three independent experiments (p < 0,01). E, Western blot analysis performed on protein extracts of infected C2C7 cells with antibodies against MyHC, Pin1, and actin.
MEF2C Interacts with Pin1 in Muscle Cells
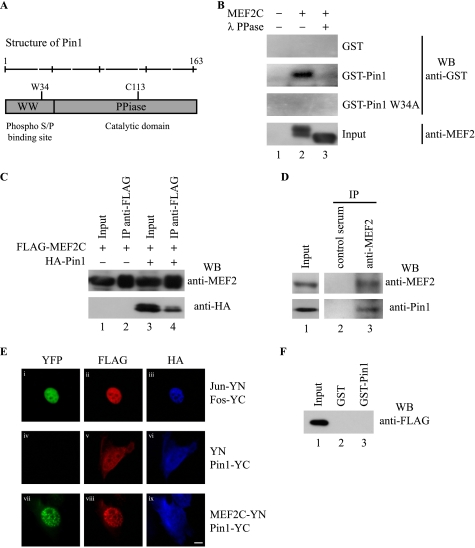
To gain insight into the molecular mechanisms underlying the observed Pin1-dependent negative regulation of skeletal muscle differentiation, we searched for targets of its catalytic activity. A large body of evidence implicates MEF2 proteins as key downstream effectors of signaling pathways that control vertebrate skeletal muscle differentiation (20). Notably, MEF2C protein is phosphorylated on several serine residues, most of which reside in Ser/Thr-Pro motifs, putative consensus sites that might be targeted by Pin1 (21–23). Pin1 contains an amino-terminal WW domain that binds to specific phospho-Ser/Thr-Pro motifs and a carboxyl-terminal enzymatic domain that catalyzes the cis-trans isomerization of the phospho-Ser/Thr-Pro bonds of target proteins (7) (Fig. 2A). To test whether Pin1 and MEF2C are able to associate directly, we performed a Far Western analysis by using GST-Pin1 or GST to detect anti-FLAG immunoprecipitates from FLAG-MEF2C or empty vector-overexpressing COS1 cells. As shown in Fig. 2B, the FLAG-tagged MEF2C protein (lane 2) was specifically recognized by GST-Pin1 but not by GST alone, indicating that Pin1 binds directly to MEF2C. We observed a lack of binding upon treating the lysates with λ-phosphatase (lane 3) or by using the GST-Pin1 W34A mutant, which is unable to recognize its substrates (19). These results suggest that this interaction is strictly dependent both on phosphorylation of MEF2C and on the integrity of the Pin1 WW domain, respectively. Moreover, Pin1 and MEF2C also associate in vivo in C2C7 skeletal muscle cells, as shown by co-immunoprecipitation of both overexpressed and endogenous proteins (Fig. 2, C and D, respectively). To demonstrate that the interaction observed in myoblasts is direct, we used the bimolecular fluorescence complementation approach (24). To this end, MEF2C and Pin1 were fused to the amino- or carboxyl-terminal fragment of YFP, respectively, and transfected into C2C7 myoblasts. In this experiment, the fusion proteins Jun-YN and Fos-YC were used as positive controls (Fig. 2E, i). Notably, co-expression of MEF2C-YN and Pin1-YC in C2C7 cells resulted in complementation of the YFP in the nucleus, indicating that MEF2C and Pin1 directly interact in C2C7 cells and that this interaction takes place in the nuclear compartment (Fig. 2E, vii). We observed no bimolecular complementation when the YN fragment alone, which does not interact with Pin1 in GST pull-down assays (Fig. 2F), was expressed in association with Pin1-YC (Fig. 2E, iv). In summary, these data demonstrate that MEF2C is a phosphorylation-dependent Pin1 target in muscle cells.
FIGURE 2.
