Abstract
Multiple interactions exist between human follicle-stimulating hormone (FSH) and the N-terminal hormone-binding fragment of the human FSH receptor (FSHR) extracellular domain (ECD). Binding of the other human glycoprotein hormones to their cognate human receptors (luteinizing hormone receptor (LHR) and thyroid-stimulating hormone receptor (TSHR)) was expected to be similar. This study focuses on amino acid residues in β-strands 2 (Lys74), 4 (Tyr124, Asn129, and Thr130), and 5 (Asp150 and Asp153) of the FSHR ECD identified in the human FSH·FSHR ECD crystal structure as contact sites with the common glycoprotein hormone α-subunit, and on noncontact residues in β-strands 2 (Ser78) and 8 (Asp224 and Ser226) as controls. These nine residues are either invariant or highly conserved in LHR and TSHR. Mutagenesis and functional characterization of these residues in all three human receptors allowed an assessment of their contribution to binding and receptor activation. Surprisingly, the six reported α-subunit contact residues of the FSHR ECD could be replaced without significant loss of FSH binding, while cAMP signaling potency was diminished significantly with several replacements. Comparative studies of the homologous residues in LHR and TSHR revealed both similarities and differences. The results for FSH/FSHR were analyzed on the basis of the crystal structure of the FSH·FSHR ECD complex, and comparative modeling was used to generate structures for domains, proteins, and complexes for which no structures were available. Although structural information of hormone-receptor interaction allowed the identification of hormone-receptor contact sites, functional analysis of each contact site was necessary to assess its contribution to hormone binding and receptor activation.
Keywords: G Protein-coupled Receptors (GPCR), Glycoprotein Hormones, 7-Helix Receptor, Ligand-binding Protein, Protein-Protein Interactions, Receptor Structure-Function, Signal Transduction, Extracellular Domain, Glycoprotein Hormone Receptors
Introduction
The heterodimeric glycoprotein hormones (luteinizing hormone (LH),3 CG, FSH, and TSH), each consisting of a common α-subunit and a hormone-specific β-subunit, regulate a number of developmental, reproductive, and metabolic processes by activating their cognate GpHRs, LHR, FSHR, and TSHR (1, 2). GpHRs are composed of two approximately equally sized but functionally distinct modules as follows: an N-terminal ECD, responsible for ligand recognition and binding (3–5), linked to a prototypical GPCR domain (6), responsible for signaling.
GpHRs belong to the subfamily of LRR-containing GPCRs (7). Within the ECD of a given GpHR, the LRR region that is flanked by N- and C-terminal cysteine-rich domains is responsible for ligand-binding specificity and affinity (8–17). The ECDs of the three GpHRs display a lower sequence identity (∼40%), reflecting the observation that hormone specificity is completely encoded within the LRR region, compared with their transmembrane helix portions (∼70%).
Structural information related to the transmembrane helix moiety of GpHRs has become available for several GPCRs as follows: rhodopsin in the dark and constitutively active forms (18–21), the β1- and β2-adrenergic receptors (22–25), and the A2A-adenosine receptor (26). The crystallographic structure of bovine rhodopsin, in particular, has been used for homology modeling of many other GPCRs, including the GPCR moiety of GpHRs (27).
Based on models of GpHR ECDs, it was suggested that the LRR region consists of successive β-strands and α-helices that organize themselves into a cusp-shaped structure (8, 28, 29). Isolated ECDs are capable of binding the hormones with high affinity, as demonstrated by the co-purification of FSH with part of the ECD of the FSHR (30). Studies on chimeric glycoproteins and their receptors have proven highly useful in elucidating structure-function relationships of GpHRs, and detailed mutagenesis studies allowed the identification of a relatively small number of positive and negative key hormone-selective amino acid residues in the ECDs of GpHRs (4, 14–17, 31).
The crystallographic structure of an N-terminal hormone-binding fragment of the FSHR ECD complexed with single-chain FSH provided, for the first time, a crystal structure of a glycoprotein hormone bound to its cognate receptor (30). In the complex, electron densities were determined for amino acid residues 18–259 of the ECD, and both subunits of FSH were found to make multiple contacts with several LRRs of the FSHR ECD fragment in a clasped hand arrangement. This structure has aided enormously in interpreting mutagenesis data (14–17) and vice versa. Complementing the FSHR ECD structure, the structure of the TSHR ECD complexed with a monoclonal antibody was found to be very similar to that of the FSHR ECD (32).
The goal of this study was to analyze several invariant and highly conserved amino acid residues in the ECDs of human FSHR, LHR, and TSHR (Fig. 1) that have been identified as hormone-receptor contact sites in the FSH·FSHR ECD structure (30). By comparing Ala replacements of these amino acid residues in the three receptors as well as multiple replacements of some, it is possible to obtain a measure of the generality of the FSHR ECD structure to that of LHR and TSHR and to ascertain the functional aspects of the reported contacts. The FSHR ECD-FSH α-subunit contacts were chosen because the α-subunit is common in the glycoprotein hormones, and a comparison with the other two receptors will show if they behave in a similar or dissimilar manner. Based on their contact with the FSH α-subunit, the following six amino acid residues were chosen in the FSHR ECD (30): Lys74 in β-strand 2; Tyr124, Asn129, and Thr130 in β-strand 4; and Asp150 and Asp153 in β-strand 5. The following three amino acid residues were chosen as controls as they have no interaction with either subunit of FSH: Ser78 in β-strand 2; Asp224 and Ser226 in β-strand 8. The corresponding residues in the other two receptors are as follows: Lys77, Ser81, Tyr127, Asn132, Thr133, Glu154, Asp157, Asp228, and Ser230 in LHR; and Arg80, Ser84, Phe130, Asn135, Thr136, Glu157, Asp160, Asp232, and Ser234 in TSHR. Comparative modeling and docking studies were also done to complement the experimental results and to aid in their interpretation.
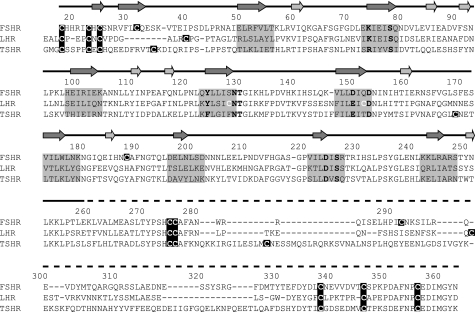
FIGURE 1.
Alignment of the three human GpHR ECDs. The numbering given above the sequence is based on FSHR. The arrows, positioned on the solid line (above the numbering) that represents the part of the FSHR of which the structure is known (30), indicate the β-strands on the concave (dark gray) or convex (light gray) side of the FSHR ECD. The dashed line (above the numbering) indicates the part of the FSHR ECD of which no structural information is known yet. The invariant and highly conserved amino acid residues that were studied are shown in boldface, and all cysteine residues are presented in white on a darker background. Residues on a gray background are forming the β-strands facing the hormone in FSHR and are presumed to do the same in LHR and TSHR.
Although replacements have been made of several of the above amino acid residues in the three human GpHRs (FSHR/D224V (33); LHR K77A and S81A (11), LHR Y127A, N132A, and T133A (12); TSHR R80A,R80D (32), R80K (14), and E157A (13, 34)), there has not been a systematic investigation, and comparison between FSHR, LHR, and TSHR at either of the sites chosen. Some related information is also available for rat LHR/K81D,K81R (35), which corresponds to human LHR/Lys77, and rat LHR/D232E,D232K,D232N (8), which corresponds to human LHR/Asp228. Our results establish that invariant and highly similar amino acid residues in homologous positions of these three similar human GPCRs have distinct functional roles when comparing FSH/FSHR, CG/LHR, and TSH/TSHR binding and hormone-mediated signaling. Whether these differences reflect distinct binding modalities, microenvironments, and/or localized conformational changes remains to be established.
EXPERIMENTAL PROCEDURES
Materials
Dulbecco's modified Eagle's medium (DMEM) was purchased from Cellgro Mediatech, Inc. (Herndon, VA) and Sigma. NCS, antibiotics, Waymouth's media, and trypsin-EDTA were purchased from Invitrogen, and TransFectin lipid reagent was from Bio-Rad. BSA, isobutylmethylxanthine, a goat anti-rat IgG peroxidase conjugate, and the recombinant human hormones CG and FSH and bovine TSH were purchased from Sigma. Also, human CG, LH, and FSH were obtained from NV Organon (Oss, The Netherlands, courtesy of Dr. W. G. E. J. Schoonen); human TSH was from Sigma and from Dr. A. F. Parlow, National Institutes of Health, Torrance, CA. The high affinity (monoclonal) anti-HA antibody was from Roche Diagnostics. 125I-CG and 125I-FSH were iodinated using the IODO-GEN iodination reagent following the standard iodination protocol by Pierce. 125I-cAMP RIA kits and [125I]NaI were obtained from PerkinElmer Life Sciences and Amersham Biosciences, respectively.
Construction of Mutant FSHRs, LHRs, and TSHRs
The cDNAs encoding human FSHR and LHR were kindly provided by Dr. T. Minegishi (Gunma University School of Medicine, Maebashi, Japan) and Dr. E. Milgrom (INSERM, Paris, France), respectively, and that for human TSHR was obtained from the Missouri S&T cDNA Resource Center. Following subcloning of the cDNAs into the pcDNA3.1/V5-His-TOPO expression vector (Invitrogen), PCR-based mutagenesis was used with the FSHR, LHR, and TSHR templates to obtain the desired replacements. An epitope (YPYDVPDYA) from the influenza virus hemagglutinin (HA) was inserted between the C terminus of the signal peptide and the N terminus of the mature receptor, i.e. between Gly17–Cys18 of FSHR, Leu27–Arg28 of LHR, and Gly20–Gly21 of TSHR.
Cell Culture and Transient Transfection of Cells
HEK 293 cells (obtained from the American Type Culture Collection, Manassas, VA) and HEK 293T cells (36) were grown at 37 °C in humidified air containing 5% CO2 in DMEM supplemented with 10% (v/v) newborn calf serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 125 ng/ml amphotericin B, and 10 mm HEPES, pH 7.4 (HEK 293 cells), or with 10% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B, pH 7.0–7.2 (HEK 293T cells) and transiently transfected as described elsewhere (16, 17, 37, 38). Briefly, the HEK 293 cells were transiently transfected (1 or 5 μg of wild type (WT) cDNA and 5 μg of mutant cDNA) using TransFectin, and the HEK 293T cells were transiently transfected (1 μg of WT and mutant cDNAs) with 10 μg of pCRE/β-gal plasmid (39) using the modified bovine serum transfection method (15, 16). To achieve comparable receptor densities between WT and mutant receptors in the HEK 293 cells, each mutant receptor was compared with a WT control in which the Bmax values were within a 2–3-fold range of each other. Hence, in some cases, WT receptor controls, transfected with 1 μg of cDNA, were compared with mutant receptors transfected with 5 μg of cDNA.
