Abstract
Our recent studies established essential and distinct roles for RalA and RalB small GTPase activation in K-Ras mutant pancreatic ductal adenocarcinoma (PDAC) cell line tumorigencity, invasion, and metastasis. However, the mechanism of Ral GTPase activation in PDAC has not been determined. There are four highly related mammalian RalGEFs (RalGDS, Rgl1, Rgl2, and Rgl3) that can serve as Ras effectors. Whether or not they share distinct or overlapping functions in K-Ras-mediated growth transformation has not been explored. We found that plasma membrane targeting to mimic persistent Ras activation enhanced the growth-transforming activities of RalGEFs. Unexpectedly, transforming activity did not correlate directly with total cell steady-state levels of Ral activation. Next, we observed elevated Rgl2 expression in PDAC tumor tissue and cell lines. Expression of dominant negative Ral, which blocks RalGEF function, as well as interfering RNA suppression of Rgl2, reduced PDAC cell line steady-state Ral activity, growth in soft agar, and Matrigel invasion. Surprisingly, the effect of Rgl2 on anchorage-independent growth could not be rescued by constitutively activated RalA, suggesting a novel Ral-independent function for Rgl2 in transformation. Finally, we determined that Rgl2 and RalB both localized to the leading edge, and this localization of RalB was dependent on endogenous Rgl2 expression. In summary, our observations support nonredundant roles for RalGEFs in Ras-mediated oncogenesis and a key role for Rgl2 in Ral activation and Ral-independent PDAC growth.
Keywords: Carcinogenesis, Cell Motility, Cellular Regulation, G Proteins, Oncogene, Pancreas, Ras, Signal Transduction
Introduction
Mutational activation of the KRAS oncogene is the most prevalent genetic alteration associated with essentially 100% of pancreatic ductal adenocarcinomas (PDACs)3 (1). Therefore, considerable effort has been made to develop inhibitors of K-Ras for PDAC treatment. Currently, most of these efforts are focused on inhibitors of K-Ras downstream effector signaling, with numerous inhibitors of the Raf-MEK-ERK mitogen-activated protein kinase (MAPK) or phosphatidylinositol 3-kinase (PI3K)-AKT pathway currently under clinical evaluation (2). However, recent studies suggest that additional Ras effector pathways may also play significant roles in Ras-mediated oncogenesis. In particular, there is increasing evidence for the importance of Ral guanine nucleotide exchange factors (RalGEFs), activators of the Ral (Ras-like) small GTPases (3), in cancer growth (4).
Ral GTPases function as GDP/GTP-regulated binary switches that regulate extracellular stimulus-regulated signaling networks that control a diversity of cellular processes, including exocytosis, endocytosis, activation of transcription factors, and actin cytoskeletal reorganization (4). Surprisingly, despite their significant sequence identity (82%) and interaction with a common set of effectors, the related RalA and RalB isoforms serve very distinct functions in oncogenesis. Importantly, Chien and White (5) found that RalA is necessary for tumor cell anchorage-independent growth, whereas RalB is necessary for tumor but not normal cell survival. Our studies determined that RalA but not RalB was essential for PDAC cell line anchorage-independent growth and tumorigenic growth in vivo (6). In contrast, RalB and, to a lesser degree, RalA were essential for PDAC cell Matrigel invasion and lung colony metastatic tumor growth in a lung colonization model. Distinct roles for RalA and RalB have also been described in bladder (7) and prostate (8) cancer growth. These distinct biological roles are due, in part, to their distinct subcellular localization, with RalA found at the plasma membrane and endosomes, whereas RalB is found only at the plasma membrane (9). The critical importance of subcellular localization in Ral function was also demonstrated by our recent observation that Aurora-A phosphorylation of RalA resulted in relocation from the plasma membrane to endosomes, causing a change in effector utilization required to promote PDAC anchorage-independent and tumorigenic growth (10).
Despite the importance of Ral activation in cancer growth, the mechanisms by which Ral GTPases are aberrantly activated remain poorly understood. In our analyses of PDAC tumors and cell lines, we observed elevated levels of GTP-bound active RalA and RalB (6, 11). At least six Ral-specific mammalian guanine nucleotide exchange factors (RalGEFs) (4) and two Ral-specific GTPase-activating proteins (RalGAPs) (12) have been identified. The RalGEFs activate the Ral GTPases by catalyzing the exchange of GDP for GTP, whereas the RalGAPs inactivate Ral GTPases by increasing hydrolysis of bound GTP to GDP. Four of the RalGEFs contain Ras association (RA) domains that can function as Ras effectors: RalGDS, Rgl1, Rgl2/Rlf, and Rgl3 (13–16). In light of the high frequency of KRAS mutations in PDAC, a logical hypothesis is that one or more RA domain-containing RalGEFs are required for Ral activation in PDAC. However, the two pleckstrin homology domain-containing RalGEFs may also be activated, perhaps indirectly by K-Ras activation of their phosphotidylinositol 3-kinase effectors (17). Moreover, although no mechanism for regulation of RalGAPs has been described, another possible mechanism of Ral activation in PDAC could involve loss of RalGAP activity, similar to the hyperactivation of Ras due to loss of the neurofibromin RasGAP in glioblastoma (18) and hyperactivation of Rho due to loss of the RhoGAP DLC1 in multiple cancer types (19).
The four RA domain-containing RalGEFs share an identical domain organization (Fig. 1). However, overall sequence identity within these domains is limited (17–33%), with very little identity in the sequences flanking the functional domains. This sequence divergence may result in important functional differences between the GEFs. For example, a RalGEF-independent function of RalGDS has been described involving activation of AKT (20). Although ectopic overexpression of activated Ras proteins can activate the different RalGEFs in vivo (21–24), there are currently no data implicating a specific RalGEF in endogenous mutant KRAS-mediated growth transformation. Mice lacking RalGDS have a deficiency in oncogenic H-Ras-induced skin tumor formation (25). However, it is unclear whether RalGDS is the only RalGEF expressed in mouse skin or whether it possesses a unique function in tumorigenesis. We found that a constitutively activated mutant of Rgl2/Rlf was sufficient to phenocopy activated H-Ras to cause anchorage-independent growth transformation of immortalized human embryonic kidney (HEK-HT) cells, although whether activated forms of other RalGEFs were also transforming was not evaluated (26). Interestingly, we found that RalA and RalB were not activated concurrently in the same PDAC cell lines (11), indicating distinct mechanisms of regulation. All four family members are widely expressed in many tissues, but their relative abilities to activate RalA and RalB have not been investigated. The different RalGEFs may also exhibit distinct subcellular localization, leading to spatially distinct Ral activation and engagement of different effector populations (27).
FIGURE 1.
Domain structure and sequence identity of RA domain-containing RalGEFs. The four RalGEFs that can serve as effectors of Ras share the same domain topology, with an N-terminal Ras exchange motif (REM), followed by the CDC25 homology RalGEF catalytic domain and a C-terminal RA domain. The sequence identities to RalGDS are indicated below each domain. The precise function of the Ras exchange motif domain is not known, but deletion analysis of RalGDS indicates that it is dispensable for RalGEF catalytic activity in vivo. Unlike other CDC25 homology domain-containing GEFs, the RalGEFs are selective for the two Ral isoforms. The RA domain binds to GTP-bound Ral. Expression vectors encoding full-length mouse RalGDS (NM_009058), Rgl1 (NM_016846), Rgl2 (NM_009059), and Rgl3 (NM_023622) were generated that included either N-terminal HA epitope or GFP sequences. To generate plasma membrane-targeted versions that mimic constitutive association with activated K-Ras, the C-terminal 20 residues of human K-Ras4B were added to the C terminus of each RalGEF; this sequence includes the CAAX prenylation signal sequence and the polybasic second signal required for plasma membrane association. Rlf-CAAX contains an N-terminal HA epitope tag (MAYPYDVPDYASTD) followed by residues 2–532 of mouse RGL2 and then 12 vector-encoded residues (HAPGRHGRGIVN) and then terminating with 20 residues that include the K-Ras4B sequence (KMSKDGKKKKKKSKTKCVIM). The C-terminal 246 residues of Rlf, including the RA domain, are deleted.
The essential requirement for Ral GTPase activation in PDAC growth has stimulated our interest in evaluating candidate anti-Ral approaches for PDAC treatment (10, 28). Establishing a role for RalGEFs in activation of Ral GTPases in PDAC may validate targeting mechanisms of RalGEF activation for pharmacologic inhibition of Ral GTPases (29). In this study, we first compared the biochemical and biological functions of the four RA domain-containing RalGEFs and determined that they equally activated RalA and RalB but nevertheless exhibited widely variable plasma membrane association-dependent transforming potencies. We identified Rgl2 protein overexpression in PDAC tumors and determined that Rgl2 was essential for PDAC cell line anchorage-independent growth and invasion in PDAC lines in vitro, in part through a Ral-independent mechanism(s) independent of AKT activation. Finally, we demonstrated that RalB subcellular localization to the plasma membrane at the leading edge was dependent on Rgl2 expression, suggesting that RalGEFs may also regulate spatially restricted Ral activation.
