Abstract
The small intestinal BB Na+/H+ antiporter NHE3 accounts for the majority of intestinal sodium and water absorption. It is highly regulated with both postprandial inhibition and stimulation sequentially occurring. Phosphatidylinositide 4,5-bisphosphate (PI(4,5)P2) and phosphatidylinositide 3,4,5-trisphosphate (PI(3,4,5)P3) binding is involved with regulation of multiple transporters. We tested the hypothesis that phosphoinositides bind NHE3 under basal conditions and are necessary for its acute regulation. His6 proteins were made from the NHE3 C-terminal region divided into four parts as follows: F1 (amino acids 475–589), F2 (amino acids 590–667), F3 (amino acids 668–747), and F4 (amino acids 748–832) and purified by a nickel column. Mutations were made in the F1 region of NHE3 and cloned in pet30a and pcDNA3.1 vectors. PI(4,5)P2 and PI(3,4,5)P3 bound only to the NHE3 F1 fusion protein (amino acids 475–589) on liposomal pulldown assays. Mutations were made in the putative lipid binding region of the F1 domain and studied for alterations in lipid binding and Na+/H+ exchange as follows: Y501A/R503A/K505A; F509A/R511A/R512A; R511L/R512L; R520/FR527F; and R551L/R552L. Our results indicate the following. 1) The F1 domain of the NHE3 C terminus has phosphoinositide binding regions. 2) Mutations of these regions alter PI(4,5)P2 and PI(3,4,5)P3 binding and basal NHE3 activity. 3) The magnitude of serum stimulation of NHE3 correlates with PI(4,5)P2 and PI(3,4,5)P3 binding of NHE3. 4) Wortmannin inhibition of PI3K did not correlate with PI(4,5)P2 or PI(3,4,5)P3 binding of NHE3. Two functionally distinct phosphoinositide binding regions (Tyr501–Arg512 and Arg520–Arg552) are present in the NHE3 F1 domain; both regions are important for serum stimulation, but they display differences in phosphoinositide binding, and the latter but not the former alters NHE3 surface expression.
Keywords: Phospholipid, Signal Transduction, Sodium Transport, Sodium Proton Exchange, Trafficking
Introduction
Many transport proteins, including pumps, channels, and transporters, are regulated by phosphoinositides. This rapidly expanding list includes voltage-gated K+ channels, inwardly rectifying K+ channels, and members of the TRP channel family, ENaC, NHE1, and NBCe1 (1–8). This regulation has been explained on the basis of two contrasting mechanisms. (i) There is direct phosphoinositide interaction with specific amino acids in the transport protein, explained either on the basis of charge (8–13) or presence of canonical lipid binding domains such as pleckstrin homology domains (1, 8, 14). Common to these direct interaction studies has been the demonstration that mutagenesis of specific amino acids leads to changes in molecular interactions with phosphoinositides that lead to subsequent changes in physiologic channel or transporter activity. (ii) An indirect mechanism involves an additional intermediate, either protein or lipids, that binds via a phosphoinositide-dependent mechanism (5, 11).
Sodium/hydrogen exchangers (SLC9a family) are ubiquitous transporters serving many functions in the cell, including regulation of intracellular pH, cell volume, and sodium absorption (15, 16). Grinstein and co-workers (17) have demonstrated that NHE1, the ubiquitous member of the sodium hydrogen exchanger gene family, is regulated by PI(4,5)P2, which binds to its C terminus. Using a unique whole cell patch pipette technique, Fuster et al. (18) showed that NHE3 is rapidly stimulated in opossum kidney cells by intracellular application of PI(3,4,5)P3.2 However, the mechanism of this stimulation is unknown. We hypothesized that the epithelial brush border Na+/H+ antiporter NHE3 binds phosphoinositides based on the recognition that gene families have similar structural/functional organization (19). The aim of this study was to understand the mechanism of NHE3 regulation by phosphoinositides by the following: (i) investigating whether NHE3 can directly bind phosphoinositides; (ii) identifying regions and amino acids that are necessary for this interaction, and (iii) studying the physiologic relevance of this interaction.
EXPERIMENTAL PROCEDURES
Materials
Lipids, phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, PI(3,4,5)P3, and PI(4,5)P2 were from Avanti Polar Lipids. QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA). EZ-link sulfo-NHS-SS-biotin was from Thermo Scientific (Rockford, IL). Nigericin and 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein were from Invitrogen. DNA primers were from Operon Biotechnologies (Huntsville, AL). Unless specified, all other chemicals and materials were from Sigma.
Antibodies
Monoclonal mouse antibodies to the hemagglutinin (HA) epitope (MMS 101-R) were from Covance Research Products (Princeton, NJ). Monoclonal mouse anti-polyhistidine antibodies (H1029) and monoclonal mouse anti-VSV-glycoprotein antibodies (A1970) were purchased from Sigma.
Construction of Expression Vectors for NHE3 C-terminal His6 Fusion Proteins and NHE3 C-terminal Point Mutations
Four His6-tagged cDNAs together spanning nearly the entire rabbit NHE3 C terminus were engineered by PCR to encode F1(amino acids 475–589), F2 (amino acids 590–667), F3 (amino acids 668–747), and F4 (amino acids 748–832). Fragments were ligated into pET 30a vector (Novagen) with N-terminal His6 tag using HindIII and EcoRI restriction sites. A 2-amino acid linker (LL) was placed at the C terminus, and a stop codon was inserted at the 3′ end for all inserts just after the linker region. Point mutations in full-length NHE3 were prepared using QuikChange site-directed mutagenesis kit according to the manufacturer's protocol (Table 1). All the cDNAs were fully sequenced to ensure proper sequence, orientation, and reading frame.
TABLE 1.
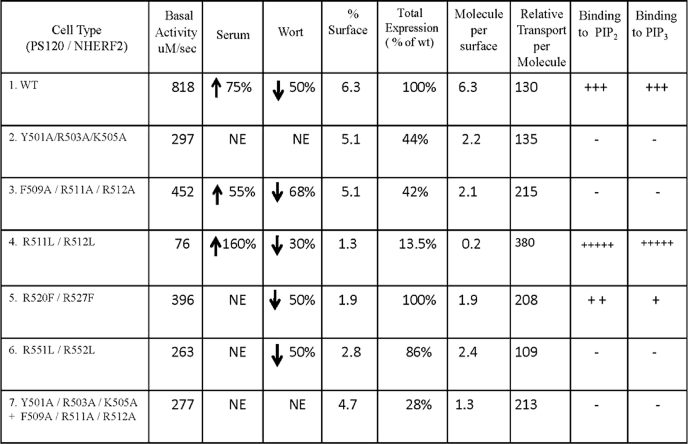
Summary of Na+/H+ exchange rates, surface biotinylation, and phosphoinositide binding studies for WT and NHE3 F1 point mutations
1st column, PS120/NHE2 cells stably transfected with cDNAs are as listed. 2nd column, transport activity of NHE3 WT and mutant proteins under basal conditions as μm/s and serum (3rd column) and wortmannin (Wort) (4th column) conditions are as percentage increase/decrease of basal. 5th column, percentage of NHE3 on surface are as calculated in Fig. 6. 6th column, total expression of proteins are standardized to WT (at 100%). 7th column, molecule per surface is calculated from product of % surface and total expression (normalized to wild type). 8th column, relative transport per molecule was computed by dividing the basal Vmax activity by molecule per surface (2nd column/7th column). 9th and 10th columns, in vitro binding of NHE3 fusion proteins to PI(4,5)P2 and PI(3,4,5)P3 liposome, respectively, is represented as + for presence and − for absence of binding with WT NHE3 binding set to +++. NE means no effect.