Cell Surface Expression
Receptor levels at the cell surface were measured by specific hormone binding (described below) and ELISA measurements (15–17). For the latter, the HEK 293T cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 30 min and then blocked with 1% dried nonfat milk in 0.1 m NaHCO3 for at least 4 h, all at room temperature. The cells were then incubated at 4 °C overnight with a 1:200 dilution of anti-HA high affinity antibodies in Tris-buffered saline containing 0.1% bovine serum albumin. The following day, the cells were incubated with a 1:500 dilution of peroxidase-conjugated goat anti-rat IgG in 1% dried nonfat milk in 0.1 m NaHCO3 at room temperature for 2 h. A 3,3′,5,5′-tetramethylbenzidine liquid substrate system (Sigma) was used to visualize the peroxidase activity of the WT and mutant receptors. Absorbance values (at 450 nm) of mock transfected cells were subtracted, and mutant receptor expression values were expressed as the percentage of WT receptor expression. Experiments were repeated a minimum of three times from independent transfections, each performed in duplicate (HEK 293) or triplicate (HEK 293T), and the results are presented as mean ± S.E.
Hormone Binding
Binding assays with HEK 293 cells were performed as given elsewhere (37, 38, 40, 41). Briefly, about 16–18 h after transfection, the cells were replated (1 × 105 cells per well) into 12-well tissue culture plates, and 24 h later the cells were assayed for 125I-labeled human hormone (CG or FSH) competitive and saturation binding. Competitive binding experiments were performed with a fixed concentration of labeled hormone (50–100 pm) and increasing concentrations of unlabeled hormone, whereas the saturation binding studies were conducted with various concentrations of labeled hormone (50–5000 pm). All binding studies involved incubation of the hormone and cells at 37 °C for 6 h in the presence of binding buffer (278 mm sucrose, 0.1% glucose, 5 mm HEPES, pH 7.4, 5 mm KCl, 1.2 mm MgSO4, 1 mm NaHCO3, 1 mm CaCl2·2H2O, 1.2 mm KH2PO4) and 0.1% bovine serum albumin (42). Nonspecific binding was determined by addition of a 1000-fold excess of unlabeled hormone. The experiments were repeated 3–5 times, each in duplicate, using independent transfections, and the results are presented as Kd and Bmax values relative to the WT receptor. As discussed above, transfection conditions were chosen to minimize significant differences between WT and mutant receptors in the HEK 293 cells in which both binding and signaling studies were conducted. Binding studies were not performed for TSH and TSHR because iodination of the human hormone effectively abolishes specific binding to the receptor, and we prefer not to compare data using a nonhomologous system, e.g. bovine TSH.
Cyclic AMP Assays
The transfected HEK 293 cells were prepared as described above and incubated with increasing concentrations of the cognate hormone, FSH, CG, and TSH, at 37 °C for 30 min in the presence of 0.8 mm 3-isobutyl-1-methylxanthine (37, 38). The incubation medium was removed, and the cells were lysed in 100% ethanol at −20 °C overnight. The extract was collected, dried under vacuum, and resuspended in the buffer of the 125I-cAMP assay kit. Cyclic AMP concentrations were determined by RIA as recommended by PerkinElmer Life Sciences. All experiments were repeated 3–5 times, each in duplicate, and the results for EC50 and Rmax are given as means ± S.E., relative to the WT receptor. In separate experiments, signaling was measured with the HEK 293T cells as described elsewhere (15–17). Briefly, 2 days after transfection, the cells were cultured in HEPES-modified DMEM containing 0.1% bovine serum albumin and 0.1 mm 3-isobutyl-1-methylxanthine and then incubated for 6 h with the cognate hormone, including human LH and LHR. The conversion of o-nitrophenyl-β-d-galactopyranoside into o-nitrophenol was measured at 405 nm, reflecting the changes in β-galactosidase activity mediated by the gonadotropins. The β-galactosidase gene used to transfect the HEK 293T cells was under the control of a human vasoactive intestinal peptide promoter containing five cAMP-response elements (39). The collected data were related to 10 μm forskolin-induced changes and were thus expressed in arbitrary units. Experiments were repeated three times, each in triplicate, and the results are given as means ± S.E.
Data Analysis
Both binding and cAMP data were analyzed by Prism software (Graph Pad Software, San Diego) using nonlinear regression analysis. Whenever possible, the Bmax and Kd values, obtained from saturation binding, and the EC50 and Rmax values, obtained from RIA measurements, were used to determine the coupling efficiency (43), Q = {(1/2)(1 + (Kd/EC50))/(Rmax/Bmax)} of receptors expressed in HEK 293 cells. Relative values of Bmax, EC50, and Rmax were also determined independently with the HEK 293T cells. In addition to standard statistical analysis to determine significance (p < 0.05), differences between expression, binding, and signaling of mutant receptors had to exceed that of WT receptor by ≥3- or ≤3-fold. This rather stringent criterion was imposed to correct for interassay variability and to help ensure that stated differences are functionally significant.
Molecular Modeling
Models of complexes of human LH, CG, and TSH bound to the LRR domain of their respective human receptors were built using the available structures of FSH·FSHRHB (Protein Data Bank code 1XWD) (30) and M22·TSHR (Protein Data Bank code 3G04) (32). The high sequence identity between the three GpHRs, the identical α-subunit, and the high sequence identity shared by the three β-subunits of the hormones contributed to this process. The sequences of CG, LHR, TSH, and TSHR were aligned to the stretch of residues present in the crystal structure of the FSH·FSHR ECD complex taking into account sequence identity, the position of disulfide bonds, and structural features. Comparative modeling was done using MODELLER (44), which uses the satisfaction of spatial restraints, along with simultaneous optimization of CHARMM energies, using conjugate gradients and molecular dynamics with simulated annealing (45). A script with increased variable target function method and molecular dynamics optimization was used to generate a model for each complex. The models were evaluated by examination of the MODELLER output log files, their Ramachandran plots, visual inspection, and finally cross-checked with existing mutagenesis data. The programs PISA (46), PROTORP (47), PIC (48), RAMPAGE, LIGPLOT (49), HBPLUS 3.0 (50), and PyMOL (51) were used to identify the residues involved in the interaction between hormone and receptor and to calculate physical and chemical parameters of the protein interaction sites. Models of each glycoprotein hormone·receptor complex were used to generate maps of the electrostatic surface potentials and to study the hormone receptor interface. Visualization and molecular graphics were done using PyMOL and Discovery Studio Visualizer (Accelrys Software, Inc.).
RESULTS
Locations of the FSHR and LHR Mutations
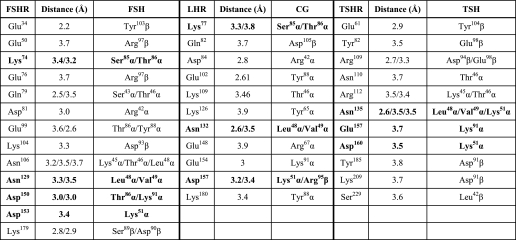
Fig. 1 shows the aligned amino acid sequences of the three GpHR ECDs and highlights the location of the nine amino acid residues replaced with Ala. Additional replacements with other amino acid residues were also made of the Lys/Arg in β-strand 2 and the Asp/Glu in β-strands 5 and 8. The structures of β-strands 2, 4, and 5 of FSHR are shown in supplemental Fig. 1, A–C, and the FSH-FSHR contacts (defined as <4 Å) are given schematically in supplemental Fig. 1D. We have determined the details of the FSH·FSHR ECD interactions (supplemental Table 1) based on the coordinates of the FSH·FSHR ECD structure as reported by Fan and Hendrickson (see supplemental material in Ref. 30). The listing in supplemental Table 1 indicates that the six residues bind to the FSH α-subunit via ion-ion, ion-dipole, H-bonding, and hydrophobic interactions. Our assessment of hormone·receptor contacts agree, in general, with those given by Fan and Hendrickson (30). A few differences, however, do exist. Of the nine amino acid residues chosen for Ala replacement, six were reported to be in contact with the FSH α-subunit or β-subunit in Fig. 2 of their paper (30). In our analysis, we find contacts with just the FSH α-subunit, as also concluded in the supplemental material by Fan and Hendrickson (30). Moreover, Ser78 was reported in Fig. 2 of the Fan and Hendrickson paper (30) to exhibit a fractional solvent accessibility (FSA) of 0.1–0.4, although a direct contact was not indicated in the supplemental material of Ref. 30. Our analysis of the structure indicated no direct contact between the FSH α-subunit and Ser78 in FSHR, and we have chosen to use this residue as a control in our studies.
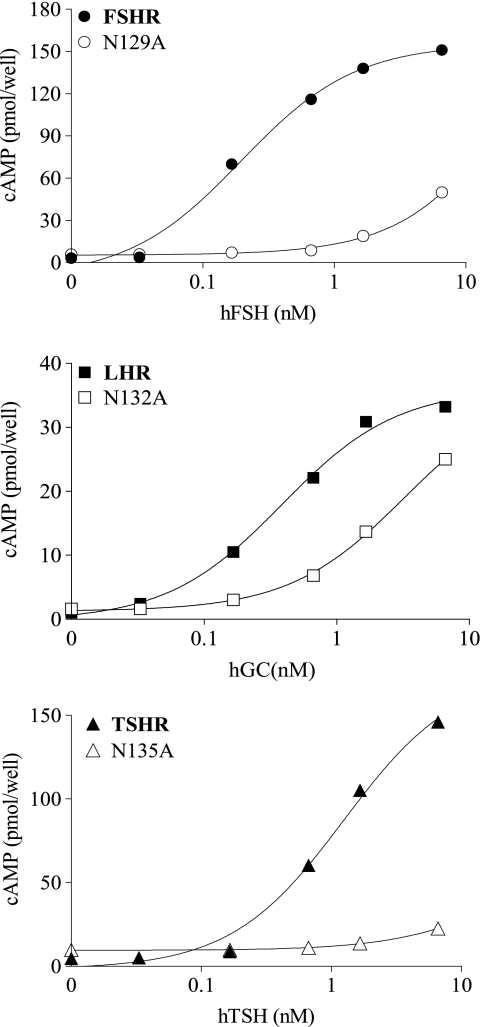
FIGURE 2.
