Abstract
Select adhesion molecules connect pre- and postsynaptic membranes and organize developing synapses. The regulation of these trans-synaptic interactions is an important neurobiological question. We have previously shown that the synaptic cell adhesion molecules (SynCAMs) 1 and 2 engage in homo- and heterophilic interactions and bridge the synaptic cleft to induce presynaptic terminals. Here, we demonstrate that site-specific N-glycosylation impacts the structure and function of adhesive SynCAM interactions. Through crystallographic analysis of SynCAM 2, we identified within the adhesive interface of its Ig1 domain an N-glycan on residue Asn60. Structural modeling of the corresponding SynCAM 1 Ig1 domain indicates that its glycosylation sites Asn70/Asn104 flank the binding interface of this domain. Mass spectrometric and mutational studies confirm and characterize the modification of these three sites. These site-specific N-glycans affect SynCAM adhesion yet act in a differential manner. Although glycosylation of SynCAM 2 at Asn60 reduces adhesion, N-glycans at Asn70/Asn104 of SynCAM 1 increase its interactions. The modification of SynCAM 1 with sialic acids contributes to the glycan-dependent strengthening of its binding. Functionally, N-glycosylation promotes the trans-synaptic interactions of SynCAM 1 and is required for synapse induction. These results demonstrate that N-glycosylation of SynCAM proteins differentially affects their binding interface and implicate post-translational modification as a mechanism to regulate trans-synaptic adhesion.
Keywords: Carbohydrate Function, Carbohydrate Structure, Cell Adhesion, Neurobiology, Neurodevelopment, Synapses, CADM, SynCAM, Nectin-like
Introduction
Synapses in the central nervous system are highly specialized sites of neuronal adhesion. They are morphologically defined by a presynaptic terminal filled with synaptic vesicles, an apposed postsynaptic specialization that contains neurotransmitter receptors, and a synaptic cleft of 20-nm width that separates pre- and postsynaptic sites (1, 2). This cleft is filled with proteinaceous material (3).
The proteins spanning the synaptic cleft not only tie pre- and postsynaptic membranes together. Select synaptic surface molecules can also instruct the organization of nascent synapses (4–6). This was first demonstrated for neuroligins, postsynaptic membrane proteins that bind the presynaptic neurexins (7–9). Adhesion molecules of the Ig superfamily and proteins containing leucine-rich repeats additionally mediate synaptic differentiation (10–14). Similarly, receptor tyrosine kinases, including EphB receptors, instruct synaptogenesis through trans-synaptic signaling (15, 16). These synapse-inducing proteins act in conjunction with N-cadherins that set the pace of synaptic maturation (17, 18).
Among these synapse-organizing proteins, SynCAMs4 form a family of four Ig superfamily members that are single-spanning membrane proteins with three extracellular Ig-like domains (19). SynCAMs, also known as nectin-like molecules, are prominently expressed throughout the brain and are enriched in synaptic plasma membranes (10, 20). They are N-glycosylated proteins, consistent with the presence of multiple predicted N-glycosylation sites in their extracellular Ig domains (19, 20). SynCAMs 1, 2, and 3 can bind themselves through homophilic binding, but SynCAMs 1/2 and 3/4 preferentially engage each other in specific heterophilic interactions (20, 21). SynCAM 1 and 2 form a trans-synaptic adhesion complex and promote the number of functional excitatory synapses (20).
Consistent with the critical importance of synapse organization for brain functions, trans-synaptic adhesion molecules need to be regulated. Mechanisms include the control of their sorting by intracellular interactions as shown for neuroligins (22) and alternative splicing within sequences encoding extracellular domains, which specifies neurexin-neuroligin interactions and has been analyzed at atomic resolution (23–26). Post-translational modifications regulating synaptic adhesion molecules are less understood, but a negative effect of glycosylation on neuroligin 1 binding has been reported (27).
Here, we address the molecular properties that underlie and regulate SynCAM adhesion and function. Our crystallographic, mass spectrometric, and biochemical analyses of SynCAM 1 and 2 demonstrate that they carry N-glycans adjacent to and within the first Ig domain that provides their extracellular binding interface. Unexpectedly, the glycosylation of these two SynCAM family members serves different roles. Although N-glycans within the Ig1 binding interface of SynCAM 2 reduce its binding, glycosylation at the SynCAM 1 Ig1 domain promotes its adhesion. Consequently, the ability to N-glycosylate SynCAM 1 increases its trans-synaptic interactions and synapse inducing activity. Together, these results identify glycosylation as a novel mechanism for positively and negatively regulating the trans-synaptic SynCAM adhesion complex in the brain.
EXPERIMENTAL PROCEDURES
Antibodies
Specific antibodies against SynCAM 1 (YUC8) and SynCAM 2 (YU524) were described previously (20). For immunostaining of SynCAMs 1–3, we utilized a pleio-SynCAM antibody (T2412) raised against the SynCAM 1 C terminus that equally recognizes this conserved sequence in full-length SynCAM 2 and 3 (20) but not the GPI-anchored SynCAM constructs used in this study. Monoclonal antibodies to synaptophysin (7.2) and GDI (81.2) were obtained from Synaptic Systems (Göttingen, Germany), monoclonal antibodies to CASK were from Millipore (Billerica, MA), and monoclonal antibodies to FLAG (M2) were from Sigma. Monoclonal antibodies to SV2 (developed by Kathleen Buckley) were obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa.
Expression Vectors
A pCMV5 GPI vector backbone was generated by amplifying the GPI targeting sequence from GPI-VAMP2 (a gift from Dr. James Rothman, Department of Cell Biology, Yale University) with 5′ SalI and 3′ BamHI sites and subcloning into pCMV5. pCMV5 FLAG-GPI was generated analogously, adding the FLAG epitope DYKDDDDK N-terminal of the GPI anchoring sequence. Full-length SynCAM extracellular sequences or sequences lacking select Ig domains were amplified from pCMV IG9 vectors described previously (10) and subcloned into pCMV5 GPI or pCMV5 FLAG-GPI vectors. Point mutants were generated using site-directed PCR mutagenesis (QuikChange; Stratagene, La Jolla, CA). Expression vectors for full-length SynCAM 1 carrying an extracellular FLAG epitope N-terminal of the transmembrane region and for extracellular SynCAM sequences fused to a thrombin cleavage and IgG1-Fc sequence were described previously (20). GFP was expressed from pCAG GFP, a gift from Dr. Nenad Sestan (Department of Neurobiology, Yale University).
