Abstract
Reduced bone mass is a common complication in chronic inflammatory diseases, although the mechanisms are not completely understood. The PHEX gene encodes a zinc endopeptidase expressed in osteoblasts and contributes to bone mineralization. The aim of this study was to determine the molecular mechanism involved in TNF-mediated down-regulation of Phex gene transcription. We demonstrate down-regulation of the Phex gene in two models of colitis: naive T-cell transfer and in gnotobiotic IL-10−/− mice. In vitro, TNF decreased expression of Phex in UMR106 cells and did not require de novo synthesis of a transrepressor. Transfecting UMR-106 cells with a series of deletion constructs of the proximal Phex promoter identified a region located within −74 nucleotides containing NF-κB and AP-1 binding sites. After TNF treatment, the RelA/p50 NF-κB complex interacted with two cis-elements at positions −70/−66 and −29/−25 nucleotides in the proximal Phex promoter. Inhibition of NF-κB signaling increased the basal level of Phex transcription and abrogated the effects of TNF, whereas overexpression of RelA mimicked the effect of TNF. We identified poly(ADP-ribose) polymerase 1 (PARP-1) binding immediately upstream of the NF-κB sites and showed that TNF induced poly(ADP-ribosyl)ation of RelA when bound to the Phex promoter. TNF-mediated Phex down-regulation was completely abrogated in vitro by PARP-1 inhibitor and overexpression of poly(ADP-ribose) glucohydrolase (PARG) and in vivo in PARP-1−/− mice. Our results suggest that NF-κB signaling and PARP-1 enzymatic activity cooperatively contribute to the constitutive and inducible suppression of Phex. The described phenomenon likely contributes to the loss of bone mass density in chronic inflammatory diseases, such as inflammatory bowel disease.
Keywords: Bone, Cytokine Action, Inflammation, NF-κB, Tumor Necrosis Factor (TNF), Inflammatory Bowel Disease, Colitis, p65
Introduction
Bones have several physiological functions including maintaining organism structure, serving as a hematopoietic niche, calcium and inorganic phosphate reservoir, and regulating and feeding back to other organs via hormone release. To balance both normal serum mineral concentrations and bone mineral density, bones have a dynamic homeostasis that involve bone-forming osteoblasts and bone-resorbing osteoclasts. This balance is influenced by hormones, neurotransmitters, and cytokines that act directly on osteoblasts and osteoclasts to affect mineral deposition and release (1).
Although normal bone structure is the result of the balance of these two opposite processes, certain genetic disorders can skew this balance leading to low bone mineralization. In vitamin D-resistant, X-linked hypophosphatemic rickets, an inactivating mutation of PHEX3 (phosphate-regulating gene with homologies to endopeptidases on the X chromosome) leads to hypophosphatemia, aberrant vitamin D levels, high serum alkaline phosphatase, and osteomalacia (2, 3). Although the Phex gene encodes a membrane-bound, zinc metallopeptidase expressed only in osteoblasts and odontoblasts, the effects of its mutation lead to phosphate wasting in the kidney by decreasing the expression and activity of the Na+/Pi cotransporter, NaPi-IIa (NPT2; SLC24A1) in proximal convoluted tubules (4, 5). Phex inactivation was postulated to indirectly affect the kidney through bone-released, phosphaturic factors known as phosphatonins. Fibroblast growth factor 23 (FGF23) was one of the leading phosphatonin candidates due to its potent negative effects on renal phosphate reabsorption and its highly elevated expression in X-linked hypophosphatemic ricket patients (6). Although initial studies seemed to confirm this mechanism (7), it was later shown that Phex mutations lead to increased FGF23 expression rather than processing (8), and that FGF23 is cleaved by subtilisin-like proprotein convertases and not PHEX (9).
In addition to the renal abnormalities in X-linked hypophosphatemic rickets, PHEX inactivation also leads to osteoblast mineralization deficits. This is exemplified by the inability of immortalized osteoblasts from Phex-deficient Hyp mice to mineralize in vitro (10). Although the target(s) of PHEX proteolytic activity remains uncertain, PHEX protein may influence bone metabolism by binding and stabilizing matrix extracellular phosphoglycoprotein, dentine matrix protein 1 (DMP1) (11), and osteopontin (12). Specifically, when PHEX binds to these substrates, it prevents their cleavage and release of a small, acidic protease-resistant ASARM peptide (acidic serine-aspartate-rich matrix extracellular phosphoglycoprotein-associated motif). These ASARM peptides have been shown to inhibit mineralization in vivo and in vitro, and most likely function by directly binding to hydroxyapatite crystals and by decreasing the expression of Phex (13).
PHEX itself is regulated by several hormones and cytokines important for skeletal homeostasis. Phex is up-regulated after treatment with insulin-like growth factor 1, growth hormone (14), and glucocorticoids (15). Alternatively, Phex was found to be down-regulated by parathyroid hormone (16), parathyroid hormone-related peptide (17), and vitamin D (18).
We have shown that the proinflammatory cytokine TNF decreases Phex expression in vivo and in vitro (19), an observation with significant pathophysiological implications. During chronic inflammation such as in inflammatory bowel diseases (IBD), circulating and/or infiltrating lymphocytes and other mononuclear cells produce cytokines that can influence bone metabolism by altering the balance of bone mineral deposition and resorption. Decreased bone mineral density is a common outcome of IBD. Indeed, 31 to 59% of adult IBD patients are classified as osteopenic, whereas 5 to 41% are actually diagnosed with osteoporosis, although rates of up to 70% of adult and pediatric IBD patients with low bone mineral density have been reported (20). In our earlier study (19), we showed that TNF treatment and chemically induced colitis decrease Phex mRNA expression via a transcriptional mechanism, and that the polyadenine (poly(A)) region located −116 to −110 bp upstream of the transcriptional start site was necessary for the TNF-mediated inhibition. This decrease in Phex expression correlated with decreased mineral deposition in osteoblast-like UMR-106 cells.
In the present work, we extend our previous observations to demonstrate the down-regulation of Phex in two more representative models of human IBD: microflora-induced colitis in gnotobiotic IL-10−/− mice and adoptive CD4+CD45RBhigh T-cell transfer into Rag2−/− recipients. Phex gene promoter analysis showed that the polyadenine (poly(A)) region in the murine Phex promoter constitutively binds PARP-1, whereas TNF induces binding of the NF-κB complex proximally to this element. RelA is then PARylated to increase its activity as a transrepressor. PARP-1 activity was indispensable for the effects of TNF as demonstrated by a blunted response to the cytokine in the presence of a PARP-1 inhibitor or overexpressed poly(ADP-ribose) glycohydrolase (PARG), and by a complete abrogation of the response to TNF in PARP-1 knock-out mice.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
10× TBE, 20× SSC, 100 mmol/liter of sodium pyruvate, 100× antibiotic-antimycotic, restriction enzymes, TRIzol reagent, DMEM high glucose medium, gels, and T4 DNA ligase were purchased from Invitrogen. Fetal bovine serum was from Mediatech, Inc. and Irvine Scientific (Santa Ana, CA), [γ-32P]deoxyadenosine triphosphate was from PerkinElmer Life Sciences (Boston, MA), ToxiLight BioAssay Kit was purchased from Lonza (Rockland, ME), TransIT-LT1 reagent from Mirus Bio LLC (Madison, WI), Galacto-Star β-galactosidase Reporter Gene Assay System and TaqMan primer sets for real time RT-PCR were purchased from Applied Biosystems (Foster City, CA). DNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). qScript cDNA Synthesis Kit and PerfeCTa qPCR SuperMix were from Quanta Biosciences Inc. (Gaithersburg, MD). Recombinant murine (mTNF) and human (hTNF) TNF were purchased from PeproTech (Rocky Hill, NJ). Human NF-κB expression plasmids pFLAG-CMV2-RelA and pFLAG-CMV2-p50 were kindly provided by Dr. Christian Jobin (University of North Carolina, Chapel Hill, NC). Human poly(ADP-ribose) polymerase 1 (pCMV6-PARP-1), human poly(ADP-ribose) glucohydrolase (pCMV6-PARG) expression plasmids, and control vector pCMV6-null were from OriGene Technologies Inc. (Rockville, MD). Control expression vector pTarget, Caspase-Glo 3/7 Assay Systems, and Streptavidin Magna-Sphere® Paramagnetic Particles (SA-PMPsm) were from Promega (Madison, WI). All other reagents, unless otherwise indicated, were purchased from Fisher Scientific (Pittsburgh, PA) or Sigma.