Pin1 interacts with MEF2C in a phosphorylation-dependent manner. A, schematic representation of the functional domains of Pin1. WW domain accounts for the phosphoprotein binding, whereas the peptidyl-prolyl cis/trans-isomerase domain is involved in substrate isomerization. Also shown are critical amino acids that, when mutated, hamper the binding (W34A) or the catalytic (C113A) activity of Pin1, respectively. B, far Western assay. COS1 cells were transiently transfected with the empty FLAG vector (lane 1) or the FLAG MEF2C expression vector (lanes 2 and 3). Cells were then lysed and were treated with λ-phosphatase (λ PPase) where indicated (lane 3). Anti-FLAG immunoprecipitation was performed on the lysates, and immunoprecipitated proteins were resolved on SDS-PAGE; blotted onto a PVDF membrane; incubated with bacterially purified GST, GST-Pin1, or GST-Pin1 W34A; and then analyzed by Western blot (WB) with the anti-GST antibody. About 5% of the input immunoprecipitated proteins were analyzed using the anti-MEF2 antibody. C, co-immunoprecipitation of overexpressed MEF2C and Pin1 proteins from muscle cells. C2C7 cells ectopically expressing FLAG-MEF2C alone (lanes 1 and 2) or in association with HA-Pin1 (lanes 3 and 4) were cross-linked with DSP and then lysed and immunoprecipitated (IP) with anti-FLAG antibody (lanes 2 and 4). Western blot was performed with anti-MEF2 (polyclonal) and anti-HA antibodies. D, co-immunoprecipitation of endogenous MEF2 and Pin1 proteins from muscle cells. C2C7 cells were cross-linked with DSP and then lysed and immunoprecipitated with anti-MEF2 (polyclonal) antibody (lane 3) or with control serum (lane 2). Western blot was performed with anti-MEF2 (monoclonal) and anti-Pin1 antibodies. E, BiFC assay. C2C7 cells were transfected with FLAG-Jun-YN and HA-Fos-YC-positive control (i–iii), FLAG-YN- and HA-Pin1-YC-negative control (iv–vi); and FLAG-MEF2C-YN- and HA-Pin1-YC (vii–ix). After 36 h from transfection, cells were fixed and immunostained using the antibodies against FLAG (red stain, second column) to detect YN alone and the Jun-YN and MEF2C-YN fusion proteins and against HA (blue stain, third column) to detect Fos-YC and Pin1-YC fusion proteins. The first column represents the fluorescence of the YFP (green stain) resulting from the bimolecular complementation assay. Scale bar, 10 μm. F, total protein extracts of COS1 cells expressing FLAG-YN were subjected to a GST (lane 2) or GST-Pin1 (lane 3) pull-down assay and immunoblotted with the anti-FLAG antibody. FLAG-YN protein input was also checked (lane 1).
Identification of the Relevant Pin1 Binding Sites on MEF2C in Muscle Cells
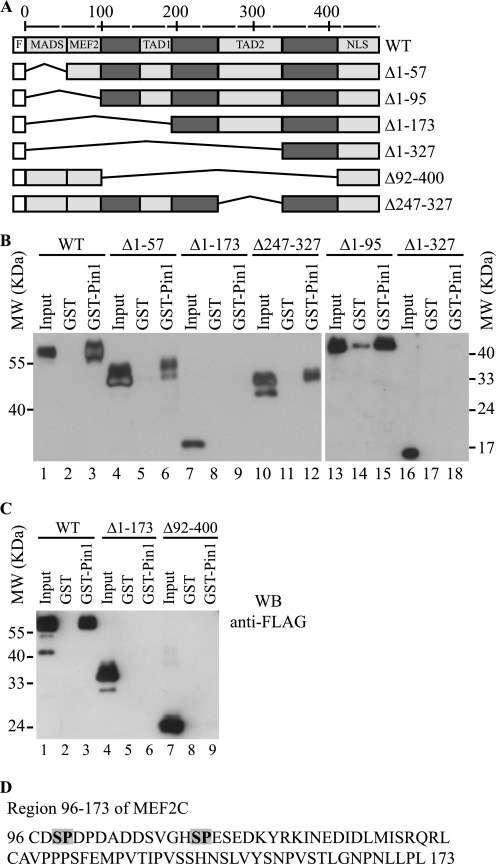
To identify the Pin1 binding domain on MEF2C, we first generated a series of FLAG-MEF2C deletion mutants (Fig. 3A) and assayed their ability to bind GST-Pin1 in a GST pull-down assay. As shown in Fig. 3, B and C, the full-length MEF2C protein and the deletion mutants Δ1–57, Δ1–95, and Δ247–327 exhibited similar affinity for Pin1 (Fig. 3, B, lanes 3, 6, 15, and 12, respectively) and C, lane 3. In contrast, the deleted MEF2C proteins Δ1–173, Δ1–327, and Δ92–400 did not bind to Pin1 (Fig. 3, B, lanes 9 and 18) and C, lanes 6 and 9. This suggests that the minimal region of MEF2C required for binding Pin1 lies between amino acids 96 and 173.
FIGURE 3.
Region spanning amino acids 96–173 of MEF2C is involved in the binding of Pin1. A, schematic representation of the deletion mutants of FLAG-MEF2C used to identify the binding sites of Pin1. The light boxes indicate the FLAG tag (F) and the following functional domains: MADS (DNA binding domain), MEF2 (dimerization domain), transcriptional activation domains 1 and 2 (TAD1 and TAD2, respectively), and nuclear localization signal (NLS). The numbers reported indicate the positions of the amino acid residues of MEF2C. B and C, total extracts of COS1 cells expressing the full-length FLAG-MEF2C (WT) or its deletion mutants were subjected to GST and GST-Pin1 pull-down followed by Western blot with anti-FLAG antibody. D, amino acid sequence of the region of MEF2C spanning the residues 96–173. Serines 98 and 110 are shown in boldface type and shaded; they represent the only serine-proline motifs in this region.
