Abstract
One of the proliferating cell nuclear antigen loader complexes, Ctf18-replication factor C (RFC), is involved in sister chromatid cohesion. To examine its relationship with factors involved in DNA replication, we performed a proteomics analysis of Ctf18-interacting proteins. We found that Ctf18 interacts with a replicative DNA polymerase, DNA polymerase ϵ (pol ϵ). Co-immunoprecipitation with recombinant Ctf18-RFC and pol ϵ demonstrated that their binding is direct and mediated by two distinct interactions, one weak and one stable. Three subunits that are specifically required for cohesion in yeast, Ctf18, Dcc1, and Ctf8, formed a trimeric complex (18-1-8) and together enabled stable binding with pol ϵ. The C-terminal 23-amino acid stretch of Ctf18 was necessary for the trimeric association of 18-1-8 and was required for the stable interaction. The weak interaction was observed with alternative loader complexes including Ctf18-RFC(5), which lacks Dcc1 and Ctf8, suggesting that the common loader structures, including the RFC small subunits (RFC2–5), are responsible for the weak interaction. The two interaction modes, mediated through distinguishable structures of Ctf18-RFC, both occurred through the N-terminal half of pol ϵ, which includes the catalytic domain. The addition of Ctf18-RFC or Ctf18-RFC(5) to the DNA synthesis reaction caused partial inhibition and stimulation, respectively. Thus, Ctf18-RFC has multiple interactions with pol ϵ that promote polymorphic modulation of DNA synthesis. We propose that their interaction alters the DNA synthesis mode to enable the replication fork to cooperate with the establishment of cohesion.
Keywords: DNA Polymerase, DNA Replication, DNA Synthesis, Protein Assembly, Protein-Protein Interactions
Introduction
The eukaryotic loader complex replication factor C (RFC)2 consists of one large and four small subunits (RFC1 and RFC2–5, respectively) and loads the clamp complex proliferating cell nuclear antigen (PCNA) onto a primer/template DNA through its ATPase activity. PCNA is a homotrimeric complex functioning as the processivity factor for replicative DNA polymerase δ (pol δ). Three RFC1 paralogues, Ctf18, Rad17, and Elg1, exist in eukaryotes and function as the largest subunits of three alternative loader complexes, Ctf18-RFC, Rad17-RFC, and Elg1-RFC, respectively, in association with RFC2–5 (for review, see Refs. 1, 2). These loader complexes target PCNA or the alternative clamp, the Rad9-Hus1-Rad1 (9-1-1) complex, and contribute to various reactions tightly related to DNA replication, such as chromosome cohesion, DNA damage responses, and maintenance of genome stability (3, 4).
Ctf18-RFC forms a heteroheptameric complex combining the pentameric core loader complex, Ctf18 and RFC2–5 (hereafter referred to as Ctf18-RFC(5)), and two additional cohesion-specific factors, Dcc1 and Ctf8 (5, 6), whose yeast orthologues are required for faithful sister chromatid cohesion (7–9). Ctf18-RFC loads functional PCNA onto DNA and stimulates pol δ (6, 10). Its PCNA-unloading activity depends on replication protein A concentration (11), but its functional significance in vivo has not been elucidated. We have reported that Ctf18-RFC physically interacts with a Y-family DNA polymerase, pol η, and stimulates its activity (12).
Sister chromatid cohesion is achieved by the tethering of the cohesin complex of SMC1, SMC3, SCC1, and SCC2 on DNA to ensure precise segregation of sister chromosomes in M phase (13, 14). Because establishment of sister chromatin cohesion occurs concomitantly with DNA replication, a mechanism for cross-talk between them is necessary. Several lines of evidence from yeast genetics have demonstrated interactions between these processes. First, PCNA and the cohesion factor Ctf7/acetyltransferase interact genetically (15–17). Second, the presence of one allele of pol2, a mutant form of the catalytic subunit of pol ϵ, causes sister chromatid cohesion defects, and POL2 also interacts genetically with four genes involved in sister chromatid cohesion, SMC1, ECO1/CTF7, and TRF4 and TRF5, which redundantly encode a nuclear nucleotidyl transferase (18). Third, the cohesion factor Ctf4 interacts genetically and physically with another replicative DNA polymerase, pol α (19, 20). Recent work with Dcc1-deficient human cells has demonstrated that human Ctf18-RFC is required for acetylation of SMC3 and controls replication fork progression (21).
Pol ϵ is a eukaryotic replicative DNA polymerase involved mainly in leading strand synthesis (22). Pol ϵ is composed of a catalytic subunit, p261 (POLE1), and three associated subunits: p59 (POLE2), p17 (POLE3), and p12 (POLE4) (23, 24). Pol2 (p261) and Dpb2 (p59) are essential for the viability of yeast cells (25–27), and although neither Dpb3 (p17) nor Dpb4 (p12) is essential in Saccharomyces cerevisae, Dpb3 is essential in Schizosaccharomyces pombe (28–30). The functions of these small subunits are not fully understood, but they form a heterodimer complex of Dpb3/Dpb4 and contain histone-fold motifs involved in protein-protein and protein-DNA interactions (31). p261 consists of 140 kDa of a catalytically active, N-terminal region containing six polymerase and five exonuclease motifs and 120 kDa of a noncatalytic, C-terminal region, which is required for interactions with other pol ϵ subunits (32). p261 is involved in several cellular functions, such as initiation of DNA replication, DNA repair, DNA recombination, S phase checkpoint, gene silencing, and sister chromatid cohesion in yeast (18, 33, 34). The C-terminal region, but not the catalytic N-terminal region, is essential for cell growth in yeast (32, 35, 36).
In this study, we detected pol ϵ as a Ctf18-interacting protein in human cells. We studied the interactions of pol ϵ and Ctf18 in detail and found that they interact in two distinct modes, weak and stable. Interestingly, both interactions occurred through the N-terminal half of pol ϵ, which includes the catalytic domain and regulates the DNA synthesis activity. Thus, our results provide a novel molecular link between the DNA replication fork components and the chromosome cohesion apparatus in human cells. This link may directly modulate fork progression in human cells.