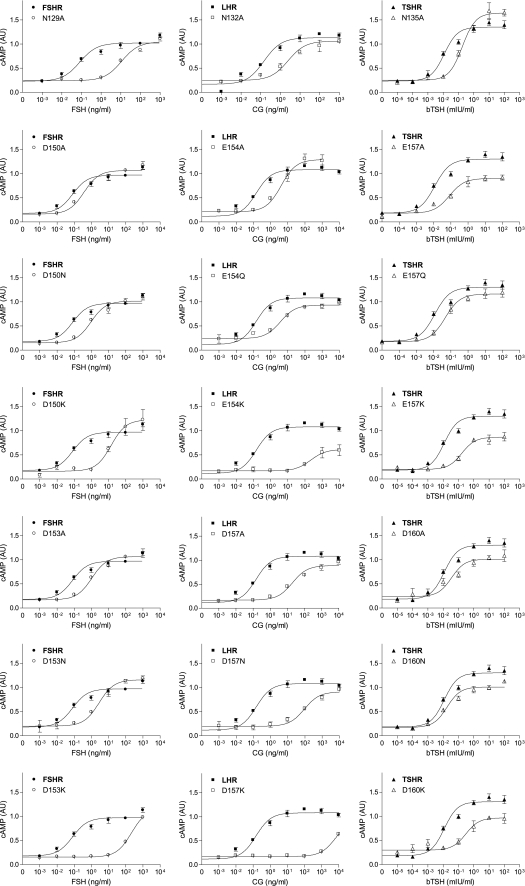
Dose-response signaling curves of the three GpHRs, WT, and mutant forms in β-strand 4. Representative cAMP responses are shown for the WT receptors and Ala replacements at Asn129 (FSHR), Asn132 (LHR), and Asn135 (TSHR) upon receptor activation with the cognate hormones (hCG being used to activate LHR; h is human). All assays of intracellular cAMP were done by RIA with HEK 293 cells.
Measured Experimental Parameters (Tables 1–6)
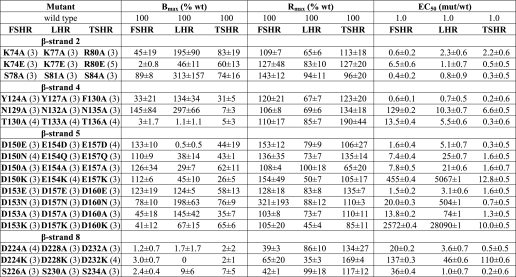
TABLE 1.
Summary of expression, binding, and signaling results for β-strand 2 mutants in HEK 293 cells
| Mutant | Bmax | Kd (mutant/WT) | EC50 (mutant/WT) | Rmax | Q (mutant/WT) |
|---|---|---|---|---|---|
| % WT | % WT | ||||
| FSHR | |||||
| K74A (n = 5) | 87 ± 23 | 0.8 ± 0.2 | 1.0 ± 0.2 | 84 ± 11 | 0.9 ± 0.2 |
| K74E (n = 2) | 102 ± 24 | 0.8 ± 0.3 | 0.7 ± 0.2 | 210 ± 60 | 2.4 ± 0.1 |
| S78A (n = 5) | 66 ± 26 | 0.9 ± 0.3 | 1.4 ± 0.3 | 89 ± 25 | 0.9 ± 0.1 |
| LHR | |||||
| K77A (n = 4) | 143 ± 56 | 1.1 ± 0.2 | 1.0 ± 0.1 | 143 ± 51 | 1.0 ± 0.04 |
| K77E (n = 4) | 159 ± 52 | 1.3 ± 0.5 | 1.7 ± 0.3 | 228 ± 42 | 0.9 ± 0.1 |
| S81A (n = 4) | 217 ± 23 | 1.4 ± 0.3 | 1.3 ± 0.3 | 165 ± 42 | 0.9 ± 0.03 |
| TSHR | |||||
| R80A (n = 3) | a | b | 0.8 ± 0.1 | 70 ± 18 | b |
| R80E (n = 3) | a | b | 0.9 ± 0.3 | 104 ± 23 | b |
| S84A (n = 3) | a | b | 0.5 ± 0.1 | 145 ± 18 | b |
a Data were not determined by binding (see Table 6).
b Data were not determined.
TABLE 2.
Summary of expression, binding, and signaling results for β-strand 4 mutants in HEK 293 cells
| Mutant | Bmax | Kd (mutant/WT) | EC50 (mutant/WT) | Rmax | Q (mutant/wt) |
|---|---|---|---|---|---|
| % WT | % WT | ||||
| FSHR | |||||
| Y124A (n = 5) | 52 ± 15 | 0.7 ± 0.2 | 1.1 ± 0.2 | 72 ± 14 | 1.1 ± 0.2 |
| N129A (n = 3) | 45 ± 23 | 4.8 ± 2.8 | 226 ± 59 | a | a |
| T130A (n = 3) | 310 ± 52 | 3.2 ± 0.6 | 1.8 ± 0.1 | 258 ± 47 | 1.2 ± 0.3 |
| LHR | |||||
| Y127A (n = 4) | 82 ± 9 | 1.3 ± 0.2 | 2.1 ± 0.5 | 83 ± 10 | 0.9 ± 0.06 |
| N132A (n = 3) | 112 ± 34 | 3.4 ± 1.5 | 10.3 ± 1.6 | 126 ± 34 | 0.9 ± 0.06 |
| T133A (n = 3) | 103 ± 36 | 3.3 ± 1.1 | 2.8 ± 0.6 | 93 ± 5 | 1.0 ± 0.07 |
| TSHR | |||||
| F130A (n = 3) | b | c | 3.3 ± 0.5 | 71 ± 7 | c |
| N135A (n = 3) | b | c | 10.5 ± 0.7 | a | c |
| T136A (n = 3) | b | c | 2.2 ± 0.2 | 75 ± 11 | c |
a The EC50 value was too high to permit accurate measurement of Rmax and Q.
b Data were not determined by binding (see Table 6).
c Data were not determined.
TABLE 3.
Summary of expression, binding, and signaling results for β-strand 5 mutants in HEK 293 cells
| Mutant | Bmax | Kd (mutant/WT) | EC50 (mutant/WT) | Rmax | Q (mutant/WT) |
|---|---|---|---|---|---|
| % WT | % WT | ||||
| FSHR | |||||
| D150E (n = 2) | 165 ± 28 | 2.1 ± 0.4 | 0.8 ± 0.2 | 224 ± 2 | 3.0 ± 0.6 |
| D150N (n = 3) | 249 ± 65 | 2.8 ± 0.6 | 3.0 ± 0.5 | 231 ± 72 | 0.8 ± 0.03 |
| D150A (n = 3) | 60 ± 24 | 1.2 ± 0.4 | 6.4 ± 1.1 | 115 ± 9 | 0.8 ± 0.1 |
| D150K (n = 2) | 105 ± 2 | 1.0 ± 0.1 | >200 | a | a |
| LHR | |||||
| E154D (n = 4) | 36 ± 8 | 1.9 ± 0.9 | 5.6 ± 0.9 | 80 ± 13 | 1.1 ± 0.2 |
| E154Q (n = 4) | 62 ± 25 | 12 ± 2 | 27 ± 2 | a | a |
| E154A (n = 4) | 33 ± 6 | 10 ± 4 | >33 | a | a |
| E154K (n = 4) | 20 ± 6 | 2.5 ± 0.6 | a | 0 | a |
| TSHR | |||||
| E157D (n = 3) | b | c | 0.8 ± 0.1 | 119 ± 27 | c |
| E157Q (n = 3) | b | c | 1.4 ± 0.2 | 84 ± 5 | c |
| E157A (n = 3) | b | c | 1.0 ± 0.1 | 77 ± 2 | c |
| E157K (n = 3) | b | c | 6.6 ± 0.4 | a | c |
a The EC50 values was too high to permit accurate measurement of Rmax and Q.
b Data were not determined by binding (see Table 6).
c Data were not determined.
TABLE 4.
Summary of expression, binding and signaling results for β-strand 5 mutants in HEK 293 cells
| Mutant | Bmax | Kd (mutant/WT) | EC50 (mutant/WT) | Rmax | Q (mutant/WT) |
|---|---|---|---|---|---|
| % WT | % WT | ||||
| FSHR | |||||
| D153E (n = 2) | 90 ± 22 | 1.2 ± 0.4 | 1.5 ± 0.4 | 203 ± 1 | 1.8 ± 0.05 |
| D153N (n = 3) | 210 ± 51 | 3.2 ± 0.6 | 6.5 ± 1.8 | 130 ± 60 | 0.3 ± 0.06 |
| D153A (n = 4) | 49 ± 7 | 1.1 ± 0.2 | 25.0 ± 3.0 | 106 ± 9 | 0.5 ± 0.1 |
| D153K (n = 3) | 47 ± 1 | 0.6 ± 0.1 | >200 | ∼0 | a |
| LHR | |||||
| D157E (n = 5) | 234 ± 90 | 2.0 ± 0.5 | 2.4 ± 0.3 | 215 ± 76 | 1.1 ± 0.2 |
| D157N (n = 4) | 40 ± 8 | 3.9 ± 1.0 | >400 | a | a |
| D157A (n = 4) | 95 ± 35 | 13 ± 1 | >95 | a | a |
| D157K (n = 2) | 22 ± 8 | 2.4 ± 0.7 | a | ∼0 | a |
| TSHR | |||||
| D160E (n = 3) | b | c | 3.3 ± 0.5 | 56 ± 15 | c |
| D160N (n = 2) | b | c | 3.3 ± 1.2 | 56 ± 12 | c |
| D160A (n = 3) | b | c | 1.7 ± 0.1 | 68 ± 13 | c |
| D160K (n = 3) | b | c | 6.6 ± 0.4 | a | c |
a The EC50 was too high to permit accurate measurement of Rmax and Q.
b Data were not determined by binding (see Table 6).
c Data were not determined.
TABLE 5.