Cell Culture and cDNA Transfection of NHE3 C-terminal Point Mutations
The cDNAs of wild type (WT) rabbit (HA) NHE3 and point mutants Y501A/R503A/K505A, F509A/R511A/R512A, R511L/R512L, and R551L/R552L in pcDNA3.1/ G418 (Invitrogen) consisting of an N-terminal triple HA tag, as described previously (20), were transfected into plasma membrane Na+/H+ exchanger-deficient PS120 fibroblasts (stably expressing NHERF2 with a hygromycin-resistant selectable marker) at 70% confluence, using ∼10 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Similarly, point mutation R520F/R527F with a C-terminal VSV-G epitope tag in pcDNA3.1/hygromycin was transfected into PS120 fibroblasts stably expressing NHERF2 (21). Cells stably transfected with NHE3 (NHE3 wild type and mutants) were selected by an H+ selection procedure consisting of exposure to 50 mm NH4Cl/saline solution for 1 h, followed by an isotonic 2 mm Na+ solution for 1 h, as described previously (22). Surviving cells were placed in normal culture medium (Dulbecco's modified Eagle's medium with 4.5 g/liter glucose, 584 mg/ml l-glutamine, 110 mg/ml sodium pyruvate, and 10% FBS) with hygromycin and G418 to maintain selection pressure and allowed to reach 30–50% confluence. The H+ selection process was initially repeated every 2–3 days until more than 50% of the cells survived and was then repeated weekly thereafter.
Expression and Purification of Escherichia coli Fusion Proteins
Rosetta 2 (DHE-3) cells grown in Luria broth (LB) medium were used to express His6-tagged fusion proteins using 1 mm isopropyl 1-thio-β-d-galactopyranoside for induction. Crude cell extracts were prepared from culture pellets by breaking the cells in a microfluidizer. Lysates were loaded on columns packed with a Ni2+-nitrilotriacetic acid resin. The column was washed five times with 50 mm Tris, pH 7.4, and 300 mm NaCl and eluted in 50 mm Tris, pH 7.4, 300 mm NaCl and 250 mm imidazole (protocol modified from Qiagen, Valencia, CA). The proteins were subjected to SDS-PAGE and stained with Coomassie Blue demonstrating a single predominant band for each protein (supplemental Fig. 2).
In Vitro Liposomal Binding Assays
PI(4,5)P2, PI(3,4,5)P3, or PI lipid vesicles were prepared with 30 mol % phosphatidylcholine, 17 mol % phosphatidylserine, 20 mol % phosphatidylethanolamine, 24.75 mol % phosphatidylinositol (PI) and 8.25 mol % PI(3,4,5)P3, PI(4,5)P2 or additional PI (in control experiments). Vesicles with serial dilutions of PI(4,5)P2, PI(3,4,5)P3, or PI were prepared by altering molar concentrations of PI with respect to PI(4,5)P2 or PI(3,4,5)P3, keeping the total molar concentration at 100 mol %. The lipids were dried in a speed vacuum and rehydrated using bath sonication in 250 μl of 0.2 m sucrose, 20 mm KCl, 20 mm HEPES, pH 7.4, 0.01% sodium azide. 150 of ng of test proteins (F1, F2, F3, F4, and point mutations) were mixed with 100 μl of PI(3,4,5)P3, PI(4,5)P2, or PI vesicles in 900 μl of liposomal reaction buffer (containing 0.12 m NaCl, 1 mm EGTA, 0.2 mm CaCl2, 1 mm MgCl2, 5 mm KCl, 20 mm HEPES, pH 7.4, 1 mg/ml BSA, and 0.75 mm fresh DTT) and incubated for 15 min at 37 °C. The 1000-μl mixture was then ultracentrifuged at 100,000 × g for 45 min in a Beckman Ti70.1 rotor. The supernatant was removed by fine tip vacuum suction, and the pellet thus obtained was resuspended in NuPAGE LDS sample buffer (Invitrogen) preheated at 60 °C and bath-sonicated for 10 min at room temperature. 35 μl of sample was separated on 14% SDS-PAGE and transferred onto nitrocellulose for immunoblot analysis.
Immunoblot Analysis
The membranes were blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS) (10 mm potassium phosphate buffer, pH 7.4, and 0.15 m NaCl) for 1 h. Blots were then incubated with primary antibodies (where indicated: anti-His (1:5000); anti-HA (1:1000); anti VSVG (1:100)) in 5% milk/PBS for 1 h at room temperature and then washed with 0.05% Tween/PBS three times for 10 min each. The blots were incubated with fluorescently labeled secondary antibody goat anti-mouse IRDye 800 at 1:15,000 (Rockland Immunochemicals, Gilbertsville, PA) for 1 h. Finally, blots were washed three times for 10 min each with 0.05% Tween/PBS, and bands were visualized by the Odyssey system (LI-COR, Lincoln, NE).
Measurement of Na+/H+ Exchange Activity
The transfected PS120 cells were grown to a confluence of ∼70% on glass coverslips and then placed in serum-free medium for 4–5 h to arrest cell division. The Na+/H+ exchanger activity of these cells was measured fluorometrically by using the intracellular pH-sensitive dye 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester (21). The experiments were performed under basal, stimulated (10% dialyzed serum), and inhibited conditions (100 nm wortmannin). To study the effect of serum, PS120 cells were incubated with NH4Cl medium with 10% serum for 20 min at 37 °C. This was followed by sequential perfusion with tetramethylammonium and Na+ media containing 10% serum as described previously (22). Study of the wortmannin effect was by adding 100 nm wortmannin to NH4Cl medium and incubating for 45 min at 37 °C followed by perfusion with tetramethylammonium and Na+ media. Na+/H+ exchange rates (H+ efflux) were calculated as the product of buffering capacity at each pHi and Na+-dependent change in pHi and were analyzed using data analysis software Origin (Microcal), by which data are fit to a general allosteric model as described by the Hill equation. In all experiments, Vmax and K′ (H+)i values were calculated. Means ± S.E. were calculated from kinetic parameters from at least five different experiments for each condition.
Cell Surface Biotinylation
The transfected PS120 cells were grown to 70–80% confluence in 10-cm Petri dishes. The cells were then serum-starved for 4–5 h. Cells were washed with ice-cold phosphate-buffered saline three times (150 mm NaCl and 20 mm Na2HPO4, pH 7.4) and subsequently once in borate buffer (1 mm boric acid, 154 mm NaCl, 7.2 mm KCl, and 1.8 mm CaCl2, pH 9.0). The surface labeling of NHE3 was done by incubating cells with 0.5 mg/ml sulfo-NHS-SS biotin (21) for 20 min and repeated once. Post-labeling, the cells were washed three times with quenching buffer (20 mm Tris, pH 7.4, and 120 mm NaCl) to scavenge the unreacted biotin. Cells were washed three times with ice-cold phosphate-buffered saline and solubilized with N+ lysis buffer (60 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm KCl, 5 mm Na3EDTA, 3 mm EGTA, and 1% Triton X-100). The lysates were then incubated with streptavidin-agarose beads for 4 h. After precipitation of the streptavidin-agarose complex, the supernatant was recovered and called the intracellular fraction. The avidin-agarose beads were washed in N+ buffer six times. For recovering the plasma membrane NHE3, the streptavidin-agarose beads were resuspended in loading buffer (5 mm Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, and 1% 2-mercaptoethanol), boiled for 5 min at 70 °C, and separated on 10% SDS-PAGE. The size-fractionated gel was then transferred electrophoretically to nitrocellulose. After blocking for 1 h in 5% non-fat milk, the blots were probed with monoclonal anti-HA antibody. Western analysis and quantification of the surface fraction were performed using Odyssey software (LI-COR, Lincoln, NE), as described previously (23). As shown in Table 1, in our studies with WT and mutant NHE3, the total expression of mutations was normalized to the WT expression level, which was set at 100% in each experiment.
Statistical Analysis
All numerical data are expressed as means ± S.E., and Student's t tests were used to calculate significance of difference between experimental groups.