EXPERIMENTAL PROCEDURES
Purification of FLAG-tagged Ctf18 from 293 Cells and Mass Spectrometric Analysis of the Co-purified Proteins
A human 293 cell line expressing FLAG-tagged Ctf18 was constructed by transfection of a pcDNA3 plasmid DNA harboring the FLAG-Ctf18 cDNA sequence. A cell lysate from 1.0 × 109 cells was successively loaded onto IgG-Sepharose Fast Flow (GE Healthcare) and anti-FLAG M2 antibody affinity gel (anti-FLAG beads; Sigma) columns in buffer H (25 mm HEPES, pH 7.8, 1 mm EDTA, 0.01% Nonidet P-40, 20 μg/ml leupeptin, 0.1 mm PMSF, and 10% glycerol) containing 0.1 m NaCl, and the bound proteins were eluted with the same buffer containing 100 μg/ml FLAG peptide (Sigma). The eluted proteins were precipitated with 10% trichloroacetate and electrophoresed in 7.5–17.5% SDS-polyacrylamide gels. The area of the gel corresponding to molecular mass from approximately 300 kDa to 10 kDa was cut into 30 pieces at 2-mm intervals, and the proteins in each gel slice were analyzed with LC/MS/MS at the Medical Institute of Bioregulation at Kyushu University. The obtained raw data were further processed with the Mascot program as described in Ref. 37.
Preparation of Recombinant Human Proteins with Baculoviruses
Baculoviruses expressing human proteins with or without a FLAG peptide tag (FLAG-RFC1, FLAG-Ctf18, FLAG-Rad17, Dcc1, Ctf8, and RFCs2–5) have been described (10). Baculoviruses for human Elg1, pol ϵ p261, p59, p17, and p12 were prepared by inserting PCR-amplified human cDNA sequences into pBacPAK8 and 9 (Clontech) or pFastbac1 (Invitrogen) plasmids following the manufacturers' protocols. Histidine (His), FLAG, T7 peptide, and GFP tags were inserted at the N termini of these cDNA sequences as indicated. pFastbacGFP-Ctf18C400 was prepared by inserting the Ctf18 C-terminal fragment into the pEGFPC1 plasmid (Invitrogen) and transferring the EGFP-C400-fused sequence to pFastbac1. DNA fragments carrying Ctf18 with further C-terminal truncations (C302, C100, C23, AD4, AD6, and AD7) were amplified from pFastbacFLAGCtf18 DNA with primers carrying these target sequences and substituted for the corresponding fragment of pFastbacGFP-Ctf18C400. Recombinant human proteins were expressed by infection of Sf9 or High5 cells with baculoviruses at 27 °C in Grace's insect medium supplemented with 10% FBS or Express Five medium (Invitrogen).
Preparation of a Lysate from HeLa Cells
HeLa cells were grown in Dulbecco's minimal essential medium (DMEM; Sigma) with 10% FBS at 37 °C supplemented with 5% CO2. Cells (1.5 × 107) in a 15-cm dish were dispersed, washed with 2 ml of phosphate-buffered saline (PBS), and suspended with 50 μl of buffer B (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 10% glycerol, 20 μg/ml leupeptin, and 0.1 mm PMSF) containing 150 mm KCl. Fifty microliters of buffer B containing 850 mm KCl and 1% Nonidet P-40 was added to the sample, which was then left at 0 °C for 10 min. The supernatant was collected after centrifugation at 6.6 × 104 g for 15 min at 4 °C.
Electrophoresis and Immunoblotting
Proteins were separated in SDS-polyacrylamide gels and stained with Coomassie Brilliant Blue or silver. For immunoblotting, proteins in electrophoresis gels were transferred to Hybond-P PVDF membranes (GE Healthcare), incubated with antibodies in Can Get Signal (Toyobo) solution, and visualized with an LAS-3000mini (Fuji Film) after treatment with ECL Detection Reagent (GE Healthcare). The antibodies used were a monoclonal anti-pol ϵ p261 antibody (ATCC, CRL-2284), rabbit anti-Ctf18 polyclonal antibody (prepared with the C-terminal peptide), monoclonal anti-Ctf18 antibody (M01-1F5; Abnova), rabbit anti-Dcc1 polyclonal antibody (prepared with the C-terminal peptide), anti-Ctf8 rabbit polyclonal antibody (prepared with the C-terminal peptide), rabbit anti-RFC2 polyclonal antibody (sc-20996; Santa Cruz Biotechnology), anti-FLAG monoclonal antibody (M2; Sigma), goat anti-mouse IgG antibody conjugated with HRP (Bio-Rad), goat anti-rabbit IgG antibody conjugated with HRP (Bio-Rad), and rabbit anti-goat IgG antibody conjugated with HRP (Zymed Laboratories).
Purification of Recombinant Proteins
7.5 × 107 High5 cells in 5 × 15-cm dishes were co-infected with baculoviruses of human pol ϵ His-p261, FLAG-p59, p17, and p12 subunits for 48 h at 27 °C. The infected cells were suspended with 5 ml of buffer B and lysed by the addition of final 0.5% Nonidet P-40 and 0.5 m NaCl. The cell lysate, recovered by centrifugation at 6.6 × 104 g for 15 min at 4 °C, was loaded onto a DEAE-Sephacel (0.5 ml; GE Healthcare) in buffer B with 0.5 m NaCl, and the flow-through fraction was loaded onto an anti-FLAG antibody affinity gel column (1 ml) in buffer H containing 0.1 m NaCl. After an extensive wash of the column with the same buffer, the bound proteins were eluted with the same buffer containing 100 μg/ml FLAG peptide and successively loaded onto a Ni-nitrilotriacetic acid-Sepharose High Performance column (0.2 ml; GE Healthcare) in His tag buffer (50 mm sodium phosphate, pH 7.5, 10% glycerol, 0.01% Nonidet P-40, 20 μg/ml leupeptin, and 0.1 mm PMSF). The bound proteins were eluted successively with 20 and 100 mm imidazole in the same buffer. Pol ϵ holoenzyme recovered in the latter condition was further fractionated with a 5-ml gradient of 15–35% glycerol in buffer H containing 0.1 m NaCl by sedimentation with a SW50.1 rotor (Beckman) at 2.2 × 105 g for 11 h at 4 °C. For purification of His-p261Nter, High5 cells expressing His-p261Nter were treated as above, except for the omission of the anti-FLAG antibody column step. FLAG-tagged human Ctf18-RFC and Ctf18-RFC(5) were purified from insect cell lysates expressing these complexes as described in Ref. 10. These purified protein profiles are shown in supplemental Fig. 3A.
Pulldown Assay
Tagged recombinant loader proteins were bound with anti-FLAG beads or anti-GFP antibody beads (MBL) with buffer H containing 0.1 m NaCl. The beads were incubated with the indicated amounts of pol ϵ in 10 μl of the buffer at 4 °C for 1 h, and the bound proteins were eluted with 15 μl of 1× SDS loading buffer after five washes with 150 μl of the same buffer. Alternatively, p261 fragments prebound to 0.5 μl of Ni-nitrilotriacetic acid magnetic beads (Qiagen) were mixed with the indicated amounts of purified Ctf18-RFC or Sf9 lysates expressing Ctf18 complexes in 10 μl of buffer H with 0.1 m NaCl at 4 °C for 1 h and eluted with 15 μl of 1× SDS buffer containing 0.1 m EDTA after washes as above.