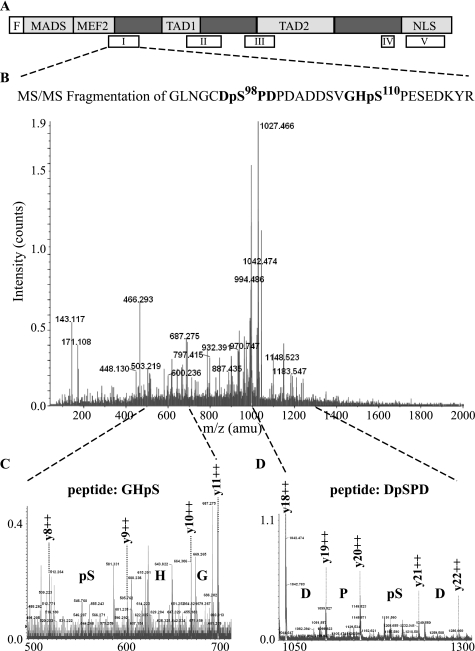
The amino acid 96–173 region, shown in Fig. 3D, contains two Ser-Pro motifs (Ser98 and Ser110), which, upon phosphorylation, could be recognized by Pin1. To determine whether these two serines are phosphorylated in muscle cells, a mass spectrometry analysis of FLAG-tagged MEF2C purified from C2C7 muscle cells was performed. The MALDI-TOF mass spectra of a tryptic digest of FLAG-MEF2C purified from myoblasts and myotubes were identical and revealed several peaks whose mass, with characteristic +80-Da shifts, is consistent with one or more phosphorylated amino acids. The positions of the functional domains of MEF2C and of the observed putative phosphopeptides (empty boxes) are summarized in Fig. 4A. Strikingly, the peptide encompassing amino acids 92–118 of MEF2C (peptide I in Fig. 4A) appeared in the MALDI-TOF spectra with a shift of +80 and +160 Da (2999.17 and 3079.17 versus 2919.17, corresponding to the unmodified peptide). Alkaline phosphatase treatment of the starting material gave rise to a MALDI-TOF spectrum of MEF2C in which the peak corresponding to the unmodified peptide (2919.17 Da) was prominent, supporting the idea that peaks m/z = 2999.17 and 3079.79 represent the mono- and diphosphorylated peptide, respectively (supplemental Fig. S2). To identify exactly the phosphorylated amino acids, peptide 92–118 carrying two phosphate moieties was fragmented using a MS/MS approach (precursor ion (MH3)3+, 1027.10 Da) (Fig. 4B). Fragmentation of the potential phosphopeptide revealed a series of y++ fragments in which the differences between y8++ and y9++ (Fig. 4C) and y20++ and y21++ (Fig. 4D) fragments can only be explained by the incorporation of a phosphate group. These fragments give high confidence in the identification of this peptide as MEF2C 92–118 phosphorylated at two previously unidentified phosphoacceptor sites: Ser98 and Ser110. Sequence alignment of the mouse MEF2C protein region encompassing amino acids 61–120 with related regions of MEF2A and MEF2D from different vertebrate species revealed a high degree of conservation of the two serine residues Ser98 and Ser110 (supplemental Fig. S3), suggesting that phosphorylation of these sites could be important for MEF2 protein function and/or regulation.
FIGURE 4.
Identification of two novel phosphorylations in MEF2C. A, schematic representation of the functional domains of MEF2C (MADS box, MEF2 domain, transcriptional activation domains 1 and 2 (TAD1 and TAD2), and nuclear localization signal (NLS) below the empty blocks representing the relative positions of the tryptic peptides analyzed by mass spectrometry (I–V). B, MS/MS analysis of the peptide I spanning amino acids 92–118 of MEF2C purified from C2C7 muscle cells. The fragmentation of the peptide generated several y fragments, which allowed us to exactly identify the phosphorylated aminoacids. Magnifications (C and D) show that both serine 98 and 110 are phosphorylated.
Concerning the other putative phosphopeptides, we identified two additional phosphoacceptor sites (Ser240 and Ser388) that have been described previously in MEF2 proteins (22, 23, 25–27). However, we failed to sequence other fragments, possibly due to a lower efficiency of nanospray ionization with respect to the MALDI source used for peptide mass fingerprinting.
Phosphorylation of Multiple SP Sites of MEF2C Is Necessary for the Interaction with Pin1
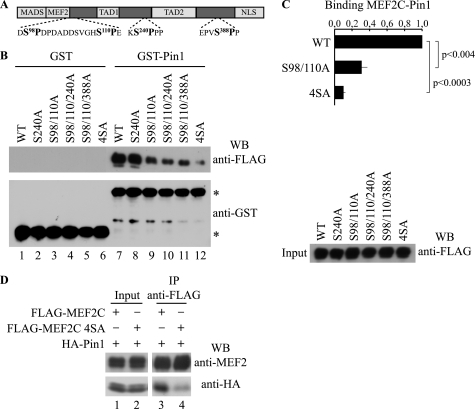
To demonstrate that Pin1/MEF2C association relied on the identified phospho-Ser-Pro motifs, mutants of Ser98, Ser110, Ser240, and Ser388 phosphoacceptor sites (Fig. 5A) were generated by Ser → Ala substitution, ectopically expressed in COS1 cells, and assayed in a GST-Pin1 pull-down experiment. As shown in Fig. 5B, wild type FLAG-MEF2C and the S240A mutant exhibited similar affinity for Pin1 (lanes 7 and 8), whereas double mutant FLAG-MEF2C S98/110A showed an about 70% reduced affinity (lane 9), as judged by densitometric analysis of the protein bands (Fig. 5C). The addition of one more mutation in triple mutants (S98A/S110A/S240A or S98A/S110A/S388A) did not significantly affect their ability to interact with Pin1 in comparison with S98A/S110A MEF2C double mutant protein (Fig. 5B, lanes 10 and 11). Substitution of all four putative Ser-Pro sites in the quadruple mutant FLAG-MEF2C S98A/S110A/S240A/S388A (indicated as 4SA in Fig. 5B, lane 12) almost completely abrogated binding capability to GST-Pin1 (about 10% of wild-type FLAG-MEF2C binding; Fig. 5C). These results were confirmed by co-immunoprecipitation of overexpressed HA-Pin1 and FLAG-MEF2C wild type or the 4SA mutant in COS1 cells (Fig. 5D).