EXPERIMENTAL PROCEDURES
Cell Lines
Immortalized human embryonic kidney cells (HEK-HT) expressing SV40 T-Ag and t-Ag as well as the catalytic subunit of telomerase (hTERT) were described previously (26). Human PDAC cell lines were obtained from ATCC or were provided by Dr. Murray Korc (Dartmouth). To establish stably infected polyclonal cell populations, the indicated parental cell lines were infected with retroviruses generated from the retroviral vectors encoding the indicated proteins, followed by selection with puromycin. To establish stably infected mass populations for stable Rgl2 knockdown cells, PDAC lines were infected with lentivirus-based vectors encoding the nonspecific negative control or sequences designed against human Rgl2 (see below for details), followed by selection with puromycin. Lentivirus was established by co-transfection of the pLKO.1 shRNA vectors described below with the helper plasmids psPAX2 and pMD2.G in HEK-293T cells with the calcium phosphate precipitation method (30).
PDAC Patient Samples
Deidentified matched normal and tumor pancreas samples were collected from the University of North Carolina Tissue Procurement Facility after approval by the University of North Carolina institutional review board. Samples were flash frozen in liquid nitrogen at the time of operation and subsequently harvested using an Nonidet P-40-based lysis buffer with 100× phosphatase inhibitors (phosphatase inhibitor mixtures 1 and 2; Sigma) and 1× protease inhibitor (Complete protease inhibitor mixture tablets; Roche Applied Science).
RalGEF Expression Vectors
The construction of the pBabe HAII-puro Rgl2-CAAX vector was described previously (26). Full-length, wild-type mouse RalGDS, Rgl1, Rgl2, and Rgl3 open reading frames were isolated from a mouse cDNA library by PCR with primers to incorporate the following flanking restriction sites: RalGDS, BamHI and EcoRI; Rgl1, BamHI and BamHI; Rgl2 and Rgl3, EcoRI and SalI. These fragments were then subcloned into the pBabe HAII-puro retrovirus expression vector, which adds an in-frame sequence to encode an N-terminal hemagglutinin (HA) epitope tag sequence. In order to add sequences to encode for the K-Ras4B C-terminal sequence to the C terminus of the RalGEFS, we ligated annealed oligonucleotides representing the 20 C-terminal amino acids of human K-ras4B and restriction site overhangs to the above RalGEF open reading frames in the pBabe-puro constructs. The primers used were as follows: for RalGDS-CAAX, Rgl1-CAAX, and Rgl3-CAAX (MluI and SalI restriction sites), 5′-CGCGTCTAAGCAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAATTATGTGAG-3′ (top) and 5′-TCGACTCACATAATTACACACTTTGTCTTTGACTTCTTTTTCTTCTTTTTACCATCTTTGCTTAGA-3′ (bottom); for Rgl3-CAAX (MluI and MluI restriction sites), 5′-CGCGTCTAAGCAAAGATGGTAAAAAGAAGAAAAAGAAGTCAAAGACAAAGTGTGTAATTATGTGAG-3′ (top) and 5′-CGCGCTCACATAATTACACACTTTGTCTTTGACTTCTTTTTCTTCTTTTTACCATCTTTGCTTAGA (bottom). RalA (S31N) and RalB (S28N) were generated by site-directed mutagenesis from pBabe-puro RalA and RalB (11). Note that the same analogous residue was mutated in both Ral isoforms, but the RalA isoform used is a splice variant that contains three additional N-terminal residues. A hygromycin-resistant expression vector for RalAQ75L was constructed subcloning the cDNA sequence from pBabe-puro-RalAQ75L (11) into the pBabe-hygro vector. Lentiviral shRNA vectors designed to target human Rgl2 (accession number NM_004761) in the pLKO.1 backbone were obtained from Open Biosystems. The target sequences are as follows: RNAi 1, 5′-GCAGTGTCTATAAGAGCATTT-3′; RNAi 2, 5′-CCATTCTGAATGGTGGCAATT-3′. The nonspecific vector is the Mission Non-Target shRNA control vector (Sigma-Aldrich) that does not target human genes.
Immunoblotting
Lysates were obtained from the indicated exponentially growing cell lines and immunoblotted with α-RalA (Transduction Laboratories), α-RalB (Upstate Biotechnology, Inc.), α-β-actin (Sigma), 12CA5 α-HA (hemagglutinin tag; Roche Applied Science), Rgl2 (Abnova), phospho-AKT (Ser473; Cell Signaling), AKT (Cell Signaling), phospho-ERK (Thr202/Tyr204; Cell Signaling), ERK (Cell Signaling), and vinculin (Sigma) antibodies. RalA-GTP and RalB-GTP levels were determined by pull-down with a bacterially expressed RalBP1 RalBD-GST, followed by immunoblot with the RalA or RalB antibodies above, as described previously (31, 32).
Immunofluorescence
The stably infected HEK-HT cells or CFPAC-I cells were plated on glass slides and 24 h later were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100. For the HEK-HT analysis, cells were incubated with the mouse 12CA5 α-HA primary antibody (Roche Applied Science) for 1 h at room temperature, followed by the Alexa Fluor 488-conjugated α-mouse secondary antibody (Molecular Probes) for 1 h. For the endogenous Rgl2 analysis, CFPAC-I cells were incubated with the mouse α-Rgl2 primary antibody (Abnova) for 1 h at room temperature followed by the Alexa Fluor 488-conjugated α-mouse secondary antibody (Molecular Probes) for 1 h. For the endogenous Rlg2, RalB, and RalB analysis, cells were incubated with 1) the mouse α-RalA (Transduction Laboratories), 2) α-Rgl2 and rabbit α-cortactin (Cell Signaling), or 3) α-RalB and rabbit α-cortactin for 1 h at room temperature, followed by both the Alexa Fluor 488-conjugated α-mouse secondary (Molecular Probes) and the Alexa Fluor 568-conjugated α-rabbit secondary antibodies (Molecular Probes) for 1 h at room temperature. Cells were mounted with Fluorosave (Calbiochem) and visualized with a Zeiss 510 LSM confocal microscope. Digital images were processed and adjusted for contrast and brightness with the LSM 5 Image browser software (Zeiss, Thornwood, NY).
Anchorage-independent Growth Assays
Anchorage-independent growth assays consisted of suspending cells in soft agar as described previously (33). Briefly, cells were trypsinized, and 4 × 104 cells (HEK-HT) or 1 × 104 cells (PDAC lines) were resuspended in growth medium containing 0.4% agar in 6-well plates. Cells were maintained at 37 °C for 4 weeks (HEK-HT cells) or 2 weeks (PDAC lines). After this time, viable colonies were stained with the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide viability stain. The total number per plate of viable colonies of >10 cells was quantified by counting the number of colonies in five representative fields of view within each plate. Results are expressed as mean ± S.D.
Invasion Assays
Invasion assays were performed with growth factor-reduced Matrigel invasion chambers (BD Biosciences) according to the manufacturer's protocol. Briefly, cells were dissociated from the plates with TrypLE Express (Invitrogen), and 1 × 105 cells were resuspended in triplicate in serum-free RPMI 1640 containing 1% bovine serum albumin into the upper chamber. RPMI 1640 containing 3% fetal bovine serum as a chemoattractant was added to the bottom well. Cells were allowed to invade for 22 h at 37 °C, and then non-invaders were removed. Invading cells were fixed and stained with the Diff-Quik stain set (Dade Behring Inc., Newark, DE). Five fields were counted for each chamber, and the total number of cells counted per chamber was used for calculating the average number of invading cells. Results are expressed as mean ± S.D.
Reverse Transcription (RT)-PCR Analysis
Total mRNA was extracted from the indicated PDAC cancer cell lines with an RNeasy kit (Qiagen). RNA was quantified by measuring the absorbance at 260 nm. cDNA libraries were then formed by reverse transcription using a PE Applied Biosystems high capacity cDNA archive kit. 10 μg of RNA was used for the reverse transcription in all cases. An RNase inhibitor (RNasin; Promega) was used during the reverse transcription. Specific primer sets were then used to amplify the respective genes by PCR amplification. PCR amplification without cDNA was performed in parallel as a negative control. Primers used were designed with the help of the Primer3 software (available on the World Wide Web) and are as follows, with forward primer followed by reverse: RalGDS, 5′-GTCTCAGGGCTCTGCAACTC-3′ and 5′-TCTTCAGCTTCCGGTCATCT-3′; Rgl1, 5′-AACCACTCAGAGGCTGAGGA-3′ and 5′-AGACAGAGCGCTTGTGGATT-3′; Rgl2, 5′-GCCTCTGATTGCCGTATCAT-3′ and 5′-CTCCATCCATGGCGTAGAAT-3′; Rgl3, 5′-CCCCCTCAAGTCCTAGAAGC-3′ and 5′-TACAGGTTCCCGTGGTCATT-3′; β-actin, 5′-GCGGGAAATCGTGCGTGACATT-3′ and 5′-GATGGAGTTGAAGGTAGTTTCGTG-3′.