Glycerol Gradient Sedimentation Analysis
HeLa cell lysates (60 μl) prepared as above were loaded onto a 2.2-ml gradient of 15–35% glycerol in buffer H containing 0.1 m NaCl and centrifuged with a TLS55 rotor (Beckman Coulter) at 1.6 × 105 g for 13 h at 4 °C. Proteins were recovered from the bottom and analyzed by immunoblotting. Sedimentation markers run in parallel were ferritin (430 kDa), alcohol dehydrogenase (50 kDa), bovine serum albumin (BSA, 66 kDa), and cytochrome c (17 kDa).
DNA Polymerase Assay
DNA synthesis was assayed as described previously (12) by measuring [32P]TMP counts adsorbed to DE81 paper after incubation of the reaction mixture at 37 °C for 30 min.
RESULTS
Analysis of Ctf18-interacting Proteins in Human Cells
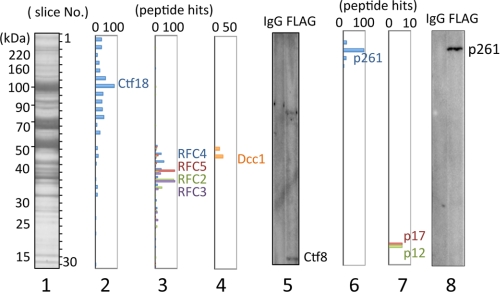
Exogenously expressed FLAG-tagged Ctf18 was purified directly from a 293 cell lysate with an anti-FLAG bead column. Co-purified proteins were identified by LC/MS/MS (Fig. 1). The Ctf18-RFC components RFC2–5 and Dcc1 were specifically identified with Ctf18 in the eluate from an anti-FLAG bead column at the gel slices of expected molecular masses (Fig. 1, panels 2–4). Fragments of the remaining component, Ctf8, were undetectable by Mascot Search among the identified peptide fragments. However, Ctf8 was detected by immunoblotting of the eluate from the anti-FLAG beads (panel 5). Therefore, FLAG-tagged Ctf18 in human cells was recovered as part of the Ctf18-RFC heptameric complex.
FIGURE 1.
Analyses of Ctf18-interacting proteins from lysates of 293 cells expressing FLAG-Ctf18. Proteins co-eluted with FLAG-Ctf18 from an anti-FLAG column were separated in a 7.5–17.5% SDS-acrylamide gel followed by Coomassie Brilliant Blue staining (lane 1). The gel was sliced into 30 pieces (numbered from top to bottom as indicated in lane 1), and included proteins were identified by LC/MS/MS analysis. Panels 2, 3, 4, 6, and 7 indicate hit numbers of detected peptides for Ctf18, RFC2–5, Dcc1, pol ϵ p261, and p17/p12, respectively. Comparable immunoblotting of Ctf8 (panel 5) and p261 (panel 8) with eluates from control IgG-Sepharose (IgG) and from the anti-FLAG column (FLAG) are shown in each panel. In addition to the Ctf8-specific band, a background band appears in lane 5 in the high molecular mass area.
In the same proteomics analysis, we found substantial peptide hits for all pol ϵ subunits except p59 in the predicted molecular mass gel slices. For example, p261 fragments were identified around No. 3, and p17 and p12 were around No. 28 (panels 6 and 7). Indeed, p261 was detected specifically by immunoblotting from the eluate of the anti-FLAG column but not from the control IgG-Sepharose eluate (panel 8). No fragments of pol ϵ p59 were detectable by LC/MS/MS analysis, although the second subunit of pol ϵ (Dpb2) corresponding to human p59 is essential for yeast viability (26). We were also unable to detect any signals by immunoblotting with an antibody for p59 under conditions sufficient to detect p59 present in the expected stoichiometric ratio with the amount of detected p261.
Ctf18-RFC and Ctf18-RFC(5) Interact Differently with pol ϵ
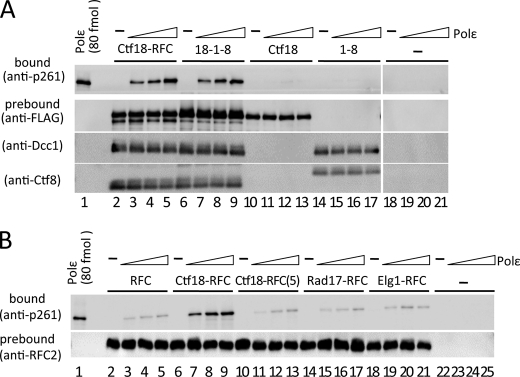
We examined whether the interaction between purified recombinant human Ctf18-RFC and pol ϵ was direct or indirect by co-immunoprecipitation. Pol ϵ efficiently co-precipitated with Ctf18-RFC, and more than half of the input pol ϵ was recovered, as judged by the p261 band intensities (Fig. 2A, lanes 2–5). Interestingly, the pentameric complex Ctf18-RFC(5), which does not contain Dcc1 or Ctf8, exhibited only weak binding, corresponding to about one-fifth of the Ctf18-RFC binding (Fig. 2B, lanes 6–13). The reverse immunoprecipitation experiments with Ni-nitrilotriacetic acid magnetic beads prebound with pol ϵ reproduced the weak and stable binding with Ctf18-RFC(5) and Ctf18-RFC, respectively (Fig. 3E, lanes 1–6). These results indicate that Ctf18-RFC interacts with pol ϵ in two different modes, one with weak binding and the other with stable binding.
FIGURE 2.
Co-immunoprecipitation of pol ϵ with loader complexes. A, approximately 0.7 pmol of Ctf18-RFC (lanes 2–5), 18-1-8 (lanes 6–9), Ctf18 (lanes 10–13), and 1-8 (with FLAG-Ctf8, lanes 14–17) prebound to anti-FLAG beads was co-immunoprecipitated with 0 (−), 0.2, 0.4, and 0.6 pmol (triangles) of pol ϵ in 10-μl reaction mixtures. Lanes 18–21 are the negative control without Ctf18 complexes. Input pol ϵ (80 fmol) was applied as the loading standard (lane 1). One-fifth of each precipitate was analyzed by immunoblotting with an anti-p261 monoclonal antibody (top panels). Prebound proteins (FLAG-Ctf18, Dcc1, and Ctf8 or FLAG-Ctf8) were compared by immunoblotting. B, 0.7 pmol of RFC (lanes 2–5), Ctf18-RFC (lanes 6–9), Ctf18-RFC(5) (lanes 10–13), Rad17-RFC (lanes 14–17), and Elg1-RFC (lanes 18–21) prebound to anti-FLAG beads was incubated with increasing amounts of pol ϵ and analyzed by immunoblotting as in A. Lanes 22–25 are the negative control without loaders. Immunoblotting of RFC2 shows the uniformity of the bound loader proteins.