FIGURE 5.
Phosphorylated Ser98, Ser110, Ser240, and Ser388 of MEF2C are required for the interaction with Pin1. A, representation of MEF2C together with the putative Pin1 consensus motifs (boldface type). B, different Ser-Ala point mutants of FLAG MEF2C were assayed for interaction with GST (lanes 1–6) and GST Pin1 (lanes 7–12) upon overexpression in COS1 cells followed by Western blot (WB) with anti-FLAG antibody (top) and anti-GST antibody (bottom); the positions of the GST and GST-Pin proteins are indicated with asterisks. The FLAG-tagged MEF2C protein inputs were also checked by anti-FLAG Western blot shown to the right. C, quantification of the ability of the MEF2C mutants to interact with Pin1. The intensities of the signals shown in B were quantified by densitometric scanning; in the histogram they are expressed relative to the value obtained with FLAG-MEF2C wild type (WT), which was assigned a value of 1 (mean ± S.D. (error bars) of three independent experiments). D, co-immunoprecipitation. COS1 cells expressing FLAG-MEF2C (lanes 1 and 3) or FLAG-MEF2C 4SA (lanes 2 and 4) in association with HA-Pin1 were cross-linked with DSP, the proteins were extracted, and the lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibody and Western blotted for anti-MEF2 (top) or anti-HA (bottom); FLAG-tagged and HA-tagged protein inputs were also checked (lanes 1 and 2).
Collectively, these data suggest that phosphorylation of Ser98 and Ser110 is necessary for the interaction of MEF2C with Pin1. However, phosphorylated Ser240 and Ser388 also contribute to the binding although only in combination with Ser98 and Ser110, thus indicating that Pin1 binds MEF2C at multiple Ser-Pro motifs to modulate its function.
Pin1 Regulates the Stability and Activity of MEF2C
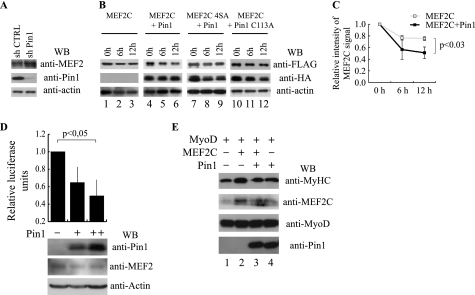
It has been reported that phosphorylation of MEF2A and MEF2D on the cognate residues of Ser240 and Ser388 of MEF2C regulates their stability (22, 25), and several reports indicated that Pin1-mediated cis/trans isomerization of the phospho-Ser/Thr-Pro peptide bond is able to alter substrate protein stability (7, 28, 29). Therefore, to characterize the mechanism responsible for Pin1-mediated repression of myogenic differentiation, we investigated whether Pin1 would influence MEF2 protein accumulation. As shown in Fig. 6A, a marked increase of MEF2 protein levels was detectable upon Pin1 knockdown in C2C7 muscle cells. Next, we tested whether the stability of MEF2C was affected by Pin1. To achieve this, we analyzed the levels of overexpressed FLAG-MEF2C alone or in association with HA-Pin1 at different time points following cycloheximide treatment. As shown in Fig. 6B, co-expression of Pin1 substantially reduced MEF2C stability, and we found MEF2C to be about 2-fold less stable in Pin1-overexpressing cells (Fig. 6C). On the contrary, this effect was not observed when Pin1 was co-expressed with the phosphorylation-defective mutant MEF2C 4SA (Fig. 6B, lane 9) nor by co-expression of the catalytically inactive mutant HA-Pin1 C113A (Fig. 6B, lane 12), demonstrating that the phosphorylation of specific serine residues and the catalytic activity of Pin1 are necessary for Pin1-dependent reduction of MEF2C stability.
FIGURE 6.