Cell Culture
COS7 cells were maintained using standard procedures and transfected with FuGENE 6 (Roche Applied Science) for transient expression. HEK293 cell lines stably expressing the SynCAM 1 or 2 extracellular domains were selected in the presence of geneticin (American Bioanalytical, Natick, MA) after transfection of HEK293 cells with the vector pcDNA3.1 SynCAM 1 extracellular domain IgG1 linearized with BglII. Dissociated cultures of hippocampal neurons were prepared as described (28).
Expression and Purification of SynCAM 1 and SynCAM 2 Extracellular Sequences
The extracellular sequences of mouse SynCAM 1 or SynCAM 2 were purified as described previously (20). Briefly, HEK293 cell lines stably expressing the full-length SynCAM 1 or SynCAM 2 extracellular sequences fused to IgG1-Fc were grown in DMEM low glucose medium supplemented with 5% FBS and 50 mg/ml geneticin. The culture supernatant was collected and replaced with fresh medium every 72 h. 2 liters of culture supernatant were filtered, concentrated, and then applied to 2 ml of protein A-agarose resin (Invitrogen) equilibrated in buffer A (50 mm Tris, pH 8.0, 150 mm NaCl, 2 mm β-mercaptoethanol). SynCAM extracellular domains were eluted from the resin by the addition of bovine α-thrombin protease (Hematologic Technologies, Essex Junction, VT) in a 1:300 molar ratio at 16 °C overnight to cleave the resin-bound N-terminal human IgG1-Fc tag. The proteins were further purified by size exclusion chromatography on a Superdex 200 column (GE Healthcare) in buffer A.
Isothermal Titration Calorimetry
The binding of SynCAM 1 to SynCAM 2 was studied by isothermal titration calorimetry in 20 mm Tris, pH 8.0, 50 mm NaCl, at 25 °C, using an iTC200 system (MicroCal, Piscataway, NJ). The sample cell contained the purified SynCAM 2 extracellular domain protein at 5 μm, and the syringe contained the SynCAM 1 extracellular domain at 50 μm, with the IgG1-Fc tags cleaved off. Typically, one initial injection of 1.5 μl and 19 serial injections of 2.0 μl of SynCAM 1 were performed at 180-s intervals. The stirring speed was maintained at 1000 rpm, and the reference power was kept constant at 5 μcal/s. The heat associated with each injection of SynCAM 1 was integrated and plotted against the molar ratio of SynCAM 1 to SynCAM 2. Thermodynamic parameters were extracted from a curve fit to the data using the Origin 7.0 software provided by MicroCal. The experiments were performed in triplicate with excellent reproducibility (<10% variation in thermodynamic parameters).
Preparation of SynCAM Extracellular Domain Complexes
SynCAM 2 was first expressed and applied to protein A-agarose resin as described above. The protein was then eluted from the resin with 0.2 m glycine, pH 3.0, and dialyzed into 20 mm Tris, pH 8.0, 50 mm NaCl, 2 mm β-mercaptoethanol. A 2-fold molar excess of the purified SynCAM 1 extracellular domain with the IgG1-Fc tag cleaved off was then added to obtain heteromeric SynCAM 1-SynCAM 2 complexes in addition to the homomeric SynCAM 2 complexes present in this preparation. The resulting mixture was incubated at 16 °C for 3 h and applied to 2 ml of protein A-agarose resin in buffer A. The SynCAM complexes were eluted from the resin with thrombin protease (1:300 molar ratio, 16 °C overnight) and were purified on a Superdex 200 column in buffer A. To aid in the subsequent crystallization step, the samples were partially deglycosylated under native conditions with PNGase F and neuraminidase (New England Biolabs) at 37 °C for 48 h and separated from the endoglycosidases and cleaved glycans on a Superdex 200 column in 50 mm Tris, pH 8.0, 50 mm NaCl, 2 mm β-mercaptoethanol.
Crystallization of the Ig1 Domain of SynCAM 2
Crystals were grown at 20 °C using the hanging drop vapor diffusion technique. The preparation of the SynCAM extracellular domain complex was concentrated to 9.0 mg/ml in 20 mm Tris, pH 8.0, 50 mm NaCl, 2 mm β-mercaptoethanol. The protein solution was mixed with an equal volume of well solution (0.1 m HEPES, pH 6.5–7.0, 21% PEG5000 monomethyl ether). Irregular bulky crystals grew after 3 months. The crystals were triclinic (space group P1) with unit cell dimensions a = 42.8 Å, b = 50.5 Å, c = 79.9 Å, α = 75.9°, β = 77.0°, and γ = 65.2°. With four molecules/asymmetric unit, the Matthews co-efficient VM is 3.36 Å3 Da−1, which corresponds to a solvent content of 63.4% (29). For data collection, the crystals were transferred to a cryoprotectant containing 0.1 m HEPES, pH 6.8, 21% PEG5000 monomethyl ether and 18% glycerol (v/v) and immediately frozen in liquid nitrogen.
Data Collection and Processing
Crystallographic data were collected at 100 K on Beamline X29A of the National Synchrotron Light Source at Brookhaven National Laboratory. The data were indexed, integrated, and scaled using the HKL2000 program suite (30). The data collection statistics are summarized in supplemental Table S1.
Structure Determination and Refinement
The crystal structure of SynCAM 2 Ig1 was determined by molecular replacement using a monomer of human SynCAM 3 (nectin-like molecule 1) Ig1, Protein Data Bank Code 1Z9M (31), as the search model in the program PHASER 2.1 (32). The details of structure determination and refinement are described in the supplemental materials.
Glycopeptide Mapping
The purified SynCAM 1 extracellular domain with the IgG1-Fc cleaved off was digested with the combined endoproteinases Lys C and trypsin. The samples were C18 RP ZipTip-cleaned and desalted prior to collecting MS data on a 9.4T Apex Qe FT-ICR MS instrument. Eluted peptides were directly infused into the mass spectrometer via nanoelectrospray at 250 nL/min into an Apollo II dual ion funnel ESI source. The spray shield voltage was set at 3500, and a 4000-V potential was applied on the glass capillary end cap. The instrument (running Compass Software with APEX control acquisition component (v.1.2) is set up to acquire single free induction decay signal (512,000) data with a mass range (m/z) from 450 to 2000. Enrichment of glycopeptides was confirmed using albumin, ovalbumin, β-casein, and RNase B as standard proteins. All of the data were processed utilizing DA analysis software v. 3.4, online GlycoMod (Expasy), and MASCOT search engine.