Summary of expression, binding, and signaling results for β-strand 8 mutants in HEK 293 cells
| Mutant | Bmax | Kd (mutant/WT) | EC50 (mutant/WT) | Rmax | Q (mutant/WT) |
|---|---|---|---|---|---|
| % WT | % WT | ||||
| FSHR | |||||
| D224A (n = 4) | 39 ± 22 | 1.8 ± 0.8 | 3.1 ± 0.4 | 41 ± 30 | 0.7 ± 0.7 |
| D224K (n = 2) | 84 ± 8 | 1.3 ± 0.1 | a | a | a |
| S226A (n = 3) | 5 ± 1 | b | b | b | b |
| LHR | |||||
| D228A (n = 3) | 37 ± 8 | 4.0 ± 1.5 | 3.9 ± 1.3 | 91 ± 13 | 2.6 ± 0.3 |
| D228K (n = 3) | 80 ± 24 | 5.3 ± 1.8 | >20 | a | a |
| S230A (n = 4) | 95 ± 19 | 0.7 ± 0.1 | 1.5 ± 0.3 | 244 ± 49 | 2.1 ± 0.1 |
| TSHR | |||||
| D232A (n = 3) | c | d | 4.0 ± 1.0 | 52 ± 20 | d |
| D232K (n = 2) | c | d | a | a | d |
| S234A (n = 3) | c | d | 1.3 ± 0.1 | 90 ± 15 | d |
a Signaling was too low to permit measurement of EC50 and Q.
b Expression was too low to permit accurate measurements.
c Data were not determined (see Table 6).
d Data were not determined.
TABLE 6.
Summary of expression and signaling results in HEK 293T cells
a Determined by ELISA.
b Data were determined from changes in β-galactosidase activity.
To fully characterize the mutant receptors and facilitate comparison with the appropriate WT receptor, the following two independent approaches were used to determine cell surface expression (Bmax) of HA-tagged FSHR, LHR, and TSHR: ELISA measurements to determine total receptor numbers (in HEK 293T cells) and saturation binding to determine functional receptor numbers (in HEK 293 cells), i.e. capable of specific binding of cognate hormone. Signaling was also based on two independent approaches as follows: a colorimetric assessment of cAMP-induced β-galactosidase activity (in HEK 293T cells) and RIA of intracellular cAMP (in HEK 293 cells). Both methods permit a quantitative comparison between WT and mutant receptors of the EC50 (potency) and Rmax (efficacy) values, i.e. the maximal cAMP produced at high ligand concentrations, corrected for the basal concentration. With all receptor mutants investigated, the basal cAMP values were within ∼2-fold that of the WT receptor; thus, there is no compelling evidence of constitutive activation in any of the mutants. Each parameter provides information about the mutant receptors, and whenever possible, coupling efficiencies were determined. To facilitate comparisons, the results for Kd, EC50, and Q values are presented as a ratio of the value of the mutant receptor to that of the WT receptor; Bmax and Rmax, as per convention, are given as a percentage of that for each WT receptor. Typical results for WT FSHR and LHR (HEK 293 cells transfected with 1 μg of cDNA) are, respectively, as follows for Bmax, Kd, EC50, and Rmax: 18 ± 4 fmol/well, 2.7 ± 0.5 nm, 0.48 ± 0.07 nm, and 51 ± 13 pmol/well for FSHR (n = 17) and 25 ± 9 fmol/well, 1.2 ± 0.2 nm, 0.49 ± 0.06 nm, and 34 ± 12 pmol/well for LHR (n = 19). For TSHR (HEK 293 cells transfected with 5 μg of cDNA), the EC50 and Rmax values were 1.5 ± 0.2 and 91 ± 17 pmol/well, respectively (n = 15).
Functional Characterization of the GpHR Mutants (Tables 1–6 and Figs. 2–6)
FIGURE 3.
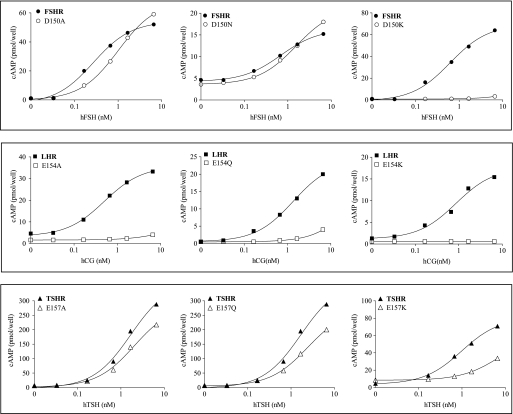
Dose-response signaling curves of the three GpHRs, WT, and mutant forms in β-strand 5. Representative cAMP responses are shown for the WT receptors and mutant forms harboring replacements of Asp150 (FSHR), Glu154 (LHR), and Glu157 (TSHR) upon receptor activation with the cognate hormones (hCG being used to activate LHR; h is human). All assays of intracellular cAMP were done by RIA with HEK 293 cells.
FIGURE 4.
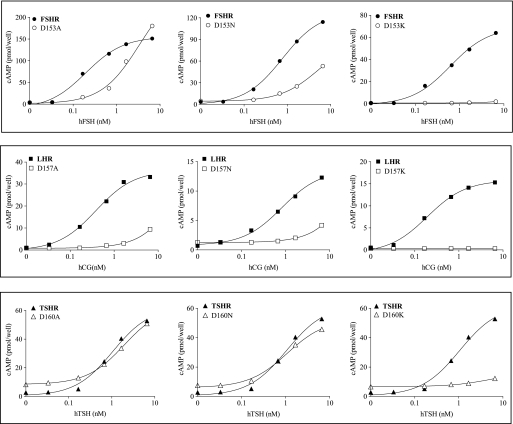
Dose-response signaling curves of the three GpHRs, WT, and mutant forms in β-strand 5. Representative cAMP responses are shown for the WT receptors and mutant forms harboring replacements at Asp153 (FSHR), Asp157 (LHR), and Asp160 (TSHR) upon receptor activation with the cognate hormones (hCG being used to activate LHR; h is human). All assays of intracellular cAMP were done by RIA with HEK 293 cells.
FIGURE 5.
Representative dose-response curves of the three WT GpHRs and selected mutants. These studies were done in HEK 293T cells, and cAMP was measured by a colorimetric assay at 405 nm based on the conversion of o-nitrophenyl-β-d-galactopyranoside into o-nitrophenol. The reaction is catalyzed by cAMP activation of a β-galactosidase gene under the control of a human vasoactive intestinal peptide promoter containing five cAMP-response elements. The results were compared with the activation achieved by 10 μm forskolin and are expressed in arbitrary units (AU). b is bovine.
FIGURE 6.
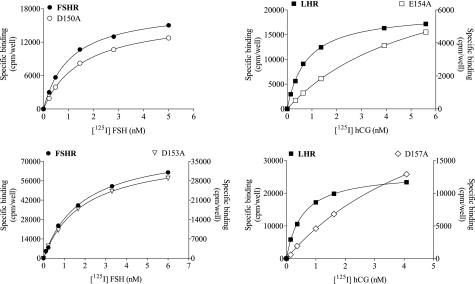
Saturation binding curves of WT and mutant forms of FSHR and LHR with Ala replacements at two positions in β-strand 5. Representative binding curves are shown for the WT receptors and mutant FSHR (D150A and D153A) and LHR (E154A and D157A) forms. All binding studies were done using 125I-labeled FSH and 125I-labeled CG with HEK 293 cells.
Tables 1–6 summarize the results obtained for the mutants in β-strands 2, 4, 5, and 8 of the three receptors. Figs. 2–5 show signaling dose-response curves for several of the more interesting mutants, and representative binding data for the two aspartic acid residues in β-strand 5 of FSHR and LHR are presented in Fig. 6. The data in Tables 1–5 and Figs. 2–4 are from the studies using HEK 293 cells; the results on expression and signaling with HEK 293T cells, based on different methods of analysis, are given in Table 6 and Fig. 5. Results were also obtained for LH and LHR, but for brevity, these are not presented. They do, however, parallel the results obtained with CG and LHR. Although there are some differences in the expression and signaling results obtained on the two cell types with different methodologies, the agreement between the two methods is good overall.
Mutants in β-Strand 2 (Tables 1 and 6 and Fig. 6)
We chose Lys74 in FSHR and the corresponding Lys and Arg in LHR and TSHR, respectively, as a contact site to study because it binds to Ser85 and Thr86 of the FSH α-subunit with an FSA ≥0.4. The K74A,K74E mutants in FSHR, the K77A,K77E mutants in LHR, and the R80A,R80E mutants in TSHR express well, and all exhibit signaling parameters like those of WT receptor. The Kd values of the FSHR and LHR mutants are also comparable with WT receptors. Thus, even though Lys74 is a contact site for the FSH α-subunit, it can be replaced with Ala or even an oppositely charged residue, Glu, without having an adverse effect on binding affinity. With the exception of the K74E mutant, the Q values are similar to that of WT receptor. Ala replacements of the Ser78 in β-strand 2, a noncontact site in FSHR, also behaved like the WT receptors. In our model of CG·LHR (see below), we find that LHR Lys77 interacts in a very similar way as FSHR Lys74, forming a hydrogen bond with Ser85 and Thr86 in the α-subunit. In our model of TSH·TSHR, we do not observe such an interaction for Arg80, which is not close to any TSH residue within a 4-Å range and has an FSA of 0.67.
Mutants in β-Strand 4 (Tables 2 and 6 and Figs. 2 and 6)
The three following side chains were selected for replacement in β-strand 4: Tyr124, Asn129, and Thr130 in FSHR as well as the corresponding residues in LHR (identical in LHR) and TSHR (Phe, Asn, and Thr, respectively, in TSHR). In the FSH·FSHR ECD structure, Tyr124, Asn129, and Thr130 contact FSH α-subunit residues Tyr88, Leu48/Val49, and Leu48 and have FSAs of 0.1–0.4, 0.1–0.4, and <0.1, respectively. With the exception of the three TSHR mutants, those for FSHR and LHR expressed well. Interestingly, the aromatic side chain could be replaced with a methyl group (on Ala) without any measurable functional change in FSHR and LHR. Likewise, the signaling parameters were similar to those of WT receptor, although the EC50 value for TSHR was slightly elevated. Replacement of the invariant Asn in the three receptors with Ala resulted in a reduction in the binding affinities (measured for FSHR and LHR) and a much larger decrease in the signaling potency, particularly for FSHR. At high concentrations of hormone (Table 6), the Rmax values are like those of the WT receptor; however, the EC50 values are greatly increased. Replacement of the invariant Thr with Ala in the three receptors led to a slight increase in the Kd values of FSHR and LHR, but the EC50 values of the three receptors were <3-fold higher than that of the WT receptor. The increase in Rmax of FSHR/T130A may reflect, at least in part, the high level of expression. Despite the low level of expression, TSHR/T136A signals similarly to the WT receptor. With the exception of FSHR/N129A, the coupling efficiencies of the FSHR and LHR mutants are like those of the WT receptors. The conserved Asn in β-strand 4 of FSHR, LHR, and TSHR is important in hormone-mediated signaling, but it seems to have a much larger role in FSHR than in the other two GpHRs. In our models, we also see that the aromatic side chains of Tyr127 (LHR) and Phe130 (TSHR) are involved in hydrophobic interactions with Tyr88 of the α-subunit, as is seen for Tyr124 in FSHR. We also see that the conserved asparagines (Asn129 in FSHR, Asn132 in LHR, and Asn135 in TSHR) are involved in multiple interactions with equivalent residues in the α-subunit. The FSA values for FSHR Asn129, LHR Asn132, and TSHR Asn135 are 0.13, 0.08, and 0.07, respectively.