Pin1 controls MEF2C stability and activity. A, Western blot analysis of MEF2, Pin1, and actin protein levels performed on protein extracts of C2C7 myoblasts infected with lentiviruses encoding short hairpin RNAs, a scramble control sequence (shCTRL) or a Pin1 silencing sequence (sh Pin1), respectively. B, protein stability assay. FLAG MEF2C wild type or mutated on the four phosphoacceptor sites (4SA) were overexpressed in COS1 cells either alone or with HA-tagged Pin1 as indicated. After 36 h, protein synthesis was blocked with cycloheximide, and the amount of FLAG MEF2C and HA Pin1 remaining at different times was checked by Western blot. Protein loading was controlled by anti-actin staining. C, the intensity of the wild type MEF2C bands of the experiment in B was quantified by densitometric scanning. The chart reports the average of the relative intensity of MEF2C signals for three independent experiments (p < 0.03). D, effect of Pin1 on the transcriptional activity of endogenous MEF2 proteins. C2C7 cells were transfected with 1 μg (+) or 3 μg (++) of the HA-Pin1 expression plasmid together with the pGL3(desMEF2)3 luciferase and the pRSVβ-gal reporter plasmids. Twenty-four hours after transfection, cells were shifted to low serum medium and kept in culture for an additional 24 h. Luciferase activity was determined and normalized to the β-gal activity. Statistical analysis was performed on data obtained from four independent experiments. The normalized luciferase activities are expressed relative to the sample transfected with the empty expression vector. The amounts of transfected HA-Pin1, endogenous MEF2 proteins, and actin were monitored by Western blot analysis using the anti-Pin1, anti-MEF2, and anti-actin antibodies, respectively. E, Pin1 blocks cooperative activation of myogenic conversion by MEF2C and MyoD. C3H10T1/2 fibroblasts were transiently transfected with the indicated plasmids. After 48 h in growth medium, cells were induced to differentiate in differentiation medium for 6 days and then lysed. Whole cell extracts were subjected to immunoblotting for MyHC, MEF2C, MyoD, and Pin1. Error bars, S.D.
To determine whether the interaction between Pin1 and MEF2C plays a functional role, we examined the effects of Pin1 on MEF2-dependent transcription in muscle cells by a reporter assay. A plasmid encoding the firefly luciferase gene under the control of three MEF2 sites derived from the desmin gene was transfected into C2C7 muscle cells, either with or without increasing amounts of the expression vector encoding HA-Pin1 (31). As summarized in Fig. 6D, MEF2-dependent trans-activation decreased upon Pin1 overexpression in a dose-dependent fashion. A similar behavior was observed in C3H 10T1/2 cells (supplemental Fig. S4). In these latter cells, Pin1 overexpression caused a 30% (p < 0,05) decrease of MEF2C-mediated transactivation of the reporter gene. Thus, Pin1 appears to regulate the trans-activating activity of ectopically expressed MEF2C in fibroblasts as well as of endogenous MEF2 proteins in muscle cells. Finally, we used a myogenic conversion assay to test whether the Pin1-dependent repression of muscle differentiation is at least partially mediated by a modulation of the synergy between MyoD and MEF2C. C3H 10T1/2 cells were transiently transfected with expression vectors for MyoD together with MEF2C alone, MEF2C and Pin1, or Pin1 alone, and the efficiency of myogenic conversion of these cells was evaluated by Western blot analysis of skeletal sarcomeric MyHC. As already reported (30), expression of MEF2C resulted in synergistic activation of myogenesis (Fig. 6E, lane 2) compared with MyoD alone (Fig. 6E, lane 1). Interestingly, when Pin1 was overexpressed, the same doses of transfected MEF2C failed to synergize with MyoD to induce myogenic conversion (Fig. 6E, compare lane 2 versus lane 3), whereas Pin1 itself did not affect the myogenic activity of MyoD alone (Fig. 6E, compare lane 1 versus lane 4). These results suggest a critical role of Pin1 in modulating MEF2C functions during myogenic differentiation.
DISCUSSION
In this work, we show for the first time that Pin1 modulates the skeletal muscle differentiation program because down-regulation of Pin1 or abolition of Pin1 isomerase activity markedly promotes myogenic differentiation. Pin1 protein is present in C2C7 myoblasts and myotubes. We find that a significant proportion of Pin1 in myotubes but not in myoblasts is excluded from the nucleus, suggesting that nuclear localized factors might be the targets for Pin1-mediated inhibition of terminal differentiation. It has been extensively reported that the prolyl isomerase Pin1 plays a key role in regulating the postphosphorylation events of its target proteins upon phosphorylation at Ser/Thr-Pro motifs in several cellular processes. Therefore, to dissect the molecular mechanisms underlying the observed Pin1-dependent negative regulation of myogenic differentiation, we investigated whether MEF2C, a myogenic transcriptional effector that serves as an end point of the several intracellular signaling pathways that control myogenesis (20, 31), might be a target for Pin1.
Our results clearly show that Pin1 binds directly to MEF2C in skeletal muscle cells and that this interaction impacts on stability and activity of the MEF2C protein. We found that the interaction between Pin1 and MEF2C is phosphospecific, and we have identified four major Pin1 binding sites.
Two of these sites, Ser240 and Ser388, correspond to the already described phosphorylation sites in MEF2A, MEF2C, and MEF2D (22, 23, 25, 32–34). Furthermore, our phosphoproteomic analysis led also to the identification of two novel phosphoacceptor sites on MEF2C (Ser98 and Ser110) that are required for Pin1 binding and that are highly conserved among other isoforms from various species. It is tempting to speculate, therefore, that Pin1 could also be a global modulator of MEF2 proteins by mediating the above mentioned phosphorylation-dependent regulatory mechanisms in other tissues.
