Abstract
The Notch signaling pathway is important for cell fate decisions in embryonic development and adult life. Defining the functional importance of the Notch pathway in these contexts requires the elucidation of essential signal transduction components that have not been fully characterized. Here, we show that Rabconnectin-3B is required for the Notch pathway in mammalian cells. siRNA-mediated silencing of Rabconnectin-3B in mammalian cells attenuated Notch signaling and disrupted the activation and nuclear accumulation of the Notch target Hes1. Rabconnectin-3B knockdown also disrupted V-ATPase activity in mammalian cells, consistent with previous observations in Drosophila. Pharmacological inhibition of the V-ATPase complex significantly reduced Notch signaling in mammalian cells. Finally, Rabconnectin-3B knockdown phenocopied functional disruption of Notch signaling during osteoclast differentiation. Collectively, these findings define an important role for Rabconnectin-3 and V-ATPase activity in the Notch signaling pathway in mammalian cells.
Keywords: Bone, Breast Cancer, Differentiation, H+-ATPase, Notch Pathway, Signal Transduction, Osteoblast, Rabconnectin-3
Introduction
Notch signaling plays an important role in the development and differentiation of many cell types in diverse organisms (1–4). Abnormal activity of the Notch pathway has been linked to several developmental disorders and malignant diseases (5, 6). Notch activity is regulated at multiple levels; in particular, intracellular trafficking and three specific cleavage events allow cells to precisely modulate Notch pathway activity (7, 8). Recent reports have shown that in Drosophila cells, the S3 cleavage of Notch, which is carried out by γ-secretase, requires the activity of the proton pump V-ATPase, a multisubunit enzyme that is highly conserved in eukaryotes and acidifies vesicles (9, 10). Moreover, it was shown that Notch signaling activity requires a novel V-ATPase regulator, Rabconnectin-3α and -3β (Rbcn-3A and -3B) (9). In Drosophila epithelial cells, in the absence of Rbcn-3A or -3B, acidification of intracellular compartments is abnormal, and Notch signaling is disrupted between the second and third cleavage step that should release the intracellular domain of Notch and allow it to enter the nucleus. The yeast homolog of Rbcn-3A is part of a RAVE complex that regulates the assembly and activity of V-ATPase (11, 12). Rbcn-3 proteins are conserved in other eukaryotic systems, including mammals. However, the biochemical functions of mammalian Rbcn-3 proteins remain uncharacterized.
These observations in Drosophila suggested that the Rbcn proteins might serve a similar role in mammalian cells. In particular, we were interested whether the Notch pathway would be sensitive to down-regulation of the Rbcn-3 proteins in distinct epithelial and bone-specific cells in which Notch signaling has been shown to play an important role in the regulation of homeostasis and differentiation. For example, the Notch pathway is known to regulate cell fate decisions in the osteoclast lineage. Osteoclasts are bone-resorbing, multinucleated cells that differentiate from monocyte or macrophage lineage precursors. Research has shown that Notch signaling at the level of receptor and transcriptional activation inhibits the differentiation of osteoclast precursors, the primary bone marrow macrophages, into mature osteoclasts (13). Bone marrow macrophages isolated from transgenic mice lacking Notch1 and Notch3 were shown to differentiate more efficiently than those derived from control mice. Osteoclast differentiation in Notch receptor-deficient osteoclast precursor cells also led to a functional increase in their capacity to resorb bone in vitro and in vivo (13). Collectively, these experiments demonstrate that loss of Notch signaling at the level of receptor and transcriptional activation promotes osteoclast differentiation.
Here, we used this model system to study the functional importance of Rbcn-3 in mammalian Notch signaling. We found that targeting of Rbcn-3B significantly affected Notch signaling in several different mammalian cell types and, in particular, had a significant effect on osteoclast differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The HaCaT skin keratinocyte cell line was maintained in Dulbecco's modified Eagle's medium, with 10% fetal bovine serum (FBS). The MCF-7 cell line was maintained in minimal essential medium with 10% FBS, nonessential amino acids, insulin, and antibiotics. The SCP28 subline was derived from the parental cell line MDA-MB-231 (American Type Culture Collection (ATCC)) (14). The subline and its genetically modified variants were maintained in DMEM supplemented with 10% FBS, penicillin/streptomycin (Invitrogen), fungizone, and appropriate selection drugs for transfected or transduced constructs. H29 cells, a packaging cell line for retrovirus production, were maintained in DMEM supplemented with 10% FBS, 2 mm l-glutamine, and antibiotics. The murine pre-osteoclast cell line MOCP5 was maintained in growth medium, α-minimal essential medium supplemented with 10% FBS and antibiotics. The murine pre-osteoclast cell line Raw 264.7 was maintained in DMEM with 10% FBS and antibiotics for regular culture and supplemented with 30 ng/ml receptor activator for nuclear factor κB ligand (PeproTech) for osteoclastogenesis assays. Bafilomycin A1 (Calbiochem) was dissolved in DMSO and used at the indicated concentrations. GSI-IX (Calbiochem) was dissolved in DMSO and used at a concentration of 1 μm unless otherwise specified.