Experimental Animals
Specific pathogen-free (SPF) wild-type (WT) 129/SvEv mice and germ-free IL-10−/− mice on the same genetic background were obtained from the National Gnotobiotic Rodent Resource Center at the University of North Carolina, Chapel Hill, NC. Germ-free IL-10−/− mice were transferred to the SPF facility and kept in sterile cages 2 days prior to being colonized with SPF fecal bacteria. 14 days after bacterial colonization allowed for the development of mild to moderate colitis in SPF-associated previously germ-free IL-10−/− mice (21). Adoptive T-cell transfer colitis was induced by intraperitoneal injection of 0.5 × 106 naive, flow-sorted CD4+CD45RBhigh lymphocytes (98% purity) into the Rag-2−/− host (both C57BL/6) (22). Control (PBS-injected) and colitic mice were sacrificed 8 weeks after transfer. Additionally, 4–5-week-old male WT and PARP-1 KO mice purchased from Jackson Labs (129s1/SvImJ and 129SParp1/J, respectively) were injected intraperitoneally once with murine recombinant TNF (PeproTech; Rocky Hill, NJ) at a dose of 150 μg/kg of body weight (22) or with an equal volume of vehicle (PBS). Mice were sacrificed 24 h after injection. All mice used for experiments described above were supplied with food and water ad libitum. At respective time points mice were sacrificed by CO2 narcosis followed by cervical dislocation. Femurs and calvarias were removed, flash frozen in liquid nitrogen, and stored at −80 °C before RNA isolation. Colitis was evaluated histologically, as well as by cytokine analysis in colonic explant culture and lymphocytes isolated from the mesenteric lymph nodes and stimulated in vitro with CD3/CD28 or with cecal antigen extract as described earlier (21, 22). Animals maintained at the University of Arizona Health Sciences Center were routinely monitored and determined as free from common murine pathogens (MHV, MPV, MVM, TMEV, Mycoplasma pulmonis, Sendai, EDIM, MNV, ecto- and endoparasites). All described animal use protocols were approved by the University of Arizona or the University of North Carolina at Chapel Hill Animal Care and Use Committee.
Cell Cultures and Sample Collection
Rat osteogenic sarcoma cells (UMR-106; ATCC number CRL-1661) were obtained from the American Type Culture Collection (Manassas, VA) and cultured in DMEM high glucose media containing 10% fetal bovine serum, 1 mmol/liter of sodium pyruvate, and 1× antibiotic-antimycotic at 37 °C with 5% CO2. For analysis of endogenous rat Phex gene expression (rPhex), UMR-106 cells were seeded on 6-well plates at 0.5 × 106 cells per well and treated with hTNF (10 ng/ml), cycloheximide (10 μm), and a combination of both or vehicle (depending on experiment). In some experiments, cells were pretreated for 30 min with THE proteasome inhibitor clasto-lactacystin-β-lactone (10 μm, cLβL). Apoptosis and cytotoxicity were evaluated with the Caspase-Glo 3/7 Assay System (Promega) and ToxiLight BioAssay kit (adenylate kinase release; Lonza Rockland, Inc., Rockland, ME), respectively, according to the manufacturer's protocols.
Real Time RT-PCR Analysis
200 ng of total RNA isolated with TRIzol were reverse transcribed using the qScript cDNA Synthesis Kit, and 2 μl of each reverse transcribed reaction (10% of total volume of real time reaction mixture) was used for real time PCR analysis using TaqMan technology and commercially available primers (Rn01455648_m1 and Mm00446973_m1 for Tbp, Rn00448130_m1 and Mm 00448123_m1 for Phex; Rn01415175_m1 for Parp-1, Rn00580158_m1 for Parg) using PerfeCTa qPCR SuperMix and the iCycler optical PCR cycler (Bio-Rad). Resulting data were analyzed by the comparative cycle threshold (Ct) method as means of relative quantitation of gene expression, normalized to an endogenous reference (TATA-box binding protein (TBP)) and relative to a calibrator (normalized Ct value obtained from control mice), and expressed as 2−ΔΔCt (Applied Biosystems User Bulletin number 2: Rev B “Relative Quantitation of Gene Expression”).
Reporter Gene Constructs and Transient Transfections
Progressive 5′-promoter deletions were generated between −522 and −74 nt of the mPhex promoter by PCR and cloned into pβGal-Basic reporter vector (Clontech, Palo Alto, CA) as previously described (18). All promoter/reporter gene constructs were sequenced to confirm fidelity. UMR-106 cells were seeded in 6- or 24-well plates (0.5 × 106 or 0.5 × 105 cells/well, respectively), and cells were transfected with mPhex reporter vector DNA (0.5–1 μg/ml of medium) by using TransIT-LT1 transfection reagent. In some experiments mPhex reporter vector DNA was co-transfected with the following expression vectors: pCMV6-null, pFLAG-CMV2-RelA, pFLAG-CMV2-p50, pCMV6-PARP-1, or pCMV6-PARG. 20–24 h after transfection, cells were treated as described under “Results.” Promoter activity was expressed as β-galactosidase activity (β-Gal, relative light units) per microgram of protein. Cells from 6-well plates were used for total RNA isolation.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear protein (NP) for EMSA was prepared from UMR-106 cells as previously described (23). Double-stranded, synthetic oligonucleotides were end-labeled with [γ-32P]dATP. For each reaction, 30,000 cpm of the probe were incubated at room temperature for 20 min with 1–2.5 μg of NP, 4 μl of 5× binding buffer (Promega), 1 μg of poly(d(I-C)) (Sigma), and H2O to a final volume of 20 μl. For competition studies, ×100 excess of unlabeled probe was added to the reaction. Antibodies for supershift assays (p50, p52, RelA, cRel, RelB, and PARP-1) were purchased from Santa Cruz Biotechnology (sc-114, sc-84, sc-372, sc-272, sc-226, and sc-74469, respectively). Binding reactions were loaded on a 6% DNA retardation gel (Invitrogen) and separated at 250 volts in 0.5× TBE. Gels were dried and then exposed to an x-ray film. Oligonucleotides used for EMSA are depicted under “Results.”
Southwestern Blot Analysis
NP was prepared as described above for EMSA, separated on 8% Tris glycine SDS-polyacrylamide gels (Invitrogen), and transferred to nitrocellulose membranes. Immobilized proteins were then re-natured and blocked overnight at room temperature in 1× EMSA buffer containing 5% nonfat dried milk. Blots were then incubated with 1 × 107 cpm of 32P end-labeled DNA probe overnight at room temperature. Following hybridization, blots were washed three times for 15 min in 1× EMSA buffer and then exposed to x-ray film (18).
DNA Affinity Precipitation Assays (DAPA)
A double-stranded probe corresponding to −133 to +1 bp of the PHEX promoter region was generated by annealing a 5′ biotin-labeled −133/+1 GS oligonucleotide (see “Results”) with an unlabeled complementary oligonucleotide. 37 pmol of biotinylated probe were mixed with: 75–100 μg of UMR-106 NP, 50 μl of 5× EMSA buffer (Promega), 1 mm DTT, phosphatase inhibitor (Sigma), protease inhibitor (Pierce), 1 μg of poly(d(I-C)) (Sigma) and incubated 2 h at 4 °C with end to end rotation (final volume 250 μl). For competition studies, 100× excess of unlabeled probe (competitor) was added to the reaction 30 min prior to addition of the biotinylated probe. 100 μl of Streptavidin Magna-Sphere® (Promega, Madison, WI) paramagnetic particles (SA-PMPs) suspended in complete 1× EMSA buffer were added 2 h later followed by a 30-min incubation at 4 °C with rotation. SA-PMPs were separated from the supernatants in a magnetic field and washed 3 times with 1× EMSA buffer. SA-PMPs were resuspended in 70 μl of Laemmli sample buffer with 5% α-mercaptoethanol, boiled for 5 min, and the supernatant was separated on 8% Tris glycine SDS-polyacrylamide gels and immunoblotted, silver stained, or used with the Experion Automated Electrophoresis System. In some studies, samples from DAPA were used for proteomic identification of the poly(A) element-binding protein, as described below and under the supplemental data.