FIGURE 3.
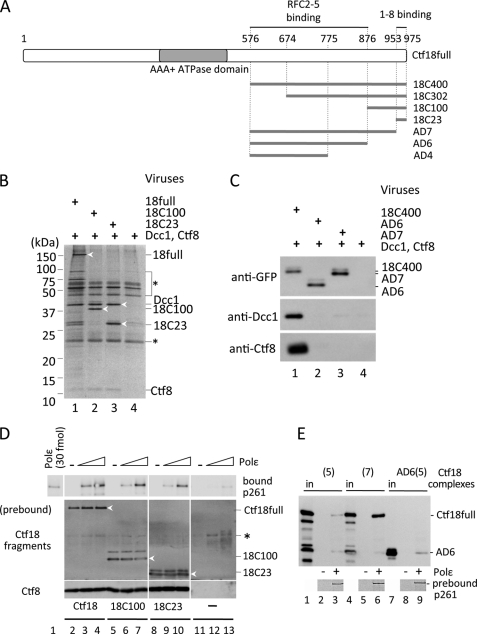
C-terminal region of Ctf18 required for association with 1-8 and the stable interaction with pol ϵ. A, truncated Ctf18 fragments associated with RFC2–5 and 1-8. Full-length Ctf18 (Ctf18full; residues 1–975) with the AAA+ ATPase domain (gray) and its truncated fragments, 18C400, 18C302, 18C100, and 18C23 carrying 400, 302, 100, and 23 residues from the C terminus, respectively, and 200-, 100-, and 23-residue C-terminal deletions from 18C400 (AD4, -6, -7) are indicated. These fragments were tagged with GFP at their N termini. The Ctf18 regions necessary for interactions with RFC2–5 and 1-8 are indicated above the map (see supplemental. Fig. 2). B and C, association of Ctf18 fragments with 1-8. GFP-tagged Ctf18 fragments 18full, 18C100, and 18C23 (B, lanes 1–3), or 18C400, AD6, and AD7 (C, lanes 1–3) were co-expressed with Dcc1 and Ctf8 in insect cells and precipitated with anti-GFP beads. Lane 4 in each panel is the negative control without GFP-Ctf18 fragments. The bound fractions were separated in 4–20% or 12.5% polyacrylamide gels and analyzed by silver staining (B) or immunoblotting with the indicated antibodies (C). Protein bands for analyses are indicated at the right of the panels and with white arrowheads in B. A bracket and a line with an asterisk (*) in B show nonspecific proteins or IgG chains from anti-GFP beads. D, interaction of pol ϵ with 18-1-8 (lanes 2–4), 18C100-1-8 (lanes 5–7), or 18C23-1-8 (lanes 8–10) is shown. Approximately 0.7 pmol of trimeric complexes were prebound to anti-FLAG beads and incubated with 0 (−), 0.2, or 0.6 pmol of pol ϵ (triangles). The bound proteins were analyzed by immunoblotting with anti-p261 (top), anti-Ctf18 (middle), or anti-Ctf8 (bottom) antibodies. Three Ctf18 fragments are indicated with white arrowheads. Lanes 11–13 are the control experiments without Ctf18 fragments, and an asterisk (*) indicates a nonspecific band. E, co-immunoprecipitation of Ctf18 complexes with Ni-nitrilotriacetic acid magnetic beads prebound with pol ϵ complex is indicated in the bottom panels (lanes 3, 6, and 9). Sf9 lysates expressing roughly equal amounts of GFP-tagged Ctf18-RFC(5) ((5) lanes 1–3), Ctf18-RFC ((7) lanes 4–6), or AD6-RFC2–5 (AD6(5) lanes 7–9) were mixed with the beads, and proteins in the 50% bound fractions were detected with an anti-GFP antibody. Lanes 1, 4, and 7 are 5% input controls of the respective fragments, and lanes 2, 5, and 8 are negative controls without pol ϵ. Lower bands in lanes 1 and 4 are degradation products of GFP-Ctf18.
Next, we asked whether the binding to pol ϵ is limited to specific loader complexes. All alternative pentameric loader complexes were reconstituted by co-expression of baculoviruses carrying RFC small subunits (RFC2–5) with either FLAG-tagged RFC1, Rad17, or Elg1, followed by co-immunoprecipitation with pol ϵ (Fig. 2B). None of the pentameric loader complexes exhibited binding as stable as that obtained with Ctf18-RFC, but substantial amounts of pol ϵ, as similar as Ctf18-RFC(5), were detected in the bound fractions. This result indicated that all loader complexes, including Ctf18-RFC(5), can interact with pol ϵ to a certain extent, probably through their common structure of the large subunits and small RFC2–5 subunits. These results strongly suggest that specific subunits of Ctf18-RFC, Dcc1, and Ctf8 lead to its stable binding. However, Ctf18 alone or a dimeric Dcc1-Ctf8 complex (which we designated 1-8) formed by co-expression of Dcc1 and Ctf8 as reported in (6) did not show any significant binding to pol ϵ (Fig. 2A, lanes 10–17). When we reconstituted the trimeric complex Ctf18-Dcc1-Ctf8 (designated 18-1-8) by co-expressing the three proteins, the complex did interact with pol ϵ at almost the same level as Ctf18-RFC (Fig. 2A, lanes 6–9). Thus, the two distinguishable weak and stable interactions with pol ϵ were with the Ctf18-RFC(5) and 18-1-8 complex, respectively.