Generation of Overexpression Cell Line and Coculture System
For stable overexpression, human Jagged1 cDNA was PCR-amplified using the primer pair 5′-ATCCTCGAGAGCACCAGCGCGAACAGCAG-3′ (sense) and 5′-ATCGAATTCCCCGCGGTCTGCTATACGAT-3′ (antisense) and cloned into the retroviral expression vector pMSCVpuro using XhoI and EcoRI restriction sites (underline indicates restriction sites). cDNA retroviral vectors were transfected into the packaging cell line H29. After 48 h, viruses were collected, filtered, and used to infect the SCP28 subline in the presence of 5 μg/ml Polybrene. The infected cells were selected with 1 μg/ml puromycin. Control cell lines were derived from parental vectors alone. To avoid clonal variations, a pooled population of at least 500 independent clones of each transfection/transduction was used to generate each stable cell line used in this study.
These cells were used to study Notch signaling activation in breast cancer cells using a coculture system. First, 1 × 105 vector control SCP28 breast cancer cells were plated into each well of a 12-well plate. The following day, these cells were transfected with the Notch reporter using Lipofectamine 2000. After 4–6 h, the transfection media were exchanged with fresh media containing 1 × 105 cells/well of Jagged1-overexpressing SCP28 breast cancer cells. As a result, the Notch pathway was activated in the control tumor cells containing the Notch reporter by the cocultured Jagged1-overexpressing tumor cells. These cocultures were subsequently treated with bafilomycin to study its effects of Notch signaling activity in tumor cells.
Quantitative RT-PCR
RNA from in vitro cultured cells was collected in RLT lysis buffer and extracted with the RNeasy mini kit (Qiagen). cDNA synthesis of RNA was performed using Superscript III first strand (Invitrogen). Quantitative RT-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) with the ABI Prism 7900HT thermocycler (Applied Biosystems) according to the manufacturer's protocol. A standard curve for each gene was generated by serial dilutions of a standard. Values were then normalized by the amount of GAPDH in each sample. Primer sequences are as follows: Rbcn-3B, primer pair 1, F4 5′-TGCTGATCACTCTGGCTC-3′ and R 5′-GCTTCTCTCACCTCCAAGCA-3′; primer pair 2, F 5′-TCACTCTGGCTCTGACCCTC-3′ and R 5′-GCTTCTCTCACCTCCAAGCA-3′; Hes1, F 5′-GTGCATGAACGAGGTGACCC-3′ and R 5′-GTATTAACGCCCTCGCACGT-3′; Hey1, F 5′-AGCCGAGATCCTGCAGATGA-3′ and R 5′-GCCGTATGCAGCATTTTCAG-3′; Hes2, F 5′-AGAACTCCAACTGCTCGAAGCT-3′ and R 5′-CGGTCATTTCCAGGACGTCT; Hey2, F 5′-AGATGCTTCAGGCAACAGGG-3′ and R 5′-CAAGAGCGTGTGCGTCAAAG-3′; and GAPDH, F 5′-GGAGTCAACGGATTTGGTCGTA-3′ and R 5′-GGCAACAATATCCACTTTACCAGAGT-3′.
Notch Reporter Assays
Mammalian cells were seeded at 2 × 105 cells/well in 12-well plates and grown to 95% confluence. The cells were then transfected with the firefly luciferase Notch reporter (15) and cytomegalovirus (CMV)-Renilla luciferase control (Promega) plasmids using Lipofectamine 2000. For siRNA experiments, the cells were also cotransfected with siRNAs according to the manufacturer's instructions. After 4 h, the transfection media were changed to regular media or media supplemented with the indicated drugs. Following 2 days, the mammalian cells were lysed and subjected to a luciferase assay as described below. Quantitative results are shown as firefly luciferase counts (Notch reporter activity) divided by Renilla luciferase counts to normalize for transfection efficiency.
Luciferase Assay
After completing treatments and/or transfections, cells were lysed for 1 h with 200 μl of lysis buffer (pH 7.8, 10 mm EDTA, 100 mm DTT, 50% glycerol, 5% Triton X-100, 125 mm Tris base) diluted 1:5 in deionized water. 30 μl of each lysate was pipetted into an opaque 96-well plate in duplicate. Half of the lysates were treated with 100 μl of firefly-luciferase assay buffer (25 mm glycylglycine, pH 7.8, 15 mm potassium phosphate, pH 7.8, 15 mm MgSO4, 4 mm EGTA, plus 2 mm ATP, 10 mm DTT, and 1 mm d-luciferin added just before use), and the other half were treated with 100 μl of Renilla-luciferase assay buffer (0.5 m NaCl, 1 mm EDTA, 0.1 m potassium phosphate, pH 7.4, plus 0.04% BSA and 2 μm coelenterazine added just before use). Firefly and Renilla-luciferase activity were measured using a GloMax 96 microplate luminometer (Promega, Madison, WI). For reporter assays, results were normalized by dividing firefly luciferase activity readings by Renilla-luciferase activity readings. Significant differences between various treatment conditions were determined by Student's t tests.