Mutants in β-Strand 5 (Tables 3, 4 and 6 and Figs. 3, 4, and 6)
The two acidic residues in β-strand 5 were chosen for replacement in the three receptors. In the FSH·FSHR ECD structure, FSHR/Asp150 and FSHR/Asp153 show FSAs of <0.1 and 0.1–0.4, respectively. In addition to the Ala replacements made in the two acidic residues, we also investigated the effects of replacements with charge retention, e.g. Asp → Glu and Glu → Asp, a polar but nonionizable residue, e.g. Asp → Asn and Glu → Gln, and a positively charged residue, Lys. With the exception of LHR/E154K and TSHR/E157K, which expressed less than 30% that of the WT receptor, the expression levels of the other mutants were reasonably good.
For FSHR, the mutants D150E and D153E have properties comparable with those of WT FSHR. Although the Kd values for D150A,D150K and D153A,D153K are similar to that of WT FSHR, the EC50 values are greatly increased, especially for the Lys replacements. The Kd and EC50 values of the D150N and D153N mutants are elevated some 3–6-fold over those of WT FSHR. The two Asp residues in β-strand 5 can thus be interchanged with Glu with no major change in functionality, but replacement with Ala, Asn, or Lys greatly reduces the signaling potency. The coupling efficiency was somewhat elevated for the D150E mutant, reflecting to a large extent the increased expression and Rmax; the Q values for the D150N,D150A and D153E,D153A mutants are similar to the WT receptor (that for D153N is some 3-fold lower than WT FSHR). The poor signaling of the Lys replacements prohibited an estimate of coupling efficiencies for those two mutants.
For LHR, the mutants E154D and D157E exhibit binding similar to that of WT LHR, but the EC50 is increased some 6-fold in the former. The Kd values of the E154A,E154Q and D157A,D157N mutants are increased some 4–13-fold over WT LHR, and the EC50 values are increased even more so. Interestingly, the E154K and D157K mutants have Kd values similar to that of WT LHR, although in both cases signaling is nearly abolished. Some of the increase in the EC50 values of the E154Q,E154A and D157N,D157A replacements can be attributed to the decreased hormone binding affinity. The coupling efficiencies of the E154D and D157E replacements are similar to WT LHR, but Q values could not be determined for the other replacements at these two positions in β-strand 5 due to the diminished signaling. For TSHR, the EC50 values of the E157D,E157Q,E157A and D160A mutants are like that of WT receptor, although those for the D160E,D160N mutants are slightly elevated. Signaling is diminished in both the E157K and D160K mutants.
The two conserved acidic residues in β-strand 5 of the gonadotropin receptors seem to make more of a contribution to binding in LHR than in FSHR. Moreover, these acidic side chains in the three GpHRs appear to be of less importance in hormone-mediated signaling in TSHR than in the other two receptors. In our models (see below), all three complexes show similar interactions between the two acidic residues in the receptor and Lys51 of the α-subunit of the hormones. Interestingly, in our CG·LHR model, an additional hydrogen bond was between Asp157 and Arg95 of the β-subunit of CG which, with its large and positively charged side chain of Arg95, is prominently present in the interface. In the β-subunits of FSH and TSH, we find the much smaller and polar residues Ser89 and Thr90, respectively. Ser89 of the FSH β-subunit makes contact with FSHR/Lys179, a residue of crucial importance for hormone selectivity of the FSH receptor (14, 16, 52). TSH residue Thr90 is not involved in a receptor interaction. Arg95, Ser89, and Thr90 have FSA values of 0.4, 0.6, and 0.77, respectively.
Mutants in β-Strand 8 (Tables 5 and 6)
The two invariant residues chosen in β-strand 8 for controls are FSHR Asp224 and Ser226, LHR Asp228 and Ser230, and TSHR Asp232 and Ser234. FSHR/S226A and each of the TSHR mutants, D232A, D232K, and S234A, expressed poorly and could not be characterized. The Kd values of FSHR D224A and D224K are comparable with that of WT receptor, but the signaling of D224A is reduced and almost abolished in D224K. The Kd values of LHR D228A and D228K are elevated over WT LHR, and the signaling is greatly reduced in D228K (the increase in EC50 of LHR/D228A may result from the increase in Kd values compared with WT LHR). LHR/S230A behaves similarly to WT LHR, except that Rmax is increased. Although FSHR/Asp224 is not a contact site for FSH binding, the side chain evidently contributes to FSH-mediated signaling, as it also does with LHR.
Molecular Modeling of LHR (Tables 7–9 and Fig. 7)
TABLE 7.
Hydrogen bonds and salt bridges within 4 Å
Summary of the hydrogen bonds and salt bridges in the FSH·FSHR ECD structure (30) and those predicted from the models generated in this study for the CG·LHR ECD and TSH·TSHR ECD complexes. Comparative modeling was based on the structures of FSH·FSHR ECD (30), the TSHR ECD (32) and CG (55, 56).
TABLE 8.
Hydrophobic contacts within 5 Å
Summary of the hydrophobic contacts in the FSH·FSHR ECD complex (30) and those predicted from the models generated in this study for CG·LHR ECD and TSH·TSHR ECD. The structures of FSH-FSHR ECD (30), TSHR ECD (32), and CG (55, 56) were used for comparative modeling.
| FSHR | FSH | LHR | CG | TSHR | TSH |
|---|---|---|---|---|---|
| Val54 | Leuβ99 | Tyr127 | Tyrα88 | Phe130 | Tyrα88 |
| Leu55 | Leuβ99/Tyrβ103 | Ile152 | Tyrα88/Tyrα89 | Pro162 | Leuα48 |
| Tyr124 | Tyrα88 | Leu159 | Valα49 | Tyr163 | Leuα48 |
| Leu148 | Tyrα88/Tyrα89 | ||||
| Ile222 | Proβ45 | ||||
| Ile174 | Tyrα89 | ||||
| Trp176 | Tyrα89 | ||||
| Val221 | Proβ42/Alaβ43 | ||||
| Ile155 | Leuα48 |
TABLE 9.
Root mean square error after superposition of Cα atoms (in Å)
FSH1 and FSH2 indicate hormone from complexes 1 and 2, respectively, in Protein Data Bank code 1XWD (30); 1FL71 and 1FL72 indicate the two independent FSH molecules present in Protein Data Bank code 1FL7 (57); Protein Data Bank code 1HCN is the structure of human CG at 2.6 Å resolution (56); LH indicates modeled structure of LH (this study); TSH indicates the modeled structure of TSH (this paper). 1XWD1 and 1XWD2 indicate ECD from complexes 1 and 2, respectively, in Protein Data Bank code 1XWD (30); LHR indicates the modeled structure of the LHR ECD (this study); Protein Data Bank code 3G04 indicates TSHR ECD from 3 Protein Data Bank code G04 (32).
| FSH1 | FSH2 | 1FL71 | 1FL72 | 1HCN | LH | TSH | CG | |
|---|---|---|---|---|---|---|---|---|
| GpHs | ||||||||
| FSH1 | X | 0.7 | 1.0 | 1.6 | 1.5 | 0.4 | 0.3 | 0.7 |
| FSH2 | 0.7 | X | 1.0 | 1.5 | 1.6 | 0.7 | 0.7 | 0.8 |
| 1FL71 | 1.0 | 1.0 | X | 1.2 | 1.7 | 1.2 | 1.0 | 0.8 |
| 1FL72 | 1.6 | 1.5 | 1.2 | X | 1.6 | 1.7 | 1.6 | 1.3 |
| 1HCN | 1.5 | 1.6 | 1.7 | 1.6 | X | 1.5 | 1.6 | 1.8 |
| LH | 0.4 | 0.7 | 1.2 | 1.7 | 1.5 | X | 0.4 | 0.7 |
| TSH | 0.3 | 0.7 | 1.0 | 1.6 | 1.6 | 0.4 | X | 0.6 |
| CG | 0.7 | 0.8 | 0.8 | 1.3 | 1.8 | 0.7 | 0.6 | X |
| ECDs | 1XWD1 | 1XWD2 | LHR | 3G04 | ||||
| 1XWD1 | X | 0.7 | 0.4 | 1.1 | ||||
| 1XWD2 | 0.7 | X | 0.5 | 1.2 | ||||
| LHR | 0.4 | 0.5 | X | 1.1 | ||||
| 3G04 | 1.1 | 1.2 | 1.1 | X | ||||
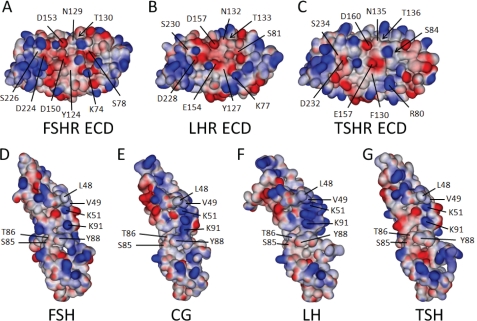
FIGURE 7.