RESULTS
Identification of a Phosphoinositide-binding Site in NHE3 by Bioinformatic Analysis
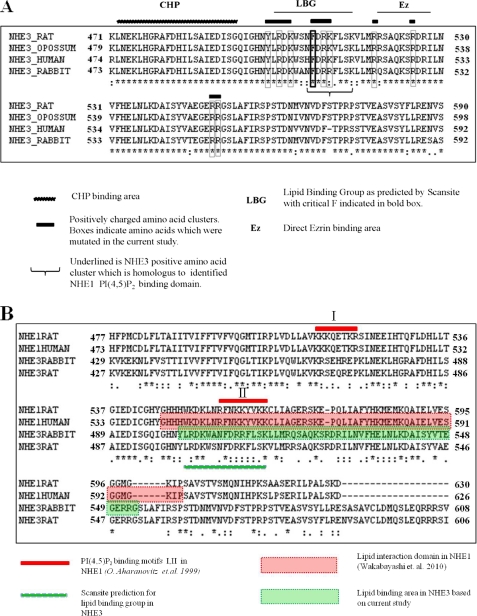
We hypothesized that the NHE3 C-terminal regulatory domain binds phosphoinositides, and we performed a bioinformatics analysis of the amino acid composition of the NHE3 C terminus using Scansite. A region in the NHE3 C-terminal domain, consisting of amino acids 502–516, was predicted by Scansite to be a candidate for lipid binding. This was not predicted by several other bioinformatics programs. This was conserved across species of NHE3. This region (Fig. 1A) has an estimated isoelectric point of 10 and contains clusters of positively charged and aromatic amino acids. The bioinformatics programs PFAM, ProSite, and Smart data bases did not identify any putative lipid binding domains in the NHE3 C terminus (specific searches for pleckstrin homology, Phox homology, FYVE (Fab-1, YGL023, Vps27, and EEA1), C1, C2, BAR (Bin, amphiphysin, Rvs), GLUE (GRAM-like ubiquitin-binding in EAP45), FERM (band 4.1, ezrin, radixin, moesin), CLAM (clathrin assembly lymphoid myeloid), and GRAM (glucosyltransferases, Rab-like GTPase activators, and myotubularins) domains were performed). This made it unlikely that such domains were involved in any NHE3-plasma membrane lipid interactions leaving the possibility of charge and hydrophobicity based interactions in the area predicted by Scansite. Therefore, as a first step we determined whether the NHE3 C terminus, including the amino acids predicted to bind lipids by Scansite (amino acids 502–516), bound PI(4,5)P2 or PI(3,4,5)P3.
FIGURE 1.
Bioinformatic analysis of NHE3. A, NHE3 F1 region (amino acids 475–589) is conserved across species and shown in the figure is the Scansite prediction for the presence of the lipid binding group (LBG) in this area. Phe509 is predicted to be necessary for lipid binding in Scansite analysis and is shown here in a bold box. Clusters of positively charged and aromatic amino acids are shown as bars, with mutated residues shown in boxes. Calcineurin homologous protein (CHP) binding area is shown as a jagged bar. Direct Ezrin binding area is shown as Ez. B, alignment of NHE1 rat, NHE1 human, NHE3 rabbit, and NHE3 rat. Motifs I and II indicate clusters of positively charged amino acids, previously shown to be important for phosphoinositides binding in NHE1 (17). Lipid interaction domain of NHE1 is shown in red. Lipid binding group (LBG) in NHE3 as predicted by Scansite is shown as a green bar. Lipid binding region in NHE3 as identified by this study is shown in green. * indicates identical amino acids; : indicates conserved amino acids; and . indicates semiconserved amino acids.
N-terminal Part of the Intracellular Domain of NHE3 (Amino Acids 475–589) but Not Other Parts of the NHE3 C Terminus Bind PI(4,5)P2 and PI(3,4,5)P3
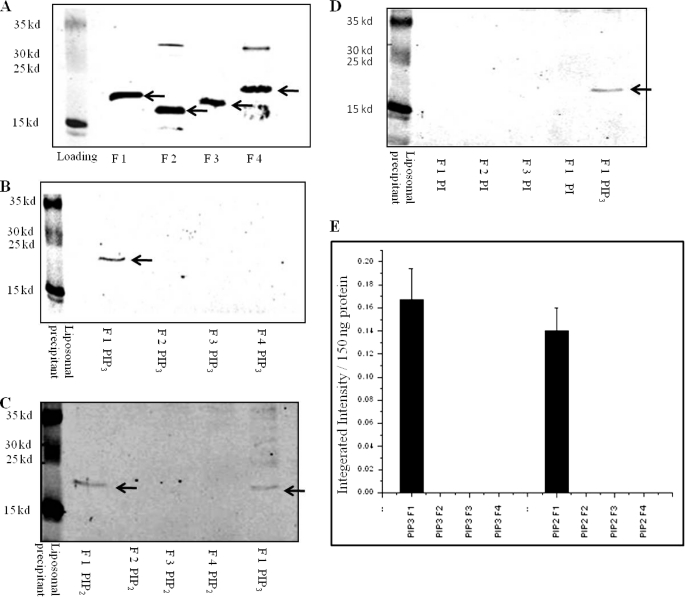
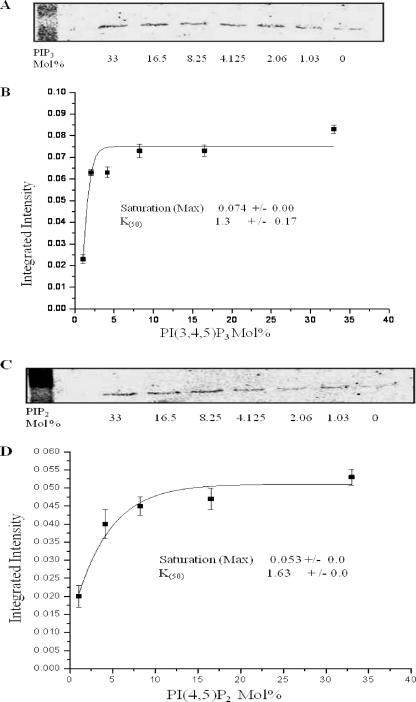
To experimentally test whether the NHE3 C terminus directly binds phosphoinositides and where that binding occurs, we used liposomal pulldown assays with 150 ng of NHE3 C-terminal His6 fusion proteins (∼10 nm) and artificial liposomes (Fig. 2A). The F1 fragment of the NHE3 C terminus (amino acids 475–589) bound to liposomes containing either PI(3,4,5)P3 (Fig. 2B) or PI(4,5)P2 (Fig. 2C). No binding to PI(4,5)P2 or PI(3,4,5)P3 occurred with 150 ng of C-terminal fragments of NHE3 F2 (amino acids 590–667), F3 (amino acids 668–747), or F4 (amino acids 748–832). Binding to F1 was saturable for PI(3,4,5)P3 and PI(4,5)P2. Both phosphoinositides had similar K50 values of 1.3 mol % for PI(3,4,5)P3 and 1.6 mol % for PI(4,5)P2 as shown in Fig. 3.
FIGURE 2.