Transfection of siRNA
Mammalian cells were transfected with a scrambled control siRNA or three distinct targeting siRNAs against Rbpj (1, GCACAGAAGUCUUACGGAAAUGAAA; 2, CCAUUACGGGCAGACUGUCAAGCUU; and 3, Rabconnectin-3, which was purchased from Invitrogen, Stealth RNAiTM siRNA WD repeat domain 7, catalogue no. 1299003, using Lipofectamine 2000). After 4 h, the transfection media were changed to regular media. For gene expression analysis, RNA from transfected cells was collected in RLT lysis buffer, extracted with RNeasy mini kit (Qiagen), and subjected to quantitative RT-PCR as described above.
Immunofluorescence and LysoTracker Assay
Cells were washed in ice-cold Dulbecco's phosphate-buffered saline (DPBS) and fixed in ice-cold methanol for 7 min. The cells were blocked in 3% milk/PBS for 1 h at room temperature and incubated with an anti-rabbit polyclonal antibody against human Hes1 (R & D Systems) (1:200 dilution in blocking solution) or an anti-rabbit monoclonal antibody against phosphorylated-Smad2 (Cell Signaling) overnight at 4 °C. After washing three times with DPBS, GFP-conjugated anti-rabbit antibodies (R & D Systems) (1:500 dilution in blocking solution) were added and incubated for 30 min at 37 °C. The cells were washed twice with DPBS. DAPI dye (1:1000 dilution in blocking solution) was added and removed immediately. The cells were washed twice with DPBS and mounted with GVA Mount (Invitrogen). Fluorescence and phase contrast images were captured on a Carl Zeiss Axiocam microscope. For LysoTracker assay, cells were washed twice with DPBS, treated with 1 μm LysoTracker Red DND-99 (Molecular Probes) for 1 min, treated with DAPI, and washed twice with DPBS. Fluorescence images were captured on a Carl Zeiss Axiocam microscope.
Western Blot Analyses
SDS lysis buffer (0.05 mm Tris-HCl, 50 mm β-mercaptoethanol, 2% SDS, 0.1% bromphenol blue, 10% glycerol) was used to collect protein from cultured cells. Heat-denatured protein was then equally loaded, separated on an SDS-polyacrylamide gel, transferred onto a pure nitrocellulose membrane (Bio-Rad), and blocked with either 5% milk or 5% BSA. Primary antibodies for immunoblotting included rabbit anti-N1ICD (1:1000 dilution, Cell Signaling) and mouse anti-β-actin (1:4000 dilution, Abcam) for loading control. Membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (1:2000 dilution, GE Healthcare) or anti-rabbit secondary antibody (1:2000 dilution, GE Healthcare) for 1 h, and chemiluminescence signals were detected by ECL substrate (GE Healthcare).
Osteoclastogenesis Assay
Murine pre-osteoclast Raw 264.7 (2 × 105 cells/well) or MOCP5 (5 × 105 cells/well) cells were cultured on 12-well plates in regular growth media on day 0. The cells were treated with control, Rbpj, Rbcn-3 siRNAs using Lipofectamine 2000 according to manufacturer's protocol on day 1. After 4 h, transfection media were changed to growth media containing 30 ng/ml recombinant murine soluble receptor activator for nuclear factor κB ligand. Media were changed every 2 days. TRAP staining was performed on day 5 using a leukocyte acid phosphatase kit from Sigma (16). TRAP-positive multinucleated cells were scored as mature osteoclasts. The average number of mature osteoclasts in five distinct fields from each well of the 12-well plate was quantified. Data are expressed as averages and standard deviations of the number of mature osteoclasts in three distinct wells from each experimental group.
Statistical Analysis
Results are presented as average ± S.D. or as average ± S.E. as indicated in the figure legends. Comparisons were analyzed by unpaired two-sided independent Student's t test without equal variance assumption unless otherwise indicated.