Proteomic Identification of the Poly(A)-element Binding Protein
DAPA was performed as described above. Biotin label was incorporated at the 5′-end of either the lead-strand (containing poly(A) sequence) or 5′-end of the reverse-complement strand (containing poly-T sequence) to avoid bias. Affinity-precipitated nuclear protein from UMR-106 cells were separated on SDS-PAGE gel, silver-stained, and band(s) that could be competed with ×100 excess of unlabeled oligo were excised, digested with trypsin, and analyzed by tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS). To independently verify the findings, affinity-precipitated nuclear protein with or without an excess of unlabeled probe was used without gel separation with tandem mass spectrometry coupled to tandem liquid chromatography (LC-LC-MS/MS). The experimental details are provided under the supplemental data. DNA interaction of the protein consistently identified with all approaches (PARP-1) was further confirmed by EMSA (supershift) and chromatin immunoprecipitation assay (ChIP) as described below.
ChIP Assay
Phex ChIP was performed with UMR-106 cells utilizing the enzymatic ChIP-IT Express Chromatin Immunoprecipitation Kit according to the manufacturer's protocol (Active Motif, Carlsbad, CA) with the following antibodies: RelA, PARP-1, RNA polymerase II (RNAP II), and the respective IgG (Santa Cruz and Active Motif). PCR were performed with primers specific for the proximal rat Phex gene promoter or 500, 1300, 3000, and 4100 bp downstream from the transcription start site (RNAP II walking), or with control primers for rat β-actin. Sequences of primers and the conditions of PCR are depicted under “Results.”
Immunoprecipitation
TNF-treated or control UMR-106 cells were lysed with PBS with 1% Nonidet P-40 (IP buffer) and 500 μg of protein extract was rotated with 2 μg of RelA (Santa Cruz; sc-372x) or control IgG (Santa Cruz; sc-2027) for 2 h at 4 °C in a total volume of 0.5 ml. 75 μl of Protein A (Sigma) was then added to the samples and rotated overnight at 4 °C. Samples were centrifuged (3,000 × g for 2 min), beads were washed 3 times with 750 μl of the IP buffer, and resuspended and boiled in 75 μl of 5× Laemmli buffer with β-mercaptoethanol. The eluted protein was used for Western blotting with antibodies against RelA, poly(ADP-ribose) chain (Calbiochem/EMD Chemicals, Gibbstown, NJ), or PARP-1 (Santa Cruz).
Statistical Analysis
Statistical significance was determined by the Student's t test or analysis of variance followed by followed by Fisher's protected least significant difference test, using Statview software package version 4.53 (SAS Institute, Cary, NC). Data are expressed as mean ± S.E.
RESULTS
Chronic T-cell-mediated Colitis Suppresses Phex Expression in Mouse Osteoblasts in Vivo
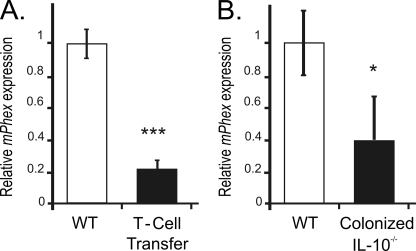
Adoptive transfer of CD4+CD45RBhigh T-cells (naive T-cells) from healthy wild-type mice into syngeneic Rag2−/− recipients that lack T- and B-cells induce pancolitis and small bowel inflammation within 5–8 weeks with symptoms resembling human Crohn's disease. The CD45RBhigh T-cell transfer led to 80% reduction in the bone Phex mRNA compared with PBS-injected mice (Fig. 1A), coinciding with moderate-to-severe chronic colitis, characterized by increased mucosal thickness and transmural infiltration of lymphocytes, monocytes, macrophages, and granulocytes, affecting both the proximal and distal colon as we described earlier (22). Production of selected cytokines (IFN-γ, TNF, IL-1β, and IL-17) was significantly elevated in the supernatants obtained from unstimulated colonic explants cultures as well as from CD3/CD28-stimulated MLN lymphocyte culture (Ref. 22, and data not shown). A similar decrease in Phex expression was observed in germ-free IL-10−/− mice 14 days following colonization with specific pathogen-free microflora (Fig. 1B). In this model, milder colitis (as determined by histology and cytokine production; Ref. 21, and data not shown) was associated with comparatively smaller (50%) reduction in Phex mRNA expression compared with the more severe reaction in T-cell transfer studies.
FIGURE 1.
Chronic colonic inflammation suppresses bone mPhex mRNA expression. Real time PCR analysis of mPhex mRNA expression in (A) femurs harvested from control Rag2−/− mice or Rag2−/− mice adoptively transferred with naive CD4CD45RBHi T-cells, and (B) from calvarias of wild-type (WT) mice and germ-free IL-10−/− mice collected 14 days after colonization with SPF microflora. Values are mean ± S.E. of 4–6 mice per group: *, p < 0.05; ***, p < 0.001 (Student's t test).
Inhibition of Phex Gene Expression by TNF May Not Require de Novo Synthesis of a Transrepressor and Is Not Related to Pro-apoptotic or Cytotoxic Effects of the Cytokine
Our previous studies demonstrated that neutralizing anti-TNF antibodies reversed the effects of colitis on Phex expression and that TNF inhibits Phex gene transcription and osteoblast mineralization in vitro (19). To determine whether the TNF-induced repression of the Phex gene requires de novo synthesis of a transrepressor, we tested the effects of TNF on Phex mRNA expression in cycloheximide (CHX)-treated UMR-106 cells. TNF time dependently inhibited Phex expression (supplemental Fig. S1A). CHX alone also significantly diminished the Phex transcript, suggesting the need for sustained translation of transactivator(s) and/or related signaling proteins (supplemental Fig. S1A). However, in the presence of CHX, TNF failed to further down-regulate Phex expression or to accelerate the decline of the steady-state Phex transcript levels (supplemental Fig. S1A).
We observed a transient increase in apoptosis in UMR-106 cells treated with TNF reaching statistical significance after 8 h of treatment (supplemental Fig. S1B). At later time points (14 and 24 h), coinciding with the greatest inhibition of Phex mRNA expression, there was no significant change in caspase 3/7 activation (supplemental Fig. S1B). We documented a small (∼20%), albeit significant increase in TNF-induced cytotoxicity as verified by adenylate kinase release into the medium (supplemental Fig. S1C). To test if under these experimental conditions UMR-106 were still capable of positively respond to TNF, we analyzed the expression of IκBα (NFKBAI; nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor, α), IL-6, and ICAM-1 mRNA 8 and 24 h into the treatment. All three genes (exemplified by IκBα shown in supplemental Fig. S1D) were significantly induced by TNF, CHX, and to a higher degree with the TNF/CHX combination. These data confirm the viability of the cells, and a strong positive response in the absence of protein synthesis-dependent negative feedback (TNF + CHX). Collectively, these findings suggest that under the selected experimental settings, TNF inhibits Phex expression independent of de novo protein synthesis. It is also possible, however, that in CHX-treated cells, TNF is unable to further inhibit PHEX transcription rate. In either case, the observed effects are not likely to be secondary to the minor changes in cell viability.