Statistical Analysis
Data were analyzed using an unpaired t test. Values are shown as means ± S.D. A p value of <0.05 was considered significant.
RESULTS
Although our studies identified Ral activation in PDAC and we established critical and distinct roles for RalA and RalB in PDAC cell line growth (6, 11), a mechanism for Ral activation in PDAC remains to be determined. Because mutationally activated K-Ras is found in essentially all PDACs, a logical mechanism would involve one or more of the four RA domain-containing RalGEFs (RalGDS, Rgl1, Rgl2, and Rgl3) that function as effectors of Ras (Fig. 1). The demonstration that a RalGDS deficiency alone impaired mutant H-Ras-induced skin carcinoma formation suggests non-redundant roles for RA domain-containing RalGEFs Ras-mediated oncogenesis (25). Therefore, we compared the activities of the four RalGEFs. Because antibodies were not available for recognition of all four RalGEFs, we introduced an N-terminal HA epitope tag to each RalGEF to monitor cellular expression.
Activated Ras promotes RalGEF activation by promoting RalGEF association with the plasma membrane, and previous studies showed that the addition of a Ras C-terminal plasma membrane targeting sequence results in constitutively activated RalGDS (34), Rgl2 (24), and Rgl3 (13). These previous studies verified that the addition of the Ras plasma membrane targeting sequence promoted association with membranes. In the case of RalGDS and Rgl3, highly increased membrane association was demonstrated by cell compartment fractionation, and Rgl2 membrane localization was demonstrated with microscopy. We therefore generated chimeric RalGEF proteins that terminated with the C-terminal plasma membrane targeting sequence of K-Ras4B (Fig. 1). This sequence includes the CAAX tetrapeptide motif that signals for posttranslational modification by farnesyl isoprenoid addition as well as the polybasic sequence required for plasma membrane association.
We showed previously that the RalGEF-Ral effector pathway is necessary and sufficient for activated H-Ras growth transformation of immortalized HEK-HT human embryonic kidney epithelial cells (26). Therefore, we utilized HEK-HT cells to evaluate the transforming activity of the four Ras-binding RalGEFs. We established mass populations of HEK-HT cells stably expressing wild type (WT) and membrane-targeted (designated CAAX, for cysteine, aliphatic, and terminal amino acids) versions of each RalGEF. As a positive control, we also established cells expressing the activated K-Ras4B(G12V) protein.
Wild-type RalGEFs Exhibit Similar Cytoplasmic Subcellular Localization
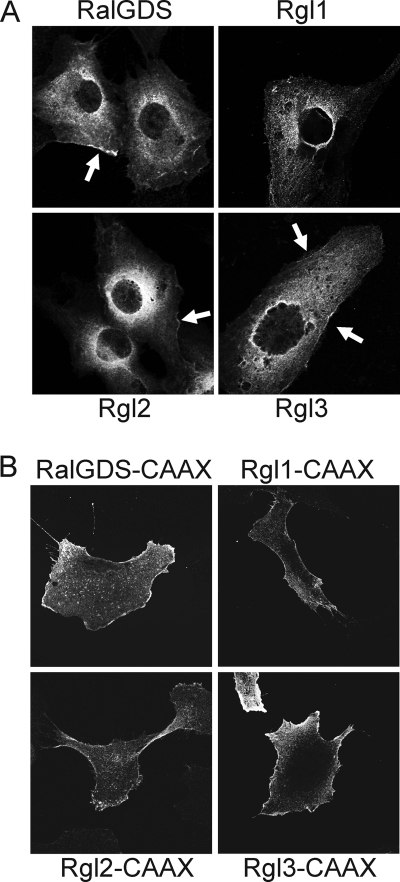
We first compared the subcellular localization of the wild-type RalGEFs and found that they showed similar diffuse cytoplasmic localization, with a concentration in the perinuclear region, with nuclear exclusion (Fig. 2). Additionally, each exhibited limited and restricted plasma membrane association. We also saw similar localizations of all four RalGEFs when GFP-tagged versions of the wild type RalGEFs were expressed transiently in COS-7 cells (data not shown). This subcellular distribution is similar to the punctuate endosome localization described previously for Rgl2 (23, 24, 26). In contrast, all of the CAAX motif-terminating RalGEFs were localized primarily and evenly along the plasma membrane in the HEK cells, with a significant loss of perinuclear staining (Fig. 2B). This distribution is similar to that seen with K-Ras4B (35). Thus, these membrane-targeted variants are expected to mimic the activity of authentic RalGEFs when associated with plasma membrane-associated activated K-Ras.
FIGURE 2.
RalGEFs exhibit similar cytosolic subcellular localization and plasma membrane association is enhanced by the addition of the K-Ras4B plasma membrane targeting sequence. A, wild type RalGEFs exhibit a cytosolic subcellular localization. Mass populations of HEK-HT cells were stably infected with pBabe-puro expression vectors expressing the indicated HA-tagged RalGEFs and were grown on glass coverslips. After labeling with anti-HA antibody followed by Alexa Fluor 488-conjugated secondary antibody, the cells were visualized by confocal microscopy. Arrows, plasma distinct plasma membrane localization. B, the addition of Ras membrane targeting sequence enhances RalGEF plasma membrane association. Analyses were done as described in A with chimeric RalGEFs terminating with the K-Ras4B plasma membrane targeting sequence. Data shown are representative of at least two independent experiments.
Plasma Membrane-targeting Promotes RalGEF Transforming Activity
We next evaluated the transforming abilities of wild type and membrane-targeted RalGEFs using HEK-HT cells. For these analyses, we included a previously described constitutively activated variant of Rgl2, designated Rlf-CAAX, which contains a similar K-Ras4B C terminus but additionally contains a deletion of the C-terminal RA domain (Fig. 1) (24). In contrast to the previously used Rlf-CAAX, our constructs retained the RA domain (Fig. 1), so that they may presumably retain their full regulation by Ras. We found previously that Rlf-CAAX can phenocopy activated H-Ras(12V) and promote the anchorage-independent growth of HEK-HT cells (26). Therefore, we utilized HEK-HT cells to evaluate the transforming activity of the four Ras-binding RalGEFs. We first verified expression of the ectopically introduced RalGEFs by Western blot analyses of stably infected HEK-HT cells (supplemental Fig. 1). The expression of all WT RalGEFs was detected readily, although Rgl2 and Rgl3 showed higher expression levels. In contrast, although Rgl3-CAAX and Rlf-CAAX (ΔRA) expression was also seen at comparable levels, we consistently found very low levels of RalGDS-CAAX, Rgl1-CAAX, and Rgl2-CAAX. Although the basis for this lower steady-state expression was not clear, it was reproducibly seen in independently established HEK-HT cell lines and when transiently expressed in 293T cells (data not shown).
We then assessed the ability of the RalGEF-expressing cells to proliferate in an anchorage-independent environment by quantitation of colony formation in soft agar (Fig. 3). Similar to our previous observations with activated H-Ras(12V) (26), ectopic expression of activated K-Ras(12V) also stimulated HEK-HT soft agar growth. As we observed previously (11, 26), cells expressing Rlf-CAAX also showed a comparable enhancement in the frequency of soft agar colony formation. In contrast, ectopic overexpression of each of the four WT RalGEFs did not cause a significant increase in colony formation above that seen for the vector control cells. However, ectopic expression of the plasma membrane-targeted versions of Rgl1, Rgl2, and Rgl3, but not RalGDS, did promote soft agar growth. When considered with the significantly lower level of steady-state expression of Rgl1-CAAX and Rgl2-CAAX when compared with their authentic counterparts, this suggests that membrane targeting greatly enhanced Rgl1 and Rgl2 transforming activity, whereas membrane targeting caused a moderate increase in Rgl3 transforming activity. Because RalGDS-CAAX expression was much lower than that of WT RalGDS, we cannot conclude that membrane targeting did not enhance RalGDS transforming activity. Finally, because the RA domain-deleted version of Rgl2 (Rlf-CAAX) was expressed at a significantly higher steady state level than the full-length counterpart (Rgl2-CAAX), we suspect that deletion of the RA domain did not significantly alter Rgl2 activity in vivo. This would also be consistent with the previous determination that deletion of the RA domain did not alter Rgl2 RalGEF catalytic activity in vitro (24).