RESULTS
Rabconnectin-3 Knockdown Reduces Notch Signaling Activity
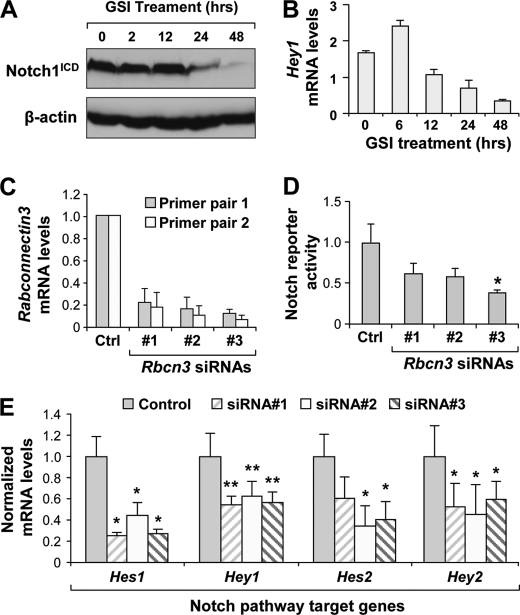
To investigate the requirement of Rabconnectin3 (Rbcn-3) in Notch signaling, we employed the immortalized human keratinocyte HaCaT cell line, which demonstrates robust Notch signaling activity as shown by high expression levels of Notch1 intracellular domain (N1ICD) protein and Hey1 mRNA. Importantly, these indicators of constitutive Notch pathway activity are susceptible to suppression by a GSI (Fig. 1, A and B), a known inhibitor of the Notch pathway. These results suggest that the HaCaT cells can be used to test whether silencing of a novel component of the Notch pathway can lead to a decrease in the base-line Notch signaling readout. Using this line of reasoning, we tested whether siRNA-mediated knockdown of Rbcn-3 in HaCaT cells disrupts Notch signaling activity. We first confirmed that Rbcn-3 mRNA expression levels were effectively knocked down by targeting siRNAs, which showed a greater than 83% reduction in Rbcn-3 siRNA-treated HaCaT cells relative to controls (Fig. 1C). We used a Notch reporter construct containing six CSL-binding elements upstream of a firefly luciferase reporter to measure Notch pathway activity. Loss of Rbcn-3 showed a modest reduction in Notch pathway reporter activity in HaCaT cells by luciferase assay (Fig. 1D). We next profiled mRNA expression levels of Notch downstream targets as an endogenous readout of Notch pathway activity. As expected, HaCaT cells treated with siRNAs against Rbcn-3 demonstrated a significant reduction in several Notch target genes of the Hes and Hey families (Fig. 1E). These results indicate that Rbcn-3 is necessary for proper Notch signaling in mammalian keratinocytes.
FIGURE 1.
siRNA-mediated knockdown of Rbcn-3 disrupts Notch signaling in HaCaT cells. A, γ-secretase inhibitor treatment disrupts Notch signaling in the HaCaT cell line as shown by Western blot analysis of Notch1 intracellular domain (Notch1ICD) protein levels. B, qRT-PCR mRNA expression levels of Hey1 in HaCaT cells treated with different durations of GSI-IX. C, qRT-PCR mRNA levels of Rbcn-3 in HaCaT cells treated with a control or three Rbcn-3 siRNAs. D, Notch reporter levels in HaCaT cells treated with a control or three Rbcn-3 siRNAs by luciferase assay. Notch reporter firefly luciferase counts were normalized to Renilla luciferase counts. E, qRT-PCR mRNA levels of several Notch target genes in HaCaT cells treated with a control or three Rbcn-3 siRNAs. Data represent average ± S.D. *, p < 0.05; **, p < 0.01 by Student's t test.
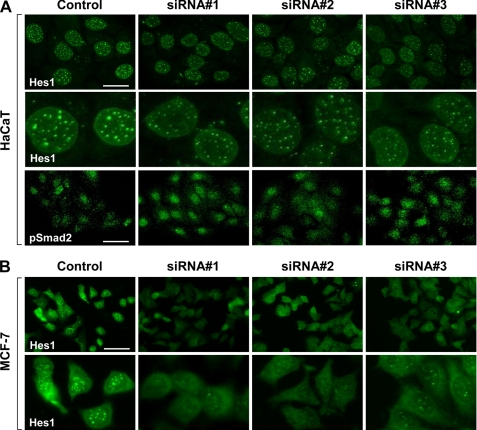
Based on these results, we decided to examine the activity of the Notch pathway downstream mediators in response to Rbcn-3 siRNA treatment. We reasoned that if the Notch pathway is compromised by silencing Rbcn-3 expression, activation of the nuclear transcriptional factor Hes1, a prominent downstream target of the Notch signaling pathway, should concomitantly be disrupted. The Notch target Hes1 stained brightly and localized as punctate clusters in the nucleus of control HaCaT cells, consistent with its role as a transcriptional regulator. In contrast, Rbcn-3 siRNA-treated HaCaT cells demonstrated a modest decrease in staining intensity and fewer punctate clusters in the nucleus (Fig. 2A, top and middle panels). To show that these results were specific to the Notch pathway, we also stained control and Rbcn-3 siRNA-treated HaCaT cells for phospho-Smad2, an active component of the TGFβ pathway. No difference was observed in phospho-Smad2 staining intensity and localization in Rbcn-3 siRNA-treated cells compared with the control (Fig. 2A, lower panel). To evaluate the generality of these results in the mammalian system and extend the implications to a breast cancer model, we tested whether Rbcn-3 is necessary for proper Hes1 localization in the human MCF-7 breast cancer cell line. siRNA-mediated knockdown of Rbcn-3 in MCF-7 cells showed a pronounced reduction in Hes1 staining intensity and almost complete absence of nuclear localization compared with control cells (Fig. 2B). These findings strongly suggest that appropriate Notch signaling activity, including localization of downstream targets, requires Rbcn-3 function.