Structures and models of the three GpHR ECDs and bound cognate hormones. Shown are the electrostatic surfaces and relevant amino acid residues (each indicated with a line; those buried with an arrow) studied herein, displayed using an orientation showing the hormone-binding interface of the FSHR (A), LHR (B), and TSHR (C). Also shown are the electrostatic surfaces of the three cognate hormones, FSH (D), CG (E), LH (F), and TSH (G), with the interacting residues highlighted. The structures of the FSHR ECD and FSH are taken from the crystal structure of the complex (30) and that for the TSHR ECD is from the M22·TSHR ECD complex (32). The predicted structures of bound LHR, CG, LH, and TSH were modeled from the published structures of free human CG (55, 56), free FSH (57), and the FSH·FSHR complex (30) and have been separated to enhance visibility of the interacting surfaces. As discussed in the text, CG has a more positive charge localization than FSH and TSH in a region predicted to be part of the binding complex. E, this localization is defined in large part by the close proximity of α-subunit residues Lys51 and Lys91 and β-subunit residue Arg95. Protorp outputs for the two forms of the FSH·FSHR ECD complex (30) are given in supplemental Table 2, along with those for our models of the CG·LHR ECD and TSH·TSHR ECD complexes. The supplemental Table 3 lists the root mean square errors and Ramachandran values for the hormones and receptors, indicating that the models generated provide good structures.
To study the possible effects of our mutations on the binding of CG to LHR and TSH to TSHR, and to compare those to the available complex structure of FSH·FSHR, models were generated using comparative modeling. The structures that were used as input are the coordinates of the hormone-binding domain of FSHR in complex with FSH (Protein Data Bank code 1XWD), the structure of the ECD of TSHR from the crystal structure of TSHR in complex with a TSHR autoantibody (Protein Data Bank code 3G04), and the structure of human CG (Protein Data Bank code 1HCN). CG·LHR and TSH·TSHR models were built using the program MODELLER version 9.7 (44), checked for a low overall energy score, the absence of large conformational errors, and the presence of biologically relevant interactions based on the large number of published mutations.
A summary of the interface contacts is presented in Tables 7 and 8; clearly, there are many similarities and a number of potentially important differences in the three complexes. Overall, the three complexes had very low overall C-α root mean square deviation values when superimposed (Table 9), with an average root mean square deviation of 1.1 Å. Fig. 7 shows the crystal structure of the FSHR ECD (Fig. 7A) and that of the structure of bound FSH (Fig. 7D), taken from Fan and Hendrickson (30), both emphasizing the electrostatic surfaces. The hormone and receptor have been separated to enable a better view of the amino acid residues studied herein and the hormone residues that contribute to the hormone·receptor interface. Also shown in Fig. 7 is the model developed in this study of the ECD of the LHR (Fig. 7B) based on the structures of the TSHR ECD (Fig. 7C) (32) and FSHR ECD, together with its modeled hormone CG (Fig. 7E) based on the structure of CG and complexed FSH (30, 55–57). In addition, models for LH (Fig. 7F) and TSH (Fig. 7G) were generated in the same way based on complexed FSH and CG structures. The supplemental Table 2 gives the PROTORP outputs for the two forms of the FSH·FSHR ECD complex (30) and the models for CG·LHR ECD and TSH·TSHR ECD complexes. All models were generated as complexes, which might explain why CG has a slightly higher overall root mean square deviation (supplemental Table 3) compared with the structure of free human CG. The root mean square errors of the various structures and models, including those for LH and TSH, are presented in supplemental Table 3, together with the Ramachandran values for each individual molecule (both experimental and modeled structures).
DISCUSSION
Differences in the β-subunits of the glycoprotein hormones (LH, CG, FSH, and TSH) are thought to contain the molecular determinants that are involved in specificity for their respective receptors. However, these hormones share the same α-subunit, suggesting that this subunit mainly contributes to receptor activation (58). The results of this study revealed that some of the identical or highly conserved amino acid residues, present in different β-strands of the LRRs of the GpHRs, do not have the same functions in the three receptors. Because these amino acid residues have been shown to be in contact with the α-subunit of FSH and, by inference, LH, CG, and TSH, these findings suggest that their effects on cell surface expression (presumably reflecting differences in protein folding, post-translational modifications, and/or trafficking), hormone binding affinity, and/or the hormone-mediated cAMP response differ between the GpHRs. The functional differences observed in binding and signaling may result from localized conformational differences in the hormones (see Refs. 55–57 for a comparison of the subtle structural differences in CG and FSH), the receptors, or the complexes, as evidenced by small induced conformational changes when FSH binds to the ECD of FSHR (30).
The amino acid residues in FSHR that were chosen for replacement include ones that bind the FSH α-subunit via hydrogen bonds and salt bridges (e.g. the Nξ on Lys74 contacts the Oγ of α-subunit residues Ser85 and Thr86; the Oγ of Asn129 interacts with the N of α-subunit residues Leu48 and Val49; the Oδ2 of Asp150 contacts the Nξ of α-subunit residue Lys91; and the Oδ2 of Asp153 interacts with the Nξ of α-subunit residue Lys51) and hydrophobic interactions (e.g. Tyr124 with α-subunit residue Tyr88, Thr130 with α-subunit residue Leu48, and Asp153 with α-subunit residue Val49) (see the supplemental Table 1a in Ref. 30 and supplemental Table 1 in this study). The fractional solvent accessibilities are ≥0.4 (Lys74), 0.1–0.4 (Tyr124 and Asp153), and <0.1 (Asn129, Thr130, and Asp150). The three residues chosen for controls, Ser78 in β-strand 2 and Asp224 and Ser226 in β-strand 8, have no detectable contact with the α-subunit.
Interestingly, replacing FSHR/Lys74 with Ala had no major effect on the functionality of the mutant receptor. Thus, the N-O interactions are not required for WT-like receptor structure and function; indeed, even reversing the charge with K74E has no measurable impact on the parameters measured. This weak interaction is consistent with the FSA of ≥0.4. Likewise, the hydrophobic interaction of Tyr124 with α-subunit residue Tyr88 can be reduced with no major change in functionality as evidenced by the WT-like properties of the Y124A mutant. In contrast, the side chain interaction of the O on Asn129 is important for WT-like function as indicated by the increase in Kd values associated with the N129A mutant and the very large increase in the EC50. This contact site has an FSA <0.1 and clearly contributes significantly to both binding and signaling. The results on the T130A mutant are somewhat enigmatic in that there were differences in the expression and signaling parameters measured on the HEK 293 and 293T cells. With the HEK 293 cells, only a slight increase was noted in the Kd value, with the signaling parameters like those of WT FSHR. The results on the HEK 293T cells suggested, however, that the EC50 value was increased in FSHR/T130A. We have no explanation for these discrepancies. For both Asp150 and Asp153, Glu can replace Asp with little if any effect on the measured values. This is somewhat surprising because the respective O-N interactions with α-subunit residues Lys91 and Lys51 exhibit FSAs of <0.1 and 0.1–0.4 for Asp150-α-subunit residue Lys91 and Asp153-α-subunit residue Lys51, respectively. Thus, there appears to be some plasticity in these interactions. Replacement of Asp150 and Asp153 with Asn, Ala, and Lys had no major effect on binding (although there was a small increase in the Kd values of FSHR/D150N and FSHR/D153N), but the EC50 values were increased in each of the three mutants of the two acidic side chains in β-strand 5, notably in D150K and D153K and less so, but still significant, in D150A and D153A. These findings indicate important roles of Asp150 and Asp153 in ligand-mediated signaling. One of the residues chosen as a control for Ala replacements, FSHR/S226A, expressed very poorly and could not be characterized. One control mutant, FSHR/S78A, exhibited WT-like properties, as did FSHR/D224A, although signaling was somewhat diminished in the latter. Signaling was greatly compromised in FSHR/D224K, suggesting that FSHR/Asp224 may be involved in a conformational change necessary for receptor activation following hormone binding.
Overall, the major change that occurs in the FSHR mutants in direct contact with the FSH α-subunit is a reduction in signaling potency for N129A, D150K, and D153K and, to a lesser extent, D150A and D153A. Of the 13 mutants examined on six contact sites, only N129A gave a significant decrease, e.g. nearly 5-fold, in the binding affinity; less, but not significant, reductions were also noted for T130A, D150N, and D153N. The noncontact controls, D224A and D224K, gave a reduction in signaling potency, suggesting that the negative charge in WT FSHR is required for WT-like signaling.
The same amino acid residue replacements in LHR and TSHR as in the FSHR contact sites led to both similarities and differences in receptor function (for TSHR expressed in HEK 293 cells the results apply to expression and signaling only). Focusing on major changes, the results can be summarized as follows. The conserved positively charged group in β-strand 2 (FSHR/Lys74, LHR/Lys77, and TSHR/Arg80) can be replaced with Ala or even Glu with no significant consequence on receptor function. In β-strand 4, replacement of the conserved aromatic group (FSHR/Tyr124, LHR/Tyr127, and TSHR/Phe130) with the smaller less hydrophobic methyl group (Ala) was also without major effect on functionality. These findings are in contrast to the reports of others who found decreases in binding affinity and signaling potency of LHR/K77A (11), LHR/Y127A (12), and TSHR/R80A and TSHR/F130A (34). However, Ala replacement of the invariant Asn (FSHR/Asn129, LHR/Asn132, and TSHR/Asn135) reduced the binding affinity in FSHR (and to a lesser degree in LHR) but greatly reduced the signaling potency in FSHR and to a lesser extent in LHR and TSHR. All three receptors are flanked on the C-terminal side of the invariant Asn by Thr. The N-terminal side of the invariant Asn is Ser in FSHR, although in LHR and TSHR it is Cys and Phe, respectively. Whether or not these localized neighboring differences impact on the binding and/or signaling remain to be established. The results with the invariant Thr (FSHR/Thr130, LHR/Thr133, and TSHR/Thr136) were less definitive, but it seems that the Ala replacement reduced binding and signaling potency somewhat in LHR consistent with that found by Song et al. (12).
