Abstract
Many organisms use sophisticated systems to acquire growth-limiting iron. Iron limitation is especially apparent in bacterial pathogens of mammalian hosts where free iron concentrations are physiologically negligible. A common strategy is to secrete low molecular weight iron chelators, termed siderophores, and express high affinity receptors for the siderophore-iron complex. Staphylococcus aureus, a widespread pathogen, produces two siderophores, staphyloferrin A (SA) and staphyloferrin B (SB). We have determined the crystal structure of the staphyloferrin B receptor, SirA, at high resolution in both the apo and Fe(III)-SB (FeSB)-bound forms. SirA, a member of the class III binding protein family of metal receptors, has N- and C-terminal domains, each composed of mainly a β-stranded core and α-helical periphery. The domains are bridged by a single α-helix and together form the FeSB binding site. SB coordinates Fe(III) through five oxygen atoms and one nitrogen atom in distorted octahedral geometry. SirA undergoes conformational change upon siderophore binding, largely securing two loops from the C-terminal domain to enclose FeSB with a low nanomolar dissociation constant. The staphyloferrin A receptor, HtsA, homologous to SirA, also encloses its cognate siderophore (FeSA); however, the largest conformational rearrangements involve a different region of the C-terminal domain. FeSB is uniquely situated in the binding pocket of SirA with few of the contacting residues being conserved with those of HtsA interacting with FeSA. Although both SirA and HtsA bind siderophores from the same α-hydroxycarboxylate class, the unique structural features of each receptor provides an explanation for their distinct specificity.
Keywords: ABC Transporter, Bacteria, Iron, Receptor Structure-Function, Transport Metals, X-ray Crystallography, Import, Siderophore, Staphylococcus aureus, Staphyloferrin
Introduction
Staphylococcus aureus is a Gram-positive bacterium that is typically a commensal of the mammalian host. S. aureus is prevalent, colonizing 10–35% of the adult human population persistently and 20–75% intermittently (1). However, it is also an opportunistic pathogen that can cause a wide range of infections from minor skin wounds to more severe invasive infections (2). S. aureus is one of the most frequent causes of hospital-acquired bacterial infection, and the frequency of drug resistance has surged over recent years (3, 4). The resulting decrease in treatment options has highlighted the need to understand fundamental biological processes of S. aureus and develop new anti-infective therapeutics (3).
Iron is a limiting nutrient to most organisms, especially bacterial pathogens. In the human body, free iron levels are extremely low (5, 6), and to overcome this growth limitation, S. aureus acquires iron by sequestering it from numerous host sources (7–9). Mammalian iron is primarily found intracellularly in heme and heme proteins, ferritins, or a labile iron compartment. Extracellular iron is predominantly found in protein carriers, such as transferrin and lactoferrin (6). S. aureus encodes several iron uptake systems to use most host iron sources for growth. For example, the Isd uptake system removes heme from hemoglobin and transfers it through the cell wall to a membrane transporter for uptake into the cell followed by degradation (7, 10, 11).
Another prominent microbial iron acquisition strategy is to use siderophores, low molecular weight, high affinity iron chelators that are secreted to scavenge iron (12). S. aureus produces at least two siderophores, staphyloferrin A (SA)4 and staphyloferrin B (SB), both belonging to the hydroxycarboxylate siderophore class (13–16). In addition, S. aureus can use exogenously produced hydroxamate type siderophores through the ferric hydroxamate (Fhu) uptake system (17–19). The enzymatic pathways for SA and SB synthesis have been identified and shown to be encoded by the sfa and sbn operons, respectively (20–23). The sfa operon is present in all sequenced staphylococcal genomes, whereas the sbn operon is unique to S. aureus strains, although the original characterization of SB was from Staphylococcus hyicus culture supernatants (13, 14, 21). The only other documented occurrence of the SB biosynthetic operons in sequenced genomes are in the Gram-negative plant pathogen Ralstonia solanacearum, which was shown to synthesize SB, and the soil bacterium Cupriavidus metallidurans (23, 24). The reception and import systems for Fe(III)-SA (FeSA) and Fe(III)-SB (FeSB) are composed of a lipoprotein surface receptor (HtsA for FeSA and SirA for FeSB) and the heterodimeric permeases (HtsBC and SirBC) (20, 25). In concert with the multifunctional ATPase, FhuC, these systems drive the energy-dependent import of ferric loaded siderophore (26).
HtsA binds FeSA within a electropositive pocket, followed by conformational change in three C-terminal loops to enclose FeSA with a dissociation constant in the low nanomolar range (27). The converse strategy is employed in the hydroxamate uptake system, where affinity (mid to high nanomolar dissociation constants) is sacrificed for broader specificity (28, 29). Because S. aureus does not produce hydroxamate siderophores, this import pathway is solely parasitic and probably provides additional growth advantage to S. aureus in complex communities. SB is from the same siderophore class as SA, but the respective transporters are highly specific (20), although the mechanism of specificity has been unclear.
To understand the molecular basis of SB specific recognition by SirA, we have determined the x-ray crystal structures of the SB receptor in its apo and FeSB-bound forms to 2.2 and 1.7 Å resolution, respectively.
EXPERIMENTAL PROCEDURES
Cloning and Protein Expression
Analysis of the SirA primary structure reveals a predicted N-terminal secretion signal with a Cys lipidation target at position 21. The SirA primary structure was analyzed for regions of predicted disorder using the DISOPRED2 disorder prediction server (30, 31). The two constructs used in this study consist of residues 37–330 (SirAT37) and 54–330 (SirAK54). The coding regions for both constructs were cloned into pET28a to incorporate a thrombin-cleavable His6 tag at the N terminus. Expression was from Escherichia coli BL21(DE3) inoculated into 2× YT medium and grown at 30 °C to an optical density of ∼0.8 at 600 nm. Protein expression was induced with the addition of 0.3 mm isopropyl β-d-thiogalactosidase and continued incubation at 25 °C. Cells were pelleted by centrifugation, resuspended in 50 mm Tris, pH 8.0, 100 mm NaCl, and lysed using an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, Canada). Insoluble cell debris was removed by centrifugation followed by filtration through an 0.8-μm syringe filter. His-tagged-SirA was purified using a Ni2+-loaded HisTrap HP Column (GE Healthcare). Purified protein was dialyzed into 40 mm Hepes, pH 7.8, and digested with thrombin (1:300, SirA/thrombin, w/w) overnight at 4 °C. SirA was separated using Source 15S resin with 40 mm Hepes, pH 7.8, and a linear salt gradient to 500 mm NaCl. Pure SirA (>98%, as determined by SDS-PAGE) was dialyzed into 20 mm Tris, pH 8.0, and concentrated to ∼20–25 mg/ml for crystallization. Selenomethionine-labeled SirAK54 was expressed as previously described (32) and purified as described for unlabeled SirA.
Recombinant SbnC, SbnE, SbnF (SB synthetases), and SbnH (decarboxylase) were required to produce staphyloferrin B in vitro. Proteins were cloned, overexpressed, and purified as described (21). Overexpression and purification of recombinant HtsA as well as the in vitro synthesis of staphyloferrin A have been described (27).