FIGURE 2.
siRNA-mediated knockdown of Rbcn-3 reduces expression levels and disrupts localization of the Notch target Hes1 in HaCaT skin and MCF-7 breast cancer cells. A, localization of Hes1 and pSmad2 in HaCaT cells treated with a control or three Rbcn-3 siRNAs by immunofluorescence. Scale bar, 100 μm. B, localization of Hes1 in MCF-7 breast cancer cells treated with a control or three Rbcn-3 siRNAs by immunofluorescence. Scale bar, 100 μm.
Inhibition of V-ATPase Reduces Notch Signaling Activity
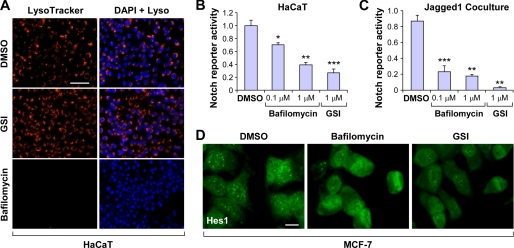
It has been previously proposed that Rbcn-3 and its homologs function to regulate the V-ATPase activity and are therefore important for the acidification of intracellular compartments (9, 11). Based on this understanding, we decided to test whether Rbcn-3 is functioning through the V-ATPase complex to regulate Notch activity (18, 19). To accomplish this, we first tested whether Rbcn-3 is required for V-ATPase activity in mammalian cells. Consistent with previously published studies in Drosophila, siRNA-mediated silencing of Rbcn-3 disrupted V-ATPase activity in HaCaT keratinocytes and MCF-7 breast cancer cells as shown by staining cells with LysoTracker, a dye that specifically identifies the acidic intracellular compartment of cells (Fig. 3). We next examined whether V-ATPase activity was necessary for Notch signaling by employing bafilomycin A1, a macrolide antibiotic and potent V-ATPase inhibitor isolated from Streptomyces species. We first confirmed that bafilomycin disrupted V-ATPase function as shown by the significant reduction in LysoTracker staining, whereas γ-secretase inhibitor treatment had no effect (Fig. 4A). Next, we transiently transfected HaCaT cells with the Notch reporter and subsequently treated these cells with bafilomycin to test whether disruption of the V-ATPase complex influenced Notch signaling activity. Bafilomycin treatment reduced Notch activity in HaCaT cells in a dose-dependent fashion, approaching levels in response to γ-secretase inhibitor treatment, which served as a positive control (Fig. 4B). In addition, we employed a mammalian coculture system in which Jagged1-expressing tumor cells activate the Notch pathway in cocultured isogenic control tumor cells that contain the Notch reporter to test whether bafilomycin treatment disrupted Notch signaling activation in breast cancer cells (see under “Experimental Procedures” for details). Consistently, inhibition of the V-ATPase complex led to a dose-dependent reduction in Notch reporter activity in the signaling receiving tumor cells by luciferase assay (Fig. 4C). Furthermore, we showed that bafilomycin treatment disrupted the nuclear accumulation of Hes1, similar to the phenotype achieved by γ-secretase inhibitor treatment (Fig. 4D). These findings demonstrate that V-ATPase activity is required for the Notch pathway and further suggest that Rbcn-3 is most likely functioning through the V-ATPase complex to regulate Notch activity.
FIGURE 3.
Rbcn-3 is required for ATPase activity in HaCaT keratinocytes and MCF-7 breast cancer cells. A, LysoTracker staining of acidic intracellular compartments in HaCaT cells treated with a control or three Rbcn-3 siRNAs by immunofluorescence. Scale bar, 100 μm. B, Lysotracker staining of acidic intracellular compartments in MCF-7 cells treated with a control or three Rbcn-3 siRNAs by immunofluorescence. Scale bar, 100 μm.
FIGURE 4.
Bafilomycin A1 treatment disrupts Notch signaling in mammalian cells. A, LysoTracker staining of acidic intracellular compartments in HaCaT cells treated with DMSO, 1 μm GSI, or 0.5 μm bafilomycin A1. Scale bar, 100 μm. B, Notch reporter activity in HaCaT cells treated with the indicated concentrations of bafilomycin A1 and GSI IX. C, Notch reporter activity in Jagged1-overexpressing tumor cell coculture system treated with the indicated concentrations of bafilomycin A1 and GSI. Notch reporter firefly luciferase counts were normalized to Renilla luciferase counts. B and C, data represent average ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test. D, localization of Hes1 in MCF-7 breast cancer cells treated with DMSO, bafilomycin A1, or GSI IX by immunofluorescence. Scale bar, 30 μm.