Proximal Phex Promoter Region Is Involved in TNF-mediated Inhibition of Phex Transcription
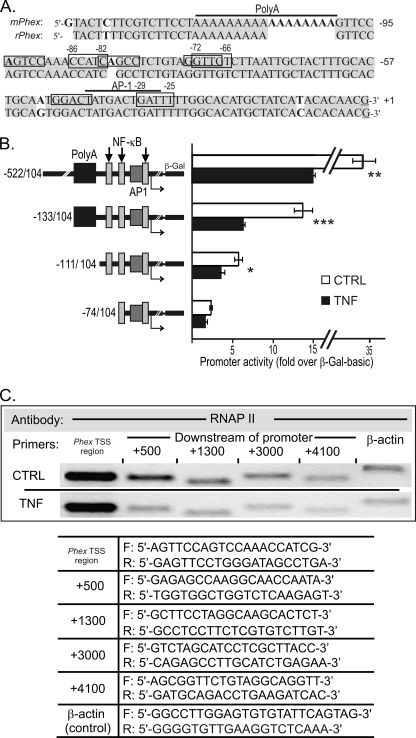
The proximal regulatory regions of the mouse and rat Phex genes are highly homologous (Fig. 2A). The rPhex poly(A) region is shorter than in the mPhex gene, although prediction analysis of putative cis-elements by Match® software (24), including relatively weak NF-κB and AP-1 binding sites, are very similar. To characterize the mechanism of the transcriptional response of the murine Phex gene (mPhex) to TNF, we tested a series of 5′ progressive deletions of its promoter from −542 to −74 for TNF responsiveness in transiently transfected UMR-106 cells (Fig. 2B). TNF treatment reduced the activity of constructs spanning −522/+104 and −133/+104nt by ∼50%. A construct truncated to remove the poly(A) region (−111/+104 nt) was inhibited to a lesser degree (35%), whereas the shortest construct (mPhex −74/+104) containing a putative AP-1 and two NFκB binding sites was not significantly inhibited by TNF. These observations indicated that cis-element(s) required for TNF-mediated repression of the Phex promoter activity are located within −111/−74 nt, whereas the immediately upstream poly(A) region is necessary for the maximal inhibition.
FIGURE 2.
Functional analysis of the murine Phex gene promoter and its response to TNF. A, alignment and schematic representation of the −133/+1 Phex promoter region of mouse (mPhex) and rat (rPhex) genes with the depicted polyadenine region (poly(A)) and predicted NF-κB (rectangles) and AP-1 cis-elements (line). B, effects of TNF (10 ng/ml, 24 h) on mPhex promoter activity in UMR-106 cells transiently transfected with reported constructs containing progressive promoter deletions. Promoter activity is expressed as fold-increase over β-galactosidase activity in cells transfected with promoter-less vector pβ-Gal basic. The poly(A) element, and NF-κB and AP-1 binding sites are depicted by black or gray rectangles. Values are mean ± S.E. from 3 independent experiments; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student's t test). C, ChIP analysis in control (CTRL) and TNF-treated (10 ng/ml, 24 h) UMR-106 cells using RNA polymerase II antibody (RNAP II walking) and primers specific for the mPhex TSS vicinity, and primers designed for downstream regions of the gene (+500, +1300, +3000, +4100 bp; see table below). Primers specific for the rat β-actin were used as a control (CTRL).
TNF Inhibits Phex Transcript Elongation, but Not RNA Polymerase II (RNAP II) Recruitment
To address the possible effect of TNF on the RNAP II recruitment to the Phex gene transcriptional start site (TSS), we performed RNAP II walking using ChIP with an RNAP II antibody followed by PCR with primers specific for the TSS vicinity, and primers designed for downstream regions of the gene (+500, +1300, +3000, and +4100 bp). Although TNF had no influence on the interaction of RNAP II with the Phex TSS region, significantly less PCR product was associated with RNAP II in downstream regions of the gene (Fig. 2C). These results were confirmed in three independent experiments and suggest that TNF-induced changes within the proximal Phex promoter result in impaired promoter clearance by RNAP II rather than its altered recruitment to the TSS.
Identification of Poly(A) Element-binding Protein
We have previously described the poly(A) element in the proximal Phex promoter and postulated a transcriptional modulator of an approximate molecular mass of 110 kDa, tentatively termed PAP110 (PHEX-activating protein 110) as an important element in basal as well as 1,25-(OH)2 vitamin D-mediated transcriptional control of the Phex gene (18). DNA affinity enrichment using biotinylated double-stranded probe spanning the poly(A) region (5′-CTAAAAAAAAAAAAAAAAAGT-3′) followed by tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS) identified PARP-1 as a likely candidate with a matching molecular weight (supplemental Table S1 and Fig. S2). Another protein fulfilling the search criteria was SP120. The latter rat protein has been deposited to GenBankTM by others (BAA031336.1), but not published as a peer-reviewed report. SP120 is a putative rat nuclear scaffold protein binding to the matrix attachment regions of DNA, with high homology to human scaffold attachment factor A. The presence of PARP-1, but not SP120, in the precipitated DNA-protein complex was independently confirmed without gel separation with MudPIT (Multidimensional Protein Identification Technology) using LC-LC-MS/MS and double-stranded probes spanning the poly(A) region of the Phex promoter, with a biotin label either on poly(A) or poly(T) strands (data not shown).
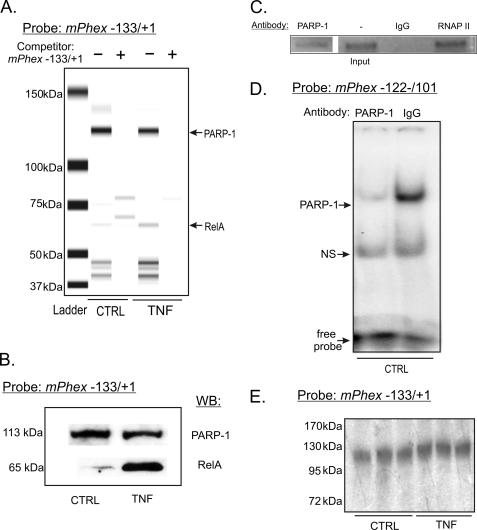
DAPA using the sequence depicted in Fig. 2A and followed by microfluidics-based automated protein electrophoresis (Experion Automated Electrophoresis System; Bio-Rad) identified a strong and constitutive (not altered by TNF treatment) association of a protein of approximately 110 kDa (consistent with PARP-1), and a TNF-inducible protein of approximately 65 kDa. Both were competed by an excess of unlabeled probe (Fig. 3A). Western blot analysis of the same samples confirmed PARP-1 and RelA (p65) as two proteins binding to the proximal Phex promoter (Fig. 3B). ChIP assay, EMSA with PARP-1-specific antibodies, as well as Southwestern blotting further confirmed the ∼110-kDa protein as PARP-1 (Fig. 3, C–E).
FIGURE 3.
PARP-1 and RelA interact with −133/+1 nt of the mPhex promoter. A, DAPA followed by automated protein electrophoresis identified a constitutively associating 110-kDa protein and inducible binding of a 65-kDa protein in TNF-treated UMR-106 cells. These DNA-protein complexes were competed with 100-fold excess of the non-biotinylated probe. B, DAPA was followed by SDS-PAGE separation and Western blot analysis of the complexes formed with −133/+1 nt of the mPhex gene and confirmed the bands to represent PARP-1 and RelA. C, ChIP analysis of PARP-1 association with the proximal mPhex promoter. Chromatin was immunoprecipitated with anti-PARP-1 or RNAP II and IgG (as a positive and negative control, respectively) and PCR amplified with primers specific for mPhex TSS (see Fig. 3 for sequences). D, neutralization of PARP-1 binding to the poly(A) element of the mPhex gene in EMSA analysis in untreated UMR-106 cells. E, Southwestern blot analysis of nuclear proteins isolated from control (CTRL) or TNF-treated UMR-106 cells with a radiolabeled probe spanning −133/+1 nt of the mPhex gene.