Staphyloferrin B Enrichment from Culture Supernatant
Staphyloferrin B was enriched from concentrated S. aureus culture supernatants as described previously (25). For co-crystallization, concentrated culture supernatants containing SB were incubated with 5 mm FeCl3 and added in ∼2-fold molar excess to SirA. The solution was incubated at room temperature for 30 min prior to buffer exchange and concentration.
Staphyloferrin B Synthesis and Purification
Using recombinant SbnC, SbnE, SbnF, and SbnH, staphyloferrin B biosynthesis reactions were set up as described previously (21) with the exception of preincubating SbnH with a 5-fold molar excess of pyridoxal phosphate followed by a 3× wash of excess cofactor from the enzyme using an Amicon® Ultra-0.5 10k filter column (Millipore Corp.) spinning at 14,000 × g for 5 min. Enzymes, substrates, and remaining cofactors were incubated for 16 h. The staphyloferrin B reaction mixture was centrifuged in an Amicon® Ultra-0.5 10k filter column (Millipore) at 14,000 × g for 15 min to remove enzymes. The filtrate was then supplemented with 3 mm FeCl3 and centrifuged at 18,000 × g to remove any precipitate. Fifty μl of the solution was then injected onto a Waters xTerra C18 reversed-phase 5-μm column (150 × 2.1 mm) fitted onto a Beckman System Gold HPLC equipped with a photodiode array detector. Samples were run at 0.2 ml/min using a step gradient as described previously (22). Solvent A was 10 mm tetrabutylammonium phosphate, pH 7.3, in HPLC grade water (Fisher), and solvent B was 100% acetonitrile (Fisher), and peaks were monitored at 340 nm. The peak corresponding to FeSB eluted at 4–6 min and was collected. Collected fractions were then vacuum-centrifuged until dry and resuspended in deionized water. The siderophore resuspension was then analyzed by Q-TOF mass spectrometry as described (21) to confirm the presence of SB. The presence of EDTA in samples analyzed by Q-TOF allowed the detection of iron-free staphyloferrin B but not the iron-loaded form. Samples run in the absence of EDTA allowed detection of a mixture of the iron-free and iron-loaded forms of the siderophore.
Other Siderophores
Staphyloferrin A was synthesized and purified as described previously (27), and desferrioxamine B (used as DesferalTM) was purchased from the London Health Sciences Centre (London, Canada).
Determination of Ferric Staphyloferrin B Concentration
Atomic absorption spectrometry was used to determine the concentration of iron in HPLC-purified FeSB samples. Therefore, the concentration of iron was used to determine the concentration of staphyloferrin B after assuming a 1:1 molar ratio of iron to siderophore in an FeSB complex. Samples were diluted in 1 m nitric acid before being drawn by an SPS five-sample preparation system (autosampler) into a Varian AA240 atomic absorption spectrometer. Absorbance was detected by an iron/manganese hallow cathode lamp, which emits at 248.3 nm specific for iron detection. Absorbance data were analyzed and compared with a linear calibration curve based on known iron standards in ppm. Iron standards were diluted in 1 m nitric acid from an atomic absorption spectrometer certified 1000 ppm ± 1% stock (Fisher). Calibration standards were separately analyzed first before FeSB samples.
Fluorescence Spectroscopy
Fluorescence titration experiments were performed at room temperature using recombinant SirA (15 nm) buffered in 50 mm HEPES, pH 7.4, with a Fluorolog-3 spectrofluorometer (ISA Instruments). The excitation and emission slits were set at 2.1 and 6.3 nm, respectively. The excitation and emission wavelengths were set at 280 and 340 nm, respectively. Dissociation constants (Kd) and relevant parameters were calculated by fitting the fluorescence titration data for FeSB (across a concentration range between 0.1 and 112 nm ligand) to a one-site binding model accounting for ligand depletion. Data were analyzed by nonlinear regression analysis as described previously (28, 29).
Siderophore Plate Bioassays
Siderophore plate bioassays involving S. aureus sirA and htsABC mutants were performed as described (20). Ten-μl aliquots of HPLC-purified FeSB were spotted onto sterile paper discs before being placed onto Tris-minimal succinate agar seeded with S. aureus transporter mutants. Growth promotion, as measured by the diameter of the growth halo around each disk, was determined after a 36-h incubation at 37 °C.
SirA Crystallization
Diffraction quality crystals of SirAT37 were only obtained in the presence of FeSB. These crystals grew in 1:1 mixtures of 25 mg/ml protein and 0.1 m Hepes, pH 7.0, 30% Jeffamine ED-2001. Crystals were flash frozen in the same buffer, supplemented with 32% Jeffamine ED-2001 and 15% glycerol. Selenomethionine-labeled apo-SirAK54 crystals were grown in a 1:1 mixture of 20 mg/ml protein solution and 0.1 m BisTris, pH 6.5, 24% polyethylene glycol 3350. Apo-SirAK54 crystals were frozen in cryoprotectant consisting of mother liquor supplemented to 0.1 m BisTris, pH 6.5, 26% polyethylene glycol 3350, 20% ethylene glycol.
SirA Structure Solution and Analysis
Selenomethionine apo-SirAK54 diffraction data were collected at the Stanford Synchrotron Radiation Lightsource on beamline 7-1. Two-wavelength multi-wavelength anomalous dispersion data were collected at 0.97882 and 0.97947 Å and were processed with HKL2000 (33). The crystals were in the space group P21 with two molecules (four selenium atoms) in the asymmetric unit. Four selenium peaks were identified using the program Solve (34) with an overall figure of merit phasing of 0.59. The program Resolve (35, 36) built 371 of the 552 expected residues as Ala, and successive rounds of ArpWarp (37) assigned 274 residues to the sequence. At this stage, the regions built were primarily from chain A and the N-terminal domain of chain B. The remainder of the structure was built manually in the program Coot (38) and refined with Refmac5 (39). The two molecules in the apo-SirAK54 asymmetric unit differ significantly. The B-factors of the C-terminal domain of Chain B are elevated, and the two chains overlay with an r.m.s. deviation of ∼0.62 Å. Two loops in chain B of apo-SirA were poorly defined (B-factors of 60–90 Å2), and several residues could not be refined with good geometry. For this reason, chain A was used for phase solution and structural comparison with holo-SirA. TLS refinement parameters (40, 41) were included in Refmac5 to account for the added flexibility in chain B, lowering Rwork to 0.185 from 0.199 and Rfree to 0.232 from 0.251.