Glycan Profiling
Glycosidase treatment of the SynCAM 1 extracellular sequence with the IgG1-Fc cleaved off was performed using the glycosidases PNGase F or endoglycosidase H. Cleaved glycans were enriched with a Carbograph column, followed by C18 RP ZipTip prior to direct infusion into a 9.4T FT-ICR MS instrument. The electrospray source was configured with a capillary (low flow) sprayer optimized for positive mode. Ions were detected in the 450–2500 m/z with 512K data points/MS scan. All of the data were processed utilizing DA analysis software v.3.4, online GlycoMod (Expasy).
Tissue Preparation
Samples from rat brain regions were prepared by rapid homogenization in 8 m urea. Protein concentrations were determined using the Pierce BCA assay.
Protein Deglycosylation
Enzymatic deglycosylation was performed using neuraminidase (sialidase; Roche Applied Science) and PNGase F (New England Biolabs) according to the manufacturers' instructions.
Affinity Chromatography
The SynCAM 1 extracellular domain was immobilized on protein A beads to serve as affinity matrix. Rat forebrain proteins were solubilized with 1% CHAPS (Roche Applied Science), and affinity chromatography and quantitative immunoblotting were performed as described (10, 20).
Surface Expression Control
COS7 cells expressing SynCAM constructs tagged with an extracellular FLAG epitope were fixed, labeled with anti-FLAG antibodies to detect surface-expressed epitopes (antibody M2; 1:1000), washed, and then permeabilized using 0.1% Triton X-100 to perform immunostaining for total SynCAM protein (antibody T2412; 1:1000). The images were acquired on a Zeiss LSM 510 META laser scanning confocal microscope.
Cell Overlay Experiments
Cell overlay assays were performed as described (21). Briefly, COS7 cells were co-transfected with expression vectors encoding extracellularly FLAG-tagged SynCAM constructs and soluble GFP or GFP alone as negative control. After 2 days, live cells were overlaid for 20 min at 25 °C with the purified SynCAM 1 extracellular domain at 2 μg/ml or the SynCAM 2 extracellular domain at 10 μg/ml. The IgG1-Fc fusion tag of these overlaid fusion proteins was directly detected by including Alexa 546-conjugated protein A (6 μg/ml; Invitrogen) in this step. Surface-expressed SynCAM proteins were detected in these live cells by simultaneously adding anti-FLAG (antibody M2; 1:1000) and secondary anti-mouse antibodies conjugated to Alexa 488 (Invitrogen) (1:1000). The medium was then replaced with DMEM without phenol red, and the cells were immediately imaged with a Hamamatsu Orca camera attached to a Nikon Eclipse TE2000-U microscope. The signal of the secondary Alexa 488 antibody detecting anti-FLAG antibodies was used to define regions of interest, within which the fluorescence from the Alexa 546-conjugated protein A was measured and normalized to the anti-FLAG signal. Signals were quantified using a custom Matlab (MathWorks) script that is available upon request.
Mixed Co-culture Assay for Synapse Induction
Co-culture assays were performed as described (28). Briefly, COS7 cells co-expressing GPI-anchored SynCAM 1 constructs and soluble GFP or GFP alone as negative control were seeded atop neurons at 6–7 days in vitro. At 8–9 days in vitro, these mixed co-cultures were fixed and immunostained for the presynaptic marker SV2 and for neuronal SynCAM proteins with the antibody T2412. The images were acquired on a Zeiss LSM 510 META laser scanning confocal microscope. The surface area of COS7 cells immunopositive for neuronal SynCAMs and SV2 was quantified using a Matlab script that is available upon request. The images were collected blind to the synaptic marker channel.
Miscellaneous Procedures
Sequence similarities were analyzed using the T-Coffee method (33). Amino acid numbers refer to the position in the protein including the signal peptide. Statistical analyses were performed using the two-tailed t test, with statistical errors corresponding to the standard errors of mean.
RESULTS
High Affinity Binding of SynCAM 1 to SynCAM 2 Requires the Ig1 Domain
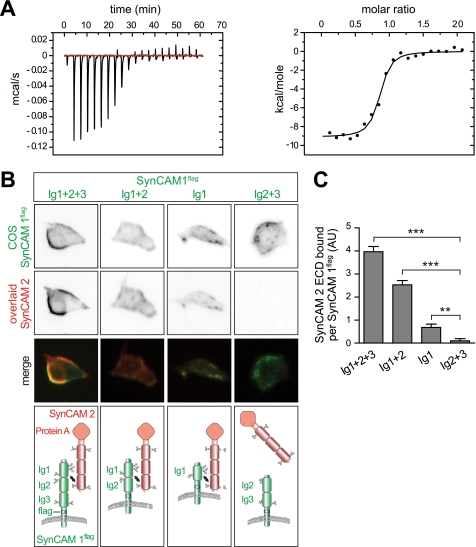
To define the molecular properties of SynCAM interactions, we measured the affinity between the SynCAM 1 and SynCAM 2 extracellular sequences by isothermal titration calorimetry. The resulting isotherm was consistent with a single binding interface between the two proteins in a 1:1 complex, with a tight apparent dissociation constant (Kd) of 78 nm (Fig. 1A). This Kd is very similar to the neuroligin 1/neurexin 1β interaction (34).
FIGURE 1.