The observation that, during myogenic conversion, Pin1 affects the synergy between MEF2C and MyoD yet leaves unaffected the myogenic activity of MyoD alone suggests that the inhibitory effect of Pin1 on muscle differentiation is due to inhibition of MEF2C function. Indeed, we show that Pin1 destabilizes MEF2C, and this effect depends on the catalytic activity of Pin1 and on the phosphorylation status of MEF2C. However, our data show that, in addition to protein destabilization, other mechanisms could contribute to the inhibitory effect of Pin1 on MEF2C function, possibly involving transcriptional regulation. In support of this, we show that Pin1 represses in a dose-dependent manner the protein level and transcriptional activity of endogenous MEF2 proteins in a muscle cellular context (Fig. 6D). The region of MEF2C that appears to be minimally required for interaction with Pin1 (residues 96–173) lies carboxyl-terminal to the minimal DNA binding and dimerization domain. Nevertheless, the interaction does not seem to interfere with the binding capability of MEF2C with DNA as assessed in gel shift and chromatin immunoprecipitation experiments (data not shown). However, we cannot rule out that this interaction could prevent the interactions between MEF2C and myogenic basic helix-loop-helix factors or other molecules, either adaptor proteins or components of the basal transcription machinery, by steric hindrance or by inducing a conformation of MEF2C that is unable to associate with co-activators.
Our findings suggest that the interaction of Pin1 with Ser98, Ser110, Ser240, and Ser388 multiphosphorylated MEF2C reduces the latter's activity and may represent a novel negative regulatory step controlling MEF2C activity in muscle cells. This finding might help explain why the expression of MEF2 transcription factors is not always accompanied by MEF2-dependent transcription (35). Unlike these phosphorylation events, MEF2C phosphorylation on other residues by the MAP kinase p38 or ERK5 (extracellular signal-regulated kinase 5) up-regulates MEF2-dependent transcription (36, 37). We suggest that diverse cellular signals can induce distinct combinations of phosphorylated sites on MEF2C, which in turn differentially regulate the function of MEF2C in promoting muscle-specific gene expression. We do not know the protein kinases that phosphorylate MEF2C on the residues whose modification is necessary for the highly efficient interaction with Pin1. It has been shown that in neurons, Cdk5 phosphorylates MEF2A and MEF2D at a residue that corresponds to the Pin1 binding site on Ser388 of MEF2C (25, 27, 32). However, although we cannot exclude a role for Cdk5 in controlling the activity of MEF2C in muscle, given the reported observation that Cdk5 stimulates muscle differentiation (38), it appears unlikely that this kinase is responsible for the phosphorylation of the Pin1 binding sites of MEF2C. Thus, our findings suggest a role for signaling through one or more protein kinases other than Cdk5. Possible kinases include members of Cdk family, like Cdk2 and Cdk4, that have been shown to repress muscle differentiation through inhibition of MyoD, myogenin, and MEF2C in proliferating muscle cells (39–41).
To monitor changes in MEF2C phosphorylation on the relevant Pin1 binding sites during C2C7 differentiation, we have recently obtained anti-phospho-Ser98 and anti-phospho-Ser110-specific antibodies that can be used in immunoblotting analyses. Preliminary results indicate a reduction of phosphorylation of these phosphoacceptor sites in lysates prepared from differentiated myocytes (data not shown), consistent with a role of these covalent modifications in Pin1-dependent repression of MEF2C activity.
The evidence that down-regulation of Pin1 promotes terminal differentiation of C2C7 cells also suggests that the function of Pin1 could be modulated during the coordinated processes of muscle differentiation to permit proper activity of MEF2C. However, we observed that although the levels of MEF2 proteins progressively increased during muscle terminal differentiation (data not shown), the levels of Pin1 remained unchanged. Intriguingly, we found that Pin1 becomes excluded from the nucleus with concomitant increase of the cytoplasmic fraction upon induction of differentiation. To our knowledge, this is the first report indicating that the function of Pin1 is regulated by subcellular localization during muscle differentiation. We do not know how Pin1 localization is determined. It was only recently discovered that Pin1 possesses a putative nuclear localization signal, and it was therefore suggested that, despite being a small protein, it may not undergo only passive diffusion through nuclear pore complexes but rather that Pin1 nuclear transport might predominantly be mediated by the importin transport system (42). Therefore, a better understanding of the mechanisms controlling the activity and nuclear transport of Pin1 protein during muscle differentiation may provide more insight into muscle differentiation. As discussed above, the total amount of Pin1 in the cells remains unaffected. The observation that Pin1 is relegated to the cytoplasm during the differentiation process, whereas MEF2C remains strictly nuclear, provides a plausible explanation to a reduced Pin1-MEF2C association and to the up-regulation and enhanced activity of MEF2 proteins.