Holo-SirAT37 data were collected at the Stanford Synchrotron Radiation Lightsource on beamline 9-2 at 0.97946 Å wavelength and processed using HKL2000 (33) to 1.7 Å resolution. Holo-SirAT37 crystallized in the space group P212121 with one molecule in the asymmetric unit, and phases were determined using MolRep (42). The model was edited in Coot (38) and refined with Refmac5 (39). Coordinates and library files for SB were generated in the program Sketcher from the CCP4 program suite (43). The FeSB iron peak was identified as a large peak in the difference map and the remainder of FeSB was clearly visible in the density prior to the addition of water. FeSB at full occupancy modeled into the holo-SirAT37 and refined to an average B-factor of 18.0 Å2. Data collection and refinement statistics are shown in Table 1. Figures were generated with PyMOL (44).
TABLE 1.
Data collection and refinement statistics
| Selenomethionine apo-SirAK54 | FeSB-SirAT37 | |
|---|---|---|
| Data collectiona | ||
| Resolution range (Å) | 40–2.1 (2.08–2.10) | 50–1.7 (1.76–1.70) |
| Space group | P21 | P212121 |
| Unit cell dimension (Å) | a = 42.1, b = 61.5, c = 114.6, β = 100.1° | a = 57.7, b = 71.6, c = 72.6 |
| Unique reflections | 34,042 | 33,855 |
| Completeness (%) | 99.7 (100.0) | 96.6 (89.6) |
| Redundancy | 4.9 (4.9) | 6.8 (6.6) |
| Average I/σI | 19.5 (3.7) | 39.7 (5.8) |
| Rmerge | 0.066 (0.406) | 0.044 (0.322) |
| Wilson B (Å2) | 27.9 | 19.4 |
| Refinement | ||
| Rwork (Rfree) | 0.192 (0.236) | 0.182 (0.206) |
| No. of atoms (average B (Å2)) | ||
| Protein | 4351 (40.4) | 2372 (19.8) |
| Water | 246 (37.5) | 188 (26.7) |
| Staphyloferrin B | 31 (18.0) | |
| Iron | 1 (12.7) | |
| r.m.s. deviation bond length | 0.013 | 0.013 |
| Ramachandran plot (%) | ||
| In most favorable | 91.6 | 91.3 |
| In disallowed | 0.0 | 0.0 |
a Data collection values in parentheses represent data for the highest resolution shell.
Sequence Alignment of HtsA and SirA
The two S. aureus α-hydroxycarboxylate type siderophore receptors, SirA (FeSB-SirAT37) and HtsA (PDB entry 3LI2, FeSA-bound, closed structure (27)), were structurally aligned using the combinatorial extension method (45) in the alignment program Strap (46). The alignment was exported from Strap and used as a profile to generate full-length alignments with ClustalX (47) using sequence accession numbers BAB41330 (SirA) and NP_375290 (HtsA) from S. aureus strain N315. The resulting full-length alignment was viewed and rendered in Jalview (48).
RESULTS
S. aureus Imports FeSB via the SirABC Transporter
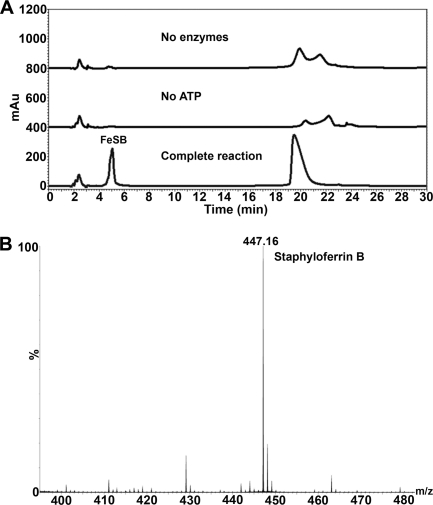
Our previous studies demonstrated that staphyloferrin B, synthesized by enzymes encoded by the sbn operon in S. aureus, is transported through the SirABC transport system (21). In that study, complete SB reaction mixtures were able to promote growth of S. aureus ΔhtsABC mutants but not sirA mutants, as demonstrated using siderophore plate bioassays. Equivalent bioassay results were obtained using HPLC purified FeSB (fractions collected at the 4–6 min window of the HPLC chromatogram, Fig. 1A). No other HPLC fractions promoted growth of S. aureus (data not shown). Q-TOF mass spectrometry on the bioactive fractions confirmed the presence of SB with mass ion [M-H]− = 447.1 (Fig. 1B). In contrast, reaction mixtures containing no enzymes or ATP did not produce the FeSB peak on HPLC; nor did they promote the growth of S. aureus (data not shown). The observation that FeSB was only able to promote the growth of S. aureus ΔhtsABC and not strains harboring a sirA mutation highlights the specificity and requirement of the SirABC transport system for the recognition and import of the FeSB complex.
FIGURE 1.
Synthesis and purification of in vitro synthesized staphyloferrin B. A, HPLC traces of in vitro SB synthesis, demonstrating that enzymes and ATP are required for SB synthesis. B, Q-TOF mass spectrometry identified the main constituent of in vitro purification as having a mass ion [M-H]− = 447.1 as expected for SB. mAu, milliabsorbance units.
FeSB-SirA Binding
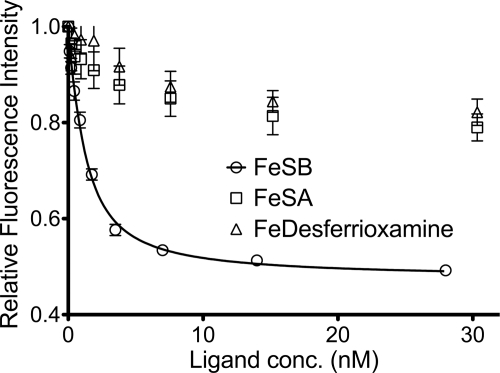
SirA is a class III substrate binding protein that is N-terminally lipidated and serves as the receptor for ferric staphyloferrin B. Fluorescence spectroscopy was used to measure the affinity of this receptor protein for its siderophore ligand. Titrations of HPLC-purified FeSB with recombinant SirA revealed a dissociation constant (Kd) that is in the low nanomolar range (Fig. 2). Due to a limitation in the fluorescence intensity, we are unable to report an accurate Kd value other than an upper limit. The affinity observed for SirA and SB resembles the affinity observed for HtsA and SA (27). The specificity of SirA for FeSB was further demonstrated by using FeSA (another α-hydroxycarboxylate siderophore) and Fe-desferrioxamine (an unrelated hydroxamate siderophore). Using either of the latter iron-siderophore complexes, some fluorescence quenching was observed but never reached the level of quenching observed for the FeSB complex (Fig. 2), suggestive of nonspecific quenching or photobleaching of the protein.
FIGURE 2.
Fluorescence quenching of SirA upon ligand binding. Fe(III)-loaded staphyloferrin B, staphyloferrin A, and desferrioxamine were titrated into SirA, and quenching of fluorescence was monitored. Titrations were preformed with concentrations of Fe(III)-siderophores between 0 and 120 nm, but only data from 0–30 nm are shown.