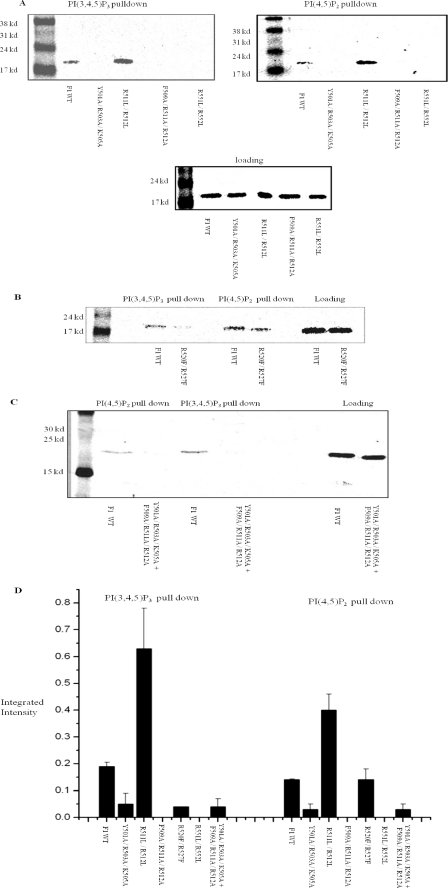
F1 region of rabbit NHE3 (amino acids 475–589) binds to liposomes containing PI(3,4,5)P3 and PI(4,5)P2 but not PI. A, Western blot showing 150 ng of His6 NHE3 C-terminal fusion proteins F1, F2, F3, and F4. These represent input for all experiments. Arrows indicate major bands detected by anti-His6 antibody, 1:5000. B, only F1 but not F2, F3, or F4 bind with PI(3,4,5)P3 in liposomal pulldown assays. Representative Western blots are shown that were repeated three times with similar results. 150 ng of each NHE3 fusion protein was used in liposomes consisting of 30 mol % phosphatidylcholine, 17 mol % phosphatidylserine, 20 mol % phosphatidylethanolamine, 24.75 mol % phosphatidylinositol, and 8.25 mol % PI(3,4,5)P3. C, only F1 but not F2, F3, or F4 binds with PI(4,5)P2. Liposomes as in Fig. 1B, except 8.25 mol % PI(4,5)P2, were used instead of PI(3,4,5)P3. A representative Western blot is shown, which was repeated four times with similar results. Right lane is positive control showing F1 fragment binding with PI(3,4,5)P3 liposomes. D, no binding to PI. 150 ng of each NHE3 fusion protein was used with liposomes as in B and C but with 33 mol % phosphatidylinositol. A PI(3,4,5)P3 liposomal pulldown with F1 is shown in the right lane as a positive control. A representative Western blot is shown, which was repeated three times with similar results. E, quantification for binding from B and C was done by measuring integrated signal intensity of bands by Odyssey system (LI-COR, Lincoln, NE) normalized to 150 ng of fusion protein. Integrated intensity of bands shown as means ± S.E. for PI(3,4,5)P3, n = 3, and PI(4,5)P2, n = 4, is plotted on the y axis. Only the F1 fragment bound PI(3,4,5)P3 and PI(4,5)P2. Arrows in Fig. 2, B–D indicate F1 fragment in liposomal pulldowns.
FIGURE 3.
NHE3-F1 fusion protein binds to liposomes containing PI(3,4,5)P3 and PI(4,5)P2 in a saturable manner. A, liposomal pulldowns assays performed with 150 ng of F1 protein and liposomes containing varied amounts of PI(3,4,5)P3. The makeup of the liposomes was set at 100 mol % by replacing mol % of PI(3,4,5)P3 by the same mol % PI. His6-tagged NHE3 F1 was identified by Western blot analysis of the pulldown by anti-His6 antibody, and the intensity of bands was quantified by Odyssey system (LI-COR). B, binding curve based on intensity values (y axis) versus PI(3,4,5)P3 in mol % (x axis). Background intensity due to F1 protein binding in absence of PI(3,4,5)P3 was subtracted from all data points in calculating kinetics of binding. All experiments were done three times, and the curve representing means ± S.E. calculated from data points of three different experiments is shown. Saturation (max) and K50 were calculated using Origin software. C, liposomal pulldowns done with 150 ng of F1 protein and liposomes containing varied amounts of PI(4,5)P2; PI(4,5)P2 was replaced by the same mol % PI in PI(4,5)P2 liposomes. Intensity of bands was quantified by Odyssey system (LI-COR). D, binding curve based on intensity values (y axis) versus PI(4,5)P2 in mol % (x axis). Background intensity due to F1 protein binding in the absence of PI(4,5)P2 was subtracted from all data points in calculating kinetics of binding. All experiments were done three times, and the binding curve representing means ± S.E. calculated from data point of three different experiments is shown. Saturation (max) and K50 values (mol %) were calculated using Origin software.
As a negative control, binding of the F1 fragment did not occur with liposomes with 33 mol % of phosphatidylinositol, an unphosphorylated phospholipid (Fig. 2D). Further studies explored how the F1 domain bound to PI(3,4,5)P3 and PI(4,5)P2 and the functional consequences of that binding.
Mutation of Positively Charged and Hydrophobic Amino Acids in the NHE3 C-terminal F1 Region Alters PI(3,4,5)P3 and PI(4,5)P2 Binding
Phosphoinositides bind to sites containing positively charged and aromatic amino acids (1). By sequence analysis, we identified four putative binding motifs in the F1 region of NHE3 (amino acids 475–589), which contain clusters of positive charges and were conserved across species of NHE3 (Fig. 1A). As shown in Fig. 1A, these clusters were located C-terminal to the putative calcineurin homologous protein binding area of NHE3, which we predicted by homology to the NHE1 calcineurin homologous protein-binding site, the structure of which had been solved (24, 25). One of these positively charged amino acid clusters (Arg520/Arg527) has previously been shown to be involved in the direct binding of the cytoskeletal linking protein ezrin to the NHE3 C terminus (21). A second motif (Tyr501/Arg503/Lys505) is immediately adjacent to His499, which had been shown by mutagenesis to be part of the NHE3 intracellular pH sensor (26).
To determine whether any of these conserved areas of clustered positively charged amino acids contributed to the interaction of NHE3 with PI(3 ,4,5)P3 and/or PI(4,5)P2, we individually mutated at least two positive charges in each cluster. In several instances, we also mutated an aromatic amino acid in the region. His6 fusion proteins of the mutated F1 domains were studied for phosphoinositide binding, and the same mutations were made in full-length NHE3 and expressed in PS120/NHERF2 cells for Na+/H+ exchange activity measurements. Table 1 lists the mutations that were made in these domains with arginine or lysine being generally mutated to an alanine or leucine except for R520F/R527F, a mutation that had previously been shown to reduce direct ezrin binding to NHE3 (21).
Liposomal pulldown assays with purified mutant F1 domains confirmed binding of wild type NHE3 to liposomes containing PI(3,4,5)P3 and PI(4,5)P2. In contrast, the NHE3 F1 point mutants Y501A/R503A/K505A, F509A/R511A/R512A, R551L/R552L, and combination mutation Y501A/R503A/K505A + F509A/R511A/R512A showed loss of phosphoinositide binding. Mutation R520F/R527F displayed greater loss of PI(3,4,5)P3 (68.1%) binding than PI(4,5)P2 binding (23.3%). In contrast, the F1-R511L/R512L mutant exhibited increased binding to both PI(3,4,5)P3- and PI(4,5)P2-containing liposomes (Fig. 4, A–D).
FIGURE 4.
Mutagenesis of positively charged amino acids in N-terminal part of cytosolic region of NHE3 alters PI(3,4,5)P3 and PI(4,5)P2 binding. A, PI(3,4,5)P3 and PI(4,5)P2 liposomal pulldown assays were performed with 150 ng of WT F1 His6 fusion protein and point mutated fusion proteins as follows: Y501A/R503A/K505A, R511L/R512L, F509A/R511A/R512A, and R551L/R552L as described in Fig. 1A and Table 1. Y501A/R503A/K505A, F509A/R511A/R512A, and R551L/R552L do not bind to PI(4,5)P2 or PI(3,4,5)P3 containing liposomes, whereas R511L/R512L had increased binding to PI(4,5)P2 and PI(3,4,5)P3. 150 ng of NHE3 fusion proteins were loaded as shown in the lower panel. B, PI(3,4,5)P3 and PI(4,5)P2 liposomal pulldown assays were performed with 150 ng of WT F1 His6 fusion protein and point mutated fusion protein R520F/R527F. This mutant displayed greater reduction of PI(3,4,5)P3 binding than PI(4,5)P2 binding when compared with its control (68% loss versus 23%). Loading of 150 ng of proteins is shown in the right-most lanes. C, PI(3,4,5)P3 and PI(4,5)P2 liposomal pulldown assays were performed with 150 ng of WT F1 His6 fusion protein and combination mutations Y501A/R503A/K505A + F509A/R511A/R512A. The combination mutation binds minimally with PI(3,4,5)P3- and PI(4,5)P2-containing liposomes. D, quantification: integrated intensity of bands from A–C was measured by Odyssey system (LI-COR). Pulldowns were done three times, and average values of integrated intensity were plotted as means ± S.E. Standard error is too small to see in Y501A/R503A/K505A and R520F/R527F PI(3,4,5)P3, although F509A/R511A/R512A and R551L/R552L had approximately zero intensity values.