Replacement of the two acidic residues in β-strand 5 (FSHR Asp150 and Asp153, LHR Glu154 and Asp157, and TSHR Glu157 and Asp160) with a similarly charged side chain (Glu or Asp), a similar but uncharged side chain (Gln or Asn), a small hydrophobic side chain (Ala), and an oppositely charged side chain (Lys) yielded some interesting comparative results. In FSHR, replacement of either Asp with Glu was without major consequence; TSHR/E157D was also much like WT TSHR in signaling. Expression was reduced in LHR/E154D, as was the signaling potency, but in both FSHR and LHR the binding was unaffected by the respective Asp/Glu and Glu/Asp replacements. In FSHR, replacement of either of the aspartates decreased the signaling potency slightly in D150N, somewhat in D150A, and significantly in D150K. Binding was not affected in any of the mutants except for a slight reduction in D150N and D153N. In LHR, the signaling potency was reduced considerably with all replacements, although less so with E154D and D157E, and the binding affinity was decreased in E154Q and E154A, and D157N and D157A. Signaling in TSHR was only marginally altered, if at all, except in the Lys replacements of either acidic side chain, which led to decreases in the potency. Overall, signaling potency is reduced by the replacements much more in LHR than in FSHR and TSHR. In the FSH·FSHR ECD complex (30), Asp150 interacts with α-subunit residues Lys91 and Thr86, and the latter also contacts FSHR Lys74 and Glu99. Asp153, on the other hand, interacts with α-subunit residues Lys51 and Val49, and one or more of these contacts appear more critical in receptor activation. Our models of the CG·LHR and TSH·TSHR ECD complexes (Fig. 7) predict interactions of Glu154 in LHR and of Glu157 in TSHR with α-subunit residue Lys91 and of Asp157 in the LHR and Asp160 in the TSHR ECD complexes with α-subunit residue Lys51. In the CG·LHR ECD model, we predict that Asp157 interacts with β-subunit residue Arg95, whereas in the TSH·TSHR ECD complex, Asp160 also interacts with its neighboring receptor residue Thr159. The predictions on the TSH·TSHR ECD are, overall, in good agreement with those of Núñez Miguel et al. (58). Thus, disruption of the ionic bridges in the LHR ECD impacts more adversely on signaling than similar changes in the other two GpHRs.
Ala replacements of the invariant serines in the three GpHRs in β-strand 2, a noncontact site in the FSH·FSHR ECD complex, were without significant effects, although others have reported a decrease in binding affinity and signaling potency in LHR/S78A (11). The other noncontact sites in FSHR β-strand 8 that were chosen for controls, FSHR/Asp224 and FSHR/Ser226, gave interesting comparative results. FSHR/D224A and FSHR/D224K, although binding FSH normally, exhibited decreased signaling potency. FSHR/S226A did not express at sufficient levels to permit evaluation. It has been reported that the hinge region of FSHR is important in receptor activation (60, 61). Although structural information is lacking beyond residue Glu259 in FSHR (30), it appears that the last two/three strands are much shorter with fewer hydrogen bonds stabilizing the parallel strands. It is thus possible that changes in this region can more easily cause disorder and perhaps affect the way the LRR domain interacts with the C-terminal cysteine-rich domain. If so, both proper folding (and hence expression) and signaling would be affected. LHR/D228A and LHR/D228K bound hCG with a reduced affinity, and the signaling potency was also decreased, as was observed also with TSHR/D232A and TSHR/D232K. LHR/S230A and TSHR/S234A exhibited properties like those of the WT receptor. LHR/D232E, but not D232Q, was found to result in decreased binding affinity (9); likewise TSHR/D232A and D232R exhibited Kd increases compared with WT TSHR (34). Thus, the invariant Asp in β-strand 8 appears to be involved in part of the pathway leading to hormone-mediated receptor activation.
The free energy of binding FSH to WT FSHR (and most of the mutants) is estimated to be about −13 kcal/mol (ΔG = −RT lnKa, where the association constant Ka = 1/Kd), which arises from many relatively weak interactions spread over a large surface area of 2600 Å2 containing an unusually high content of electrostatic charges (30). With LHR, the hormone-receptor affinity is somewhat higher than that of FSH-FSHR leading to a free energy of binding of about −13.7 kcal/mol with hCG. The observation of FSH binding to a negatively charged domain of the ECD validates earlier reports on modeling the hormone binding regions of LHR, which suggested a strong electrostatic component to hormone-receptor binding (8, 14, 28, 59). In studies between proteins interacting with high affinity, such as FSH or hCG binding to the FSHR or LHR ECD, respectively, it is not surprising that disruption of single hydrogen bonds, ion-ion interactions, ion-dipole interactions, or even single hydrophobic interactions by Ala replacement fails to lead to a significant reduction in affinity. (Note that the binding assay used is not perfectly reversible because some hormone-receptor internalization can occur and some of the 125I-labeled hormone may be degraded over the time course of the experiment (41). The Kd values obtained, however, agree with measurements from numerous other laboratories, and thus confidence can be placed in the validity of the assay, at least when used for comparative purposes, as was done herein.) Although binding of the FSHR and LHR mutants to their cognate hormones was reduced at most 3–5-fold, much larger decreases were found in the signaling potency and efficacy. Thus, FSHR residues Asn129, Asp150, and Asp153 and LHR residues Asn132, Glu154, and Asp157 appear to be involved, in varying degrees, in receptor activation. In TSHR, Asn135 seems much more important in receptor activation following hormone binding than are Glu157 and Asp160.
In a comprehensive comparative analysis of FSH-FSHR and TSH-TSHR (putative) interactions, Núñez Miguel et al. (58) concluded that of the 43 amino acid residues making contact between the two receptors and the cognate hormones, 22 were equivalent, 13 in FSHR contact FSH, but the corresponding positions in TSHR do not contact TSH, and 8 from TSHR contact TSH, but the corresponding positions in FSHR do not contact FSH. Furthermore, they suggested that just four residues from the two receptors made contact with the respective hormones in a similar fashion as follows: FSHR/Tyr124 and TSHR/Phe130, FSHR/Asn129 and TSHR/Asn135, FSHR/Glu157 and TSHR/Asp150, and FSHR/Asp153 and TSHR/Asp160. Yet our functional studies showed that an Ala substitution of FSHR/Tyr124 had no effect on signaling potency in FSHR but led to a 3-fold reduction in that of TSHR/Phe130. We found that Ala substitution on FSHR/Tyr124 had no effect on the binding affinity, and Sanders et al. (34) reported at most a 3-fold reduction in binding affinity of porcine TSH to TSHR/F130A compared with WT receptor. With FSHR/N129A, we found a much greater reduction in signaling potency than in TSHR/N135A. Ala replacements of Glu157 and Asp160 in TSHR produce at most a 2-fold reduction in signaling potency (Tables 3 and 4) (34), whereas in FSHR the same replacements decrease signaling potency some 6–25-fold. Thus, even though the interactions may be comparable, the functional consequences can vary considerably between receptors. In general, our results on the three GpHRs support the conclusion, based on comparative studies with FSHR and TSHR, that the hormone α-subunit contacts are more involved with signaling than with binding (58).
A careful examination of the FSH·FSHR ECD structure (30) and the models predicted in this study, CG·LHR ECD and TSH·TSHR ECD (Fig. 7), reveals that the number of interface contact residues is similar, averaging 24–28 on the ECDs and 32–36 on the hormones. Furthermore, the interfaces are more polar than hydrophobic in nature. The numbers of polar nonionizable residues, charged residues, and hydrophobic residues on the hormones vary from 11 to 16, 9 to 11, and 9 to 10, respectively, whereas those on the ECDs vary from 9 to 12, 8 to 11, and 4 to 7, respectively. Of interest is a comparison of the surface H-bonds and salt bridges in the three ECDs as follows: FSHR has 7 and 20, respectively (30); LHR has 6 and 16, respectively; and TSHR has 2 and 11, respectively. In addition, in the interface nonionizable polar and charged amino acid residues are more comparable between FSH and FSHR and CG and LHR than between TSH and TSHR. For the latter, our model predicts an excess of nonionizable polar, charged, and hydrophobic residues on the hormones compared with the receptor. These predicted numbers for LHR and TSHR, coupled with the structural data on FSHR (30), suggest a less constrained structure of TSHR, consistent with its higher basal cAMP compared with FSHR and LHR. The electrostatic surfaces of the hormones and receptors appear to be very important in directing hormone binding and probably specificity. For example, a comparison of some of the hormone-receptor interactions found in the FSH·FSHR ECD complex (30) with our models of the CG·LHR and TSH·TSHR ECD complexes (Fig. 7; see Ref. 58 for TSH·TSHR) reveals a more localized positive surface on CG, arising in part from the localization of α-subunit residues Lys51 and Lys91, and β-subunit residue Arg95, interacting with a more negative surface on LHR, relative to the FSH-FSHR and TSH-TSHR interactions as can be seen in Fig. 6.
There are examples of earlier work on the hormones that support many of the predicted interactions proposed herein. For example, with human CG binding to and activating LHR, a number of reports have demonstrated the importance of the α-C-terminal region, in particular Tyr88, Tyr89, and Lys91 (62–65), as well as certain amino acid residues (Arg94, Asp99, and Lys104) on the β-subunit of CG (66–68).
In summary, these results on mutations of nine selected amino acid residues in β-strands 2, 4, 5, and 8 of the three GpHRs show that, of the six reported contact sites of the FSHR with the FSH α-subunit, replacements can be made in FSHR without reducing the binding affinity more than 3–5-fold. On the other hand, signaling is greatly reduced in most mutants at positions Asn129, Asp150, and Asp153. The functional responses of the same replacements in LHR and TSHR as in FSHR were, in many cases, quantitatively and qualitatively different from those of FSHR. In and of themselves, these findings do not necessarily argue against the published crystal structure of the FSH·(truncated) FSHR ECD complex (30), but they do raise interesting questions on the roles of hormone binding and receptor activation. These results strongly suggest that the modalities of hormone binding and hormone-mediated signaling differ to varying extents in the three homologous receptors, even in identical or highly conserved amino acid residues, a conclusion also reached by others on FSH-FSHR and TSH-TSHR interactions (58). The molecular models we have suggested for the CG·LHR ECD and the TSH·TSHR ECD also exhibit subtle differences with each other and with the FSH-FSHR ECD structure. Finally, it should be emphasized that the C-terminal cysteine-rich region of the ECD has been reported to also function in hormone binding and receptor activation (60, 61, 69–71).
Supplementary Material
Acknowledgment
We thank Arjen Jakobi for assistance in the molecular modeling.
This work was supported, in whole or in part, by National Institutes of Health Grants DK069711 and DK033793.
This paper is dedicated to the memory of Yongsheng Li.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Results, Tables 1–3, Fig. 1, and additional references.
- LH
- luteinizing hormone
- CG
- chorionic gonadotropin
- ECD
- extracellular domain (ectodomain)
- FSH
- follicle-stimulating hormone
- FSHR
- FSH receptor
- GPCR
- G protein-coupled receptor
- GpHR
- glycoprotein hormone receptor
- LHR
- LH receptor
- LRR
- leucine-rich repeat
- TSH
- thyroid-stimulating hormone
- TSHR
- TSH receptor
- FSA
- fractional solvent accessibility
- RIA
- radioimmunoassay.