SirA Overall Fold
Primary structure analysis of SirA revealed an N-terminal secretion signal with a lipidation site at Cys21. Protein disorder predictions suggested that residues 22–36 are disordered to form a flexible linker following the membrane anchor site. Expressed recombinant SirA constructs were truncated to at least residue Thr37 to exclude the predicted linker. Crystal structures were determined from two different constructs. The longer SirAT37 construct did not yield apo crystals but would only crystallize in the presence of FeSB. Good quality apo crystals were only attained with the shorter SirAK54 construct.
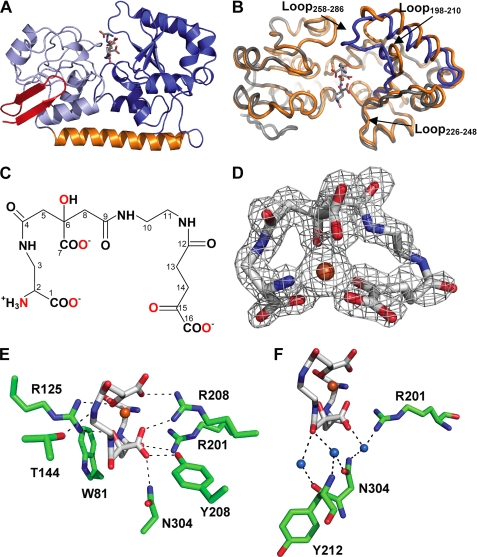
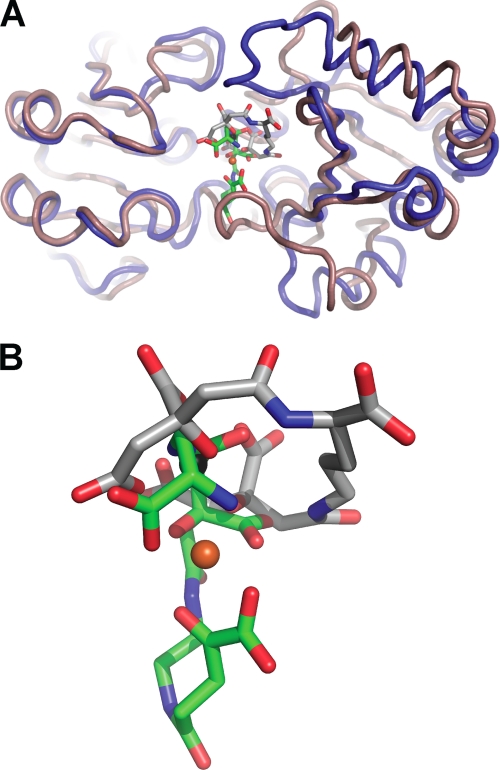
The SirA structures are of the class III binding protein fold (Fig. 3A). Compositionally similar N- (residues 37–161) and C-terminal (residues 188–329) domains contain a central parallel β-sheet surrounded by short α-helical segments. The domains are bridged by a single α-helix (residues 162–187) that runs the width of the protein. The region of SirA excluded in the SirAK54 construct corresponds to two β-strands that pack against the side of the N-terminal domain (Fig. 3A). This region is variably present in other class III binding protein structures. In S. aureus proteins, the β-strands are present in HtsA (20) but absent in the heme-binding protein IsdE (49). The excluded region encodes several hydrophilic residues that may have contributed additional flexibility to the lipid-anchored linker. The N-terminal β-strands in the FeSB-SirAT37 structure form a crystal contact that may stabilize the observed conformation. In concert with conformational changes associated with FeSB binding, the strand stabilization could enable crystallization of the holoprotein but not the apoprotein. Interestingly, the apo-SirAK54 molecules pack such that the two molecules in the asymmetric unit contact each other across the same region that would be occupied by the β-strands (not shown).
FIGURE 3.
Structure SirA-SB. A, overall structure of SirA-SB. The backbone is shown as a schematic diagram with the N-terminal domain, α-helix-bridging, and C-terminal domains shown in light blue, orange, and blue, respectively. The N-terminal β-strand extension (residues 37–53) present in the Sir-SB structure only is highlighted in red. B, structural changes upon SB binding in the open (orange) and closed (blue) SirA. Backbones are shown as threads. C, molecular structure of SB. Carbon atoms are numbered according to Drechsel et al. (13). Fe(III)-coordinating atoms are highlighted in red. D, the crystal structure of SB shown as sticks with carbon, nitrogen, oxygen, and iron in gray, blue, red, and orange. Omit difference electron density is shown as a gray mesh contoured at 3.5 σ. E, polar SirA-FeSB contacts are shown as sticks as in D, with protein carbons in green. Dashes represent H-bonds. F, SB waters and the extended water-bridged interactions. Representations and coloring are the same as for E.
There are several generously allowed residue conformations in the apo-SirAK54 structure. Asn304 is located in a highly constrained turn within the SB binding pocket with similar main chain torsional angles in both the apo and FeSB-bound structures. The other residues occur in the C-terminal domain of chain B (average B-factor of 80.7 Å2) of the apo structure, which is less well ordered than in chain A (average B-factor of 31.8 Å2). These residues are Lys188, Asp189, Ala209, Gly210, Gly211, and Asp261). Due to the disorder in some regions of chain B, chain A was used for all comparative analysis with FeSB-SirA.
Staphyloferrin B Structure
Staphyloferrin B is synthesized from l-2,3-diaminopropionic acid, citric acid, 1,2-diaminoethane, and α-ketoglutarate. The chemical structure (Fig. 3C) and biosynthetic pathways have been determined (13, 14, 21); however, the conformation of FeSB was previously unknown. Electron density for FeSB was obvious upon inspection of the SirA binding pocket. The iron peak was identified in Fo − Fc difference maps as the most prominent peak. The groups coordinating the iron and surrounding atoms are clearly defined at 1.7 Å resolution (Fig. 3D). FeSB orientation was further supported by the tetrahedral l-configuration of the l-2,3-diaminopropionate group at C2 (see numbering in Fig. 3C), which was clearly distinguishable from the planar configuration of the α-ketoglutarate group at C15 on the opposite SB terminus (Fig. 3C). FeSB is six-coordinate, with five oxygen atoms and one nitrogen atom of SB in a distorted octahedral geometry with angles ranging from 74–113° and bond distances from 2.0 to 2.2 Å. Two of the metal ligands are from the terminal carboxylate (C1) and the amino substituent of C2 derived from l-2,3-diaminopropionate, two are from the terminal carboxylate (C16) and carbonyl groups (C15) of the α-ketoglutarate-derived component, and two are from the hydroxyl substituent of C6 and carboxylate (C7) derived from citrate (Fig. 3D). FeSB is oriented in the binding pocket such that the citrate portion (C4–C9) of the siderophore is the most buried. An extended length (9 atoms) of backbone spanning the citrate, 1,2-diaminoethane and α-ketoglutarate components (C8–C14) link the functional groups that coordinate the iron. This linker makes few protein contacts, and the corresponding electron density is not well defined (Fig. 3D), resulting in slightly elevated B-factors of ∼25–40 Å2 compared with an average of 18.2 Å2 for the entire FeSB molecule. Electron density for FeSB from co-crystals of SirA-FeSB using in vitro synthesized SB closely resembled the FeSB from culture supernatants, but due to lower resolution diffraction, the data are not included.