The first Ig domain mediates tight heterophilic binding of SynCAM 1 to SynCAM 2. A, isothermal titration calorimetry analysis of the binding of the SynCAM 1 extracellular sequence to SynCAM 2. Left panel, enthalpic heat released at 25 °C during the titration of the SynCAM 1 extracellular sequence into the isothermal titration calorimetry cell containing the SynCAM 2 extracellular sequence. Right panel, integrated binding isotherms of the titration and best fit to a single-site model. The best fit yielded a dissociation constant Kd = 78.0 nm, enthalphy ΔH = −9.1 kcal/mol, and binding stoichiometry n = 1. B, analysis of adhesive SynCAM binding by cell overlay. COS7 cells expressed full-length SynCAM 1 or variants containing the indicated Ig domains, with an extracellular FLAG epitope inserted proximal to the transmembrane region. These surface-expressed proteins were detected in live cells by the addition of anti-FLAG antibodies and secondary antibodies conjugated to Alexa 488 (top row, green in the merge). The cells were simultaneously overlaid with the SynCAM 2 extracellular domain fused to IgG1-Fc together with protein A conjugated to Alexa 546 (second row, red in the merge) to label the retained protein. The first Ig domain of SynCAM 1 is required for adhesive binding to SynCAM 2 as depicted in the model below. C, quantification of the results in B. The results are expressed as a protein A signal detecting retained SynCAM 2 normalized to the signal from the indicated anti-FLAG labeled SynCAM 1 constructs expressed on COS7 cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We next mapped this single binding interface within the three extracellular Ig-like domains. Utilizing constructs comprised of subsets of SynCAM 1 Ig domains, we measured their adhesive interaction with the SynCAM 2 extracellular domain using a cell overlay approach (Fig. 1B). These experiments extended previous affinity chromatography studies (20) and allowed us to analyze SynCAM interactions as they occur on the cell surface. We expressed an array of SynCAM 1 Ig constructs carrying an extracellular FLAG epitope and labeled the expressed proteins in live COS7 cells with anti-FLAG antibodies. To quantify adhesive binding, we overlaid these cells with the soluble SynCAM 2 extracellular sequence fused to IgG1-Fc and detected retained protein using fluorophore-labeled protein A. This signal was divided by the fluorescence measured with anti-FLAG antibodies, which normalized for each cell the extent of SynCAM 2 retention to the amount of its surface-expressed SynCAM 1. All of the SynCAM 1 Ig constructs were properly N-glycosylated and sorted to the plasma membrane, with the Ig1 domain carrying N-glycans to the highest apparent extent (supplemental Fig. S1). These experiments showed that the tandem Ig1 + 2 domains of SynCAM 1 were sufficient for strong binding (Fig. 1, B and C). Moreover, the SynCAM 1 Ig1 domain was required for binding because the SynCAM 1 Ig2 + 3 construct did not retain SynCAM 2. The SynCAM 1 Ig1 domain alone was sufficient for SynCAM 2 binding, albeit at a lower strength. This reduced interaction of the SynCAM 1 Ig1 domain in the absence of the Ig2 and Ig3 domains is possibly due to a role of these domains in conferring a steric orientation to SynCAM 1 Ig1 that is favorable for its interaction with SynCAM 2. Together, these results show that the first Ig domain of SynCAM 1 provides its binding interface.
Crystal Structure of the SynCAM 2 Ig1 Domain
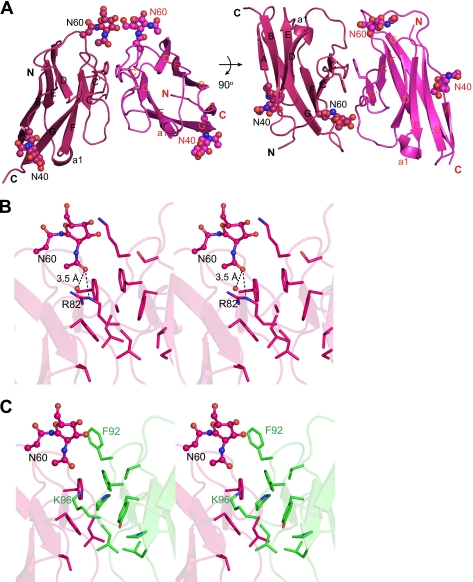
Aiming to characterize the extracellular SynCAM interactions at atomic resolution, we performed crystallization trials of the SynCAM 1/2 extracellular domain complex. The crystal structure, which was refined at 2.21 Å resolution (r = 0.197, Rfree = 0.245), showed that the crystals contained only the Ig1 domain of SynCAM 2. This was consistent with the presence of a major protein band at 17 kDa in these crystals (data not shown), corresponding to the size of one Ig domain. Upon closer examination of the drop that produced the crystals, fungal growth was observed. This suggested that secreted fungal proteases may have cleaved the SynCAM 1/2 extracellular domain complex, allowing SynCAM 2 Ig1 to crystallize by itself. Several other proteins have been crystallized as a result of either intentional proteolytic cleavage or serendipitous cleavage by secreted fungal proteases (35, 36). The crystal structure of the SynCAM 2 Ig1 monomer (residues 35–131) showed that it adopts an Ig-like fold of the variable type (37) as predicted by sequence analysis (19), comprising two β-sheets with nine antiparallel β-strands (denoted A to G; Fig. 2A). Hydrophobic interactions between the two sheets form the core of the domain. A disulfide bridge between Cys53 and Cys113 links β-strands B and G, further stabilizing the domain. Two N-linked N-acetylglucosamine residues are visible in the structure on Asn40 and Asn60, respectively. The remainder of these N-glycans had been removed during sample preparation to aid crystallization (see “Experimental Procedures”).
FIGURE 2.
Structure of the SynCAM Ig1 domain interface. A, crystallographic results show that the SynCAM 2 Ig1 domain forms a dimer, characterized by mostly hydrophobic interactions across a noncrystallographic 2-fold axis, as shown in this ribbon representation. The N and C termini are marked. The N-acetylglucosamine residues on Asn40 and Asn60 are marked as spheres. B, stereodiagram of the SynCAM 2 Ig1 trans-homodimer interface, with the hydrogen bonding interactions of the N-acetylglucosamine on Asn60 shown as dashed lines. Side chains in the interfaces are shown in stick representation. Water molecules are highlighted as red spheres. C, theoretical model of the trans-heterodimer interface of SynCAM 1 Ig1/SynCAM 2 Ig1 shown as stereodiagram. Residues in the interface are depicted in stick representation, with SynCAM 1 and SynCAM 2 in green and magenta, respectively. Interfaces are displayed in the same orientation as in B. The N-linked glycan on Asn60 of SynCAM 2 participates in the SynCAM 1 Ig1/SynCAM 2 Ig1 trans-heterodimer interface.