C-terminal Region of Ctf18 Is Necessary for the Formation of the 18-1-8 Subcomplex and Stable Binding to pol ϵ
Because both Ctf18 and 1-8 were required for stable binding, the formation of the 18-1-8 subcomplex must be tightly linked with the interaction. When we prepared lysates co-expressing any two of GFP-Ctf18, Dcc1, and Ctf8 and precipitated them with anti-GFP beads, about half of the amount of Dcc1 was recovered with Ctf18 as when 18-1-8 was expressed, and only a limited amount of Ctf8 was recovered with Ctf18 (supplemental Fig. 1). Thus, both Dcc1 and Ctf8 can interact independently with Ctf18 to some extent, but their mutual association with Ctf18 is necessary for the stable formation of the 18-1-8 complex. It has been reported that the 82-amino acid stretch of the C terminus of yeast Ctf18 is necessary for its interaction with Dcc1 and Ctf8 (11). To identify the minimum region of Ctf18 that interacts with 1–8, we constructed a series of GFP-tagged deletion fragments carrying the 400, 302, 100, and 23 amino acids from the C terminus (18C400, 18C302, 18C100, and 18C23, respectively; Fig. 3A). The C-terminal-most 23-residue stretch was chosen because its sequence is highly conserved from yeast to human. The deletion constructs were co-expressed with Dcc1 and Ctf8 in insect cells and precipitated with anti-GFP beads. All of the Ctf18 fragments precipitated both Dcc1 and Ctf8 efficiently (representative data for 18C100 and 18C23 are shown in Fig. 3, B and C), indicating that 1-8 could form a complex with Ctf18 through these 23 residues at the C terminus. Conversely, deletions of the C-terminal 100 and 23 residues from 18C400 (AD6 and AD7, respectively) abolished the association with both Dcc1 and Ctf8 (Fig. 3C). Thus, the 23 residues are sufficient for the formation of the trimeric complex. Then we studied the interaction of 18C100-1-8 and 18C23-1-8 with pol ϵ by immunoprecipitation (Fig. 3D). The result clearly indicated that the C-terminal 23 residues of Ctf18 are sufficient for the stable interaction with pol ϵ as well as for the formation of the 18-1-8 complex.
RFC2–5 also interact with Ctf18 independently of Dcc1 and Ctf8, resulting in the Ctf18-RFC(5) complex, which exhibited weak binding to pol ϵ (Fig. 2B). We also demonstrated that the 18-1-8 complex bound stably with pol ϵ without RFC2–5. Thus, Ctf18 formed two separate structures for the two distinguishable forms of binding with pol ϵ. We then addressed the question of whether the region of Ctf18 responsible for the weak interaction through association with RFC2–5 would be separate from the C-terminal region. RFC1 interacts with RFC2–5 at a region proximal to the C terminus of the AAA+ ATPase domain (38); we hypothesized that Ctf18 could use the same region to form the Ctf18-RFC(5) subcomplex. To identify the region of Ctf18 necessary for the subcomplex formation, the C-terminal fragments fused with a GFP tag were co-expressed with RFC2–5. Their immunoprecipitation with anti-GFP antibody beads indicated that Ctf18full and 18C400, but not 18C302, could form complexes with RFC2–5 (supplemental Fig. 2). Similarly, AD6 and AD7, but not AD4 (with a deletion of 200 residues from 18C400), could associate with RFC2–5. These results indicate that the minimum region necessary for the complex formation with RFC2–5 was residues 576–876. Furthermore, the pentameric complex of AD6 and RFC2–5 exhibited weak binding with pol ϵ, similar to Ctf18-RFC(5) (Fig. 3E). These data demonstrate that the two functional regions of Ctf18 for interaction with pol ϵ are structurally separable from each other.
N-terminal Half of the pol ϵ p261 Subunit Interacts with Ctf18-RFC Complexes
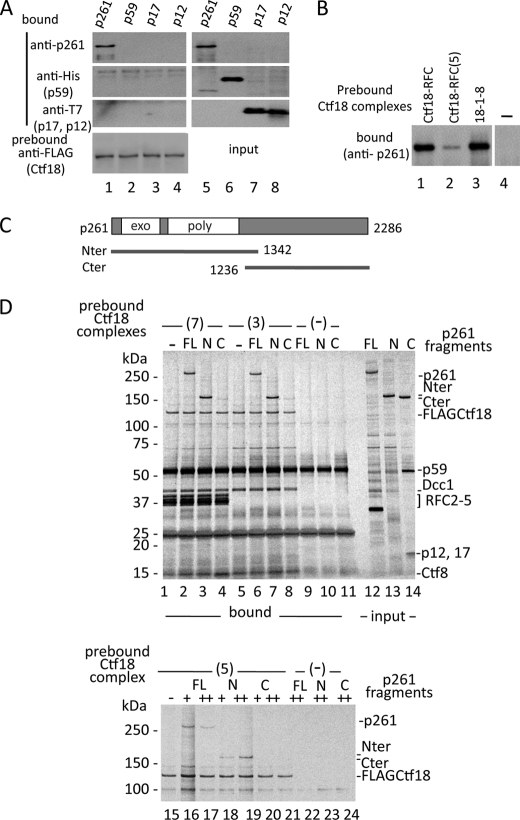
To distinguish which subunits of pol ϵ interact with Ctf18-RFC, we prepared insect cell lysates expressing four human pol ϵ subunits separately and co-precipitated them with beads prebound with Ctf18-RFC (Fig. 4A). Only p261, the catalytic subunit, bound to Ctf18-RFC. Further experiments demonstrated that p261 exhibited the same binding modes as the pol ϵ complex in terms of affinity and specificity. p261 bound stably with Ctf18-RFC and 18-1-8 and weakly with Ctf18-RFC(5) (Fig. 4B). Thus, our observed interactions of Ctf18-RFC with pol ϵ were mediated only by p261. As demonstrated before with yeast, the catalytic subunit consists of two functionally separable regions; the N-terminal half contains an exonuclease and DNA polymerase, and the C-terminal half interacts with factors involved in various cellular functions (35, 36). Only the C-terminal half is essential for cell viability. We expressed the N- and C-terminal half fragments (Nter and Cter, respectively; Fig. 4C) and tested their interaction with the Ctf18 complexes. The results demonstrated that Nter was responsible for both the weak binding with Ctf18RFC(5) and the stable binding with Ctf18-RFC and 18-1-8 (Fig. 4D).
FIGURE 4.