Conformational Change upon FeSB Binding
A variable amount of interdomain hinge motion upon ligand binding is observed in members of the class III binding protein family (50–52). Our recent structure of the FeSA-HtsA ternary complex revealed isolated conformational change to specific loops rather than large interdomain hinged motions (27). Comparison of the apo and FeSB-SirA structures reveals localized structural reorganization while maintaining the same rigid interdomain orientation (hinged motion of <2°). Structural alignment of the apo- and holo-SirA reveals an r.m.s. deviation of ∼1.09 Å2 over all Cα (Fig. 3B). However, the N terminus of SirA is largely unaltered by ligand binding (residues 54–160 Cα atoms overlay with r.m.s. deviation of ∼0.51 Å2). The majority of the intradomain conformational change is isolated to loops in the C-terminal domain (residues 190–329 Cα atoms overlay with r.m.s. deviation of ∼1.59 Å2) (Fig. 3B). The conformational differences are maximal in two loops, which refold into the binding groove of the ligand (Fig. 3B). The region containing residues 258–286 (Loop258–286) has a maximal translation of 11.3 Å between Asn263 Cα in the apo and holo structures. The other loop comprises residues 198–210 (Loop198–210), with a maximal displacement of Ala202 Cα of ∼2.1 Å. Loop198–210 forms part of the FeSB binding site, and the conformational change establishes several direct FeSB-SirA contacts. A third loop composed of residues 226–248 (Loop226–248) undergoes a modest shift (<1.6 Å) toward the pocket following FeSB binding. The conformational changes occlude FeSB such that only 23.6% of the surface area of the siderophore is exposed to bulk solvent (as calculated in AREAIMOL (43). The percentage of exposure is intermediate to FeSA exposure in the open and closed forms of FeSA-HtsA (∼33 and 14.5%, respectively) (27).
SirA Recognition of Staphyloferrin B
Fe(III) binding to SB neutralizes its net −3 charge, and remaining electronegative regions of SB are neutralized by three Arg residues in the SirA binding site (Fig. 3E). One Arg (Arg125) supplied by the N-terminal domain forms two salt bridges, one from each NH group, to the iron-coordinating carbonyl of the α-ketoglutarate component (C15) and the coordinating carbonyl (C9) from the citrate group (see Table 2 for a list of FeSB–SirA contact distances). Arg125 is stabilized by water-mediated H-bonds to Trp81 Nϵ and Glu103 Oϵ2. Thr144 forms an H-bond to the amide linkage between C11 and C12. Two Arg residues within the C-terminal domain form interactions with FeSB. Arg201 Nη forms an H-bond with the carbonyl oxygen linking the l-2,3-diaminopropionic acid and citrate groups. Arg201 is in turn stabilized by H-bonds through its Nη and Nϵ atoms to Tyr208 Oϵ and Asn199 Oδ, respectively. The side chain of Arg206 forms salt bridges to the carboxylate groups of the citrate and α-ketoglutarate components. Arg206 lies across the iron site opposite to Arg125 and is part of a network of H-bonds with His204 Nϵ, Asn240 Oϵ, and Tyr208 OH, respectively. Tyr208 OH also forms a direct H-bond to the citrate carboxylate group (C7). The final polar FeSB-SirA contact is between Asn304 Nδ and the carboxylate of the citrate group (C7).
TABLE 2.
FeSB–SirA hydrogen bonds
| SirA atom–FeSB atom | Distance |
|---|---|
| Å | |
| Trp81 π–C2a amide N | 3.3 |
| Arg125 NH1–C6 hydroxyl | 2.8 |
| Arg125 NH2–C15 carbonyl | 3.0 |
| Thr144 Oγ–C9-C10 amide N | 3.2 |
| Arg201 NH–C3-C4 carbonyl | 3.0 |
| Arg206 NH1–C7 carboxyl | 2.8 |
| Arg206 NH2–C16 carboxyl | 3.0 |
| Tyr208 OΗ–C7 carboxyl | 2.6 |
| Asn304 Nδ–C6 carbonyl | 2.8 |
| Water–C9 carbonyl | 2.9 |
| Water–C9 carbonyl | 2.9 |
| Water–C4 carbonyl | 3.0 |
a Components of SB numbered according to carbon atoms in Fig. 3C.
Trp81 is located near the base of FeSB, close to the loop that undergoes the largest conformational change (Fig. 3E). The indole ring is ∼3.3 Å from amide nitrogen that links the l-2,3-diaminopropionic acid and citrate groups (C3-C4), such that the Trp81 π system forms an amide-π interaction. Of three buried water molecules in the FeSB pocket, two are at the base of the pocket and form H-bonds to the carbonyl oxygen (C9) between citrate and 1,2-diaminoethane (Fig. 3F). The first water in turn forms H-bonds with the Asn304 Oδ and Tyr212 backbone nitrogen. The second water forms an H-bond with the Asn304 main chain oxygen. A third water beneath the siderophore in the pocket is within H-bonding distance to the carbonyl oxygen (C4) between l-2,3-diaminopropionic acid and citrate and mediates interactions with residues Arg201 and Asn304 (Fig. 3F). The remainder of water molecules modeled into the structure that interact with FeSB are located in solvent-exposed surface regions.
Comparison with FeSA-HtsA Complex
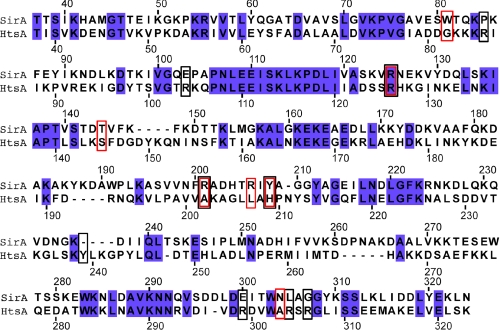
A search of the Dali server (53) with SirA as the query revealed the staphyloferrin A transporter HtsA (PDB entries 3LI2, 3LHS, 3EIW, and 3EIX) as the top hit (z score = 25.5, r.m.s. deviation = 2.4, 32% sequence identity over 250 residues) (20, 27). Despite sharing ∼32% sequence identity and the highest structural similarity, few residues implicated in siderophore binding are conserved between SirA and HtsA (Fig. 4). The next most similar structures in the data base (z scores = 21–23.5, r.m.s. deviation = 2.6–3.3, 22–29% sequence identity over 243–257 residues) are E. coli FitE (PDB entries 3BE5 and 3BE6) (52), Bacillus subtilis BSU3320 (PDB entry 3G9Q), B. subtilis FeuA (PDB entries 2WI8 and 2PHZ) (51), and S. aureus IsdE (PDB entries 2Q8P and 2Q8Q) (49).
FIGURE 4.
Structure-based sequence alignment of SirA and HtsA (PDB entry 3LI2). Numbering is according to SirA (Entrez ID SA0111) and HtsA (Entrez ID SAV2177) sequences. Red and black boxes indicate SirA and HtsA residues forming direct contacts with FeSB and FeSA, respectively. Blue-filled boxes highlight sequence identities.