FIGURE 3.
Membrane association promotes RalGEF transformation of HEK-HT cells. HEK-HT cells were suspended in soft agar as described (57), and the number of proliferating viable colonies of >30 cells were quantitated after 4 weeks. Data shown are the average ± S.D. (error bars) of triplicate plates and are representative of three independent experiments. *, p < 0.005 versus vector control.
Wild-type and Activated RalGEFs Activate RalA and RalB
Our previous finding that RalA and RalB activities varied widely when evaluated in PDAC cell lines suggested that the different RalGEFs may exhibit preferential abilities to activate RalA and RalB. This possibility was supported by RNAi suppression studies, where depletion of RalGDS caused accumulation of binucleate cells as seen with RalA depletion in HeLa cells (36). In contrast, depletion of Rgl1 caused bridged cells, as was seen with RalB depletion in HeLa cells. No effect was seen with Rgl2 or Rgl3 depletion. To address this possibility, we did pull-down analyses to assess RalA and RalB activation by ectopic expression of WT or membrane-targeted RalGEFs in HEK-HT cells (supplemental Fig. 1). As expected, K-Ras(12V) cells showed elevated RalA and RalB GTP-bound protein levels. Ectopic overexpression of WT Rgl2 in particular caused a strong increase in both RalA and RalB activity. A lesser increase was seen with Rgl3-expressing cells, whereas no significant increase was seen for RalGDS- or Rgl1-expressing cells. Surprisingly, for the membrane-targeted RalGEFs, only significant RalA and RalB activation was seen with Rgl3-CAAX and Rlf-CAAX. However, due to the much lower expression levels of the CAAX versions of RalGDS, Rgl, and Rgl2, it was difficult to determine whether their inability to activate RalA and RalB was due to any inherent differences between the GEFs. Furthermore, despite the stronger transforming activity of Rgl2-CAAX, it showed a reduced ability to increase Ral-GTP levels when compared with WT Rgl2. Thus, we saw that the different RalGEFs did not differentially regulate RalA and RalB activity and that the Ral-GTP levels did not correlate with transforming activity.
Rgl2 Overexpression in PDAC Tumors
The RalGEFs are all widely expressed in different human tissues, with RalGDS and Rgl2 showing the widest tissue distribution (15, 37–39). UniGene analysis (see the NCBI Web site) identified RalGDS, Rgl2, and, to a lesser extent, Rgl1 mRNA in pancreas and pancreatic cancer tissue (Rgl3 was not in the data base). The Oncomine data base identified mRNA expression of all four GEFs in normal and pancreatic cancer tissue by microarray analysis (40), although their relative abundances were not determined. Interestingly, the Oncomine data base also shows that Rgl2 but not any of the other GEFs was overexpressed in pancreatic cancer versus normal tissue. To more directly compare the expression levels of the RalGEFs in PDAC, we performed RT-PCR to determine the presence of mRNA of the four RalGEFs in several PDAC cell lines (Fig. 4A). We readily detected both RalGDS and Rgl2 mRNA in all of the PDAC lines. In contrast, Rgl1 and Rgl3 transcription was readily detected only in CFPAC-I cells. Rgl1 and Rgl3 mRNA have been detected previously in lung tissue (15, 39). Thus, as a positive control for Rgl1 and Rgl3 expression, we also readily detected Rgl1 and Rgl3 mRNA in several NSCLC lines (data not shown). Based on these observations and on the transforming activity of membrane-targeted Rgl2, we focused on Rgl2.
FIGURE 4.
Pancreatic carcinoma cells express multiple RalGEFs. A, RT-PCR detection of RalGEF mRNA colorectal and pancreatic ductal adenocarcinoma cell lines, with β-actin as a control. Total cellular RNA was isolated from the indicated cell lines. After reverse transcription, PCR was performed with RalGEF-specific and β-actin-specific primers. B, Rgl2 protein expression in PDAC cell lines. Lysates from the cell lines were separated by SDS-PAGE and transferred to a PVDF membrane and blotted for Rgl2 protein expression. A parallel blot for β-actin was done to ensure equivalent loading of total cellular protein. C, overexpression of Rgl2 protein in tumor (T) versus normal (N) matched PDAC patient samples, as analyzed by Western blot analysis, with γ-tubulin monoclonal antibody (Sigma) as a control for equivalent total protein.
To determine the protein levels of Rgl2 in PDAC lines, we have performed Western blot analysis (Fig. 4B). Consistent with the RT-PCR analysis, we detected Rgl2 protein in all PDAC cell lines. However, its expression level was highly variable between lines. We attempted to generate rabbit antisera to detect RalGDS, Rgl1, and Rgl3; however, these antisera were not able to recognize endogenous proteins. We also tested various commercial RalGDS antibodies, and we could not detect even overexpressed RalGDS in HEK-HT lysates. Therefore, we could not perform similar analyses of endogenous RalGDS, Rgl1, and Rgl3 protein. Additionally, we used Western blot analyses and determined that Rgl2 protein was overexpressed in three of the five PDAC patient samples tested when compared with normal tissue (Fig. 4C), suggesting that Rgl2 expression is also elevated in PDAC cell lines.
RalGEFs Promote Ral Activation and PDAC Anchorage-independent Growth and Invasion through Matrigel
To determine if Ral activation and PDAC growth are dependent on RalGEF function, we utilized dominant-negative mutants of Ral that antagonize RalGEF function (41). The RalA(31N) and RalB(28N) mutants are analogous to the Ras(17N) dominant negative and should form nonproductive complexes with both RA and pleckstrin homology domain RalGEFs, thus preventing RalGEF-dependent Ral activation (42); however, they should not impair Ral activation due to loss of RalGAP function. Because the RalGEFs contain other domains not necessarily involved in Ral activation, the use of dominant negatives may block RalGEF functions besides Ral activation. For these analyses, we utilized three PDAC cell lines that we determined previously to be strongly dependent on RalA and RalB function for anchorage-independent growth and invasion through Matrigel (6). We established SW-1990, MIA PaCa-2, and CFPAC-I PDAC cells stably expressing dominant-negative RalA(31N) or RalB(28N) (Fig. 5A). First, we found that dominant negative Ral protein expression was well tolerated and expressed severalfold above endogenous Ral protein expression. Moreover, we did not observe significant alteration in anchorage-dependent growth on plastic. Ectopic expression of either RalA(31N) and RalB(28N) significantly reduced the steady-state levels of both RalA-GTP and RalB-GTP in SW-1990 and CFPAC-I cells, suggesting that RalGEF activity is a key basis for Ral activation in these two PDAC lines (Fig. 5A). In contrast, in MIA PaCA-2 cells, although RalB(28N) reduced RalB-GTP levels, RalA(31N) expression did not significantly reduce RalA-GTP levels. Thus, Ral activation in MIA PaCa-2 cells appears to be less dependent on RalGEF function.
FIGURE 5.
Dominant negative Ral mutant inhibitors of RalGEFs impair PDAC soft agar growth and invasion through Matrigel. A, effect on the RalGEFs in RalA- and RalB-GTP levels in pancreatic ductal adenocarcinoma cell lines. The indicated cell lines were stably infected with retroviral pBabe empty vector or Ral dominant negatives (RalA(31N) and RalB(28N)) and selected with puromycin. RalA- and RalB-GTP were detected as previously described (29) by pull-down from the indicated PDAC lines, followed by immunoblot analysis with the RalA or RalB antibody. Total RalA and RalB were derived from immunoblotting total lysate and show stable expression of RalA(31N) or RalB(28N), respectively. Blot analysis with anti-β-actin antibody was done to verify equivalent total protein. B, role of the RalGEFs in PDAC anchorage-independent growth. The number of proliferating viable colonies of >30 cells were quantitated after 2 weeks. Data shown are the average ± S.D. (error bars) of triplicate plates and are representative of at least two independent experiments. C, role of the RalGEFs in PDAC invasion. The indicated cells were dissociated and resuspended in serum-free growth medium containing 1% BSA and incubated at 37 °C in the upper chamber of a Matrigel invasion chamber. Data shown are the percentage of invaded cells relative to vehicle and are the average ± S.D. of triplicate chambers and are representative of at least two independent experiments. *, p < 0.05 versus pBabe vector control. **, p < 0.005 versus pBabe vector control.