Interestingly, SynCAM 2 Ig1 forms homodimers in the crystals, with each asymmetric unit containing two dimers. The SynCAM 2 Ig1 homodimer has approximate dimensions of 60 × 42 × 33 Å (Fig. 2A). Dimer formation buries a total of 698 Å2 (11.5%) of solvent-accessible surface/monomer. The N and C termini of each subunit in the dimer are antiparallel, indicating that the dimer corresponds to the trans-adhesion complex. The dimer interface is mostly hydrophobic (35% of the residues are nonpolar) and closely resembles the dimer interface of the Ig1 domain of SynCAM 3, also known as nectin-like molecule 1 (31), which is its closest structural homolog. SynCAM 3 also participates in cell adhesion (20, 38). The Ig1 domains of SynCAM 2 and SynCAM 3 have high sequence identity (63%) and structural similarity (root mean square deviation, 0.7 Å over 96 equivalent Cα atoms). However, although the SynCAM 3 structure lacks glycans, in SynCAM 2 the N-acetylglucosamine on Asn60 forms a weak intersubunit contact in the trans-dimer interface of SynCAM 2 Ig1 (Fig. 2, A and B). Specifically, the carbonyl oxygen atom in the acetyl moiety of the first residue of the glycan is within 3.5 Å of the Nϵ atom of the Arg82 side chain in the other monomer and of a structured water molecule located in the dimer interface. The location of Asn60 at the dimer interface leaves little room for a bulky glycan, however, suggesting that full glycosylation at Asn60 may interfere with adhesive dimer formation.
Homology Model of the SynCAM 1/2 Ig1 trans-Heterodimer
Like SynCAM 2, SynCAM 1 engages in both homo- and heterophilic adhesion complexes (20, 21). The high sequence identity of 44% between the Ig1 domains of SynCAM 1 and SynCAM 2 enabled us to build a homology model for SynCAM 1 Ig1 based on our crystal structure of SynCAM 2 Ig1. Our model predicts that the trans-dimeric interface of the SynCAM 1 Ig1 homodimer is more hydrophobic than that of SynCAM 2 Ig1 (supplemental Fig. S2A). The residues that form additional hydrophobic contacts in the SynCAM 1 model are Val76, Phe92, and Pro94.
Although capable of homophilic binding, SynCAMs preferentially assemble into specific heterophilic complexes, and SynCAM 1 strongly binds SynCAM 2 (20, 21, 38–40). To better understand dimerization specificities, we modeled SynCAM 1 Ig1/SynCAM 2 Ig1 trans-heterodimers using the homodimeric crystal structure of SynCAM 2 Ig1 as template. Interestingly, a glycan-mediated contact occurs in the heterodimer model between the N-linked glycan on Asn60 of SynCAM 2 and the side chain of Lys96 of SynCAM 1 (Fig. 2C). The Asn60 site of SynCAM 2 is not conserved in SynCAM 1, and this glycan may contribute to regulating the heterophilic binding of SynCAM 2 to SynCAM 1. Conversely, two N-glycosylation sites of the Ig1 domain of SynCAM 1, Asn70 and Asn104, are located on one face of the Ig1 domain, in the loop between strands B and C and in the middle of strand E, respectively (supplemental Fig. S2B). Residues 70 and 104 are both ∼20 Å from the trans-dimer interface and face away from the interface.
N-Glycosylation of the SynCAM 1 Ig1 Domain
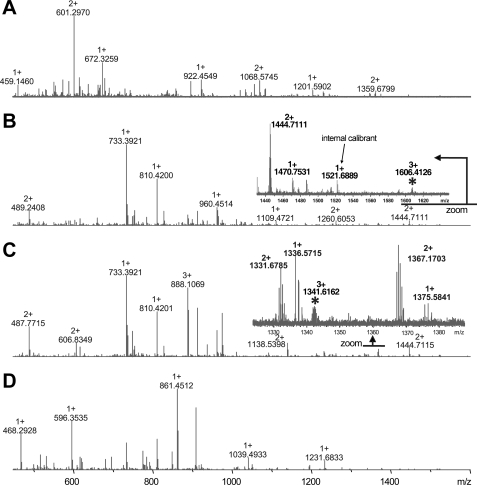
These crystallographic results map glycans to different surfaces of the Ig1 domain in SynCAM 1 and SynCAM 2. To examine SynCAM 1 N-glycosylation, we performed a mass spectrometry analysis of the SynCAM 1 extracellular sequence purified from HEK293 cells, which glycosylate SynCAM 1 to the same apparent extent as found in brain (20). Using several different enzymes or combinations thereof, we observed a very high number of extracellular SynCAM 1 peptides/glycopeptides, which provide >70% sequence coverage of the protein. Glycopeptide mapping identified the asparagines Asn70 and Asn104 in the first Ig1 domain as potential N-glycosylation sites based on FT-ICR high mass accuracy and GlycoMod prediction (41) (Fig. 3). The deconvoluted mass list of each spectrum was entered into GlycoMod to predict possible glycosylations sites along with their potential glycan composition based on mass accuracy and the consensus sequence for N-glycosylation (42). The GlycoMod output identifies the Asn104 site as glycosylated (in the CNBr + trypsin and LysC + trypsin digest conditions; Fig. 3, B and C), with several glycopeptides showing sialylated glycan (NeuAc) modifications at that site. N-Glycosylation at Asn70 was predicted from the mass list of SynCAM 1 digested with CNBr + trypsin (Fig. 3B). To obtain a profile of these N-glycan structures, we subjected SynCAM 1 to PNGase F or endoglycosidase H to cleave the glycans. Glycan masses observed by FT-ICR suggested the presence of Hex, HexNac, and NeuAc carbohydrates as predicted by GlycoMod (data not shown).
FIGURE 3.
Glycopeptide mapping and glycan profiling of the SynCAM 1 extracellular sequence. Broadband FT-ICR MS mass spectrum of four different enzymatic digestion of the purified, glycosylated SynCAM 1 extracellular sequence. 10 μg of SynCAM 1 was utilized for each digestion with CNBr (A), CNBr + trypsin (B), Lys C + trypsin (C), and protease type XIII (D). The asterisks indicate potential glycosylation sites based on exact mass measurements and GlycoMod prediction (41). The inset in B shows an enlarged region illustrating a predicted glycopeptide at m/z 1606.413 (3+) that corresponds to a modification at the Asn70 position. Note that internal calibrations were utilized to obtain mass accuracy of <5 ppm. The inset in C shows an enlarged region with a glycopeptide that corresponds to the modified Asn104 residue of SynCAM 1 at m/z 1341.616 (3+).