N-terminal half of pol ϵ p261 interacts with Ctf18-RFC. A, pol ϵ subunits (p261, His-tagged p59, T7-tagged p17, and T7-tagged p12; 1.4 pmol each) incubated with anti-FLAG beads prebound with 1.8 pmol of Ctf18-RFC (lanes 1–4). One-third of the bound samples was used for immunoblotting with the indicated antibodies. Lanes 5–8 are the input controls (one-tenth of the total samples). B, binding of p261 with Ctf18 subcomplexes. 1 pmol of p261 was incubated with anti-FLAG beads prebound with 0.8 pmol of Ctf18-RFC (lane 1), Ctf18-RFC(5) (lane 2), or 18-1-8 (lane 3), and bound p261 was detected with an anti-p261 antibody. Lane 4 is the control without Ctf18 complexes. C, schematic of p261, including the exonuclease (exo) and DNA polymerase (poly) regions. The N- and C-terminal fragments (Nter and Cter) are indicated. Numbers represent the length of p261 in residues and the ends of Nter and Cter. D, upper panel, binding of p261 full-length (FL; lanes 2, 6, and 9), Nter (N; lanes 3, 7, and 10), and Cter (C; lanes 4, 8, and 11) with Ctf18-RFC ((7) lanes 1–4) or 18-1-8 ((3) lanes 5–8) in a pulldown assay with anti-FLAG antibody beads. Lanes 1, 5, and 9–11 are negative controls without p261 fragments or Ctf18 complexes (−). Partially purified p261, Nter, and Cter (1 pmol each) were mixed with beads prebound with about 0.5 pmol of Ctf18-RFC or 18-1-8, and 50% of the bound fractions were used for silver staining after 4–20% SDS-PAGE. Lanes 12–14 are 6% of the input as a control. A prominent band of about 35 kDa in lane 12 is a degradation product from p261. Cter was complexed with the p59, p17, and p12 subunits. These subunits were coexpressed in the insect cells to facilitate solubility of Cter in the lysate. Lower panel, same experiment with Ctf18-RFC(5). 0 (−), 0.5 (+), or 1 pmol (++) of p261 fragments were mixed with beads prebound with 0 (lanes 22–24) or 0.5 pmol (lanes 15–21) of Ctf18-RFC(5), and 50% of the bound fractions were analyzed as above. Lane 16 has slightly more sample than the others judging from the recovered FLAG-Ctf18. Only the upper half of the gel image is shown.
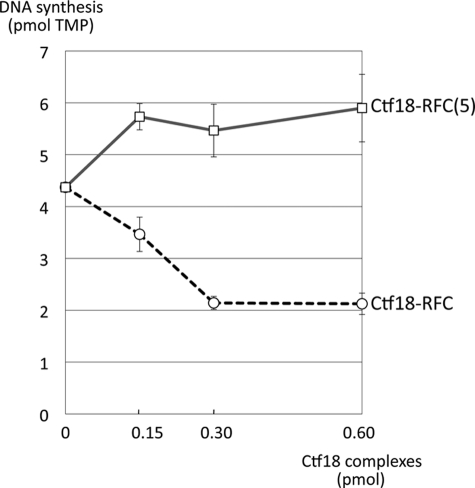
Because the interaction is mediated by the DNA polymerase region, we asked whether the interaction might affect the polymerase activity. The N-terminal half alone has polymerase activity in yeast (35, 36). Indeed, Nter alone exhibited rather higher polymerase activity than the pol ϵ complex as reported with yeast pol ϵ (39) (supplemental Fig. 3B). When we added Ctf18-RFC complexes to Nter, its polymerase activity was stimulated by Ctf18-RFC(5) and inhibited by Ctf18-RFC (Fig. 5). Both effects were saturated with the loader complexes at 3-fold molar excess to the input Nter and reached about 120 and 50% of the initial activity with Ctf18-RFC(5) and Ctf18-RFC, respectively.
FIGURE 5.
Effect of Ctf18-RFC and Ctf18-RFC(5) on the DNA synthesis activity of Nter. Purified Nter (0.1 pmol) was incubated with poly(dA)·oligo(dT) and [α-32P]TTP in the presence of 0.15, 0.3, or 0.6 pmol of Ctf18-RFC or Ctf18-RFC(5) at 37 °C for 30 min. The mean of two independent results of incorporated TMP are shown.
pol ϵ and Ctf18-RFC in Human Cells Exist in High Molecular Mass Complexes
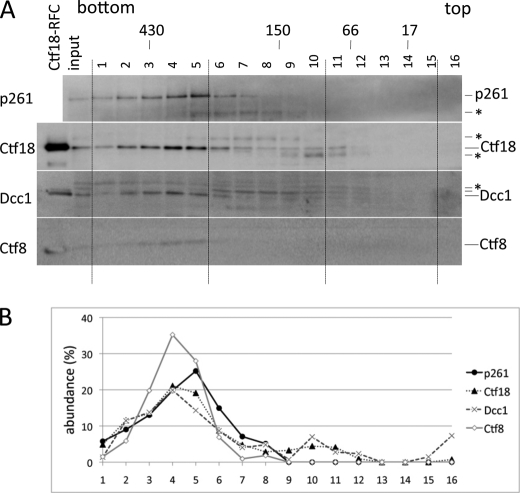
To study the status of native complexes of pol ϵ and Ctf18-RFC in human cells, a HeLa cell lysate was subjected to glycerol gradient sedimentation (Fig. 6). The majority of Ctf18, Dcc1, and Ctf8 were detected in fractions 2–5, corresponding to a molecular mass of about 400 kDa. A substantial fraction of pol ϵ p261 co-sedimented in the same high molecular mass fractions. Because recombinant pol ϵ and the Ctf18-RFC complexes each sedimented at around 250 kDa in a glycerol gradient sedimentation in the same conditions (supplemental Fig. 4), these two complexes should interact with other components in HeLa cells, and the coincidence of their sedimented fractions at higher molecular mass strongly suggested that they would interact with each other in vivo. Most Dcc1 (45 kDa) and Ctf8 (13 kDa) were recovered in the same fractions as Ctf18, indicating that most were present as a complex with Ctf18, and their free or dimeric forms as 1-8 were minimal in HeLa cells.
FIGURE 6.
Fractionation of HeLa lysate by glycerol gradient sedimentation. A, HeLa lysate was fractionated by glycerol gradient sedimentation in 2.2 ml of 15–35% glycerol gradient in buffer H containing 0.1 m NaCl. Proteins were fractionated into 16 tubes, separated in a 15% acrylamide gel, and analyzed by immunoblotting with antibodies against pol ϵ p261, Ctf18, Dcc1, or Ctf8. Sedimentation positions of marker proteins with their molecular masses are shown at top. *, nonspecific proteins. B, abundances of four proteins in the fractions are indicated as percent of band intensities (band intensity/sum of band intensities ×100).
DISCUSSION
Proteomics Analysis for Ctf18-RFC-binding Proteins
Ctf18 is partially required for chromosomal cohesion in yeast (7, 8) and behaves as a component of an alternative PCNA loader complex among eukaryotes (1, 2). As indicated in Fig. 1, FLAG-Ctf18 in 293 cells was eluted from an anti-FLAG antibody column along with RFC2–5, Dcc1, and Ctf8, its assembly partners for Ctf18-RFC, and various interacting proteins. Several replication proteins in addition to pol ϵ were identified in the co-eluting fractions, but not in the fractions obtained from a control IgG-Sepharose column. For example, the pol δ p125, replication protein A p70 and p34, PCNA, and MCM subunits were detected in an LC/MS/MS analysis and further confirmed by immunoblotting (supplemental Fig. 5). Some of these interactions with Ctf18 have been reported previously (11, 40, 41), indicating the reliability of our results. We previously reported that translesion DNA polymerase pol η also associates with Ctf18-RFC and stimulates its activity (12); however, pol η was not detected in this analysis. Because pol η is involved in the DNA damage response pathway, some functional conditions, such as the induction of DNA damage, may be necessary to observe the stable interactions of pol η with Cft18-RFC in human cells. Indeed, we have observed their co-immunoprecipitation when they are overexpressed in HeLa cells (12).