Recruitment of NF-κB, but Not AP-1 Complex to the Proximal Phex Promoter Is Induced in TNF-treated UMR-106 Cells
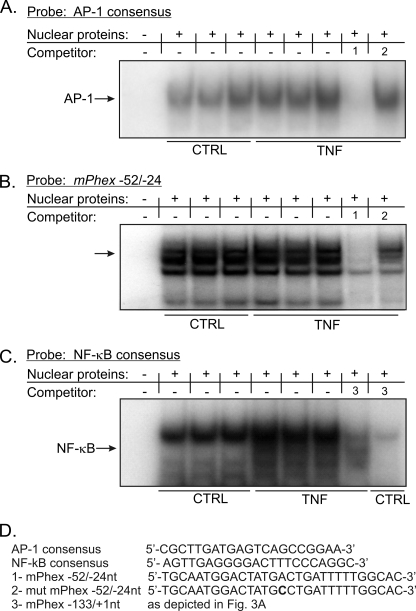
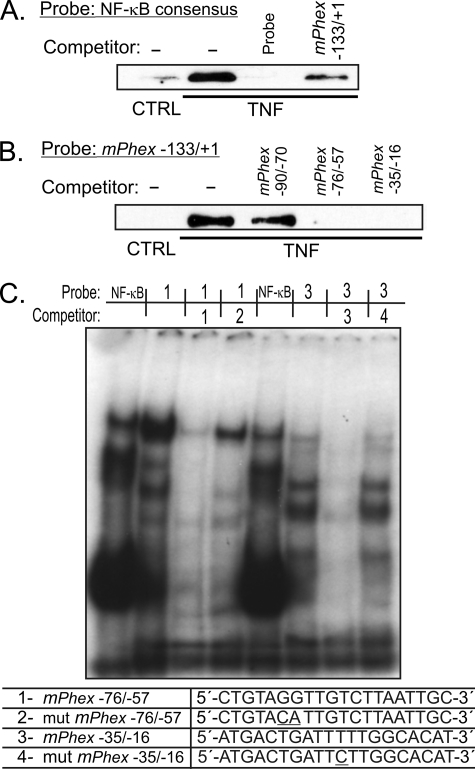
Prediction analyses indicated the presence of putative AP-1 and NF-κB binding sites between the poly(A) element and the transcription start site (Fig. 2B). EMSA with consensus AP-1 oligonucleotide as a probe showed no significant induction of this transcription factor complex (Fig. 4A). AP-1 binding to this consensus probe could be competed with a probe spanning the putative AP-1 site in the Phex promoter (−52/−24 nt; Fig. 4A, competitor 1), but not by the probe with a mutated AP-1 cis-element (Fig. 4A, competitor 2). This indicates that the AP-1 complex can bind to the proximal Phex promoter, despite the lack of detectable induction by TNF. Indeed, in EMSA with the −52/−24-nt fragment used as the labeled probe, strong, but not TNF-inducible binding was observed (Fig. 4B). On the other hand, TNF activated NF-κB, as demonstrated in EMSA with a NF-κB consensus probe (Fig. 4C). Moreover, this binding could be competed with a probe spanning −133/+1 nt of the Phex gene, thus confirming the observations from DAPA assays (Fig. 3, A and B) and suggesting TNF-inducible binding of the NF-κB complex to the proximal Phex promoter.
FIGURE 4.
NF-κB and AP-1 complexes interact with the proximal mPhex promoter. A, EMSA analysis with nuclear protein isolated from control and TNF-treated UMR-106 cells with a radiolabeled consensus AP-1 probe. No detectable increase in AP-1 DNA binding was reported in response to TNF, although binding could be competed with an excess of the unlabeled mPhex −52/−24-nt probe spanning the AP-1 consensus element but not its mutated form. B, consistently, only constitutive AP-1 binding was observed with the −52/−24-nt sequence used as a probe. C, TNF treatment increases NF-κB binding to a consensus NF-κB element. This interaction can be effectively competed with an excess of the unlabeled −133/+1 mPhex probe. D, probe sequences used in EMSA analysis. An excess of the free probe is not demonstrated due to the necessary complex separation and for figure clarity, but was confirmed in an independent assay (not shown).
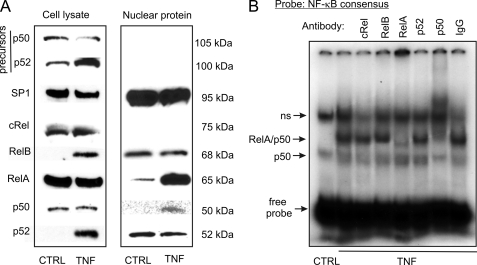
A subsequent Western blot study showed that in UMR-106 cells, TNF induces translocation of RelA (p65) and p50 to the cell nucleus (Fig. 5A). Although TNF induced p100 processing and increased total p52 levels, Western blotting with nuclear proteins did not indicate increased nuclear translocation of this subunit (Fig. 5A). EMSA with supershifting or blocking antibodies verified inducible DNA binding of RelA (p65) and p50 in TNF-treated UMR-106 cells (Fig. 5B). DAPA with the biotinylated NF-κB consensus probe also demonstrated inducible binding of RelA, which could be competed with excess of both the unlabeled consensus probe or the unlabeled −133/+1-nt fragment of the Phex promoter (Fig. 6A). When the −133/+1-nt fragment was used as a probe, competitor spanning −90/−70 nt of the promoter was not effective, whereas both the −76/−57-nt and the −35/−16-nt probes containing putative NF-κB binding sites competed for RelA binding (Fig. 6B). This was further verified in EMSA with wild-type or mutated −76/−57-nt and −35/−16-nt probes (Fig. 6C). Collectively, the data indicated the involvement of two relatively weak NF-κB binding sites located in proximity to the TSS in TNF-mediated Phex inhibition.
FIGURE 5.
Nuclear translocation of the NF-κB complex components and their DNA association in TNF-treated UMR-106 osteoblasts. A, representative Western blot analysis of NF-κB proteins in whole cell lysate or in the nuclear fraction of control and TNF-treated UMR-106 cells (10 ng/ml, 24 h). Only RelA (p65) and p50 demonstrated detectable and inducible nuclear translocation in response to TNF. B, supershift/blocking EMSA analysis of NF-κB/DNA complexes with nuclear protein from TNF-treated UMR-106 cells confirming the predominant involvement of the p50/p65 heterodimer.
FIGURE 6.
RelA (p65) protein binds to the proximal region of the mPhex gene promoter. DAPA was performed with nuclear proteins isolated from control or TNF-treated UMR-106 cells with: A, biotinylated NF-κB consensus DNA probe, or B, biotinylated double-stranded DNA probe corresponding to the −133/+1 region of the mPhex promoter. The Western blot was performed with antibodies specific to RelA. TNF-inducible binding could be competed with an excess of unlabeled NF-κB consensus probe, a probe spanning −133/+1 nt and the two putative NF-κB elements located at −76/−57 nt and −35/−16 nt of the mPhex promoter, but not another putative NF-κB site located at −90/−70 nt. C, EMSA analysis with radiolabeled consensus NF-κB probe or probes spanning −76/−57 nt (probe 1) or −35/−16 nt of the mPhex promoter. Competing probes were designed to have intact or mutated core elements of the putative NF-κB sites (underlined nucleotides). All binding reactions were performed with nuclear extracts from UMR-106 cells treated with TNF. An excess of the free probe is not demonstrated due to the necessary complex separation and for figure clarity, but was confirmed in an independent assay (not shown).
Inhibition of NF-κB Signaling Reverses whereas Overexpression of RelA Mimics TNF-mediated Inhibition of Phex Expression
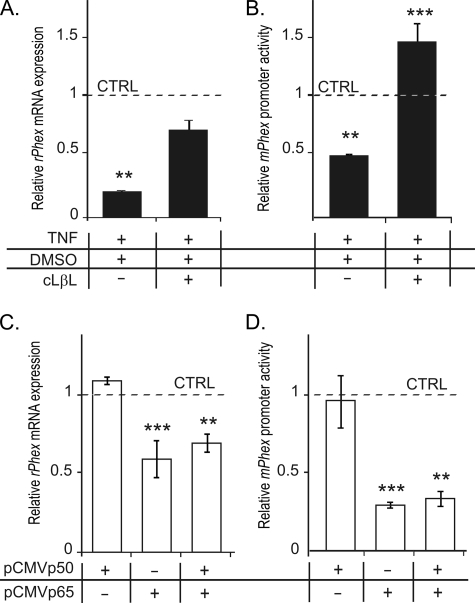
To better define the role of NF-κB signaling in the regulation of Phex gene transcription, we used decoy peptides (IMG-2000, IMG-2001, and IMG-2002), pyrrolidine dithiocarbamate, BAY 11-7082, proteasome inhibitor clasto-lactacystin-β-lactone, as well as RelA siRNA. Although the decoy peptides pyrrolidine dithiocarbamate, BAY, and p65 siRNA were able to decrease the level of RelA in the nuclei of UMR-106 cells, their combination with TNF treatment was associated with severe cytotoxicity, therefore yielding data difficult to interpret. Pretreatment with the proteasome inhibitor cLβL, at a concentration resulting in no discernable cytotoxicity (10 μm), partially (but significantly) reversed the inhibitory effects of TNF on the rPhex mRNA level (Fig. 7A). It also fully reversed the negative effects of TNF on the mPhex promoter activity (Fig. 7B). Overexpression of RelA in UMR-106 cells transiently transfected with pFLAG-CMV2-RelA mimicked the effects of TNF on both the expression of endogenous rPhex mRNA (Fig. 7C) and gene promoter activity (Fig. 7D). Overexpression of p50 alone or in combination of RelA did not result in a further decrease of Phex expression or promoter activity (Fig. 7, C and D).