Both SirA and HtsA bind α-hxdroxycarboxylate type siderophores that possess a net negative charge. Without iron bound, SA carries a −5 net charge, whereas SB has a −3 net charge. Because both siderophores are synthesized and utilized by S. aureus, we wanted to understand the differences in siderophore iron binding and receptor recognition that allow discrimination by the SirA and HtsA receptors. Aside from the common citrate constituent in both siderophores, Fe(III) coordination in SA and SB is distinct. In FeSA, all six Fe(III)-coordinating oxygen atoms are located at the termini of SA derived from two citrate molecules. The SA backbone wraps around Fe(III) as a hand holding iron with the palm edge in the HtsA binding pocket (Fig. 5) (27). In contrast, the iron coordinating groups in FeSB are located throughout the molecule. Four of the coordinating atoms are localized in pairs at each siderophore terminus: an α-hydroxy carboxylate and one α-amino carboxylate group. The remaining two atoms are from the midchain citrate α-hydroxycarboxylate, such that SB binds Fe(III) with the palm facing up and outward from the SirA binding pocket (Fig. 5). Most polar siderophore-protein interactions in SirA and HtsA are formed directly with the Fe(III)-coordinating functional groups. The lateral placement of siderophore in the binding pocket also differs between HtsA and SirA.
FIGURE 5.
Comparison of SirA and HtsA structures. A, SirA-SB (blue) and HtsA-SA (tan) overlay with an r.m.s. deviation of ∼2.4 Å. B, a close-up view of SB (green) and SA (gray) in the binding pocket.
In a FeSA-HtsA and FeSB-SirA overlay, the iron atoms are ∼3 Å apart, and the siderophores are related by an ∼90° rotation such that the long axis is either perpendicular (FeHtsA) or parallel (FeSB) to the plane of the protein domain interface (Fig. 5A). The terminal carboxylates from the citrate (FeSA) and l-2,3-diamonipropionate (FeSB) groups are the only chemically similar portions derived from the two siderophores that are bound in similar positions. Both are anchored by an H-bond to the conserved Arg125 (Fig. 5). FeSB makes extensive contacts with residues in Loop198–210, none of which are conserved with FeSA-HtsA contacts (Fig. 4). For example, Tyr208 at the base of the SirA loop forms an H-bond with the FeSB citrate carboxylate group, whereas the FeSA backbone carbonyl oxygen located ∼2.8 Å away forms an H-bond with His209 in HtsA. Alternatively, a unique FeSA-HtsA interface is formed with three Arg residues located at the base of the HtsA binding pocket (Lys299, Lys304, and Lys306), whereas the similar region in FeSB-SirA forms only a single H-bond through Asn304 (Fig. 4). In addition, the higher net negative charge of SA is reflected in a higher number of neutralizing positive charges in HtsA. Five or six Arg residues interact with FeSA in the HtsA-FeSA structure in the open and closed forms (27). In contrast, FeSB forms only three direct contacts with positively charged arginine residues. Instead, the contacts are made with a higher proportion of polar coordinating residues. Thus, the overall structural features of both receptors are similar, but the three main C-terminal loops involved in siderophore binding make unique interactions with the cognate siderophores.
DISCUSSION
High affinity receptors are essential for acquiring limiting nutrients, and it has become apparent that the class III binding proteins recognize their ligands by diverse means. The endogenously synthesized S. aureus siderophores are of the negatively charged α-hydroxycarboxylate type (9). Given the similarities between SA and SB and the prevalence of broad specificity hydroxamate transporters, the utility and mechanism of specialized uptake systems for chemically similar siderophores was unclear (17, 18). Comparison of the FeSB-SirA and FeSA-HtsA structures explains specificity of the receptors because the siderophore-protein contacts are almost entirely different, and siderophore binding induces distinct conformational changes. The conformational changes in each receptor probably maximize both the affinity and specificity by expanding the contact area between the receptor and siderophore. These high affinity, high specificity uptake systems may enable S. aureus to outcompete other organisms attempting to parasitize the SA or SB systems. This specificity is not seen in the S. aureus ferric hydroxamate receptor (FhuD), which binds a broad range of exogenously synthesized ferric hydroxamate complexes, suggesting that this uptake system would have utility in environments with complex microbial communities (17, 18, 28, 29). The combination of iron acquisition systems reflects the broad range of environments S. aureus encounters and its success as a pathogen.
The SirA-FeSB crystal structure reveals that SB coordinates Fe(III) through five oxygen atoms and a single nitrogen atom in strained octahedral geometry. Although siderophores generally use oxygen atoms to coordinate Fe(III), several examples of nitrogen and sulfur ligands exist (for a review of siderophore chemistry, see Ref. 54). The Pseudomonas aeruginosa siderophore, pyochelin, coordinates iron by nitrogen atoms from the thiazolin and thiazolidin rings as well as by carboxylate oxygen atoms (55, 56). Other examples are mycobactins and exochelins, both mixed siderophores that satisfy octahedral coordination geometry with five oxygen atoms from hydroxamate groups and a nitrogen atom from β-hydroxyhistidine (57, 58). Primary amines function as iron ligands in several heme proteins, such as Lys coordination in cytochrome c nitrite reductase (59). However, to our knowledge, this is the first crystal structure of a siderophore using a primary amine as an Fe(III) ligand.
The high affinities of S. aureus siderophore receptors compared with their Gram-negative homologs probably reflect their location in the process of ligand transport. The highly specific HtsA and SirA receptors bind ligands with low nanomolar affinities, and the FhuD1 and FhuD2 receptors have dissociation constants reported in the midnanomolar range (27–29). Alternatively, the Gram-negative counterparts, such as E. coli FhuD, bind hydroxamates with high nanomolar to low micromolar dissociation constants (60). E. coli FhuD was recently shown to interact with TonB, suggesting that it is localized near the outer membrane receptor FhuA, thereby removing the need for high affinity binding (61). Instead, the binding affinities of Gram-positive lipoproteins for siderophores are more in line with the affinities measured for outer membrane siderophore receptors of Gram-negative uptake systems (62), ultimately resulting in systems with the same affinity for ligands at the cell surface.
Conformational changes in class III binding proteins are variable. Siderophore receptors, such as E. coli FitE and B. subtilis FeuA, undergo large hinged motions that probably allow them to clamp around their ligands (51, 52). E. coli FhuD displays modest hinged motions (50, 63, 64); also, small angle x-ray scattering data for S. aureus FhuD1 and FhuD2 suggest that they undergo only modest interdomain conformation change upon hydroxamate binding (28, 29). Crystal structures of E. coli BtuF, Shigella dysenteriae ShuT, and P. aeruginosa PhuT do not differ greatly between apo and holo forms (65–67); however, molecular dynamics simulations of ShuT, PhuT, and BtuF suggest that both hinged movements and twisting of the domains along the axis defined by the domain-bridging α-helix occur to a greater extent than observed in the crystal structures (68–70). Despite the focus on hinged ligand binding, other conformational changes can be associated with ligand binding.