Ctf18 has been shown to interact with pol2 and Dpb2 in S. cerevisiae by proteome analysis (42), and pol ϵ has been shown to be involved in the cohesion pathway by yeast genetics (18). Thus, the interaction of Ctf18-RFC and pol ϵ is highly conserved among eukaryotes. We were not able to identify the p59 subunit corresponding to Dpb2 in our LC/MS/MS analysis with a human cell lysate, although the other three subunits were detected. This may be because of the lower abundance of p59 than p261 in the 293 cells. When we purified the pol ϵ complex from human cells exogenously expressing recombinant FLAG-tagged p59 by the same method described here, we were able to co-purify all components of Ctf18-RFC with the p59-containing pol ϵ complex (data not shown). Thus, the presence of p59 in the pol ϵ complex and the association of pol ϵ with Ctf18-RFC are not necessarily exclusive events. However, these results also suggest that p59 is not required in vivo for the interaction of pol ϵ and Ctf18-RFC.
Two Interaction Modes
Our results clearly indicated that Ctf18-RFC and pol ϵ interact by two different mechanisms. The weak interactions, observed in common among the clamp loaders, might be derived mainly from their common subunits, RFC2–5. This property suggests that all clamp loaders have the potential to be tethered to pol ϵ, probably at DNA synthesis sites, and to function cooperatively with the DNA replication fork complex. Indeed, the Escherichia coli loader subunit γ complex interacts with the pol III complex to form the replisome complex (for review, see Ref. 43). However, the observed stability of eukaryotic loaders with pol ϵ is much lower than in E. coli, in which their interaction functions as a framework for the replisome. Because not all loaders are necessarily prerequisite components of the replisome, their unstable but clear interaction with pol ϵ may represent a mechanism to recruit them transiently to the eukaryotic replisome through an unknown regulatory mechanism, which may change the functions of the replisome according to its upcoming template structures. Further studies will be necessary to elucidate the possible regulatory mechanism of the replisome through the association of loader complexes.
It is of interest that, among loader complexes, Ctf18-RFC specifically and stably interacts with pol ϵ, and the additional cohesion factors Dcc1 and Ctf8 are responsible for the stable interaction. Studies with truncated Ctf18 fragments showed that 18C23 is sufficient to form the 18-1-8 complex and to interact stably with pol ϵ (Fig. 3D, lanes 8–10). Because Dcc1 and Ctf8 formed a stable dimer that did not exhibit any interaction with pol ϵ (Fig. 2A), and the interactions of Ctf18 with Dcc1 or Ctf8 separately are less stable than with the 1-8 complex (supplemental Fig. 1), the interfaces of the trimeric complex of 18C23, Dcc1, and Ctf8 likely form a robust structure to lock in the interaction target of p261.
Functional Relevance of pol ϵ Binding and PCNA Loading of Ctf18-RFC
Ctf18-RFC(5) bound PCNA and loaded it to DNA with activity that was indistinguishable from that of Ctf18-RFC (6, 10), indicating that the two additional subunits, Dcc1 and Ctf8, have no roles in PCNA binding and loading. The region from residues 576–876 of Ctf18 was important for the association with RFC2–5 (supplemental Fig. 2A), whose assembly is necessary for PCNA loading. The obvious special separation of the PCNA loading structure and the stable pol ϵ association structure reflects the functional independence of the two events, loading of PCNA and binding to pol ϵ. Our previous report indicated that Ctf18-RFC interacts with pol η and stimulates its activity (12). In this case, Ctf18-RFC(5) was sufficient for the interaction with pol η, and the same interaction occurred with RFC, resembling the weak interaction of loader complexes with pol ϵ shown in the current study. PCNA was not required for the stimulation of pol η but cooperatively stimulated the activity. Taken together, Ctf18-RFC may have two independent functions for pol ϵ activity through interactions with two separable structures; one is the PCNA loader structure, and the second is the cohesion specific subunit assembly. It will be also possible that these two functions will have to be linked together on one Ctf18 molecule.
Functional Significance of the Stable Association with pol ϵ
Our data clearly demonstrated that cohesion-specific components in Ctf18-RFC are concomitantly required for the stable interaction with pol ϵ, suggesting a strong connection of pol ϵ with cohesion activity. Involvement of pol ϵ (pol2) in cohesion reactions has been demonstrated by its genetic interaction with several cohesion proteins in S. cerevisiae, although the C terminus of pol2 interacted with those proteins (18), whereas the interaction in our work was mediated through the N-terminal catalytic half of pol ϵ p261. This suggests that pol ϵ is a platform for multiple cohesion proteins and that its DNA polymerase activity will also function in the establishment of cohesion through Ctf18-RFC. Terret et al. demonstrated that human Dcc1 is essential for cell growth, and defective Dcc1 in human cells resulted in a marked reduction of Ctf18, a decrease of SMC3 acetylation, and impaired fork progression (21). They propose that Ctf18-RFC is required for modification and remodeling of the cohesin ring to allow the replisome to pass through it. We observed a result seemingly inconsistent with their observation that the interaction of Ctf18-RFC with the pol ϵ catalytic half resulted in reduction of its DNA synthesis activity. It should be noted that the inhibitory effect of Ctf18-RFC on pol ϵ was partial, and only 50% inhibition was observed in the saturated condition. Thus, we propose that the association of Ctf18-RFC with pol ϵ will slow down the replisome rate to coordinate the progression with the passing of the ring. This function would be important to stabilize the replisome at the cohesion sites and to move it forward efficiently. Further analysis will be necessary to test this idea by analyzing the fork progression in vivo when the interaction of Ctf18-RFC and pol ϵ is blocked by mutation of target sites or introduction of the target peptides in cultured cells.
Analysis of pol ϵ and Ctf18-RFC complexes in HeLa cells demonstrated that their complex status does not seem to be dynamic because populations of free complexes are limited and most of the 1-8 subcomplex (Fig. 6), the key factor for engaging binding of pol ϵ and Ctf18-RFC, is involved in the Ctf18 complex. This indicates that the majority of these proteins are in the complex and function together in vivo. Chromosome cohesion is required not only in S phase, but also for DNA damage repair reactions throughout the cell cycle (for review, see Ref. 44), and interaction of Ctf18-RFC and pol ϵ may occur any cell cycle phase. Thus, it will also be important to study the behavior of these two complexes in cells during the cell cycle and upon DNA damage treatment, focusing on their interaction. Our biochemical evidence has revealed a novel molecular link between a cohesion-specific alternative clamp loader complex and the DNA synthesis apparatus and has brought important insights into the mechanism of coordination of replication fork progression and establishment of cohesion in S phase.