Basal Transport and Serum Stimulation of NHE3 Activity but Not Wortmannin Inhibition of Basal Activity Directly Correlates with PI(3,4,5)P3 and PI(4,5)P2 Binding of NHE3
The function of PI(4,5)P2 and PI(3,4,5)P3 binding on NHE3 transport activity was studied in PS120/NHERF2 cells transfected with wild type NHE3 and full-length NHE3 mutated to contain the same point mutations listed in Table 1. Kinetic constants of WT NHE3 activity were compared with the mutants under basal, serum-stimulated, and wortmannin-inhibited conditions (Fig. 5, A–H, Table 1, and supplemental Table 1). All mutants with altered phosphoinositide binding displayed lower basal activity. This was true for both Vmax and K′(H+)i. Those mutants with decreased phosphoinositide binding had decreased or absent serum stimulation of NHE3 activity. In contrast, the single mutant with increased PI(3,4,5)P3 and PI(4,5)P2 binding (R511L/R512L) had increased serum stimulation of NHE3 activity.
FIGURE 5.
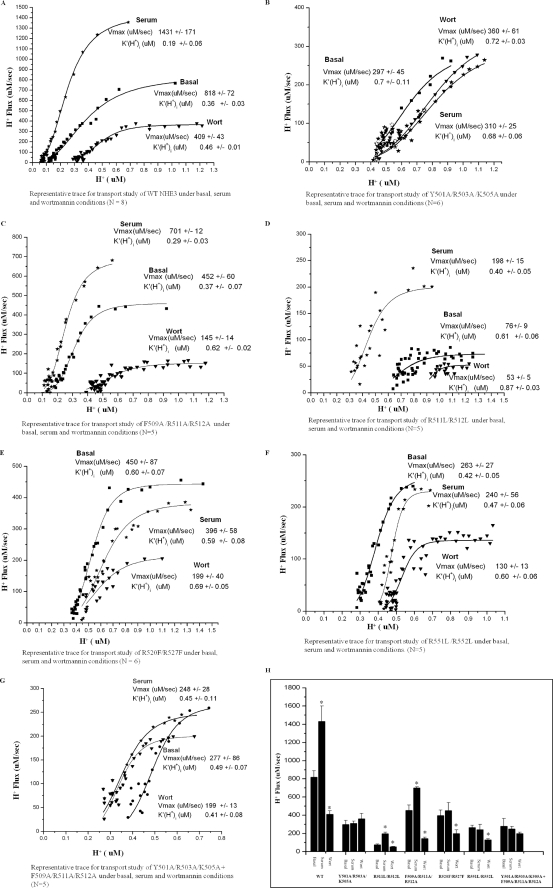
Na+/H+ exchange rates for WT and NHE3 F1 point mutants expressed in PS120 cells. A–G, basal rates (■), stimulation by serum (10%) ( ), and inhibition by wortmannin (▾) (100 nm) are shown as Vmax (μm/s) and K′(H+)i (μm) for representative experiments and means ± S.E. for the series of experiments shown in text. NHE3 mutants Y501A/R503A/K505A, R520F/R527F, and R551L/R552L and Y501A/R503A/K505A + F509A/R511A/R512A lost serum stimulation. F509A/R511A/R512A showed a decrease in serum stimulation. R511L/R512L showed increased serum stimulation. All experiments were done using mixed pairs and repeated five or more times (N shown in individual legends). H represents comparison of Vmax values of WT and NHE3 point mutations under basal, serum, and wortmannin conditions as mean ± S.E. (*, p < 0.05; for each NHE3 cell type, comparison of Na+/H+ exchange rates was done between basal/serum and basal/wortmannin).
), and inhibition by wortmannin (▾) (100 nm) are shown as Vmax (μm/s) and K′(H+)i (μm) for representative experiments and means ± S.E. for the series of experiments shown in text. NHE3 mutants Y501A/R503A/K505A, R520F/R527F, and R551L/R552L and Y501A/R503A/K505A + F509A/R511A/R512A lost serum stimulation. F509A/R511A/R512A showed a decrease in serum stimulation. R511L/R512L showed increased serum stimulation. All experiments were done using mixed pairs and repeated five or more times (N shown in individual legends). H represents comparison of Vmax values of WT and NHE3 point mutations under basal, serum, and wortmannin conditions as mean ± S.E. (*, p < 0.05; for each NHE3 cell type, comparison of Na+/H+ exchange rates was done between basal/serum and basal/wortmannin).
Under basal conditions, WT NHE3 (Fig. 5, A and H, Table 1, and supplemental Table 1) had transport activity of 818 ± 72 μm/s (Vmax) and K′ (H+)i of 0.36 ± 0.03 μm. NHE3 responded to serum with the following: (i) 75% increase in Vmax, 1431 ± 171 μm/s, p < 0.05 and (ii) K′(H+)i change to 0.19 ± 0.06 μm, p < 0.05 (increased sensitivity to H+). Wortmannin-inhibited basal NHE3 activity was as follows: 50% inhibition of Vmax, 409 ± 43 μm/s, p < 0.005, and (ii) K′(H+)i change to 0.46 ± 0.01 μm, p < 0.05. (p values are in comparison with basal activity.)
As an example of the changes in the mutants compared with wild type NHE3, Y501A/R503A/K505A, which does not bind PI(4,5)P2 or PI(3,4,5)P3, had the following: (i) lower basal activity (basal activity Vmax, 297 ± 45 μm/s, and K′(H+)i, 0.70 ± 0.11 μm), and (ii) no change in activity when exposed to serum or wortmannin (serum Vmax, 310 ± 25 μm/s, NS, K′(H+)i, 0.68 ± 0.06 μm, NS, and wortmannin Vmax, 360 ± 61 μm/s, NS, K′(H+)i, 0.72 ± 0.03 μm, NS) (Fig. 5, B and H, Table 1, and supplemental Table 1). The meaning of changes in basal transport is commented on under “Discussion.”
As the bioinformatic analysis from Scansite predicted, Phe509 and two positive charges in the same amino acid cluster were important for phosphoinositide binding. As shown in Fig. 4, mutant F509A/R511A/R512A lost PI(3,4,5)P3 and PI(4,5)P2 binding. This mutant displayed lower serum stimulation (55 ± 2.6%), which was significantly different from WT serum stimulation (75 ± 4.3% p < 0.05). In addition, this mutation had a wortmannin inhibitory response (68% inhibition ± 1.9%), which was significantly increased compared with WT wortmannin response (50 ± 5.4%, p < 0.01). Basal Vmax, 452 ± 60 μm/s, serum Vmax, 701 ± 12 μm/s, p < 0.05, wortmannin Vmax, 145 ± 14 μm/s, p < 0.05, K′(H+)i basal, 0.37 ± 0.07 μm, K′(H+)i serum 0.29 ± 0.03 μm, NS; K′(H+)i wortmannin, 0.62 ± 0.02 μm, p < 0.05 (Fig. 5, C and H, Table 1, and supplemental Table 1).
To determine whether the effect of mutations Y501A/R503A/K505A and F509A/R511A/R512A acted on similar or different pathways, a combination of both mutations was expressed in PS120 cells. This combination double mutant compared with WT had lower basal activity (Vmax, 277 ± 86 μm/s, and K′(H+)i, 0.49 ± 0.07 μm) and no change in activity when exposed to serum or wortmannin (serum Vmax, 248 ± 28 μm/s, NS, K′(H+)i 0.45 ± 0.11 μm, NS and wortmannin Vmax, 199 ± 13 μm/s, NS, K′(H+)i, 0.41 ± 0.08 μm, NS). Thus, this combination mutation mimics Y501A/R503A/K505A by itself with no evidence of additivity of effects of the two mutants (Fig. 5, G and H, Table 1, and supplemental Table 1).