Comparison of HtsA and SirA in the holo and apo crystal forms reveals little hinge motion between domains, but rather localized conformational changes of ∼12 Å are observed in loops that form part of the ligand binding site. However, the loop undergoing the largest change in HtsA is only modestly shifted in SirA. Instead, the large siderophore-encasing movement in SirA occurs in Loop258–286, located on the opposite end of the interdomain interface of the siderophore-binding pocket. The analogous region of HtsA shifts away from FeSA upon binding to accommodate other conformational changes (Fig. 5A). The medial binding loop in both structures undergo similar shifts into the pocket (Loop198–210 in SirA); however, in HtsA, the shift is accompanied by one H-bond formed from the main chain nitrogen of an Ala in the loop to SA. In SirA, the analogous loop shifts to allow three FeSB-SirA H-bonds to form, primarily to the regions immediately adjacent to the ferric binding site. The structures demonstrate that both receptors use local conformational changes to bind, but the loops involved are distinct.
Ligand binding drives conformational changes in substrate-binding protein receptors, which in turn control receptor-permease interactions. The crystal structures of BtuCD-F reveal that, in the substrate-binding protein, two conserved Glu residues interact with corresponding Arg-rich patches in the transmembrane permease (71). Further, site-directed mutagenesis studies have identified analogous Glu-Arg interactions for the E. coli Fec system and the S. aureus Fhu system, both in vitro and in vivo (28, 72). SirA possesses conserved glutamates (Glu109 and Glu245) that overlay with the permease-interacting Glu residues in BtuF when these receptors are superimposed (not shown). Through a combination of localized loop shifts and the slight hinged motion in SirA, the distance between Glu109 and Glu245 Cα atoms is decreased by ∼1.9 Å to ∼46.1 Å. A similar distance is observed in several other substrate-binding protein receptors, including the permease-docked BtuF structure (Table 3) (65, 67). This conservation of interglutamate distance in both FeSB-SirA and FeSA-HtsA suggests that the subtle Glu shifts in these receptors enable the permeases to discriminate between ligand-free and ligand-bound protein. Interestingly, the conserved interglutamate distances suggest that this spacing may be the prime determinant of permease docking whether the conformational change is a result of hinged motion or localized refolding of loops.
TABLE 3.
Distance between permease-binding conserved glutamate residues (Cα-Cα) substrate-binding proteins in ligand-free and ligand-bound forms
| Protein (organism) | Apo structure (PDB code) | Substrate-bound (PDB code) |
|---|---|---|
| Å | Å | |
| SirA (S. aureus) | 48.0 (3mwg) | 46.1 (3mwf) |
| HtsA (S. aureus) | 48.0 (3eiw) | 45.6 (3li2) |
| BtuF (E. coli) | 46.2 (1n4d) | 46.2 (1n4a) |
| FhuD (E. coli) | 45.5 (1efd) | |
| FeuA (B. subtilis) | 48.5 (2wi8) | 44.4 (2why) |
In summary, we have defined FeSB recognition by the S. aureus receptor, SirA. Binding is accompanied by structural changes isolated primarily to three surface loops that enclose FeSB in the binding pocket, contributing to its low nanomolar dissociation constant. Furthermore, we demonstrate that FeSB orientation and the SirA binding residues are largely distinct from those in the FeSA-HtsA complex, demonstrating a mechanism of siderophore specificity. Finally, we showed that the small conformational changes in SirA lead to Glu positioning primed for permease interaction.
Acknowledgments
We thank Angele Arrieta for technical assistance, John Cooper for providing staphyloferrin A, Michael Tiedemann for help with atomic absorption spectroscopy, Suya Liu for performing mass spectrometry, and Stephanie Pfaffen for critical reading of the manuscript. Portions of the research described in this paper were performed at the Stanford Synchrotron Radiation Light Source, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by the National Institutes of Health National Center for Research Resources, Biomedical Technology Program, and NIGMS.
This work is supported by Canadian Institutes of Health Research Grants MOP-102596 (to M. E. P. M.) and MOP-38002 (to D. E. H.).
The atomic coordinates and structure factors (codes 3MWG and 3MWF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SA
- staphyloferrin A
- SB
- staphyloferrin B
- FeSA
- ferric bound staphyloferrin A
- FeSB
- Fe(III)-SB
- Fhu
- ferric hydroxamate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.
- root mean square
- PDB
- Protein Data Bank
- H-bond
- hydrogen bond.
REFERENCES
- 1.Kluytmans J. A., Wertheim H. F. (2005) Infection 33, 3–8 [DOI] [PubMed] [Google Scholar]
- 2.Lowy F. D. (1998) N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 3.Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., Craig A. S., Zell E. R., Fosheim G. E., McDougal L. K., Carey R. B., Fridkin S. K. (2007) JAMA 298, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 4.Weems J. J., Jr. (2001) Postgrad. Med. 110, 24–36 [DOI] [PubMed] [Google Scholar]
- 5.Wandersman C., Delepelaire P. (2004) Annu. Rev. Microbiol. 58, 611–647 [DOI] [PubMed] [Google Scholar]
- 6.Stojiljkovic I., Perkins-Balding D. (2002) DNA Cell Biol. 21, 281–295 [DOI] [PubMed] [Google Scholar]
- 7.Grigg J. C., Ukpabi G., Gaudin C. F., Murphy M. E. (2010) J. Inorg. Biochem. 104, 341–348 [DOI] [PubMed] [Google Scholar]
- 8.Maresso A. W., Schneewind O. (2006) Biometals 19, 193–203 [DOI] [PubMed] [Google Scholar]
- 9.Beasley F. C., Heinrichs D. E. (2010) J. Inorg. Biochem. 104, 282–288 [DOI] [PubMed] [Google Scholar]
- 10.Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., Schneewind O. (2003) Science 299, 906–909 [DOI] [PubMed] [Google Scholar]
- 11.Skaar E. P., Schneewind O. (2004) Microbes Infect. 6, 390–397 [DOI] [PubMed] [Google Scholar]
- 12.Winkelmann G. (2002) Biochem. Soc. Trans. 30, 691–696 [DOI] [PubMed] [Google Scholar]
- 13.Drechsel H., Freund S., Nicholson G., Haag H., Jung O., Zähner H., Jung G. (1993) Biometals 6, 185–192 [DOI] [PubMed] [Google Scholar]