We showed previously that RalA but not RalB shRNA reduced the anchorage-independent growth of these three PDAC cell lines (6). Therefore, we determined if inhibition of RalGEF function would alter the anchorage-independent growth of PDAC cells. Expression of RalA(31N) caused a greater than 50% reduction in soft agar colony formation for all three cell lines, whereas expression of RalB(28N) reduced significantly the colony formation activity of SW-1990 (∼80% reduction) but resulted in no statistically significant reduction for MIA PaCa-2 or CFPAC-I cells (Fig. 5B). These results support the critical role of RalGEFs in PDAC anchorage-independent growth. RalB(28N) blocked total RalA-GTP levels but did not affect CFPAC-I colony formation, whereas RalA(31N) blocked RalA-GTP levels and anchorage-independent growth. This might be due to spatial differences in RalA and RalB activation. Due to the different subcellular localization of RalA and RalB (9), RalB(28N) might block a non-transforming pool of RalA, whereas RalA(31N) blocks a transforming pool. Interestingly, anchorage-independent growth of MIA PaCa-2 was reduced by RalA(31N), despite RalA-GTP levels not being lowered, suggesting that there may be Ral-independent effects of the RalGEFs in anchorage-independent growth.
We determined previously that RalA shRNA reduced SW-1990 and MIA PaCa-2 invasion through Matrigel, whereas RalB shRNA significantly reduced invasion for all three lines (6). RalB(28N) but not RalA(31N) expression statistically reduced (p < 0.05) MIA PaCa-2 and CFPAC-I invasive ability of all three PDAC lines (Fig. 5C). Taken together, these results support an important role for RalGEFs in PDAC growth and invasion, at least in part, through Ral activation. Interestingly, these results also suggest that there are RalGEFs that are somewhat specific for RalA or RalB activation, because in general, RalA(31N) more strongly blocked anchorage-independent growth, and RalB(28N) more strongly blocked invasion. However, in MIA PaCa-2 cells, there is no correlation of anchorage-independent growth with RalA-GTP. This strongly suggests that sequestration of the RalGEFs by dominant negatives may be altering anchorage-independent growth and invasion by means other than altering Ral activity. Indeed, in light of our demonstration of RalA-independent effects of Rgl2 in anchorage-independent growth, it is difficult to correlate the phenotypes of the dominant negatives directly with reductions in total cell steady-state RalA and RalB activity.
Rgl2 Promotes PDAC Ral Activation, Anchorage-independent Growth, and Invasion through Matrigel
Because we observed Rgl2 protein overexpression in PDAC patient samples (Fig. 4C), we next wanted to determine if Rgl2 was important for the RalGEF-dependent activities identified by Ral dominant negative mutant expression in SW-1990, MIA PaCa-2, and CFPAC-I PDAC cell lines. Using two different shRNA vectors that target different Rgl2 sequences, we established mass populations of each PDAC cell line with stable suppression of Rgl2 protein expression (Fig. 6A). Significant reduction in both RalA-GTP and RalB-GTP levels was seen in SW-1990 and CFPAC-I cells (Fig. 6A), which is consistent with our Rgl2 overexpression data that showed increased levels of both RalA and RalB activation in HEK-HT cells (Fig. 3A). However, Rgl2 knockdown in MIA PaCa-2 cells only slightly reduced RalB-GTP levels and did not detectably reduce RalA-GTP levels (Fig. 6A).
FIGURE 6.
The RalGEF Rgl2 plays a crucial role in PDAC. The indicated cell lines were stably infected with a nonspecific shRNA vector (NS), which does target any human genes, or two independent shRNA vectors targeting Rgl2. A, Rgl2 is responsible for RalA and RalB activation in PDAC lines. RalA- and RalB-GTP were detected as described previously (29) by pull-down from the indicated PDAC lines, followed by immunoblot analysis with the RalA or RalB antibody. Total RalA and RalB were derived from immunoblotting total lysate. Blot analysis with anti-β-actin antibody was done to verify equivalent total protein. Data shown are representative of two independent experiments. B, role of the Rgl2 in PDAC anchorage-independent growth. The number of proliferating viable colonies of >30 cells was quantitated after 2 weeks. Data shown are the average ± S.D. (error bars) of triplicate plates and are representative of at least two independent experiments. C, role of Rgl2 in PDAC invasion. The indicated cells were dissociated and resuspended in serum-free growth medium in the upper chamber of a Matrigel invasion chamber. After 22 h, the non-invaded cells were removed, and the chambers were fixed, stained, and counted under a microscope. Data shown are the percentage of invaded cells relative to vehicle and are the average ± S.D. of triplicate chambers and are representative of at least two independent experiments. *, p < 0.05 versus nonspecific shRNA control.
Rgl2 suppression was associated with strong reduction in both soft agar colony growth (Fig. 6B) and invasion through Matrigel (Fig. 6C) for all three cell lines, although the anchorage-independent growth suppression was only statistically significant for Rgl2 shRNA 1 in CFPAC-I cells. We concluded that Rgl2 is essential for Ral activation in a subset of PDAC cells and for PDAC cell anchorage-independent growth and invasion in vitro. Because RalA and RalB activation in MIA PaCa-2 cells is largely Rgl2-independent and there is high expression of RalGDS, we also sought to determine whether RalGDS is crucial for Ral activation in PDAC lines. However, we could not find any antibodies that recognized endogenous or overexpressed RalGDS. Furthermore, using the identical method for lentiviral shRNA knockdown, we were unable to knock down the expression of RalGDS, as judged by RT-PCR analysis (data not shown).
RalA-independent Effects of Rgl2 Mediate Anchorage-independent Growth
Because knockdown of Rgl2 and expression of a RalA dominant negative in MIA PaCa-2 cells caused a decrease in anchorage-independent growth but did not decrease RalA-GTP levels, we hypothesized that Rgl2 may promote MIA PaCa-2 anchorage-independent growth in part through a RalA-independent mechanism(s). To address this possibility, we determined whether ectopic expression of a constitutively activated mutant of RalA (Q75L) could rescue the effects of Rgl2 knockdown. Western blot analyses verified expression of the altered mobility form of RalA(75L) in MIA PaCa-2 cells at a level comparable with that of endogenous wild type RalA (Fig. 7A). Surprisingly, activated RalA did not rescue the anchorage-independent growth phenotype to the levels seen in the nonspecific shRNA control, with only partial rescue seen (Fig. 7B). Together with our previous studies (6, 11), these data indicate that Rgl2 promotes anchorage-independent growth through RalA-dependent and independent mechanisms.
FIGURE 7.
Activated RalA cannot rescue the anchorage-independent growth defect of Rgl2 knockdown in MIA PaCa-2 cells. The indicated cell lines were stably infected with a puromycin-resistant nonspecific shRNA vector (NS), which does target any human genes, or puromycin-resistant Rgl2 shRNA 1 vector targeting Rgl2, without and with ectopic expression of constitutively activated RalA(Q75L). A, blot analysis showing expression levels of endogenous wild type Rgl2 and ectopic RalA(Q75L) in the indicated PDAC lines as well as anti-tubulin antibody to verify equivalent total protein. Data shown are representative of two independent experiments. B, the number of proliferating viable colonies of >30 cells was quantitated after 2 weeks. Data shown are the average ± S.D. (error bars) of triplicate plates and are representative of at least two independent experiments. C, blot analysis demonstrating phosphorylated and total AKT and ERK levels upon Rgl2 knockdown in MIA PaCa-2 cells. Blot data for Rgl2 and β-actin are duplicated from Fig. 6A in this panel, where the same cell lysates were used for the AKT and ERK blot data. Data shown are representative of two independent experiments; *, p < 0.05 between Rgl2 shRNA 1 and shRNA 1 + RalAQ75L; **, p < 0.005 versus nonspecific shRNA control.
Next, we determined how Rgl2 may function in a Ral-independent manner to promote PDAC anchorage-independent growth. One possible mechanism is suggested by Rgl2 association with CNK, a scaffold protein that also interacts with Raf and may facilitate ERK activation (43). Thus, depletion of Rgl2 may disrupt the ability of CNK to promote Raf signaling. Another possible mechanism is based on the demonstrated ability of RalGDS to activate AKT in a Ral-independent manner (20). To address these mechanisms, we determined if Rgl2 depletion corresponded with reduced AKT and/or ERK activity levels. We used Western blot analyses to determine steady-state levels of phosphorylated and activated ERK and AKT and found unexpectedly that neither were lowered in MIA PaCa-2 cells, although phosphorylated ERK was elevated in Rgl2 shRNAi 2 but not shRNAi 1 (Fig. 7C). These results argue against these two possible Ral-independent signaling activities as the basis for Rgl2 depletion-associated growth inhibition.