To examine the role of Phe509 in the effect of the Phe509/Arg511/Arg512 mutant, Arg511 and Arg512 were mutated without mutating Phe509, creating mutant R511L/R512L. This mutation had an increase in binding to PI(3,4,5)P3 and PI(4,5)P2 (Fig. 4, A and D). This mutant also had the following: (i) reduced basal NHE3 activity; (ii) increased NHE3 activity when exposed acutely to serum. This increase was statistically significantly greater than that which occurred in the wild type NHE3 (basal Vmax, 76 ± 9 μm/s, serum Vmax, 198 ± 15 μm/s, p < 0.05); there was a 160 ± 12.7% increase in R511L/R512L compared with 75 ± 4.3% in WT, p < 0.05 (K′(H+)i basal, 0.61 ± 0.06 μm, K′(H+)i serum, 0.40 ± 0.05 μm, p < 0.05); and (iii) decrease in wortmannin inhibition (30 ± 3.2% compared with 50 ± 5.4% in WT, p < 0.01); wortmannin Vmax, 53 ± 5 μm/s, p < 0.05, K′(H+)i, 0.87 ± 0.03 μm, NS). Thus, the increase in PI(3,4,5)P3 and PI(4,5)P2 binding of this mutant correlated with an increase in serum stimulation of NHE3 activity (Fig. 5, D and H, Table 1, supplemental Table 1).
R520F/R527F, a mutation which previously had been shown to decrease ezrin binding to NHE3 by 80% (21), had greater loss of PI(3,4,5)P3 binding (68%) compared with PI(4,5)P2 binding (23%) (Fig. 4B). The mutation also displayed no serum response (basal Vmax, 396 ± 58 μm/s, serum Vmax, 450 ± 87 μm/s, NS) and normal wortmannin response (50% ± 6.7% inhibition; wortmannin Vmax, 199 ± 40 μm/s, p < 0.05; K′(H+)i basal 0.59 ± 0.08 μm, K′(H+)i serum, 0.60 ± 0.07 μm, NS and K′(H+)i wortmannin, 0.69 ± 0.05, p < 0.05) (Fig. 5, E and H, Table 1, supplemental Table 1).
Mutant R551L/R552L displayed loss of both PI(3,4,5)P3 and PI(4,5)P2 binding (Fig. 4, A and D) and functionally had no serum effect (Vmax basal, 263 ± 27 μm/s, serum Vmax, 240 ± 56 μm/s, NS) and a wortmannin inhibitory effect similar to wild type (50% ± 5.9% inhibition) (wortmannin Vmax, 130 ± 13 μm/s, p < 0.05, K′(H+)i basal, 0.42 ± 0.05 μm, K′(H+)i serum, 0.47 ± 0.06 μm, NS, K′(H+)i, wortmannin, 0.60 ± 0.06 μm, p < 0.05) (Fig. 5, F and H, Table 1, supplemental Table 1).
Basal Surface Expression of NHE3 and F1 Mutants Does Not Correlate with PI(4,5)P2 and PI(3,4,5)P3 Binding of NHE3
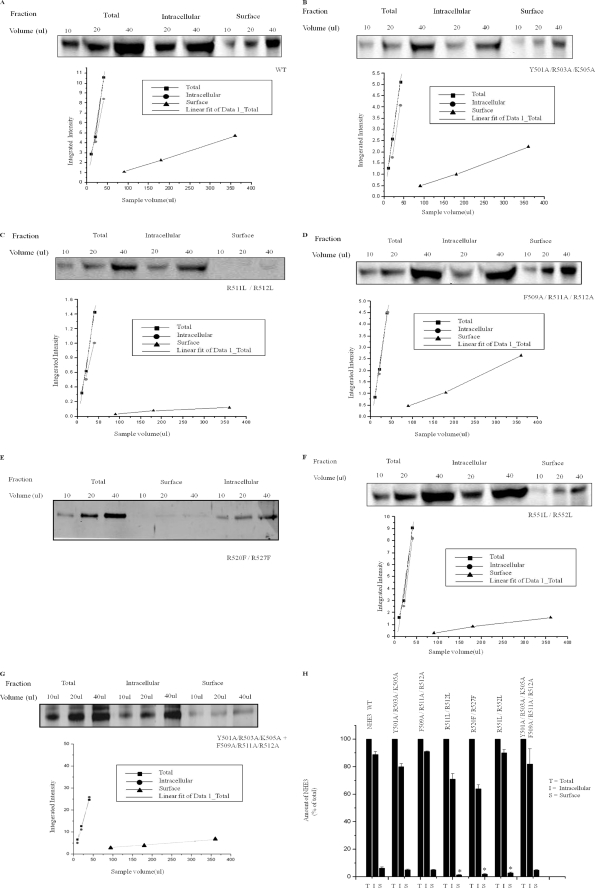
Because most regulation of NHE3 occurs by changes in regulated endocytosis or exocytosis and affects surface expression of NHE3 (15), surface expression of WT and NHE3 mutant was determined under basal conditions using cell surface biotinylation of subconfluent PS120 cells. The fraction of surface NHE3 was expressed as percent of total NHE3. The amount of protein expressed was normalized to wild type as shown in Table 1 and Fig. 6. Mutants Y501A/R503A/K505A, F509A/R511A/R512A, and Y501A/R503A/K505A + F509A/R511A/R512A had similar % surface NHE3 as compared with wild type NHE3 (5.1, 5.1, and 4.7% versus 6.3% in WT) (Fig. 5, A, B, D, and G), but all had reduced total expression. In contrast, mutant R511L/R512L had 1.3% surface expression (as shown in Fig. 5C) and also had decreased total expression. Mutant R520F/R527F had 1.9% surface expression. Mutant R551L/R55 2L also had reduced surface expression (2.8%). Both of the last two NHE3 mutants had near normal total expression of NHE3. Thus several patterns of surface and total expression were observed as shown in Table 1.
FIGURE 6.
A–H, cell surface biotinylation studies show that surface expression of WT NHE3 and F1 domain positive cluster point mutations are not different and do not correlate with PI(3,4,5)P3 and PI(4,5)P2 binding. PS120 cells stable expressing WT HA-NHE3 and NHE3 point mutants were serum-starved for 5 h and then surface-biotinylated by NHS-SS-biotin. Representative Western blots for NHE3 and mutant proteins are shown, and means ± S.E. of surface expression are shown in H and Table 1. Multiple dilutions for total, intracellular (non-avidin-precipitated), and surface (avidin-precipitated) NHE3 were probed with anti-HA antibody. Protein density was measured by Odyssey Licor Biosystems, and densities were plotted against the sample volume (μl). Fractions were compared for analysis on a single Western blot. Percentage of NHE3 on surface was calculated by the product of dilution factor of surface and total NHE3 × ratio of surface NHE3 intensity/total NHE3 intensity. Surface biotinylation for WT (A) and the following mutants showed similar percentage surface expression: Y501A/R503A/K505A (B and H), F509A/R511A/R512A (D and H), and Y501A/R503A/K505A + F509A/R511A/R512A (G and H). In contrast, R511L/R512L (C and H), R520F/R527F (E and H), and R551L/R552L (F and H) had lower surface expression than wild type. H shows the amount of surface NHE3 as percentage of total NHE3, where total NHE3 was normalized to 100% for each condition; p value (*, p < 0.05) represents comparison of percent surface NHE3 for each mutant protein compared with wild type. Experiments were done three times for each mutant protein paired with a wild type NHE3, and data shown are means ± S.E.