Loss of Rbcn-3 Function Disrupts Notch Activity in Regulating Osteoclast Differentiation
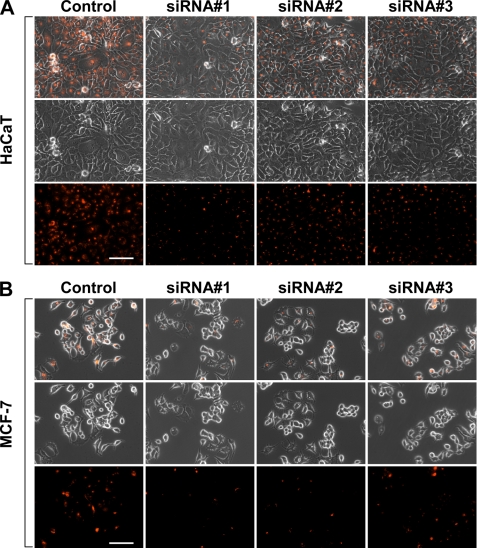
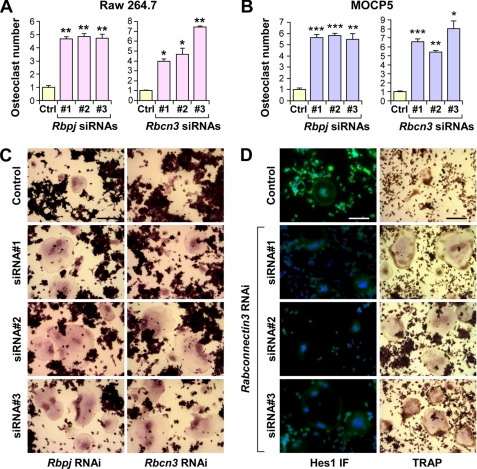
After establishing the importance of Rbcn-3 in the signaling cascade of the Notch pathway, it was important to determine whether this regulation was essential for Notch pathway functions. To investigate this, we employed a mammalian osteoclast differentiation cell culture system, because the Notch pathway has been shown to regulate the maturation status of osteoclast precursors. In particular, the loss of Notch signaling at the level of receptor and transcriptional regulation has been shown to accelerate osteoclast maturation (13). We first reproduced these results by treating the murine pre-osteoclast cell line Raw 264.7 with three distinct siRNAs against Rbpj, an essential cofactor required for transcriptional activity of the Notch pathway. Consistent with previously published data, Raw 264.7 cells treated with siRNAs against Rbpj demonstrated a greater number of differentiated, multinucleated osteoclasts compared with the control (Fig. 5, A and C, left column). We next determined whether loss of Rbcn-3 in Raw 264.7 pre-osteoclasts results in a similar phenotype. As expected, Raw 264.7 cells treated with Rbcn-3 siRNAs also showed a greater number of mature osteoclasts compared with the control (Fig. 5, A and C, right column), phenocopying the results achieved by Rbpj siRNA treatment. These results suggest that Rbcn-3 is important in the regulation of osteoclast differentiation by the Notch pathway. We additionally showed that silencing Rbcn-3 expression leads to a greater number of mature osteoclasts in an additional pre-osteoclast cell line, MOCP-5 (Fig. 5B). Moreover, we confirmed that loss of Rbcn-3 function disrupted Notch activity in the osteoclasts by examining the expression levels of Hes1 by immunofluorescence (Fig. 5D). Collectively, these results indicate that not only is Rbcn-3 required for Notch signaling activity but also is necessary for physiological functions of the Notch pathway, such as regulation of osteoclast differentiation.
FIGURE 5.
Loss of Rbcn-3 phenocopies defective Notch signaling in osteoclast differentiation. A, quantification of TRAP+ multinucleated MOCP5 osteoclasts treated with a control or three Rbpj siRNAs (left) or three Rbcn-3 siRNAs (right). B, quantification of TRAP+ multinucleated Raw 264.7 osteoclasts treated with a control or three Rbpj siRNAs (left) or three Rbcn-3 siRNAs (right). A and B, data represent average ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test. C, representative images of TRAP+ Raw 264.7 osteoclasts from the distinct experimental groups. Scale bar, 200 μm. D, left panel, localization of Hes1 in MOCP5 pre-osteoclasts treated with a control or three Rbcn-3 siRNAs by immunofluorescence. Right panel, representative images of TRAP+ MOCP5 osteoclasts treated with a control or three Rbcn-3 siRNAs, bottom panel. Scale bar, 200 μm.