FIGURE 7.
Inhibition of NF-κB signaling reverses TNF-mediated inhibition of the Phex gene expression and promoter activity, whereas overexpression of RelA (p65) mimics the TNF effect. Proteasome inhibitor cLβL reduces the negative effects of TNF on the endogenous Phex mRNA expression and mPhex promoter (−103/+104 nt) activity in transiently transfected UMR-106 osteoblasts (A and B). Overexpression of p65 alone is sufficient to inhibit the endogenous Phex mRNA expression and mPhex promoter activity in transiently transfected UMR-106 cells. All values are expressed relative to untreated/mock-transfected cells. Values are mean ± S.E.; **, p < 0.01; ***, p < 0.001 (Student's t test; n = 4).
Inhibition of Poly(ADP-ribosyl)ation (Parylation) Increases mPhex Promoter Activity and Reverses the Effects of TNF in UMR-106 Osteoblasts
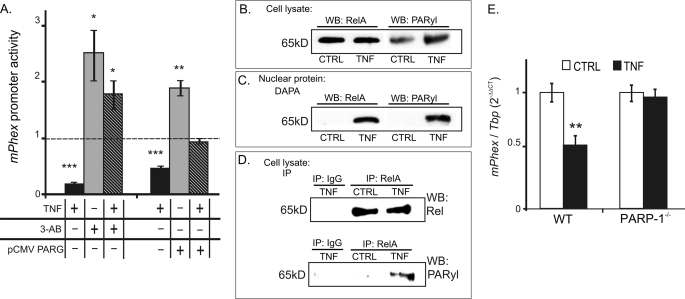
The proximity of PARP-1 binding sites to the confirmed NF-κB cis-elements in the mPhex promoter, and the reported potential interaction of these two transcriptional modulators, prompted us to investigate the effects of PARP-1 inhibition on mPhex promoter activity under basal conditions and in TNF-treated UMR-106 cells. UMR106 cells were transiently transfected with a reporter gene construct containing the mPhex-133/104 promoter region and treated for 24 h with or without TNF in the presence of poly(ADP-ribosyl)ation inhibitor 3-aminobenzamide (3-AB), or in cells overexpressing PARG. The latter enzyme catalyzes the removal of the paryl moiety from the target proteins. 3-AB and PARG overexpression caused a significant increase in Phex promoter activity over baseline in cells not exposed to TNF, and resulted in a full reversal of the inhibitory effects of TNF on Phex gene transcription (Fig. 8A).
FIGURE 8.
A, inhibition of poly(ADP-ribosyl)ation reverses TNF-mediated suppression of mPhex gene promoter activity. UMR-106 cells were transiently co-transfected with a reporter construct carrying a −133/+104-nt fragment of the mPhex gene with an “empty” or PARG expression vector. Cells were then pre-treated with or without the PARP-1 inhibitor 3-AB (1 mm) prior to a 24-h treatment with TNF (10 ng/ml). Both 3-AB and overexpression of PARG reversed the effects of TNF. Data are expressed relative to the control β-galactosidase activity in vehicle-treated cells transfected with the −133/+104 mPhex reporter construct. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (analysis of variance and Fisher protected least significant difference test; n = 4). B–D, TNF treatment results in inducible poly(ADP-ribosyl)ation (PARylation) of p65 in UMR-106 osteoblasts. B, Western blot (WB) analysis of whole protein extract with antibodies specific to poly(ADP-ribose) and RelA showed inducible PARylation of a 65-kDa protein with a migration pattern corresponding to that of p65. C, DAPA followed by Western blot detection of poly(ADP-ribose) and p65 resulted in similar findings. D, RelA was immunoprecipitated (IP) from control and TNF-treated UMR-106 cell lysates and probed for poly(ADP-ribose) to confirm poly(ADP-ribosyl)ation of this NF-κB subunit. E, TNF-mediated down-regulation of mPhex gene expression in vivo is completely abrogated in PARP-1-deficient mice. Real time PCR analysis of mPhex mRNA expression in femurs harvested from wild-type (WT) or PARP-1 KO mice (PARP-1 KO) 24 h after a single intraperitoneal injection of recombinant murine TNF (150 mg/kg). Values are mean ± S.E. (**, p < 0.01, Student's t test; n = 4).
PARP-1 Catalyzes Parylation of RelA
We next aimed to determine whether RelA is a target of PARP-1 activity. In Western blot analysis with whole cell extract of control or TNF-treated UMR-106 cells, an anti-poly(ADP-ribose) chain (PARyl) antibody detected TNF-inducible parylation of a protein with a molecular weight corresponding with that of RelA (Fig. 8B). Similar inducible binding of a parylated protein with a molecular weight consistent with RelA was observed in the DAPA with a biotinylated mPhex −133/1 probe, followed by a Western blot with an anti-PARyl antibody (Fig. 8C). When RelA was immunoprecipitated from whole cell extracts obtained from control or TNF-treated UMR-106 cells, we observed TNF-inducible parylation of this NF-κB subunit (Fig. 8D).
TNF-mediated Down-regulation of rPhex Is Completely Abrogated in PARP-1 KO Mice
To confirm the role of PARP-1 in TNF-driven and NF-κB-dependent repression of Phex gene transcription in vivo, we treated wild-type or PARP-1 null knock-out mice with recombinant mouse TNF. The TNF dose previously reported as sufficient to inhibit Phex expression in mice (150 mg/kg) reduced mPhex expression in the bone of wild-type mice by 50%, consistent with previously reported results (19). However, in PARP-1 KO mice, TNF treatment was without effect (Fig. 8E). This in vivo observation provides strong evidence for the involvement of PARP-1 as a modulator of NF-κB activity in osteoblasts exposed to elevated TNF during inflammatory conditions.
DISCUSSION
The skeletal and immune systems share numerous key players and regulatory mechanisms, an overlap that gave rise to the emerging field of osteoimmunology. A detailed understanding of the pathogenesis of bone destruction as a result of the interaction of immune cells and inflammatory mediators with bone cells is critical in designing novel strategies for the treatment of several disorders. These include rheumatoid arthritis, periodontal disease, Paget disease, osteoarthritis, multiple myeloma, metastatic bone tumors, and chronic inflammation-associated loss of bone mineral density. The latter is increasingly more recognized by the gastroenterology community due to a significant association of chronic IBD with osteopenia and osteoporosis (25). Bone loss in chronic inflammation is believed to be at least in part mediated by proinflammatory cytokines such as TNF, IL-1β, IL-6, or IFN-γ. Although TNF has been believed to affect bone primarily by causing osteoclast-driven bone erosion, newer data points to a direct effect of inflammatory mediators on the osteoblast functions as well. Defective bone formation has been reported not only in animal models of IBD (26) but also in pediatric Crohn's disease patients (27). Serum from children with Crohn's disease decreases osteoblast function including bone nodule formation in vitro (28).