Based on our results, we propose a model in which Pin1, upon binding to phosphorylated nuclear MEF2C, leads to decreased levels and transcriptional activity of MEF2C. Under this model, the presence of nuclear localized Pin1 in proliferating myoblasts would serve as a fail-safe mechanism to ensure that MEF2 would not be functional if it was promiscuously expressed in proliferating muscle cells. Upon induction of terminal differentiation, to establish a full activity of MEF2 proteins, a reduced Pin1-MEF2C association is required, possibly due to the relegation of Pin1 to the cytoplasm and to a reduced level of phosphorylation of Ser98 and Ser110.
Although our results show that Pin1 can specifically reduce the stability of MEF2C and interfere with the ability of MyoD to cooperate with MEF2C to induce muscle differentiation, we cannot exclude the possibility that Pin1 could also influence this process by acting on other myogenic regulators, such as activated Notch1, which are regulated by Pin1 in other cellular contexts (43). Previous studies suggested that signaling pathways that induce cyclin D1, a key regulator of cell cycle progression, also induce increased levels of nuclear Cdk4, which, in turn, inhibits MyoD and MEF2 function in the dividing cell (3, 40, 41). An essential role of Pin1 in regulating cyclin D1 expression and turnover through multiple mechanisms has been reported (7). Thus, it is conceivable that this Pin1 function contributes to the repression of muscle differentiation in actively proliferating myoblasts. Nevertheless, our preliminary data do not support such a hypothesis, because, as we show in supplemental Fig. S5, Pin1 depletion in C2C7 proliferating muscle cells does not correlate with a significant down-regulation of cyclin D1 or up-regulation of the general CDK inhibitor p21, whereas we detect an induction of muscle-specific gene expression (i.e. MyHC). All in all, our study raises the intriguing possibility that Pin1 might regulate cellular processes distinct from the cell cycle itself, such as terminal differentiation through a modulation of differentiation-specific gene expression.
Supplementary Material
Acknowledgments
We thank Simon M. Hughes for invaluable help in the revision of the manuscript. We thank Sara Badodi and Sandra Parenti for helping with transactivation assays. We are indebted to Sergio Ferrari, Carol Imbriano, Sandra Parenti, and Rita Perlingeiro for helpful suggestions. We are grateful to Tom Kerppola for the plasmids pBiFC-YN, pBiFC-YC, pBiFC-Jun-YN, and pBiFC-Fos-YC; Luigi Naldini for the vectors pRRL-PGK-GFP, pREV, pΔ8.74, and pVSV-G; Roberto Mantovani for the NF-YB antibody; and John McDermott for the MEF2C-specific antibody.
This work was supported by grants from MURST COFIN′06 and EC FP7-Health-2007-B Grant 223098 (OPTISTEM) (to S. F.) and by grants from Telethon and Associazione Italiana per la Ricerca sul Cancro (to G. D. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- CDK
- cyclin-dependent protein kinase
- HEK
- human embryonic kidney
- BiFC
- bimolecular fluorescence complementation
- DSP
- dithiobis(succinimidylproprionate)
- MyHC
- myosin heavy chain
- YFP
- yellow fluorescent protein
- 4SA
- S98A/S110A/S240A/S388A.
REFERENCES
- 1.Molkentin J. D., Li L., Olson E. N. (1996) J. Biol. Chem. 271, 17199–17204 [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto N., Ogashiwa M., Okumura E., Endo T., Iwashita S., Kishimoto T. (1994) FEBS Lett. 352, 236–242 [DOI] [PubMed] [Google Scholar]
- 3.Kitzmann M., Vandromme M., Schaeffer V., Carnac G., Labbé J. C., Lamb N., Fernandez A. (1999) Mol. Cell. Biol. 19, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puente L. G., Voisin S., Lee R. E., Megeney L. A. (2006) Mol. Cell. Proteomics 5, 2244–2251 [DOI] [PubMed] [Google Scholar]
- 5.Lluís F., Perdiguero E., Nebreda A. R., Muñoz-Cánoves P. (2006) Trends Cell. Biol. 16, 36–44 [DOI] [PubMed] [Google Scholar]
- 6.Suelves M., Lluís F., Ruiz V., Nebreda A. R., Muñoz-Cánoves P. (2004) EMBO J. 23, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu K. P., Zhou X. Z. (2007) Nat. Rev. Mol. Cell. Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
- 8.Yeh E. S., Means A. R. (2007) Nat. Rev. Cancer 7, 381–388 [DOI] [PubMed] [Google Scholar]
- 9.Hinits Y., Hughes S. M. (2007) Development 134, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potthoff M. J., Arnold M. A., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Mol. Cell. Biol. 27, 8143–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelelli C., Magli A., Ferrari D., Ganassi M., Matafora V., Parise F., Razzini G., Bachi A., Ferrari S., Molinari S. (2008) Nucleic Acids Res. 36, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zacchi P., Gostissa M., Uchida T., Salvagno C., Avolio F., Volinia S., Ronai Z., Blandino G., Schneider C., Del Sal G. (2002) Nature 419, 853–857 [DOI] [PubMed] [Google Scholar]