DISCUSSION
Previous genetic studies in Drosophila have shown that the homolog of Rbcn-3 proteins and the vacuolar proton pump, V-ATPase, are involved in the regulation of the Notch pathway (9, 10). In this study, we sought to investigate the importance of Rbcn-3 in mammalian Notch signaling using genetic and pharmacological approaches in several different mammalian cell types.
Distinct from its function in other tissues, the Notch pathway promotes differentiation of skin epithelial cells, maintaining strong activity throughout adult life and functioning as a tumor suppressor (20–22). In this study, we investigated whether siRNA-mediated silencing of Rbcn-3 disrupted constitutive mammalian Notch signaling in HaCaT cells. Our results indicated a requirement for Rbcn-3 in Notch signaling using reporter assays and analysis of Notch target genes. Furthermore, the strong punctate staining of Hes1 in the nucleus of HaCaT and MCF-7 cells was reduced after inhibition of Rbcn-3 expression. These results suggested an active role or Rbcn-3 in supporting the signaling functions of the Notch pathway.
Rbcn-3 has been shown to regulate V-ATPase function (9, 10), which has been shown to modulate the acidity of endocytic vesicles and cleavage of Notch receptors in Drosophila (9, 10, 23). After confirming this relationship in mammalian cells, we tested whether disrupting ATPase activity via pharmacological inhibition affects Notch signaling in human epithelial cells. As expected, we observed a reduction in Notch pathway activity in response to bafilomycin A1 treatment, a potent inhibitor of the V-ATPase complex. Acidification of intracellular compartments and protein trafficking are compromised when V-ATPase activity is inhibited, suggesting that these processes are important for Notch signaling in mammalian cells. Consistent with this notion, S3 cleavage of Notch has been found to be significantly reduced when trafficking to the early endosome is impaired (23). This can be explained by the idea that the γ-secretase complex and Notch receptors are targeted to endocytic vesicles in which γ-secretase activity is optimal and S3 cleavage occurs most efficiently (4). Moreover, there are data that suggest that γ-secretase activity is optimal in acidic endocytic vesicles (24). It is therefore conceivable that alkalization of intracellular vesicles via V-ATPase inhibition and Rbcn-3 disruption may impair γ-secretase activity and consequently reduce the efficiency of Notch signaling activation. Taken together, these results suggest that acidification of intracellular compartments, which is impaired either directly via ATPase inhibition or indirectly through disruption of Rbcn-3 function, is important for Notch signaling in mammalian cells, most likely by modifying γ-secretase activity and/or by affecting the release of NICD. Bafilomycin has been also evaluated as a potential anticancer agent because it inhibits tumor growth and chemoresistance and promotes apoptosis (25, 26). Although these anticancer effects of bafilomycin are considered to be linked to the intracellular acidosis by V-ATPase inhibition, the exact mechanism remains unclear. Our study suggests potential applications of bafilomycin in cancer therapeutics against malignancies that are dependent on Notch signaling.
Several studies have shown a context-dependent role for the Notch pathway in osteoclast maturation (13, 27). Yamada et al. (27) demonstrated that Notch signaling activity inhibited osteoclast differentiation using primary cells isolated from the bone marrow, spleen, and peritoneal cavity of mice, and a cloned macrophage-like cell line. In another study by Bai et al. (13), bone marrow macrophages isolated from transgenic mice with osteoclast-specific deletion of individual Notch receptors were examined using both in vitro and in vivo methods. The results showed that Notch1 and Notch3 collaborate in inhibiting osteoclast differentiation (13). We confirmed that attenuation of Notch signaling at the level of transcriptional regulation by silencing the expression of Rbpj in two pre-osteoclast cell lines promotes osteoclastogenesis. Similar results were obtained when Rbcn-3 was silenced in the two pre-osteoclast cell lines, which is consistent with disruption of Notch signaling downstream of receptor activation. We also confirmed that Notch signaling was compromised in Rbcn-3 siRNA-treated osteoclasts by examining the nuclear staining of Hes1.
Consistent with a context-dependent function for Notch signaling, Notch ligand Jagged1 has been shown to stimulate osteoclast differentiation. Fukushima et al. (17) demonstrated that Jagged1 signals through Notch2 to promote osteoclast maturation. Recent studies in our laboratory have similarly shown that tumor-derived or immobilized recombinant Jagged1 potently activates osteoclast differentiation in two distinct pre-osteoclast cell lines and primary bone marrow-derived osteoclast precursors.5 These findings demonstrate a unique, context-dependent role for the Notch pathway in cell fate decisions of the osteoclast lineage. We are currently pursuing further investigations in this direction to clarify the mechanism of various Notch pathway ligands, receptors, and downstream signal transducers in the regulation of osteoclast differentiation.