The Phex gene encodes a M13 family of zinc metalloendopeptidase expressed primarily in osteoblasts and odontoblasts. Phenotypically, inborn inactivating mutations in the PHEX gene result in vitamin D resistant, X-linked hypophosphatemic rickets, whereas in vitro, the expression of Phex is a prerequisite for bone matrix deposition (6). We described earlier that chemically induced colitis results in a TNF-mediated decrease in bone Phex expression, and that in vitro exposure of osteoblasts to TNF results in a corresponding decreases of Phex mRNA and protein and a mineralizing defect (19). In this report we confirm these observations in more relevant IBD models offered by gnotobiotic IL-10−/− mice and by the naive T-cell transfer, and we identify the molecular mechanism underlying this phenomenon, which likely contributes to dysfunction of osteoblast activity and bone formation in patients with chronic IBD. According to this model, TNF treatment results in the recruitment of the p65/p50 NF-κB complex to two relatively weak cis-elements located at nt −76/−57 and −35/−16 between the Phex transcription start site and a poly(A) element located upstream of the NF-κB sites. Through proteomic and molecular approaches we identified the poly(A)-binding protein as PARP-1, an enzyme participating in TNF-inducible parylation of the p65 (RelA) subunit, likely increasing the affinity of the NF-κB complex to the proximal Phex promoter, resulting in increased retention and inhibited clearance of the RNA polymerase II complex.
The obtained results highlight the importance of Phex in osteoblast function during chronic inflammatory conditions, but also the significance of NF-κB activity and the modulating role of PARP-1 in osteoblast function. It also identifies PARP-1 as a potential target in mitigating IBD-associated bone loss. Recent studies using transgenic mice bearing dominant-negative IKK-γ targeted to mature osteoblasts by the bone γ-carboxyglutamate protein-2 (Bglap2) promoter have demonstrated the anti-mineralizing effects of NF-κB (29). These mice manifested enhanced bone formation with elevated expression of bone matrix genes such as α-1 type 1 collagen, osteocalcin, secreted phosphoprotein-1, and bone sialoprotein. Studies with these transgenic mice also demonstrated that NF-κB is constitutively active, albeit to a lesser extent, under basal conditions (29). This is consistent with our results that examined mouse promoter activity in UMR-106 cells in the presence of the proteasome inhibitor cLβL. These experiments showed that cLβL alone significantly increases Phex mRNA expression. More importantly, the results from our studies and the IKK dominant-negative transgenic mouse study strongly suggest that targeting NF-κB in the treatment of osteoporosis and inflammatory bone loss will not only result in suppression of bone resorption but also in promotion of bone formation, thus facilitating the rebuilding of bone mass.
It is important to note, however, that the role of NF-κB in IBD is very complex and its pleiotropic roles in cellular proliferation, differentiation and survival, inflammation, and carcinogenesis do not make it an easy target for systemic approach (30). Identifying PARP-1 as a crucial modulator of NF-κB activity in the osteoblasts opens an alternative possibility of targeting PARP-1. Several lines of evidence suggests that PARP-1 could be a suitable pharmacological target, particularly in IBD. (a) PARP-1 has been shown to participate in triggering the NF-κB pathway, affecting both the classical pathway and the nuclear-to-cytoplasmic DNA damage-induced NF-κB pathway. In the classical pathway, PARP-1 was shown to participate in LPS-induced monocyte chemotactic protein-1 expression (31), whereas in vascular smooth muscle cells PARP-1 was critical for TNF-induced expression of ICAM-1 (but not vascular cell adhesion molecule 1) and shown to physically interact with p65 (RelA) (32). In the nuclear to cytoplasmic DNA damage-induced NF-κB pathway PARP-1 contributes to the physical assembly of the nuclear signalsome including IKKγ, PIASy (nuclear matrix-associated SUMO E3 ligase), and ATM (protein kinase ataxia telangiectasia mutated) (33). (b) Modulating PARP-1 expression or enzymatic activity has been shown in numerous studies to ameliorate the symptoms of experimental colitis (34–39).
Although PARP-1 is an abundant and ubiquitous nuclear enzyme originally identified as a key factor in the DNA repair pathway, it has now been shown to positively and negatively affect gene transcription and chromatin structure under both basal and signal-activated conditions (40). Studies examining gene expression profiles in PARP-1-deficient embryonic stem cells and liver cells from PARP-1 KO mice showed that 3.5% of the transcriptome were regulated by PARP-1, 70% of which were positively regulated (41). More notably, PARP-1 has been described as one of the major molecules involved in the propagation of inflammatory stimuli and has been proposed as a target for anti-inflammatory treatment (42). PARP-1 has been shown to affect gene transcription in several ways: as an enhancer-binding factor similar to classical sequence-specific DNA-binding activators or repressors, as a transcriptional co-regulator, or as a modifying enzyme that catalyzes the NAD+-dependent addition of ADP(ribose) polymers (PARylation) to several nuclear proteins. We demonstrate here that in TNF-treated osteoblasts, RelA is translocated to the nucleus, where it is PARylated by PARP-1. This chemical modification is critical for TNF-induced inhibition of Phex expression because this response does not occur in PARP-1-deficient mice and is reversed by the PARP-1 inhibitor 3-AB or by an overexpression of PARG. Although it is technically challenging to determine whether PARP-1 and RelA physically interact with each other in the context of the Phex gene promoter, the proximity of their binding sites and inducible recruitment of RelA suggest such a possibility.
Interestingly, we initially described the Phex poly(A) promoter region as a positive cis-element whose affinity for a 110 kDa, then unidentified binding protein was decreased in response to dihydroxy-vitamin D treatment (29). Although the exact mechanism of the effects of vitamin D on PARP-1 in osteoblasts requires further work, our findings suggest that PARP-1 may play a pleiotropic role in the regulation of Phex gene transcription, depending on the physiological or pathophysiological context.
In conclusion, our results describe a new mechanism of TNF-mediated gene regulation in osteoblasts involving NF-κB and PARP-1. Cooperatively, NF-κB signaling and PARP-1 enzymatic activity constitutively and inducibly suppress Phex gene expression.
Supplementary Material
Acknowledgments
Access to the National Gnotobiotic Rodent Resource Center at the University of North Carolina at Chapel Hill was supported by National Institutes of Health Grant 2P40RR018603 provided courtesy of Dr. Balfour R. Sartor.
This work was supported, in whole or in part, by National Institutes of Health Grant R37DK033209 (to F. K. G.). Mass spectrometric data acquired by the Arizona Proteomics Consortium was supported by National Institutes of Health Grant ES06694 from the NIEHS to The Southwest Environmental Health Sciences Center, Grant CA023074 from the NCI to the Arizona Cancer Center, and by the BIO5 Institute of the University of Arizona.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1 and S2, and Table S2.
- PHEX
- phosphate-regulating gene with homologies to endopeptidases on the X chromosome
- IBD
- inflammatory bowel disease
- PARP-1
- poly(ADP-ribose) polymerase 1
- PARG
- poly(ADP-ribose) glycohydrolase
- DAPA
- DNA affinity precipitation assay
- RNAP II
- RNA polymerase II
- TSS
- transcriptional start site
- 3-AB
- 3-aminobenzamide
- cLβL
- clasto-lactacystin-β-lactone
- nt
- nucleotide(s)
- SPF
- specific pathogen free
- NP
- nuclear protein
- CHX
- cycloheximide
- ICAM-1
- intercellular adhesion molecule 1
- PMP
- paramagnetic particles
- KO
- knock-out.