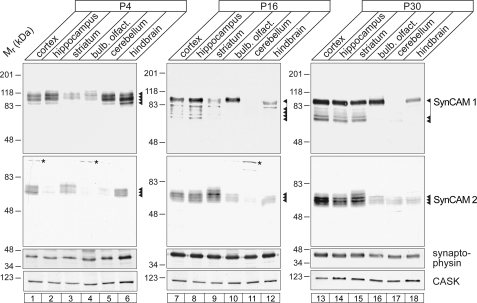
Complex Modification of SynCAM 1 and SynCAM 2 in the Brain
SynCAM 1 and 2 are heavily glycosylated in the adult brain (20). To obtain insight into the extent of post-translational SynCAM modification during early postnatal development, when most synapses form, we analyzed SynCAM 1 and 2 in several rat brain regions (Fig. 4). SynCAM modification was examined by immunoblotting prior to, during, and subsequent to the peak of synapse formation in the rodent brain at postnatal day 4 (P4), P16, and P30, respectively (43). At all stages and in all regions, the apparent molecular masses of SynCAM 1 and 2 proteins were notably higher than the 41–45 kDa predicted from their open reading frames (19). Interestingly, SynCAM 1 modifications changed during development. At P4, it was expressed as diverse species that ranged from 90 to 115 kDa. As development progressed, SynCAM 1 was detected both as an apparently uniform high molecular mass species of 100 kDa and as multiple low molecular mass species of 70–85 kDa. These changes in the modification of SynCAM 1 were accompanied by a shift of its predominant expression from hindbrain to forebrain. This is consistent with roles of SynCAM 1 in synapse formation, which progresses during brain development from the hindbrain to the forebrain. Its binding partner SynCAM 2 also followed a developmental expression increase toward forebrain, yet SynCAM 2 was expressed as the same diverse species at 62–76 kDa throughout. Other N-glycosylated proteins such as synaptophysin also did not exhibit changes in their modification (Fig. 4), consistent with the developmentally independent glycosylation of other neuronal membrane proteins.
FIGURE 4.
SynCAM post-translational modifications are regionally and developmentally regulated in the brain. The indicated brain regions were dissected from rats at P4, P16, or P30. Equal protein amounts of 30 μg were analyzed by immunoblotting using the antibodies shown. SynCAM 1 and 2 exhibited distinct expression and modification patterns as described in the text. The N-glycosylated synaptic protein synaptophysin and the scaffolding molecule CASK served as loading controls. The asterisks mark nonspecific bands.
Modification of SynCAM 2 Ig1 at Asn60 Reduces Adhesion
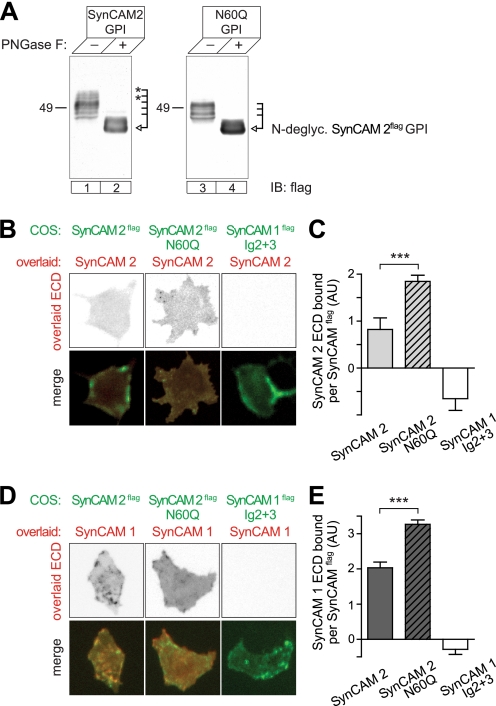
To perform a biochemical analysis of SynCAM 2 glycosylation, we changed the asparagine at position 60 to glutamine, choosing this substitution because it prevents N-glycosylation without altering immunoglobulin folds (44). Consistent with the conservative nature of this mutation, all Asn → Gln glycosylation mutants used in this study were sorted to the cell surface, indicating proper folding (see below, Fig. 5, B and D, and supplemental Fig. S3B). Furthermore, the slightly increased bulk of the glutamine residue in the N60Q SynCAM 2 mutant can be expected to be easily accommodated in the structures of its homodimer as well as the heterodimer with SynCAM 1.
FIGURE 5.
SynCAM 2 glycosylation within the Ig1 interface at Asn60 reduces adhesive binding. A, immunoblot analysis of the GPI-anchored SynCAM 2 extracellular sequence and its N60Q mutant expressed in COS7 cells. Lack of the Asn60 N-glycosylation site resulted in the absence of the higher molecular mass glycoforms marked by asterisks. Deglycosylation with PNGase F reduced both wild-type and mutant protein to the same apparent molecular mass predicted for the unmodified protein. Constructs carried a FLAG epitope for detection. B, loss of Asn60 glycosylation promotes homophilic SynCAM 2 binding. COS7 cells expressing GPI-anchored SynCAM 2 or its N60Q mutant carrying an extracellular FLAG epitope (green) were overlaid with the soluble extracellular domain of SynCAM 2 (red). Cells expressing FLAG-tagged, GPI-anchored SynCAM 1 Ig2 + 3 served as a negative control. Construct expression and SynCAM 2 retention were detected as described in Fig. 1B. C, quantification of the results in B. The results are expressed as protein A signal detecting retained SynCAM 2 normalized to the signal of COS7 surface-expressed SynCAMFLAG constructs. COS7 cells expressing GPI-anchored SynCAM 1 Ig2 + 3 or GFP alone served as negative controls. Signals are expressed relative to GFP negative control cells. Retention of SynCAM 2 on cells expressing SynCAM 1 Ig2 + 3 was lower than on GFP-expressing cells for unknown reasons (SynCAM 2, n = 24 cells; N60Q, n = 46; SynCAM 1 Ig2 + 3, n = 39; GFP = 27). ***, p < 0.001. D, loss of Asn60 glycosylation in SynCAM 2 promotes its heterophilic binding to SynCAM 1. COS7 cells expressing FLAG-tagged, GPI-anchored SynCAM 2, or its N60Q mutant (green) were overlaid with the soluble extracellular domain of SynCAM 1 (red). Construct expression and SynCAM 1 retention were detected as described in Fig. 1B. E, quantification of the results in D was performed as described for C (SynCAM 2, n = 25 cells; N60Q, n = 40; SynCAM 1 Ig2 + 3, n = 28; GFP = 28). IB, immunoblot.