In summary, our results show that Rbcn-3 is required for proper Notch signaling in mammalian cells. We also demonstrate that Rbcn-3 likely influences Notch signaling through its involvement in the acidification of intracellular compartments via the V-ATPase complex. These endocytic compartments most likely regulate the Notch pathway by either modifying the activity of γ-secretase and/or by affecting the release of NICD. Finally, using an osteoclast differentiation assay, we demonstrate that Rbcn-3 is important for carrying out Notch signaling functions. Overall, these studies support a role for Rbcn-3 in the transduction and function of Notch signaling in mammalian cells and established V-ATPase inhibitors as potential therapeutic agents to disrupt Notch signaling.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA134519 (to Y. K.). This work was also supported by Department of Defense Grant BC051647 and the New Jersey Commission on Cancer Research Grant 09-1075-CCR-EO.
N. Sethi and Y. Kang, unpublished data.
- F
- forward
- R
- reverse
- DPBS
- Dulbecco's phosphate-buffered saline
- GSI
- γ-secretase inhibitor.
REFERENCES
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2.Lai E. C. (2004) Development 131, 965–973 [DOI] [PubMed] [Google Scholar]
- 3.Tien A. C., Rajan A., Bellen H. J. (2009) J. Cell Biol. 184, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao J., Chen S., Lee B. (2010) Ann. N. Y. Acad. Sci. 1192, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizzo P., Osipo C., Foreman K., Golde T., Osborne B., Miele L. (2008) Oncogene 27, 5124–5131 [DOI] [PubMed] [Google Scholar]
- 7.Fortini M. E., Bilder D. (2009) Curr. Opin. Genet. Dev. 19, 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortini M. E. (2009) Dev. Cell 16, 633–647 [DOI] [PubMed] [Google Scholar]
- 9.Yan Y., Denef N., Schüpbach T. (2009) Dev. Cell 17, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaccari T., Duchi S., Cortese K., Tacchetti C., Bilder D. (2010) Development 137, 1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seol J. H., Shevchenko A., Shevchenko A., Deshaies R. J. (2001) Nat. Cell Biol. 3, 384–391 [DOI] [PubMed] [Google Scholar]
- 12.Smardon A. M., Tarsio M., Kane P. M. (2002) J. Biol. Chem. 277, 13831–13839 [DOI] [PubMed] [Google Scholar]
- 13.Bai S., Kopan R., Zou W., Hilton M. J., Ong C. T., Long F., Ross F. P., Teitelbaum S. L. (2008) J. Biol. Chem. 283, 6509–6518 [DOI] [PubMed] [Google Scholar]
- 14.Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 15.Zeng Q., Li S., Chepeha D. B., Giordano T. J., Li J., Zhang H., Polverini P. J., Nor J., Kitajewski J., Wang C. Y. (2005) Cancer Cell 8, 13–23 [DOI] [PubMed] [Google Scholar]
- 16.Mitchell P. G., Magna H. A., Reeves L. M., Lopresti-Morrow L. L., Yocum S. A., Rosner P. J., Geoghegan K. F., Hambor J. E. (1996) J. Clin. Invest. 97, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukushima H., Nakao A., Okamoto F., Shin M., Kajiya H., Sakano S., Bigas A., Jimi E., Okabe K. (2008) Mol. Cell. Biol. 28, 6402–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkin M. B., Carbery A. M., Fostier M., Aslam H., Mazaleyrat S. L., Higgs J., Myat A., Evans D. A., Cornell M., Baron M. (2004) Curr. Biol. 14, 2237–2244 [DOI] [PubMed] [Google Scholar]
- 19.Jékely G., Rørth P. (2003) EMBO Rep. 4, 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolas M., Wolfer A., Raj K., Kummer J. A., Mill P., van Noort M., Hui C. C., Clevers H., Dotto G. P., Radtke F. (2003) Nat. Genet. 33, 416–421 [DOI] [PubMed] [Google Scholar]
- 21.Demehri S., Turkoz A., Kopan R. (2009) Cancer Cell 16, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watt F. M., Estrach S., Ambler C. A. (2008) Curr. Opin. Cell Biol. 20, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008) J. Cell Biol. 180, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta-Rossi N., Six E., LeBail O., Logeat F., Chastagner P., Olry A., Israël A., Brou C. (2004) J. Cell Biol. 166, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y. C., Wu W. K., Li Y., Yu L., Li Z. J., Wong C. C., Li H. T., Sung J. J., Cho C. H. (2009) Biochem. Biophys. Res. Commun. 382, 451–456 [DOI] [PubMed] [Google Scholar]
- 26.Sasazawa Y., Futamura Y., Tashiro E., Imoto M. (2009) Cancer Sci. 100, 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada T., Yamazaki H., Yamane T., Yoshino M., Okuyama H., Tsuneto M., Kurino T., Hayashi S., Sakano S. (2003) Blood 101, 2227–2234 [DOI] [PubMed] [Google Scholar]