- 13.Verdel A., Curtet S., Brocard M. P., Rousseaux S., Lemercier C., Yoshida M., Khochbin S. (2000) Curr. Biol. 10, 747–749 [DOI] [PubMed] [Google Scholar]
- 14.Yaffe D., Saxel O. (1977) Differentiation 7, 159–166 [DOI] [PubMed] [Google Scholar]
- 15.Pinset C., Montarras D., Chenevert J., Minty A., Barton P., Laurent C., Gros F. (1988) Differentiation 38, 28–34 [DOI] [PubMed] [Google Scholar]
- 16.Jordan M., Schallhorn A., Wurm F. M. (1996) Nucleic Acids Res. 24, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borghi S., Molinari S., Razzini G., Parise F., Battini R., Ferrari S. (2001) J. Cell. Sci. 114, 4477–4483 [DOI] [PubMed] [Google Scholar]
- 18.Follenzi A., Ailles L. E., Bakovic S., Geuna M., Naldini L. (2000) Nat. Genet. 25, 217–222 [DOI] [PubMed] [Google Scholar]
- 19.Zhou X. Z., Kops O., Werner A., Lu P. J., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K. P. (2000) Mol. Cell. 6, 873–883 [DOI] [PubMed] [Google Scholar]
- 20.Potthoff M. J., Olson E. N. (2007) Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 21.Black B. L., Olson E. N. (1998) Annu. Rev. Cell. Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 22.Cox D. M., Du M., Marback M., Yang E. C., Chan J., Siu K. W., McDermott J. C. (2003) J. Biol. Chem. 278, 15297–15303 [DOI] [PubMed] [Google Scholar]
- 23.Zhu B., Gulick T. (2004) Mol. Cell. Biol. 24, 8264–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu C. D., Chinenov Y., Kerppola T. K. (2002) Mol. Cell. 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 25.Tang X., Wang X., Gong X., Tong M., Park D., Xia Z., Mao Z. (2005) J. Neurosci. 25, 4823–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang J., Gocke C. B., Yu H. (2006) BMC Biochem. 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grégoire S., Tremblay A. M., Xiao L., Yang Q., Ma K., Nie J., Mao Z., Wu Z., Giguère V., Yang X. J. (2006) J. Biol. Chem. 281, 4423–4433 [DOI] [PubMed] [Google Scholar]
- 28.Mantovani F., Piazza S., Gostissa M., Strano S., Zacchi P., Mantovani R., Blandino G., Del Sal G. (2004) Mol. Cell. 14, 625–636 [DOI] [PubMed] [Google Scholar]
- 29.van Drogen F., Sangfelt O., Malyukova A., Matskova L., Yeh E., Means A. R., Reed S. I. (2006) Mol. Cell. 23, 37–48 [DOI] [PubMed] [Google Scholar]
- 30.Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995) Cell 83, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 31.Naya F. J., Olson E. (1999) Curr. Opin. Cell. Biol. 11, 683–688 [DOI] [PubMed] [Google Scholar]
- 32.Gong X., Tang X., Wiedmann M., Wang X., Peng J., Zheng D., Blair L. A., Marshall J., Mao Z. (2003) Neuron 38, 33–46 [DOI] [PubMed] [Google Scholar]
- 33.Shalizi A., Gaudillière B., Yuan Z., Stegmüller J., Shirogane T., Ge Q., Tan Y., Schulman B., Harper J. W., Bonni A. (2006) Science 311, 1012–1017 [DOI] [PubMed] [Google Scholar]
- 34.Flavell S. W., Cowan C. W., Kim T. K., Greer P. L., Lin Y., Paradis S., Griffith E. C., Hu L. S., Chen C., Greenberg M. E. (2006) Science 311, 1008–1012 [DOI] [PubMed] [Google Scholar]
- 35.Naya F. J., Wu C., Richardson J. A., Overbeek P., Olson E. N. (1999) Development 126, 2045–2052 [DOI] [PubMed] [Google Scholar]
- 36.Han J., Molkentin J. D. (2000) Trends Cardiovasc. Med. 10, 19–22 [DOI] [PubMed] [Google Scholar]
- 37.Kato Y., Zhao M., Morikawa A., Sugiyama T., Chakravortty D., Koide N., Yoshida T., Tapping R. I., Yang Y., Yokochi T., Lee J. D. (2000) J. Biol. Chem. 275, 18534–18540 [DOI] [PubMed] [Google Scholar]
- 38.Lazaro J. B., Kitzmann M., Poul M. A., Vandromme M., Lamb N. J., Fernandez A. (1997) J. Cell. Sci. 110, 1251–1260 [DOI] [PubMed] [Google Scholar]
- 39.Skapek S. X., Rhee J., Kim P. S., Novitch B. G., Lassar A. B. (1996) Mol. Cell. Biol. 16, 7043–7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazaro J. B., Bailey P. J., Lassar A. B. (2002) Genes Dev. 16, 1792–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P., Wong C., Liu D., Finegold M., Harper J. W., Elledge S. J. (1999) Genes Dev. 13, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lufei C., Cao X. (2009) FEBS Lett. 583, 271–276 [DOI] [PubMed] [Google Scholar]
- 43.Rustighi A., Tiberi L., Soldano A., Napoli M., Nuciforo P., Rosato A., Kaplan F., Capobianco A., Pece S., Di Fiore P. P., Del Sal G. (2009) Nat. Cell. Biol. 11, 133–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.