To focus our analysis on extracellular interactions, we developed a GPI-anchored SynCAM 2 construct that tethered its extracellular sequence to the outer leaflet of the plasma membrane. This construct maintained a complex glycosylation pattern comparable with that seen for SynCAM 2 expressed in brain (Fig. 5A, lanes 1 and 2). The GPI construct of the SynCAM 2 N60Q mutant, however, lacked the N-glycosylated wild-type fractions above 55 kDa, consistent with selectively reduced glycosylation (Fig. 5A, lanes 3 and 4).
We next analyzed the role of modifications at Asn60 for the adhesive interactions of SynCAM 2 (Fig. 5, B–E). Using a cell overlay approach with soluble proteins, we expressed GPI-anchored SynCAM 2 carrying a FLAG epitope in COS7 cells while overlaying the cells with the soluble extracellular sequence of SynCAM 2. As described above, the COS7 cell expressed protein was labeled with anti-FLAG antibodies and the overlaid soluble protein with protein A. Notably, the absence of a glycan at amino acid 60 of SynCAM 2 strongly increased its interaction with the overlaid extracellular sequence of SynCAM 2, more than doubling its homophilic retention by 125 ± 31% (Fig. 5, B and C). Similarly, the N60Q mutation increased the heterophilic binding of SynCAM 2 to overlaid SynCAM 1 by 61 ± 10% (Fig. 5, D and E). N-Glycosylation at Asn60 of SynCAM 2 Ig1 therefore restricts its adhesive binding to both SynCAM 2 and SynCAM 1, possibly because of steric hindrance of N-glycans or charge repulsion within the binding interface.
Positive Modulation of SynCAM 1 Adhesion by N-Linked Modification of Its Ig1 Domain
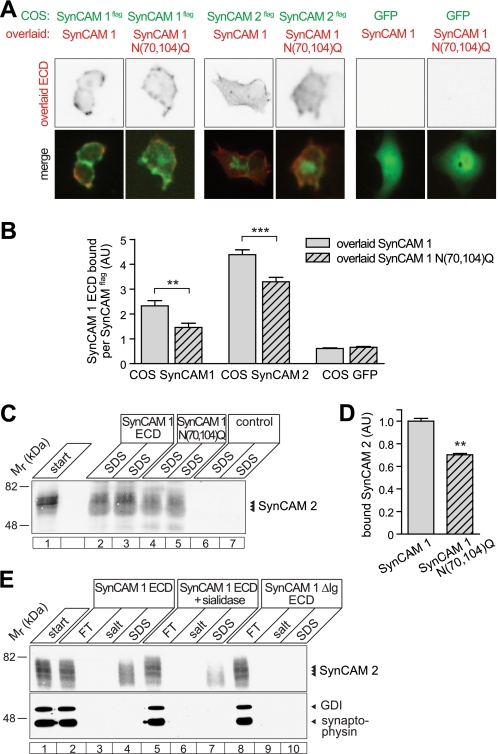
To address whether N-glycosylation within the first Ig domain of SynCAM 1 similarly regulates its adhesion, we generated a SynCAM 1 N70Q,N104Q double mutant. These two sites were selected because our structural models predicted them to flank the SynCAM 1 dimer interface (supplemental Fig. S2B) and because our mass spectrometry data suggested that they were N-glycosylated (Fig. 3). The SynCAM 1 N70Q,N104Q mutant migrated in immunoblots at a lower apparent molecular mass, consistent with reduced N-glycosylation, and was correctly sorted to the plasma membrane (supplemental Fig. S3). A SynCAM 1 N70Q,N104Q,N116Q triple mutant lacking all predicted N-glycosylation sites in the Ig1 domain could not be analyzed because it was not properly sorted to the cell surface (data not shown).
Live cell overlay assays with these GPI-anchored constructs showed that the lack of glycans at the Asn70/Asn104 sites of the Ig1 domain reduced its homophilic binding to wild-type SynCAM 1 by 51 ± 16% (Fig. 6, A and B). Similarly, the N70Q,N104Q mutations decreased the heterophilic binding of SynCAM 1 to wild-type SynCAM 2 by 30 ± 7%. This inhibitory effect of the SynCAM 1 Ig1 N70Q,N104Q mutation on its adhesive interactions contrasted with the increased binding of the SynCAM 2 Ig1 N60Q mutant. We additionally performed affinity chromatographies to analyze the effect of the SynCAM 1 N70Q,N104Q mutation on the retention of SynCAM 2 from brain (Fig. 6, C and D). Our results show that the loss of these two N-glycosylation sites reduces heterophilic binding to SynCAM 2 by 30 ± 3%, in agreement with our cell overlay data.
FIGURE 6.
N-Glycosylation of SynCAM 1 at Ig1 sites Asn70/Asn104 promotes adhesive binding. A, absence of Asn70/Asn104 glycosylation weakens the homo- and heterophilic interactions of SynCAM 1. COS7 cells expressing GPI-anchored SynCAM 1 or SynCAM 2 carrying an extracellular FLAG epitope (green) were overlaid with the soluble extracellular sequence of wild-type SynCAM 1 or its N70Q,N104Q glycosylation mutant (red). Cells expressing soluble GFP served as negative control. Construct expression and retention of soluble SynCAM 1 were detected as described in Fig. 1B. B, quantification of the results in A. The results are expressed as fluorescence intensity of retained SynCAM 1 normalized to the fluorescence intensity of COS7 surface-expressed SynCAMFLAG constructs. COS7 cells expressing GFP alone served as negative controls. **, p < 0.01; ***, p < 0.001. C, lack of SynCAM 1 glycosylation at Asn70/Asn104 reduces binding to brain SynCAM 2. The extracellular SynCAM 1 sequence or the N70Q,N104Q mutant were expressed in COS7 cells as fusions with IgG1-Fc, and equal amounts were immobilized on protein A beads. Retention of solubilized rat brain membrane proteins on the immobilized proteins was analyzed by affinity chromatography. SDS eluates obtained from two parallel affinity bindings are shown. SynCAM 2 signal was detected by quantified immunoblotting. D, quantification of results obtained as in C (n = 3). E, sialic acid modification of SynCAM 1 promotes its heterophilic binding. The SynCAM 1 extracellular sequence was expressed and immobilized as in C and treated without or with sialidase under native conditions. Retention of membrane proteins from rat brain was analyzed by affinity chromatography. Affinity matrices were first eluted with 800 mm potassium acetate and then with SDS. SynCAM 1ΔIg lacking all three Ig domains served as negative control, and the GDP dissociation inhibitor GDI and synaptophysin served as controls for nonspecific binding. SynCAM 2 signal was detected by quantified immunoblotting. FT, flow-through.