REFERENCES
- 1.Caetano-Lopes J., Canhão H., Fonseca J. E. (2009) Autoimmun. Rev. 8, 250–255 [DOI] [PubMed] [Google Scholar]
- 2.Francis F., Hennig S., Korn B., Reinhardt R., de Jong P., Poustka A., Lehrach H., Rowe P. S., Goulding J. N., Summerfield T., Mountford R., Read A. P., Popowska E., Pronicka E., Davies K. E., O'Riordan J. L., Econs M. J., Nesbitt T., Drezner M. K., Oudet C., Pannetier S., Hanauer A., Strom T. M., Meindl A., Lorenz B., Cagnoli B., Mohnike K. L., Murken J., Meitinger T. (1995) Nat. Genet. 11, 130–1367550339 [Google Scholar]
- 3.Strom T. M., Francis F., Lorenz B., Böddrich A., Econs M. J., Lehrach H., Meitinger T. (1997) Hum. Mol. Genet. 6, 165–171 [DOI] [PubMed] [Google Scholar]
- 4.Collins J. F., Scheving L. A., Ghishan F. K. (1995) Am. J. Physiol. Renal Physiol. 269, F439–F448 [DOI] [PubMed] [Google Scholar]
- 5.Tenenhouse H. S. (2005) Annu. Rev. Nutr. 25, 197–214 [DOI] [PubMed] [Google Scholar]
- 6.Kiela P. R., Ghishan F. K. (2009) Lab. Invest. 89, 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowe A. E., Finnegan R., Jan de Beur S. M., Cho J., Levine M. A., Kumar R., Schiavi S. C. (2001) Biochem. Biophys. Res. Commun. 284, 977–981 [DOI] [PubMed] [Google Scholar]
- 8.Liu S., Guo R., Simpson L. G., Xiao Z. S., Burnham C. E., Quarles L. D. (2003) J. Biol. Chem. 278, 37419–37426 [DOI] [PubMed] [Google Scholar]
- 9.Benet-Pagès A., Lorenz-Depiereux B., Zischka H., White K. E., Econs M. J., Strom T. M. (2004) Bone 35, 455–462 [DOI] [PubMed] [Google Scholar]
- 10.Xiao Z. S., Crenshaw M., Guo R., Nesbitt T., Drezner M. K., Quarles L. D. (1998) Am. J. Physiol. Endocrinol. Metab. 275, E700–E708 [DOI] [PubMed] [Google Scholar]
- 11.Martin A., David V., Laurence J. S., Schwarz P. M., Lafer E. M., Hedge A. M., Rowe P. S. (2008) Endocrinology 149, 1757–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Addison W., Masica D., Gray J., McKee M. D. (2010) J. Bone Miner. Res. 25, 695–705 [DOI] [PubMed] [Google Scholar]
- 13.Addison W. N., Nakano Y., Loisel T., Crine P., McKee M. D. (2008) J. Bone Miner Res. 23, 1638–1649 [DOI] [PubMed] [Google Scholar]
- 14.Zoidis E., Gosteli-Peter M., Ghirlanda-Keller C., Meinel L., Zapf J., Schmid C. (2002) Eur. J. Endocrinol. 146, 97–105 [DOI] [PubMed] [Google Scholar]
- 15.Hines E. R., Collins J. F., Jones M. D., Serey S. H., Ghishan F. K. (2002) Am. J. Physiol. Renal Physiol. 283, F356–363 [DOI] [PubMed] [Google Scholar]
- 16.Alos N., Ecarot B. (2005) Bone 37, 589–598 [DOI] [PubMed] [Google Scholar]
- 17.Vargas M. A., St-Louis M., Desgroseillers L., Charli J. L., Boileau G. (2003) Endocrinology 144, 4876–4885 [DOI] [PubMed] [Google Scholar]
- 18.Hines E. R., Kolek O. I., Jones M. D., Serey S. H., Sirjani N. B., Kiela P. R., Jurutka P. W., Haussler M. R., Collins J. F., Ghishan F. K. (2004) J. Biol. Chem. 279, 46406–46414 [DOI] [PubMed] [Google Scholar]
- 19.Uno J. K., Kolek O. I., Hines E. R., Xu H., Timmermann B. N., Kiela P. R., Ghishan F. K. (2006) Gastroenterology 131, 497–509 [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Bores L., Barahona-Garrido J., Yamamoto-Furusho J. K. (2007) World J. Gastroenterol. 13, 6156–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larmonier C. B., Uno J. K., Lee K. M., Karrasch T., Laubitz D., Thurston R., Midura-Kiela M. T., Ghishan F. K., Sartor R. B., Jobin C., Kiela P. R. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 295, G1079–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurston R. D., Larmonier C. B., Majewski P. M., Ramalingam R., Midura-Kiela M., Laubitz D., Vandewalle A., Besselsen D. G., Muhlbauer M., Jobin C., Kiela P. R., Ghishan F. K. (2010) Gastroenterology 138, 1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slomiany B. A., Kelly M. M., Kurtz D. T. (2000) BioTechniques 28, 938–942 [DOI] [PubMed] [Google Scholar]
- 24.Kel A. E., Gössling E., Reuter I., Cheremushkin E., Kel-Margoulis O. V., Wingender E. (2003) Nucleic Acids Res. 31, 3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilg H., Moschen A. R., Kaser A., Pines A., Dotan I. (2008) Gut 57, 684–694 [DOI] [PubMed] [Google Scholar]
- 26.Dresner-Pollak R., Gelb N., Rachmilewitz D., Karmeli F., Weinreb M. (2004) Gastroenterology 127, 792–801 [DOI] [PubMed] [Google Scholar]
- 27.Hyams J. S., Wyzga N., Kreutzer D. L., Justinich C. J., Gronowicz G. A. (1997) J. Pediatr. Gastroenterol. Nutr. 24, 289–295 [DOI] [PubMed] [Google Scholar]
- 28.Varghese S., Wyzga N., Griffiths A. M., Sylvester F. A. (2002) J. Pediatr. Gastroenterol. Nutr. 35, 641–648 [DOI] [PubMed] [Google Scholar]
- 29.Chang J., Wang Z., Tang E., Fan Z., McCauley L., Franceschi R., Guan K., Krebsbach P. H., Wang C. Y. (2009) Nat. Med. 15, 682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karrasch T., Jobin C. (2008) Inflamm. Bowel Dis. 14, 114–124 [DOI] [PubMed] [Google Scholar]
- 31.Oumouna-Benachour K., Hans C. P., Suzuki Y., Naura A., Datta R., Belmadani S., Fallon K., Woods C., Boulares A. H. (2007) Circulation 115, 2442–2450 [DOI] [PubMed] [Google Scholar]
- 32.Zerfaoui M., Suzuki Y., Naura A. S., Hans C. P., Nichols C., Boulares A. H. (2008) Cell. Signal. 20, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stilmann M., Hinz M., Arslan S. C., Zimmer A., Schreiber V., Scheidereit C. (2009) Mol. Cell 36, 365–378 [DOI] [PubMed] [Google Scholar]
- 34.Zingarelli B., Hake P. W., Burroughs T. J., Piraino G., O'Connor M., Denenberg A. (2004) Immunology 113, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez-Fidalgo S., Villegas I., Martín A., Sánchez-Hidalgo M., Alarcón de la Lastra C. (2007) Eur. J. Pharmacol. 563, 216–223 [DOI] [PubMed] [Google Scholar]
- 36.Di Paola R., Mazzon E., Xu W., Genovese T., Ferraris D., Muià C., Crisafulli C., Zhang J., Cuzzocrea S. (2005) Eur. J. Pharmacol. 527, 163–171 [DOI] [PubMed] [Google Scholar]
- 37.Zingarelli B., O'Connor M., Hake P. W. (2003) Eur. J. Pharmacol. 469, 183–194 [DOI] [PubMed] [Google Scholar]
- 38.Popoff I., Jijon H., Monia B., Tavernini M., Ma M., McKay R., Madsen K. (2002) J. Pharmacol. Exp. Ther. 303, 1145–1154 [DOI] [PubMed] [Google Scholar]
- 39.Jijon H. B., Churchill T., Malfair D., Wessler A., Jewell L. D., Parsons H. G., Madsen K. L. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G641–651 [DOI] [PubMed] [Google Scholar]
- 40.Nguewa P. A., Fuertes M. A., Valladares B., Alonso C., Pérez J. M. (2005) Prog. Biophys. Mol. Biol. 88, 143–172 [DOI] [PubMed] [Google Scholar]
- 41.Ogino H., Nozaki T., Gunji A., Maeda M., Suzuki H., Ohta T., Murakami Y., Nakagama H., Sugimura T., Masutani M. (2007) BMC Genomics 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peralta-Leal A., Rodríguez-Vargas J. M., Aguilar-Quesada R., Rodríguez M. I., Linares J. L., de Almodóvar M. R., Oliver F. J. (2009) Free Radic. Biol. Med. 47, 13–26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.