DISCUSSION
In this study, we demonstrate that the N-terminal part of the NHE3 C terminus is involved in regulation of basal and serum-stimulated NHE3 activity by interaction with the phosphoinositides PI(3,4,5)P3 and/or PI(4,5)P2. The part of NHE3 involved includes amino acids 475–552 and appears to be made up of two regions that resemble each other in effects on phosphoinositide binding and on NHE3 activity but demonstrate enough differences to make us hypothesize that they both bind phosphoinositides differently and exert different effects on NHE3 activity. These two regions (Tyr501–Arg512 and Arg520–Arg552) are both made up of two separate clusters of at least two positively charged amino acids. The first region also contains bulky, hydrophobic amino acids which when mutated alter the effects of the region on phosphoinositide binding and regulation of NHE3 activity. Mutations of each of the two positively charged clusters that make up each of the two regions behave similarly in terms of effects on NHE3 activity and phosphoinositide binding, suggesting they are acting similarly as part of a single functional domain. In fact, for the most N-terminal region, the effects on phosphoinositide binding and NHE3 activity were nonadditive when a combination made up of both mutations was studied, further supporting effects via a common mechanism. Moreover, differences in the effects of the two regions on both lipid binding and NHE3 activity are strongly suggestive that NHE3 interacts with membrane lipids using its F1 domain by more than a single mechanism. The similarities in effects of the two regions are that when mutated they all reduce basal NHE3 activity, abolish or significantly reduce serum stimulation of NHE3 activity, while also reducing liposomal binding to PI(4,5)P2 and PI(3,4,5)P3. All four mutations of NHE3, which make up the two regions, carry out basal NHE3 activity similar to that of wild type when transport activity is normalized to the total number of NHE3 molecules estimated to be on the plasma membrane. In contrast, there are significant differences in the effects of the two regions. First, for the more N-terminal region, effects of mutating either of the two positively charged clusters similarly reduced binding of the NHE3 mutant to PI(4,5)P2 and PI(3,4,5)P3, and for the more C-terminal region, mutation of R520F/R527F reduced PI(3,4,5)P3 binding more than PI(4,5)P2 binding. This is the only suggestion from these studies that binding to PI(3,4,5)P3 is more relevant for the effects of phosphoinositides on NHE3 activity than binding to PI(4,5)P2. Second, mutations in all four positively charged clusters of amino acids (Y501A/R502A/K505A, F509A/R511A/R512A, R520F/R27F, and R551L/R552L) reduced the amount of plasma membrane NHE3. The more N-terminal mutations (Y501A/R502A/K505A and F509A/R511A/R512A) have reduced total NHE3 expression but similar percent plasma membrane expression compared with wild type; we have not examined the mechanism for this reduced total expression which is likely due to decreased synthesis or increased breakdown of mutant NHE3. The latter is known to occur with misfolded proteins as is well characterized for cystic fibrosis transmembrane regulator mutants (27). The more C-terminal (R520F/R527F and R551L/R552L) mutants have total NHE3 expression similar to wild type NHE3 but reduced percent plasma membrane expression compared with wild type. This indicates that the more C-terminal region but not the more N-terminal region alters NHE3 trafficking or plasma membrane retention. Of note in interpreting these differences is that Arg520/Arg527 was previously shown to be in an α-helical domain in the NHE3 C terminus, which is where ezrin directly binds to the NHE3 C terminus and is necessary for multiple aspects of basal and regulated trafficking (21). Whether it is this direct ezrin binding or another property of this area of NHE3 that is responsible for the characteristics of this part of NHE3, including binding to phosphoinositides, is still an open question.
It is not surprising that phosphoinositide binding to NHE3 affected NHE3 activity. Detailed studies of rat NHE1 had previously shown that binding of PI(4,5)P2 to amino acids 513–520 and 556–564 is necessary for basal Na+/H+ exchange activity (17). As shown in Fig. 1B, these regions of rat NHE1 are homologous to amino acids 465–472 and 509–516 of rabbit NHE3. The second of these motifs was shown in this study to be involved in PI(4,5)P2 and PI(3,4,5)P3 binding (underlined with a brace in Fig. 1A and as motif II in Fig. 1B), although the first homologous motif (motif I in Fig. 1B; corresponding to rabbit NHE3 amino acids 465–472) was not studied here because our attempts to purify rabbit NHE3 N-terminal to amino acid 475 resulted in insoluble protein.
In addition, intracellular injection of PI(3,4,5)P3 immediately stimulated NHE3 activity in opossum kidney cells, as reported by Fuster et al. (18). Our studies are a detailed mechanistic exploration of that finding. Our previous studies of NHE3 truncation mutations expressed in PS120 fibroblasts had demonstrated that regulation of NHE3 was dependent on different subdomains of the NHE3 C terminus (22, 28); stimulation of NHE3 by okadaic acid and FGF both occurred when NHE3 was truncated at amino acid 585 (22), consistent with the role for the F1 domain in maintaining basal NHE3 and serum stimulation as demonstrated in this study. Okadaic acid, which we showed acted on the NHE3 C terminus between amino acids 509 and 543 (22), has been shown also to be an activator of PI3K and Akt (29), suggesting that its role may involve phosphoinositide binding to, in addition to, or rather than by inhibition of phosphatases (PP1, PP2A, PP6, and PP4) (22, 30); however, the mechanism of okadaic acid stimulation of NHE3 remains controversial (30).
An unexpected finding was that mutating Arg511 and Arg512 in the N-terminal subdomain without altering Phe509 increased PI(4,5)P2 and PI(3,4,5)P3 binding, which was associated with an increase in the magnitude of the serum stimulation of this NHE3 mutant. Currently, we do not understand the mechanism for this increase in binding, and we speculate on a role for allosteric interactions leading to positive enhancement and/or alteration in intramolecular interactions between the positive amino acids Arg511/Arg512 and negative amino acids such as Asp510, which might lead to an alteration in the structural conformation of this region. However, this result strongly supports the involvement of phosphoinositide binding to this area of NHE3 in serum stimulation, because this mutant established a direct correlation between magnitude of serum stimulation of NHE3 activity and extent of NHE3 C-terminal binding to PI(4,5)P2/PI(3,4,5)P3, as shown in Table 1.
In previous studies of basal and acutely stimulated NHE3 activity in all cell culture models studied, ∼50% of basal NHE3 activity was dependent on PI 3-kinase activity (15). Basal NHE3 activity was inhibited by exposure to the PI3K inhibitors, viz. LY594002 (31) and wortmannin (32, 33) at low doses, and acutely stimulated NHE3 was reduced by the same inhibitors between 50 and 100% based on the cell type expressing NHE3 and the ligand studied (FGF in PS120 cells by 50% (32); lysophosphatidic acid in opossum kidney cells by 100%) (31). These previous studies of PI3K dependence of basal and acutely stimulated NHE3 activity plus the rapid stimulation of NHE3 activity when PI(3,4,5)3 was added to the inside of opossum kidney cells (18) suggest that PI(3,4,5)P3 is the more relevant phosphoinositide for NHE3 regulation. However, this study does not separate the roles of PI(4,5)P2 and PI(3,4,5)P3, and either phosphoinositide could be relevant for physiologic or pathophysiologic regulation of NHE3. Importantly, this study did not show correlation of the extent of wortmannin inhibition of basal NHE3 activity with changes in phosphoinositide binding. This suggests that the domain of NHE3 involved in basal regulation by PI3K may be somewhat separate from the domains involved in serum-stimulated NHE3 activity. We do not rule out the possibility that mutagenesis of phosphoinositide-binding motifs can also alter association with interacting proteins in that area and contribute to alteration in NHE3 activity.
Many transport proteins have been shown to be regulated by phosphoinositides, with most emphasis being on interactions with PI(4,5)P2 rather than PI(3,4,5)P3. Recently, however, PI(3,4,5)P3 was shown to regulate TRPC6 by disrupting its association with calmodulin (11), and PI(4,5)P2 has been reported to be important for ENaC channel gating and PI(3,4,5)P3 for ENaC channel open probability and number (9, 10). These results suggest that different phosphoinositides in a cell can regulate different functional aspects of the same protein. Understanding the mechanisms by which phosphoinositides contribute to basal and regulated activities of transporters, channels and pumps have lagged behind the description of their involvement. Two aspects of the phosphoinositide interacting domains have been identified in the regulated transporters. Heo et al. (34) demonstrated a role for positively charged amino acid clusters, which appear to be important for NHE3 effects. In contrast, multiple protein/lipid interacting domains have been identified by which proteins interact with lipids, some of which are used for phosphoinositide association. Sequence searches failed to identify specific lipid binding groups in the NHE3 C terminus. Nonetheless, the possibility of the presence of additional lipid binding groups in the NHE3 C terminus cannot be excluded, as experiments with a higher concentration of C-terminal peptides (300 ng) demonstrated weak binding of the NHE3 F2 region (amino acids 590–667) to PI(3,4,5)P3 liposomes, although this binding was much less than that detected in the F1 region (supplemental Fig. 1). Thus, multiple low affinity or transiently interacting motifs capable of binding phosphoinositides might be present in the NHE3 C terminus.