Supplementary Material
Acknowledgments
We thank Dr. H. Araki (National Institute of Genetics, Japan) for critical reading of this paper and valuable comments.
This work was supported by Grants-in-aid 20370069 and 17080006 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- RFC
- replication factor C
- Cter
- C-terminal fragment
- Nter
- N-terminal fragment
- MS/MS
- tandem MS
- PCNA
- proliferating cell nuclear antigen
- pol
- polymerase.
REFERENCES
- 1.Kim J., MacNeill S. A. (2003) Curr. Biol. 13, R873–R875 [DOI] [PubMed] [Google Scholar]
- 2.Majka J., Burgers P. M. (2004) Prog. Nucleic Acids Res. Mol. Biol. 78, 227–260 [DOI] [PubMed] [Google Scholar]
- 3.Bermudez V. P., Lindsey-Boltz L. A., Cesare A. J., Maniwa Y., Griffth J. D., Hurwitz J., Sancar A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellison V., Stillman B. (2003) PLoS Biol 1, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer M. L., Gygi S. P., Aebersold R., Hieter P. (2001) Mol. Cell 7, 959–970 [DOI] [PubMed] [Google Scholar]
- 6.Bermudez V. P., Maniwa Y., Tappin I., Ozato K., Yokomori K., Hurwitz J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10237–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouprina N., Kroll E., Kirillov A., Bannikov V., Zakharyev V., Larionov V. (1994) Genetics 138, 1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer F., Gerring S. L., Connelly C., Hieter P. (1990) Genetics 124, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouprina N., Tsouladze A., Koryabin M., Hieter P., Spencer F., Larionov V. (1993) Yeast 9, 11–19 [DOI] [PubMed] [Google Scholar]
- 10.Shiomi Y., Shinozaki A., Sugimoto K., Usukura J., Obuse C., Tsurimoto T. (2004) Genes Cells 9, 279–290 [DOI] [PubMed] [Google Scholar]
- 11.Bylund G. O., Burgers P. M. (2005) Mol. Cell. Biol. 25, 5445–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiomi Y., Masutani C., Hanaoka F., Kimura H., Tsurimoto T. (2007) J. Biol. Chem. 282, 20906–20914 [DOI] [PubMed] [Google Scholar]
- 13.Haering C. H., Löwe J., Hochwagen A., Nasmyth K. (2002) Mol. Cell 9, 773–788 [DOI] [PubMed] [Google Scholar]
- 14.Nasmyth K., Haering C. H. (2005) Annu. Rev. Biochem. 74, 595–648 [DOI] [PubMed] [Google Scholar]
- 15.Ivanov D., Schleiffer A., Eisenhaber F., Mechtler K., Haering C. H., Nasmyth K. (2002) Curr. Biol. 12, 323–328 [DOI] [PubMed] [Google Scholar]
- 16.Skibbens R. V., Corson L. B., Koshland D., Hieter P. (1999) Genes Dev. 13, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. (1999) Genes Dev. 13, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards S., Li C. M., Levy D. L., Brown J., Snow P. M., Campbell J. L. (2003) Mol. Cell. Biol. 23, 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Wang T. S. (2004) Mol. Cell. Biol. 24, 9568–9579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles J., Formosa T. (1992) Mol. Cell. Biol. 12, 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terret M. E., Sherwood R., Rahman S., Qin J., Jallepalli P. V. (2009) Nature 462, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pursell Z. F., Isoz I., Lundström E. B., Johansson E., Kunkel T. A. (2007) Science 317, 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Asahara H., Patel V. S., Zhou S., Linn S. (1997) J. Biol. Chem. 272, 32337–32344 [DOI] [PubMed] [Google Scholar]
- 24.Li Y., Pursell Z. F., Linn S. (2000) J. Biol. Chem. 275, 23247–23252 [DOI] [PubMed] [Google Scholar]
- 25.Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. (1990) Cell 62, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 26.Araki H., Hamatake R. K., Johnston L. H., Sugino A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4601–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng W., Rodriguez-Menocal L., Tolun G., D'Urso G. (2003) Mol. Biol. Cell 14, 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki H., Hamatake R. K., Morrison A., Johnson A. L., Johnston L. H., Sugino A. (1991) Nucleic Acids Res. 19, 4867–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohya T., Maki S., Kawasaki Y., Sugino A. (2000) Nucleic Acids Res. 28, 3846–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiga M. G., D'Urso G. (2004) Nucleic Acids Res. 32, 4945–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson E., Macneill S. A. (2010) Trends Biochem. Sci. 35, 339–347 [DOI] [PubMed] [Google Scholar]
- 32.Dua R., Levy D. L., Campbell J. L. (1999) J. Biol. Chem. 274, 22283–22288 [DOI] [PubMed] [Google Scholar]
- 33.Sugino A. (1995) Trends Biochem. Sci. 20, 319–323 [DOI] [PubMed] [Google Scholar]
- 34.Pospiech H., Syväoja J. E. (2003) Scientific World J. 3, 87–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kesti T., Flick K., Keränen S., Syväoja J. E., Wittenberg C. (1999) Mol. Cell 3, 679–685 [DOI] [PubMed] [Google Scholar]
- 36.Feng W., D'Urso G. (2001) Mol. Cell. Biol. 21, 4495–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeishi Y., Ohashi E., Ogawa K., Masai H., Obuse C., Tsurimoto T. (2010) Genes Cells 15, 761–771 [DOI] [PubMed] [Google Scholar]
- 38.Bowman G. D., O'Donnell M., Kuriyan J. (2004) Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]
- 39.Maki S., Hashimoto K., Ohara T., Sugino A. (1998) J. Biol. Chem. 273, 21332–21341 [DOI] [PubMed] [Google Scholar]
- 40.Hanna J. S., Kroll E. S., Lundblad V., Spencer F. A. (2001) Mol. Cell. Biol. 21, 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohta S., Shiomi Y., Sugimoto K., Obuse C., Tsurimoto T. (2002) J. Biol. Chem. 277, 40362–40367 [DOI] [PubMed] [Google Scholar]
- 42.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 43.Johnson A., O'Donnell M. (2005) Annu. Rev. Biochem. 74, 283–315 [DOI] [PubMed] [Google Scholar]
- 44.Sjögren C., Ström L. (2010) Exp. Cell Res. 316, 1445–1453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.