Rgl2 Is Localized to the Leading Edge and Its Expression Is Required for RalB Association with the Leading Edge
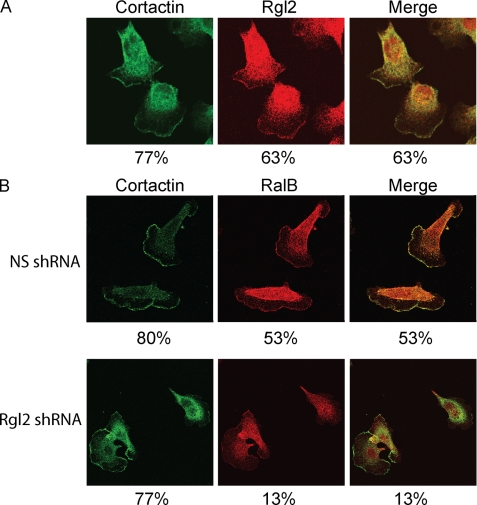
An additional function of RalGEFs may be their regulation of distinct spatial activation of their GTPase substrates (27). To further investigate the role of Rgl2 in PDAC cells, we examined the subcellular localization of endogenous Rgl2 in CFPAC-I cells. To demonstrate the specificity of the Rgl2 antibody, we used the CFPAC-I cells stably infected with either nonspecific or Rgl2 shRNAs as described earlier. The staining intensity was essentially abolished in CFPAC-I cells with knockdown of Rgl2 protein (supplemental Fig. 2), indicating the specificity of the staining. In CFPAC-I cells, Rgl2 was localized to the cytoplasm, and a specific plasma membrane localization was also observed in the majority of cells (63%; Fig. 8A), consistent with previous reports of Rgl2 localization (23, 24) and similar to the localization we observed with GFP-tagged Rgl2 (data not shown). Next, we showed that this specific membrane localization corresponded to the leading edge by co-staining with cortactin, an actin-binding protein involved in cell migration and tumor cell invasion (44) (Fig. 8A and supplemental Fig. 3A). Previous studies have validated cortactin as a marker for the leading edge of migrating cells (45–47). Importantly, in every cell that showed strong membrane localization of Rgl2, co-localization with cortactin was also observed.
FIGURE 8.
Rgl2 expression is required for RalB association with the leading edge. A, Rgl2 is localized to the leading edge. CFPAC-I cells were grown on glass coverslips. After labeling with mouse anti-Rgl2 and rabbit anti-cortactin antibodies followed by Alexa Fluor 488-conjugated and 568-conjugated secondary antibodies, cells were visualized with a confocal microscope. B, RalB localization to the leading edge is dependent on Rgl2 expression. CFPAC-I cells stably infected with either nonspecific or Rgl2 shRNA 2 were grown on glass coverslips. After labeling with mouse anti-RalB and rabbit anti-cortactin antibodies followed by Alexa Fluor 488-conjugated and 568-conjugated secondary antibodies, cells were visualized with a confocal microscope. Data shown are representative of two independent experiments. Percentages shown are percentages of 30 randomly chosen cells for each experiment that demonstrate distinct plasma membrane localization for either Rgl2, RalB, or cortactin, as shown and, in the case of the merge panel, percentages of cells that show distinct co-localization with cortactin at the leading edge.
We next addressed the possibility that Rgl2 may influence RalB subcellular localization. We determined the subcellular localization of the endogenous Ral proteins in CFPAC-I cells and found that endogenous RalB was localized strongly to a specific region on the plasma membrane and additionally a general cytoplasmic region at the opposite end of the cell in ∼50% of cells (Fig. 8B and supplemental Fig. 3B). Next, we showed that, like Rgl2, this plasma membrane region corresponded to the leading edge by co-staining with cortactin (Fig. 8B). Again, like Rgl2, in every case where plasma membrane localization of RalB was seen, this overlapped with the localization of cortactin. This leading edge localization of RalB is fully consistent with the role of RalB in migration by mobilizing the exocyst complex at the leading edge (48). Surprisingly, upon knockdown of Rgl2, the percentage of cells where RalB localized to the leading edge was dramatically lowered (Fig. 8B and supplemental Fig. 3B). Importantly, formation of the leading edge itself was not affected by Rgl2 knockdown, because a high percentage of cells still showed a distinct plasma membrane localization of cortactin upon Rgl2 depletion (Fig. 8B). Because only the active but not the total level of RalB was lowered upon knockdown of Rgl2 (Fig. 6A), this disappearance of RalB from the leading edge is explained by a redistribution away from the leading edge. We also performed the same analysis with RalA. Endogenous RalA localization in CFPAC-I cells was more cytoplasmic than RalB, with leading edge localization never seen, in contrast to RalB. Furthermore, we saw no difference in RalA localization upon Rgl2 knockdown (supplemental Fig. 4). These results demonstrate that RalB subcellular localization is not solely determined by its C-terminal membrane targeting sequences but additionally by association with Rgl2.
DISCUSSION
Evidence for a critical contribution of Ral GTPase activation in human cancer cell growth continues to accumulate at a rapid pace (4). In particular, our recent studies identified critical and distinct roles for RalA and RalB in PDAC tumorigenic and metastatic growth (6). However, the mechanisms by which Ral GTPases are activated in cancer remain to be elucidated. In light of the essentially 100% occurrence of KRAS mutations in PDAC (1), we determined if RalGEFs that can serve as K-Ras effectors are important mediators of Ral activation and PDAC growth. Additionally, because there are four RalGEFs activated by Ras, we also determined whether a specific RalGEF may be important for PDAC growth. We found that plasma membrane association was sufficient to activate RalGEF transforming potential, that all four activated RalA and RalB to comparable levels, and that spatially distinct Ral activation may be important for Ral-mediated growth transformation. We determined that RalGEF activity was important for Ral activation in PDAC cells, and in particular, Rgl2 was overexpressed in PDAC patient tumors and was essential for the anchorage-independent growth and Matrigel invasion of PDAC cell lines and for Ral activation in two of three PDAC lines.
Our results provide further support for the importance of spatial activation of Ral in promoting growth transformation. We found that all four RalGEFs exhibited a similar subcellular localization and were found predominantly in the cytoplasm with a concentration in the perinuclear region and to a lesser extent at distinct plasma membrane regions in what appear to be ruffles in cells lacking mutant Ras. As expected, plasma membrane targeting to mimic persistent association with membrane-bound K-Ras enhanced RalGEF plasma membrane association and transforming activity. However, unexpectedly, we did not find a direct correlation between the up-regulation of steady state Ral-GTP levels and transforming activity for wild type or membrane-targeted RalGEFs, although we observed previously that knockdown of RalA impaired Rgl2-CAAX-induced transformation (11). This was seen strikingly for Rgl2, where the membrane-associated form showed the same transforming potency as activated K-Ras; in contrast, wild type Rgl2 was not transforming yet did cause a significant increase in Ral-GTP levels. Thus, we show that despite being necessary for transformation in HEK-HT cells, activation of RalA alone is not sufficient for transformation and that differential localization of Ral activation may contribute to different biological effects. We suggest that the local activation of Ral at the plasma membrane, rather than the increase in total cell Ral-GTP, may be more critical to promote growth transformation. Alternatively, another possible interpretation is that RalGEFs can promote transformation via routes other than RalA activation, which is fully consistent with our data in the MIA PaCa-2 PDAC cell line, where expression of an activated RalA alone did not fully rescue the reduced anchorage-independent growth phenotype caused by Rgl2 knockdown (Fig. 7).
Interestingly, only the overexpression of wild-type Rgl2 and not the other wild-type RalGEFs in HEK-HT cells significantly activated both RalA and RalB. This result is consistent with our observation of the necessity of endogenous Rgl2 for the full activation of RalA and RalB in PDAC lines. This is especially important given that other RalGEFs, such as RAlGDS, are expressed in these PDAC lines. despite the expression of other RalGEFs, such as RalGDS. It is certainly possible that Rgl2 has higher intrinsic GEF activity than the other RalGEFs. Alternatively, although similar to the other RalGEFs in localization, a subtly different perinuclear or plasma membrane localization of Rgl2 compared with the others may result in more potent RalA and RalB activation. Taking together the observed overexpression of Rgl2 in PDAC patient samples, our demonstration that Rgl2 overexpression in HEK-HT cells causes strong RalA and RalB activation, and our observation that Rgl2 expression is necessary for full Ral activation in PDAC lines leads to a model in which Rgl2 overexpression leads to a pathological activation of RalA and RalB in pancreatic cancer.