In conclusion, NHE3 amino acids 475–589 bind phosphoinositides PI(4,5)P2 and PI(3,4,5)P3. Mutation of positively charged and hydrophobic amino acids in the NHE3 C-terminal F1 domain alters PI(3,4,5)P3 and PI(4,5)P2 binding. Thus phosphoinositide binding to NHE3 is dependent on charge-based interactions. Mutations that alter PI(3,4,5)P3 and PI(4,5)P2 binding of NHE3 all have decreased basal activity. Serum stimulation of NHE3 correlated with PI(3,4,5)P3 and PI(4,5)P2 binding of NHE3. In contrast, wortmannin inhibition of basal NHE3 activity did not correlate with PI(3,4,5)P3 or PI(4,5)P2 binding of NHE3. The NHE3 F1 domain (amino acids 475–589) has two distinct subdomains (Tyr501–Arg512 and Arg520–Arg552), both of which are important for serum stimulation, but they display different effects in total and surface expression and phosphoinositide interactions. The close proximity of two different lipid binding groups suggests the possibility of their intermolecular interaction(s) under physiologic conditions in NHE3 regulation.
Recently, Wakabayashi et al. (35) identified a lipid binding domain in the intracellular C-terminal domain of NHE1, which incorporated the area previously shown to be the PI(4,5)P2 binding domain. This domain of NHE1 physically associated with the plasma membrane under basal conditions and increased its binding with elevated diacylglycerol or phorbol ester exposure. Two subdomains in this area were tentatively identified, the N-terminal portion binding primarily to acidic phospholipids, and the C-terminal domain to diacylglycerol and phorbol esters. Shown in Fig. 1B, this domain of NHE1 overlaps almost totally with the homologous area of NHE3 we describe here, and we predict that these areas may behave similarly in terms of lipid binding and perhaps plasma membrane association. However, given that NHE1 is regulated primarily by changes in K′(H+)i and NHE3 is regulated mostly by changes in trafficking, which include changes in Vmax and to a lesser extent in K′(H+)i, indicate that these similar domains use different mechanisms for regulation of NHE1 and NHE3.
Supplementary Material
Acknowledgment
We acknowledge the expert editorial assistance of Kate Desantis.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK26523, R01-DK61765, PO1-DK72084, and R24-DK64388 (the Hopkins Basic Research Digestive Diseases Development Core Center). This work was also supported by Hopkins Center for Epithelial Disorders.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- PI(3,4,5)P3
- phosphatidylinositide 3,4,5-triphosphate
- PI(4,5,)P2
- phosphatidylinositide 4,5-bisphosphate, PI, phosphatidylinositol
- NS
- not significant.
REFERENCES
- 1.Rosenhouse-Dantsker A., Logothetis D. E. (2007) Pflugers Arch. 455, 45–53 [DOI] [PubMed] [Google Scholar]
- 2.Hilgemann D. W., Ball R. (1996) Science 273, 956–959 [DOI] [PubMed] [Google Scholar]
- 3.Hilgemann D. W., Feng S., Nasuhoglu C. (2001) Sci. STKE 2001, re19. [DOI] [PubMed] [Google Scholar]
- 4.Logothetis D. E., Petrou V. I., Adney S. K., Mahajan R. (2010) Pflugers Arch. 460, 321–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohacs T. (2009) Cell Calcium 45, 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamper N., Shapiro M. S. (2007) Nat. Rev. Neurosci. 8, 921–934 [DOI] [PubMed] [Google Scholar]
- 7.Huang C. L. (2007) Am. J. Physiol. Renal Physiol. 293, F1761–F1765 [DOI] [PubMed] [Google Scholar]
- 8.Suh B. C., Hille B. (2008) Annu. Rev. Biophys. 37, 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pochynyuk O., Bugaj V., Stockand J. D. (2008) Curr. Opin. Nephrol. Hypertens 17, 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pochynyuk O., Tong Q., Medina J., Vandewalle A., Staruschenko A., Bugaj V., Stockand J. D. (2007) J. Gen. Physiol. 130, 399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon Y., Hofmann T., Montell C. (2007) Mol. Cell 25, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohács T., Lopes C. M., Michailidis I., Logothetis D. E. (2005) Nat. Neurosci. 8, 626–634 [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin S., Murray D. (2005) Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 14.Falkenburger B. H., Jensen J. B., Hille B. (2010) J. Gen. Physiol. 135, 99–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zachos N. C., Tse M., Donowitz M. (2005) Annu. Rev. Physiol. 67, 411–443 [DOI] [PubMed] [Google Scholar]
- 16.Wakabayashi S., Shigekawa M., Pouyssegur J. (1997) Physiol. Rev. 77, 51–74 [DOI] [PubMed] [Google Scholar]
- 17.Aharonovitz O., Zaun H. C., Balla T., York J. D., Orlowski J., Grinstein S. (2000) J. Cell Biol. 150, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuster D., Moe O. W., Hilgemann D. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10482–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins M. O. (2009) Science 325, 1635–1636 [DOI] [PubMed] [Google Scholar]
- 20.Murtazina R., Kovbasnjuk O., Donowitz M., Li X. (2006) J. Biol. Chem. 281, 17845–17855 [DOI] [PubMed] [Google Scholar]
- 21.Cha B., Tse M., Yun C., Kovbasnjuk O., Mohan S., Hubbard A., Arpin M., Donowitz M. (2006) Mol. Biol. Cell 17, 2661–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine S. A., Nath S. K., Yun C. H., Yip J. W., Montrose M., Donowitz M., Tse C. M. (1995) J. Biol. Chem. 270, 13716–13725 [DOI] [PubMed] [Google Scholar]
- 23.Sarker R., Grønborg M., Cha B., Mohan S., Chen Y., Pandey A., Litchfield D., Donowitz M., Li X. (2008) Mol. Biol. Cell 19, 3859–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donowitz M., Mohan S., Zhu C. X., Chen T. E., Lin R., Cha B., Zachos N. C., Murtazina R., Sarker R., Li X. (2009) J. Exp. Biol. 212, 1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landau M., Herz K., Padan E., Ben-Tal N. (2007) J. Biol. Chem. 282, 37854–37863 [DOI] [PubMed] [Google Scholar]
- 26.Cha B., Oh S., Shanmugaratnam J., Donowitz M., Yun C. C. (2003) J. Membr. Biol. 191, 49–58 [DOI] [PubMed] [Google Scholar]
- 27.Gelman M. S., Kopito R. R. (2003) Methods Mol. Biol. 232, 27–37 [DOI] [PubMed] [Google Scholar]
- 28.Akhter S., Cavet M. E., Tse C. M., Donowitz M. (2000) Biochemistry 39, 1990–2000 [DOI] [PubMed] [Google Scholar]
- 29.Atkinson T., Whitfield J., Chakravarthy B. (2009) Biochem. Biophys. Res. Commun. 378, 394–398 [DOI] [PubMed] [Google Scholar]
- 30.Dynia D. W., Steinmetz A. G., Kocinsky H. S. (2010) Am. J. Physiol. Renal Physiol 298, F745–F753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee-Kwon W., Kawano K., Choi J. W., Kim J. H., Donowitz M. (2003) J. Biol. Chem. 278, 16494–16501 [DOI] [PubMed] [Google Scholar]
- 32.Janecki A. J., Janecki M., Akhter S., Donowitz M. (2000) J. Biol. Chem. 275, 8133–8142 [DOI] [PubMed] [Google Scholar]
- 33.Akhter S., Kovbasnjuk O., Li X., Cavet M., Noel J., Arpin M., Hubbard A. L., Donowitz M. (2002) Am. J. Physiol. Cell Physiol 283, C927–C940 [DOI] [PubMed] [Google Scholar]
- 34.Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakabayashi S., Nakamura T. Y., Kobayashi S., Hisamitsu T. (2010) J. Biol. Chem. 285, 26652–26661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.