- 14.Haag H., Fiedler H. P., Meiwes J., Drechsel H., Jung G., Zähner H. (1994) FEMS Microbiol. Lett. 115, 125–130 [DOI] [PubMed] [Google Scholar]
- 15.Konetschny-Rapp S., Jung G., Meiwes J., Zähner H. (1990) Eur. J. Biochem. 191, 65–74 [DOI] [PubMed] [Google Scholar]
- 16.Meiwes J., Fiedler H. P., Haag H., Zähner H., Konetschny-Rapp S., Jung G. (1990) FEMS Microbiol. Lett. 55, 201–205 [DOI] [PubMed] [Google Scholar]
- 17.Sebulsky M. T., Heinrichs D. E. (2001) J. Bacteriol. 183, 4994–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebulsky M. T., Hohnstein D., Hunter M. D., Heinrichs D. E. (2000) J. Bacteriol. 182, 4394–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabrera G., Xiong A., Uebel M., Singh V. K., Jayaswal R. K. (2001) Appl. Environ. Microbiol. 67, 1001–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beasley F. C., Vinés E. D., Grigg J. C., Zheng Q., Liu S., Lajoie G. A., Murphy M. E., Heinrichs D. E. (2009) Mol. Microbiol. 72, 947–963 [DOI] [PubMed] [Google Scholar]
- 21.Cheung J., Beasley F. C., Liu S., Lajoie G. A., Heinrichs D. E. (2009) Mol. Microbiol. 74, 594–608 [DOI] [PubMed] [Google Scholar]
- 22.Cotton J. L., Tao J., Balibar C. J. (2009) Biochemistry 48, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 23.Dale S. E., Doherty-Kirby A., Lajoie G., Heinrichs D. E. (2004) Infect. Immun. 72, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatt G., Denny T. P. (2004) J. Bacteriol. 186, 7896–7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale S. E., Sebulsky M. T., Heinrichs D. E. (2004) J. Bacteriol. 186, 8356–8362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speziali C. D., Dale S. E., Henderson J. A., Vinés E. D., Heinrichs D. E. (2006) J. Bacteriol. 188, 2048–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigg J. C., Cooper J. D., Cheung J., Heinrichs D. E., Murphy M. E. (2010) J. Biol. Chem. 285, 11162–11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebulsky M. T., Shilton B. H., Speziali C. D., Heinrichs D. E. (2003) J. Biol. Chem. 278, 49890–49900 [DOI] [PubMed] [Google Scholar]
- 29.Sebulsky M. T., Speziali C. D., Shilton B. H., Edgell D. R., Heinrichs D. E. (2004) J. Biol. Chem. 279, 53152–53159 [DOI] [PubMed] [Google Scholar]
- 30.Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004) Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
- 31.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 32.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 38.Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 40.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 41.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 42.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 43.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 44.DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 45.Shindyalov I. N., Bourne P. E. (1998) Protein Eng. 11, 739–747 [DOI] [PubMed] [Google Scholar]
- 46.Gille C., Frömmel C. (2001) Bioinformatics 17, 377–378 [DOI] [PubMed] [Google Scholar]
- 47.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grigg J. C., Vermeiren C. L., Heinrichs D. E., Murphy M. E. (2007) J. Biol. Chem. 282, 28815–28822 [DOI] [PubMed] [Google Scholar]
- 50.Krewulak K. D., Shepherd C. M., Vogel H. J. (2005) Biometals 18, 375–386 [DOI] [PubMed] [Google Scholar]
- 51.Peuckert F., Miethke M., Albrecht A. G., Essen L. O., Marahiel M. A. (2009) Angew. Chem. Int. Ed. Engl. 48, 7924–7927 [DOI] [PubMed] [Google Scholar]
- 52.Shi R., Proteau A., Wagner J., Cui Q., Purisima E. O., Matte A., Cygler M. (2009) Proteins 75, 598–609 [DOI] [PubMed] [Google Scholar]
- 53.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hider R. C., Kong X. (2010) Nat. Prod. Rep. 27, 637–657 [DOI] [PubMed] [Google Scholar]
- 55.Cobessi D., Celia H., Pattus F. (2005) J. Mol. Biol. 352, 893–904 [DOI] [PubMed] [Google Scholar]
- 56.Tseng C. F., Burger A., Mislin G. L., Schalk I. J., Yu S. S., Chan S. I., Abdallah M. A. (2006) J. Biol. Inorg. Chem. 11, 419–432 [DOI] [PubMed] [Google Scholar]
- 57.Gobin J., Moore C. H., Reeve J. R., Jr., Wong D. K., Gibson B. W., Horwitz M. A. (1995) Proc. Natl. Acad. Sci. 92, 5189–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hough E., Rogers D. (1974) Biochem. Biophys. Res. Commun. 57, 73–77 [DOI] [PubMed] [Google Scholar]
- 59.Einsle O., Messerschmidt A., Stach P., Bourenkov G. P., Bartunik H. D., Huber R., Kroneck P. M. (1999) Nature 400, 476–480 [DOI] [PubMed] [Google Scholar]
- 60.Rohrbach M. R., Braun V., Köster W. (1995) J. Bacteriol. 177, 7186–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carter D. M., Miousse I. R., Gagnon J. N., Martinez E., Clements A., Lee J., Hancock M. A., Gagnon H., Pawelek P. D., Coulton J. W. (2006) J. Biol. Chem. 281, 35413–35424 [DOI] [PubMed] [Google Scholar]
- 62.Hoegy F., Celia H., Mislin G. L., Vincent M., Gallay J., Schalk I. J. (2005) J. Biol. Chem. 280, 20222–20230 [DOI] [PubMed] [Google Scholar]
- 63.Clarke T. E., Braun V., Winkelmann G., Tari L. W., Vogel H. J. (2002) J. Biol. Chem. 277, 13966–13972 [DOI] [PubMed] [Google Scholar]
- 64.Clarke T. E., Ku S. Y., Dougan D. R., Vogel H. J., Tari L. W. (2000) Nat. Struct. Biol. 7, 287–291 [DOI] [PubMed] [Google Scholar]
- 65.Borths E. L., Locher K. P., Lee A. T., Rees D. C. (2002) Proc. Natl. Acad. Sci. 99, 16642–16647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho W. W., Li H., Eakanunkul S., Tong Y., Wilks A., Guo M., Poulos T. L. (2007) J. Biol. Chem. 282, 35796–35802 [DOI] [PubMed] [Google Scholar]
- 67.Karpowich N. K., Huang H. H., Smith P. C., Hunt J. F. (2003) J. Biol. Chem. 278, 8429–8434 [DOI] [PubMed] [Google Scholar]
- 68.Liu M., Su J. G., Kong R., Sun T. G., Tan J. J., Chen W. Z., Wang C. X. (2008) Biophys. Chem. 138, 42–49 [DOI] [PubMed] [Google Scholar]
- 69.Liu M., Sun T., Hu J., Chen W., Wang C. (2008) Biophys. Chem. 135, 19–24 [DOI] [PubMed] [Google Scholar]
- 70.Kandt C., Xu Z., Tieleman D. P. (2006) Biochemistry 45, 13284–13292 [DOI] [PubMed] [Google Scholar]
- 71.Hvorup R. N., Goetz B. A., Niederer M., Hollenstein K., Perozo E., Locher K. P. (2007) Science 317, 1387–1390 [DOI] [PubMed] [Google Scholar]
- 72.Braun V., Herrmann C. (2007) J. Bacteriol. 189, 6913–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]