Our observations that dominant negative Ral reduced Ral activation levels and PDAC anchorage-independent growth and Matrigel invasion support the importance of RalGEF activation, rather than RalGAP inactivation, for Ral activation in PDAC, at least in SW-1990 and CFPAC-I cells. However, the Ral dominant negative mutants are likely to block all RalGEFs, including those not known to be regulated by Ras (RalGPS1 and RalGPS2), so this approach did not establish the importance of an individual RalGEF(s). However, we did note that the RalA dominant negative preferentially impaired PDAC anchorage-independent growth, whereas the RalB dominant negative was more effective at blocking Matrigel invasion, consistent with the respective roles of RalA and RalB activity in these two PDAC growth properties (6). Thus, perhaps due to their distinct subcellular locations (9), each dominant negative mutant selectively impaired only a subset of RalGEF activities. Nevertheless, our finding that one RalGEF, Rgl2, is elevated in expression in PDAC tumors and cell lines and that suppression of Rgl2 alone was sufficient to reduce Ral activation in a subset of PDAC lines and to reduce PDAC growth in all lines demonstrates the critical role of one RalGEF in PDAC cells.
Our observation that suppression of Rgl2 alone was sufficient to suppress Ral activation in a subset of the PDAC lines that express additional RalGEFs was unexpected and suggests nonredundant roles for RalGEFs in promoting Ral activation in PDAC tumor cells. However, this is consistent with studies with other Ras effectors. For example, although Ras can interact with multiple isoforms of the p110 catalytic subunit of PI3K (α, β, δ, and γ), the inability of mutant Ras to interact with only p110α was sufficient to completely suppress H-Ras-induced skin carcinoma formation and K-Ras-induced lung carcinoma growth (49). That p110α is the key isoform for PI3K activation in cancer is also supported by the fact that only p110α is mutated in human cancers (50). Similarly, suppression of c-Raf-1 but not B-Raf reduced ERK activation in NRAS mutant melanoma cells, indicating that the c-Raf-1 isoform is the critical link to ERK activation (51).
A potential difference with individual RalGEF functions in cancer could lie in different expression levels. Consistent with this hypothesis, we demonstrated that at the mRNA level, only RalGDS and Rgl2 are expressed to detectable levels in PDAC lines, with only CFPAC-I cells, which express all four, as the exception. This is consistent with the ubiquitous expression of RalGDS and Rgl2, whereas Rgl1 and Rgl3 are more tissue-restricted (15, 37–39). Likewise, all PDAC lines tested expressed both RalGDS and Rgl2 protein. However, knockdown of Rgl2 specifically caused a large decrease in RalA and RalB activation as well as their associated phenotypes of anchorage-independent growth and invasion, respectively. Thus, the remaining presence of RalGDS in SW-1990 and MIA PaCa-2 cells and RalGDS, Rgl1, and Rgl3 in CFPAC-I cells cannot functionally compensate for the loss of Rgl2, suggesting that the RalGEFs have distinct functions in human cancer. Intriguingly, RalGDS knock-out in mice reduced the incidence and size of oncogenic H-Ras-induced skin tumors (25). Thus, the question arises as to why RalGDS is crucial in skin tumors but Rgl2 is crucial in PDAC. One potential explanation could lie in expression levels, such as the possibility that RalGDS is the only RalGEF highly expressed in mouse skin. Alternatively, this may reflect Ras isoform differences, where RalGDS is preferentially activated by H-Ras.
Another explanation for the crucial role of Rgl2 in PDAC transformation is isoform-specific Ral-independent functions, because Rgl2 has been shown to interact with the Ras scaffolding protein CNK (43), whereas RalGDS interacts with PDK1 to promote AKT activation (20, 52). In line with this hypothesis is our observations that Rgl2 knockdown and RalA31N expression strongly suppressed anchorage-independent growth in MIA PaCa-2 without suppression of RalA activity and that in MIA PaCa-2 cells, activated RalA was not able to rescue the anchorage-independent growth phenotype of Rgl2 knockdown. These results are consistent RalA-independent effects of Rgl2 that are necessary for anchorage-independent growth promotion in PDAC. This result also provides an explanation for why RalA(31N) blocked anchorage-independent growth in MIA PaCa-2 cells without lowering RalA-GTP: sequestration of Rgl2, which has RalA-independent functions in anchorage-independent growth. It is possible that knockdown of Rgl2 does not lower RalA-GTP sufficiently to cause the RalA-dependent block in anchorage-independent growth seen when RalA is knocked down (6). Because we demonstrated that Rgl2 does not activate AKT or ERK, two of the other major downstream Ras effectors, the mechanism for the promotion of anchorage-independent growth by RalA will require further investigation. There are perhaps unknown binding partners of Rgl2 that mediate a Ral-independent scaffolding function.
In addition to establishing a critical role for Rgl2 in Ral activation in PDAC, we demonstrated that this RalGEF may also dictate a distinct spatial distribution of Ral activation in the cell. Although we could not perform co-localization of Rgl2 and RalB (due to the antibodies both being of mouse origin), our observation that both co-localize with cortactin strongly suggest that Rgl2 and RalB are co-localized at the leading edge. Thus, Rgl2 may activate RalB by recruiting to and activating RalB at the leading edge. RalB can then recruit and assemble the exocyst complex to the leading edge, which is necessary for directional motility (48). This may be partially responsible for the invasion defect we saw upon knockdown of Rgl2. However, further work is needed to determine the mechanism of the demonstrated role of Rgl2 in invasion.
RalA, in contrast to RalB, did not localize to the leading edge. Likewise, unlike RalB, expression of Rgl2 did not alter RalA localization, although Rgl2 expression was responsible for the full activity of both RalA and RalB. We speculate that an internal pool of Rgl2 is responsible for the activation of RalA directly without changing its localization, whereas a leading edge pool of Rgl2 activates RalB directly and indirectly through altering its localization. Therefore, the distinct subcellular localization patterns of the four Ras proteins, as well as the distinct localization patterns of the different RalGEFs, together with the distinct subcellular localization of Rgl2 as well as RalA and RalB, provide multiple mechanisms to modulate the spatial diversity of activated Ral engagement of effectors in response to different upstream stimuli and signaling components.
In summary, our studies implicate a key role for RalGEFs, in particular Rgl2, in promoting Ral activation and PDAC growth. Presently, we have identified and are pursuing two indirect approaches to block Ral function for cancer treatment. First, we have shown that inhibitors of geranylgeranyltransferase-I cause PDAC cell growth inhibition and apoptosis, in part, by blocking RalA and RalB function (28). However, because there are more than 50 other substrates for this prenylation enzyme (53), including key Rho family small GTPases, this approach is obviously limited by considerable non-Ral activities. A second approach for a RalA-selective inhibitor is Aurora-A protein kinase inhibitors, because we found that phosphorylation of the Aurora-A site in RalA is essential for PDAC tumorigenic growth (10). However, these inhibitors will also block the function of Aurora-A in mitosis and additionally the function of other Aurora-A substrates (54). Thus, more selective approaches for blocking Ral are desired. Although GEFs may not be considered to be “druggable” targets for drug discovery, there is increasing evidence that GEFs may be tractable for small molecule inhibitor development (55, 56), and hence, there is increasing interest in developing such inhibitors (29). However, our demonstration that there are Ral-independent effects of Rgl2 suggests that inhibiting the GEF activity alone will not be sufficient for blocking the function of Rgl2 in PDAC growth. Elucidating how RalGEFs are regulated by mutant K-Ras may provide clues for the development of ways to block RalGEF function in cancer.
Supplementary Material
Acknowledgments
We thank the University of North Carolina Tissue Procurement Facility for excellent assistance. We thank Chris Counter, Donita Brady, and Adrienne Cox for helpful discussions and Lanika DeGraffenreid for assistance in manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants CA042978 and CA67771 (to C. J. D.) and National Institutes of Health, NCI, SPORE in Gastrointestinal Cancer P50 Grant CA106991 (to C. J. D. and J. J. Y.). This work was also supported by the Emerald Foundation, the Kimmel Foundation, the American College of Surgeons, and the North Carolina University Cancer Research Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- PDAC
- pancreatic ductal adenocarcinoma
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- HEK
- human embryonic kidney
- RA
- Ras association.