REFERENCES
- 1.Themmen A. P. N., Huhtaniemi I. T. (2000) Endocr. Rev. 21, 551–583 [DOI] [PubMed] [Google Scholar]
- 2.Ascoli M., Puett D. (2009) in Yen and Jaffee's Reproductive Endocrinology (Strauss J. F., 3rd, Barbieri R. eds) 6th Ed, pp. 35–55, Elsevier Publishing Co., Philadelphia [Google Scholar]
- 3.Braun T., Schofield P. R., Sprengel R. (1991) EMBO J. 10, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyle W. R., Campbell R. K., Myers R. V., Bernard M. P., Han Y., Wang X. (1994) Nature 368, 251–255 [DOI] [PubMed] [Google Scholar]
- 5.Hong S., Phang T., Ji I., Ji T. H. (1998) J. Biol. Chem. 273, 13835–13840 [DOI] [PubMed] [Google Scholar]
- 6.Ji T. H., Grossmann M., Ji I. (1998) J. Biol. Chem. 273, 17299–17302 [DOI] [PubMed] [Google Scholar]
- 7.Hsu S. Y., Kudo M., Chen T., Nakabayashi K., Bhalla A., van der Spek P. J., van Duin M., Hsueh A. J. (2000) Mol. Endocrinol. 14, 1257–1271 [DOI] [PubMed] [Google Scholar]
- 8.Bhowmick N., Huang J., Puett D., Isaacs N. W., Lapthorn A. J. (1996) Mol. Endocrinol. 10, 1147–1159 [DOI] [PubMed] [Google Scholar]
- 9.Bhowmick N., Narayan P., Puett D. (1999) Endocrinology 140, 4558–4563 [DOI] [PubMed] [Google Scholar]
- 10.Schmidt A., MacColl R., Lindau-Shepard B., Buckler D. R., Dias J. A. (2001) J. Biol. Chem. 276, 23373–23381 [DOI] [PubMed] [Google Scholar]
- 11.Song Y. S., Ji I., Beauchamp J., Isaacs N. W., Ji T. H. (2001) J. Biol. Chem. 276, 3426–3435 [DOI] [PubMed] [Google Scholar]
- 12.Song Y. S., Ji I., Beauchamp J., Isaacs N. W., Ji T. H. (2001) J. Biol. Chem. 276, 3436–3442 [DOI] [PubMed] [Google Scholar]
- 13.Smits G., Govaerts C., Nubourgh I., Pardo L., Vassart G., Costagliola S. (2002) Mol. Endocrinol. 16, 722–735 [DOI] [PubMed] [Google Scholar]
- 14.Smits G., Campillo M., Govaerts C., Janssens V., Richter C., Vassart G., Pardo L., Costagliola S. (2003) EMBO J. 22, 2692–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vischer H. F., Granneman J. C., Noordam M. J., Mosselman S., Bogerd J. (2003) J. Biol. Chem. 278, 15505–15513 [DOI] [PubMed] [Google Scholar]
- 16.Vischer H. F., Granneman J. C., Bogerd J. (2003) Mol. Endocrinol. 17, 1972–1981 [DOI] [PubMed] [Google Scholar]
- 17.Vischer H. F., Granneman J. C., Bogerd J. (2006) Mol. Endocrinol. 20, 1880–1893 [DOI] [PubMed] [Google Scholar]
- 18.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 19.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. (2004) J. Mol. Biol. 343, 1409–1438 [DOI] [PubMed] [Google Scholar]
- 20.Murakami M., Kouyama T. (2008) Nature 453, 363–367 [DOI] [PubMed] [Google Scholar]
- 21.Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 22.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen S. G., Choi H. J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. (2007) Nature 450, 383–387 [DOI] [PubMed] [Google Scholar]
- 24.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 25.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fanelli F., De Benedetti P. G. (2005) Chem. Rev. 105, 3297–3351 [DOI] [PubMed] [Google Scholar]
- 28.Jiang X., Dreano M., Buckler D. R., Cheng S., Ythier A., Wu H., Hendrickson W. A., el Tayar N. (1995) Structure 3, 1341–1353 [DOI] [PubMed] [Google Scholar]
- 29.Kajava A. V., Vassart G., Wodak S. J. (1995) Structure 3, 867–877 [DOI] [PubMed] [Google Scholar]
- 30.Fan Q. R., Hendrickson W. A. (2005) Nature 433, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dias J. A., Zhang Y., Liu X. (1994) J. Biol. Chem. 269, 25289–25294 [PubMed] [Google Scholar]
- 32.Sanders J., Chirgadze D. Y., Sanders P., Baker S., Sullivan A., Bhardwaja A., Bolton J., Reeve M., Nakatake N., Evans M., Richards T., Powell M., Miguel R. N., Blundell T. L., Furmaniak J., Smith B. R. (2007) Thyroid 17, 395–410 [DOI] [PubMed] [Google Scholar]
- 33.Touraine P., Beau I., Gougeon A., Meduri G., Desroches A., Pichard C., Detoeuf M., Paniel B., Prieur M., Zorn J. R., Milgrom E., Kuttenn F., Misrahi M. (1999) Mol. Endocrinol. 13, 1844–1854 [DOI] [PubMed] [Google Scholar]
- 34.Sanders J., Bolton J., Sanders P., Jeffreys J., Nakatake N., Richards T., Evans M., Kiddie A., Summerhayes S., Roberts E., Miguel R. N., Furmaniak J., Smith B. R. (2006) Thyroid 16, 1195–1206 [DOI] [PubMed] [Google Scholar]
- 35.Huang J., Puett D. (1995) J. Biol. Chem. 270, 30023–30028 [DOI] [PubMed] [Google Scholar]
- 36.DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., Calos M. P. (1987) Mol. Cell. Biol. 7, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angelova K., Fanelli F., Puett D. (2002) J. Biol. Chem. 277, 32202–32213 [DOI] [PubMed] [Google Scholar]
- 38.Angelova K., Fanelli F., Puett D. (2008) Mol. Endocrinol. 22, 126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W., Shields T. S., Stork P. J., Cone R. D. (1995) Anal. Biochem. 226, 349–354 [DOI] [PubMed] [Google Scholar]
- 40.Angelova K., Puett D. (2002) Endocrine 19, 147–154 [DOI] [PubMed] [Google Scholar]
- 41.Puett D., Angelova K. (2009) Methods Mol. Biol. 590, 1–20 [DOI] [PubMed] [Google Scholar]
- 42.Angelova K., Narayan P., Puett D. (2003) Mol. Cell. Endocrinol. 204, 1–9 [DOI] [PubMed] [Google Scholar]
- 43.Ballesteros J., Kitanovic S., Guarnieri F., Davies P., Fromme B. J., Konvicka K., Chi L., Millar R. P., Davidson J. S., Weinstein H., Sealfon S. C. (1998) J. Biol. Chem. 273, 10445–10453 [DOI] [PubMed] [Google Scholar]
- 44.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 45.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comput. Chem. 4, 187–217 [Google Scholar]
- 46.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 47.Reynolds C., Damerell D., Jones S. (2009) Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]
- 48.Tina K. G., Bhadra R., Srinivasan N. (2007) Nucleic Acids Res. 35, W473–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 50.McDonald I. K., Thornton J. M. (1994) J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
- 51.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 52.Bogerd J. (2007) Mol. Cell. Endocrinol. 260–262, 144–152 [DOI] [PubMed] [Google Scholar]
- 53.Laskowski R. A. (1995) J. Mol. Graph. 323–330, 307–308 [DOI] [PubMed] [Google Scholar]
- 54.Sippl M. J., Wiederstein M. (2008) Bioinformatics 24, 426–427 [DOI] [PubMed] [Google Scholar]
- 55.Lapthorn A. J., Harris D. C., Littlejohn A., Lustbader J. W., Canfield R. E., Machin K. J., Morgan F. J., Isaacs N. W. (1994) Nature 369, 455–461 [DOI] [PubMed] [Google Scholar]
- 56.Wu H., Lustbader J. W., Liu Y., Canfield R. E., Hendrickson W. A. (1994) Structure 2, 545–558 [DOI] [PubMed] [Google Scholar]
- 57.Fox K. M., Dias J. A., Van Roey P. (2001) Mol. Endocrinol. 15, 378–389 [DOI] [PubMed] [Google Scholar]
- 58.Núñez Miguel R., Sanders J., Chirgadze D. Y., Blundell T. L., Furmaniak J., Rees Smith B. (2008) J. Mol. Endocrinol. 41, 145–164 [DOI] [PubMed] [Google Scholar]
- 59.Moyle W. R., Xing Y., Lin W., Cao D., Myers R. V., Kerrigan J. E., Bernard M. P. (2004) J. Biol. Chem. 279, 44442–44459 [DOI] [PubMed] [Google Scholar]
- 60.Agrawal G., Dighe R. R. (2009) J. Biol. Chem. 284, 2636–2647 [DOI] [PubMed] [Google Scholar]
- 61.Caltabiano G., Campillo M., De Leener A., Smits G., Vassart G., Costagliola S., Pardo L. (2008) Cell. Mol. Life Sci. 65, 2484–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoo J., Ji I., Ji T. H. (1991) J. Biol. Chem. 266, 17741–17743 [PubMed] [Google Scholar]
- 63.Yoo J., Zeng H., Ji I., Murdoch W. J., Ji T. H. (1993) J. Biol. Chem. 268, 13034–13042 [PubMed] [Google Scholar]
- 64.Chen F., Wang Y., Puett D. (1992) Mol. Endocrinol. 6, 914–919 [DOI] [PubMed] [Google Scholar]
- 65.Ji I., Zeng H., Ji T. H. (1993) J. Biol. Chem. 268, 22971–22974 [PubMed] [Google Scholar]
- 66.Chen F., Puett D. (1991) Biochemistry 30, 10171–10175 [DOI] [PubMed] [Google Scholar]
- 67.Chen F., Wang Y., Puett D. (1991) J. Biol. Chem. 266, 19357–19361 [PubMed] [Google Scholar]
- 68.Xia H., Huang J., Chen T. M., Puett D. (1993) J. Mol. Endocrinol. 10, 337–343 [DOI] [PubMed] [Google Scholar]
- 69.Bruysters M., Verhoef-Post M., Themmen A. P. (2008) J. Biol. Chem. 283, 25821–25828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mueller S., Kleinau G., Jaeschke H., Paschke R., Krause G. (2008) J. Biol. Chem. 283, 18048–18055 [DOI] [PubMed] [Google Scholar]
- 71.Mizutori Y., Chen C. R., McLachlan S. M., Rapoport B. (2008) Mol. Endocrinol. 22, 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.