REFERENCES
- 1.Jones S., Zhang X., Parsons D. W., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S. M., Fu B., Lin M. T., Calhoun E. S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D. R., Hidalgo M., Leach S. D., Klein A. P., Jaffee E. M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J. R., Kern S. E., Hruban R. H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) Science 321, 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeh J. J., Der C. J. (2007) Expert Opin. Ther. Targets 11, 673–694 [DOI] [PubMed] [Google Scholar]
- 3.Wennerberg K., Rossman K. L., Der C. J. (2005) J. Cell Sci. 118, 843–846 [DOI] [PubMed] [Google Scholar]
- 4.Bodemann B. O., White M. A. (2008) Nat. Rev. Cancer 8, 133–140 [DOI] [PubMed] [Google Scholar]
- 5.Chien Y., White M. A. (2003) EMBO Rep. 4, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim K. H., O'Hayer K., Adam S. J., Kendall S. D., Campbell P. M., Der C. J., Counter C. M. (2006) Curr. Biol. 16, 2385–2394 [DOI] [PubMed] [Google Scholar]
- 7.Oxford G., Owens C. R., Titus B. J., Foreman T. L., Herlevsen M. C., Smith S. C., Theodorescu D. (2005) Cancer Res. 65, 7111–7120 [DOI] [PubMed] [Google Scholar]
- 8.Yin J., Pollock C., Tracy K., Chock M., Martin P., Oberst M., Kelly K. (2007) Mol. Cell. Biol. 27, 7538–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shipitsin M., Feig L. A. (2004) Mol. Cell. Biol. 24, 5746–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim K. H., Brady D. C., Kashatus D. F., Ancrile B. B., Der C. J., Cox A. D., Counter C. M. (2010) Mol. Cell. Biol. 30, 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim K. H., Baines A. T., Fiordalisi J. J., Shipitsin M., Feig L. A., Cox A. D., Der C. J., Counter C. M. (2005) Cancer Cell 7, 533–545 [DOI] [PubMed] [Google Scholar]
- 12.Shirakawa R., Fukai S., Kawato M., Higashi T., Kondo H., Ikeda T., Nakayama E., Okawa K., Nureki O., Kimura T., Kita T., Horiuchi H. (2009) J. Biol. Chem. 284, 21580–21588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhardt G. R., Korherr C., Wieler J. S., Knaus M., Schrader J. W. (2001) Oncogene 20, 188–197 [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi A., Demo S. D., Ye Z. H., Chen Y. W., Williams L. T. (1994) Mol. Cell. Biol. 14, 7483–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao H., Andres D. A. (2000) J. Biol. Chem. 275, 26914–26924 [DOI] [PubMed] [Google Scholar]
- 16.Wolthuis R. M., Bauer B., van 't Veer L. J., de Vries-Smits A. M., Cool R. H., Spaargaren M., Wittinghofer A., Burgering B. M., Bos J. L. (1996) Oncogene 13, 353–362 [PubMed] [Google Scholar]
- 17.Quilliam L. A., Rebhun J. F., Castro A. F. (2002) Prog. Nucleic Acid Res. Mol. Biol. 71, 391–444 [DOI] [PubMed] [Google Scholar]
- 18.McGillicuddy L. T., Fromm J. A., Hollstein P. E., Kubek S., Beroukhim R., De Raedt T., Johnson B. W., Williams S. M., Nghiemphu P., Liau L. M., Cloughesy T. F., Mischel P. S., Parret A., Seiler J., Moldenhauer G., Scheffzek K., Stemmer-Rachamimov A. O., Sawyers C. L., Brennan C., Messiaen L., Mellinghoff I. K., Cichowski K. (2009) Cancer Cell 16, 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durkin M. E., Yuan B. Z., Zhou X., Zimonjic D. B., Lowy D. R., Thorgeirsson S. S., Popescu N. C. (2007) J. Cell Mol. Med. 11, 1185–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao Y., Wong R., Feig L. A. (2008) Mol. Cell. Biol. 28, 2851–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murai H., Ikeda M., Kishida S., Ishida O., Okazaki-Kishida M., Matsuura Y., Kikuchi A. (1997) J. Biol. Chem. 272, 10483–10490 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Viciana P., Sabatier C., McCormick F. (2004) Mol. Cell. Biol. 24, 4943–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaya A., Ohba Y., Kurokawa K., Matsuda M. (2004) Mol. Biol. Cell 15, 2549–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolthuis R. M., de Ruiter N. D., Cool R. H., Bos J. L. (1997) EMBO J. 16, 6748–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.González-García A., Pritchard C. A., Paterson H. F., Mavria G., Stamp G., Marshall C. J. (2005) Cancer Cell 7, 219–226 [DOI] [PubMed] [Google Scholar]
- 26.Hamad N. M., Elconin J. H., Karnoub A. E., Bai W., Rich J. N., Abraham R. T., Der C. J., Counter C. M. (2002) Genes Dev. 16, 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaya A., Kamio T., Masuda M., Mochizuki N., Sawa H., Sato M., Nagashima K., Mizutani A., Matsuno A., Kiyokawa E., Matsuda M. (2007) Mol. Biol. Cell 18, 1850–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falsetti S. C., Wang D. A., Peng H., Carrico D., Cox A. D., Der C. J., Hamilton A. D., Sebti S. M. (2007) Mol. Cell. Biol. 27, 8003–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 30.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 31.de Rooij J., Bos J. L. (1997) Oncogene 14, 623–625 [DOI] [PubMed] [Google Scholar]
- 32.Wolthuis R. M., Franke B., van Triest M., Bauer B., Cool R. H., Camonis J. H., Akkerman J. W., Bos J. L. (1998) Mol. Cell. Biol. 18, 2486–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox A. D., Der C. J. (1994) Methods Enzymol. 238, 277–294 [DOI] [PubMed] [Google Scholar]
- 34.Matsubara K., Kishida S., Matsuura Y., Kitayama H., Noda M., Kikuchi A. (1999) Oncogene 18, 1303–1312 [DOI] [PubMed] [Google Scholar]
- 35.Roberts P. J., Mitin N., Keller P. J., Chenette E. J., Madigan J. P., Currin R. O., Cox A. D., Wilson O., Kirschmeier P., Der C. J. (2008) J. Biol. Chem. 283, 25150–25163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cascone I., Selimoglu R., Ozdemir C., Del Nery E., Yeaman C., White M., Camonis J. (2008) EMBO J. 27, 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albright C. F., Giddings B. W., Liu J., Vito M., Weinberg R. A. (1993) EMBO J. 12, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isomura M., Okui K., Fujiwara T., Shin S., Nakamura Y. (1996) Cytogenet. Cell Genet. 74, 263–265 [DOI] [PubMed] [Google Scholar]
- 39.Sood R., Makalowska I., Carpten J. D., Robbins C. M., Stephan D. A., Connors T. D., Morgenbesser S. D., Su K., Pinkett H. W., Graham C. L., Quesenberry M. I., Baxevanis A. D., Klinger K. W., Trent J. M., Bonner T. I. (2000) Biochim. Biophys. Acta 1491, 285–288 [DOI] [PubMed] [Google Scholar]
- 40.Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goi T., Rusanescu G., Urano T., Feig L. A. (1999) Mol. Cell. Biol. 19, 1731–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feig L. A. (1999) Nat. Cell Biol. 1, E25–E27 [DOI] [PubMed] [Google Scholar]
- 43.Lanigan T. M., Liu A., Huang Y. Z., Mei L., Margolis B., Guan K. L. (2003) FASEB J. 17, 2048–2060 [DOI] [PubMed] [Google Scholar]
- 44.Weaver A. M. (2008) Cancer Lett. 265, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boguslavsky S., Grosheva I., Landau E., Shtutman M., Cohen M., Arnold K., Feinstein E., Geiger B., Bershadsky A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10882–10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. (2005) Curr. Biol. 15, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 47.Kaksonen M., Peng H. B., Rauvala H. (2000) J. Cell Sci. 113, 4421–4426 [DOI] [PubMed] [Google Scholar]
- 48.Rossé C., Hatzoglou A., Parrini M. C., White M. A., Chavrier P., Camonis J. (2006) Mol. Cell. Biol. 26, 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta S., Ramjaun A. R., Haiko P., Wang Y., Warne P. H., Nicke B., Nye E., Stamp G., Alitalo K., Downward J. (2007) Cell 129, 957–968 [DOI] [PubMed] [Google Scholar]
- 50.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., Velculescu V. E. (2004) Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 51.Dumaz N., Hayward R., Martin J., Ogilvie L., Hedley D., Curtin J. A., Bastian B. C., Springer C., Marais R. (2006) Cancer Res. 66, 9483–9491 [DOI] [PubMed] [Google Scholar]
- 52.Yoshizaki H., Mochizuki N., Gotoh Y., Matsuda M. (2007) Mol. Biol. Cell 18, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid T. S., Terry K. L., Casey P. J., Beese L. S. (2004) J. Mol. Biol. 343, 417–433 [DOI] [PubMed] [Google Scholar]
- 54.Lok W., Klein R. Q., Saif M. W. (2010) Anticancer Drugs 21, 339–350 [DOI] [PubMed] [Google Scholar]
- 55.Evelyn C. R., Ferng T., Rojas R. J., Larsen M. J., Sondek J., Neubig R. R. (2009) J. Biomol. Screen. 14, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baines A. T., Lim K. H., Shields J. M., Lambert J. M., Counter C. M., Der C. J., Cox A. D. (2006) Methods Enzymol. 407, 556–574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.