These observations raised the possibility that extracellular SynCAM 1 interactions involve the participation of specific carbohydrate types. Because SynCAM 1 is modified with sialic and polysialic acid in brain (20, 45), which we confirmed in our mass spectrometry analysis of the purified protein, we tested whether sialic acids on SynCAM 1 contribute to its heterophilic SynCAM 2 binding. Affinity chromatography of SynCAM 2 extracted from brain was performed on the extracellular domain of SynCAM 1 that was either fully glycosylated (Fig. 6E, lanes 1–4) or from which sialic acids had been removed enzymatically under native conditions (lanes 5–7). A construct lacking all Ig domains served as a negative control (lanes 8–10). The removal of sialic acids from SynCAM 1 reduced its retention of SynCAM 2 by 34 ± 9% (n = 3). Although we did not determine the sites of SynCAM 1 sialylation, this result indicates that SynCAM 2 adhesion may involve specific interactions with sialic acids on SynCAM 1. Alternatively, the negative charge of sialic acids may mediate favorable electrostatic interactions across the Ig1/Ig1 trans-interface, but the observation that the SynCAM 1/2 interaction is resistant to high salt conditions does not support this (Fig. 6E).
N-Glycosylation at the Asn70/Asn104 Sites of SynCAM 1 Ig1 Promotes Synapse Induction
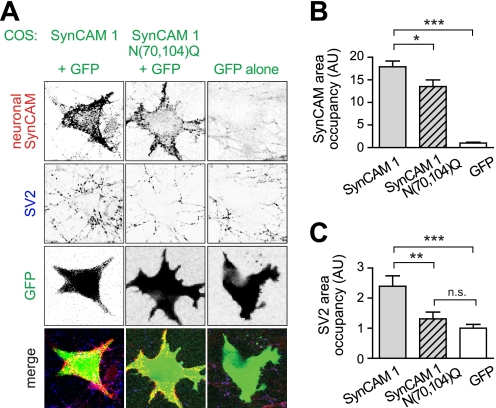
Extending our structural and biochemical analysis of SynCAM 1 N-glycosylation, we asked whether modification at the Asn70/Asn104 sites of SynCAM 1 Ig1 alters its trans-synaptic interactions and synaptogenic function. We expressed the GPI-anchored SynCAM 1 extracellular domain or the N70Q,N104Q mutant in COS7 cells and co-cultured them with hippocampal neurons (Fig. 7A). We then measured two activities, the ability of COS7-expressed SynCAM 1 to recruit neuronal SynCAM proteins upon contact and its induction of presynaptic specializations in contacted neurons (28).
FIGURE 7.
Modification of SynCAM 1 at its N-glycosylation sites Asn70/Asn104 increases trans-synaptic adhesion and synapse induction. A, wild-type SynCAM 1 recruits neuronal SynCAMs and the presynaptic marker SV2 in a mixed co-culture assay. COS7 cells co-expressing GFP with the GPI-anchored SynCAM 1 extracellular sequence or its N70Q,N104Q mutant were seeded atop dissociated hippocampal cultures at 7 days in vitro. COS7 cells expressing GFP alone served as negative control. Co-cultures were analyzed at 11 days in vitro by immunostaining for neuronally expressed SynCAM proteins (red) and the presynaptic vesicle marker SV2 (blue). GFP marked transfected COS7 cells (green). Wild-type SynCAM 1 recruits and retains neuronal SynCAMs, and SV2 puncta were detected atop COS7 cells expressing GPI-anchored SynCAM 1. Cells expressing SynCAM 1 N70Q,N104Q exhibited less SynCAM and no SV2 recruitment. B, quantification of the SynCAM recruitment shown in A (SynCAM 1, n = 30 cells; N70Q,N104Q, n = 34; GFP, n = 23; apply also to C). *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, quantification of the SV2 recruitment shown in A. n.s., not significant.
The recruitment of neuronal SynCAM proteins was determined by quantified immunostaining, taking advantage of the fact that they can be selectively detected using an antibody that does not recognize the GPI-anchored SynCAM constructs. As expected, GPI-SynCAM 1 expressed in COS7 cells efficiently recruited neuronal SynCAMs to contact sites (Fig. 7, A and B). The N70Q,N104Q mutant, however, recruited neuronal SynCAMs 24 ± 11% less, consistent with a weakening of its trans-synaptic adhesion (Fig. 7B). This weakened recruitment of neuronal SynCAMs by SynCAM 1 N70Q,N104Q correlated with its inability to induce presynaptic specializations in neuronal co-cultures (Fig. 7C). The stronger effect of this mutation on synapse induction than on SynCAM recruitment indicates that a select threshold of trans-synaptic SynCAM clustering may have to be met to induce synapses. The N60Q mutant of SynCAM 2 did not further promote the synaptogenic activity of SynCAM 2 in this mixed co-culture assay (data not shown), presumably because the activity of the wild-type protein already saturated the synapse-forming potential of neurons under the overexpression conditions of this approach. Such saturation may compromise the detection of positive modulatory effects. Together, our functional studies demonstrate that modification of the N-glycosylation sites Asn70/Asn104 of SynCAM 1 Ig1 increases both trans-synaptic adhesion and its synaptogenic activity.
DISCUSSION
Our biochemical, crystallographic, mass spectrometry, and cell biological analyses characterize N-glycosylation as a modification that modulates SynCAM adhesion. Our results further indicate roles of SynCAM 1 glycosylation in the regulation of synapse induction. Four lines of evidence support these conclusions. First, crystallographic results and structural modeling show that N-glycosylation can occur within and adjacent to the adhesive Ig1 interface of SynCAM 2 and SynCAM 1, respectively. Second, the glycosylation of these sites within the Ig1 domain differentially affects SynCAM properties, reducing the adhesion of SynCAM 2 while increasing the binding of SynCAM 1. Third, the ability to glycosylate these sites increases not only SynCAM 1 adhesion but also its synaptogenic activity. Fourth, the post-translational modification of SynCAM 1 is developmentally regulated in the brain, suggesting functional roles in vivo.
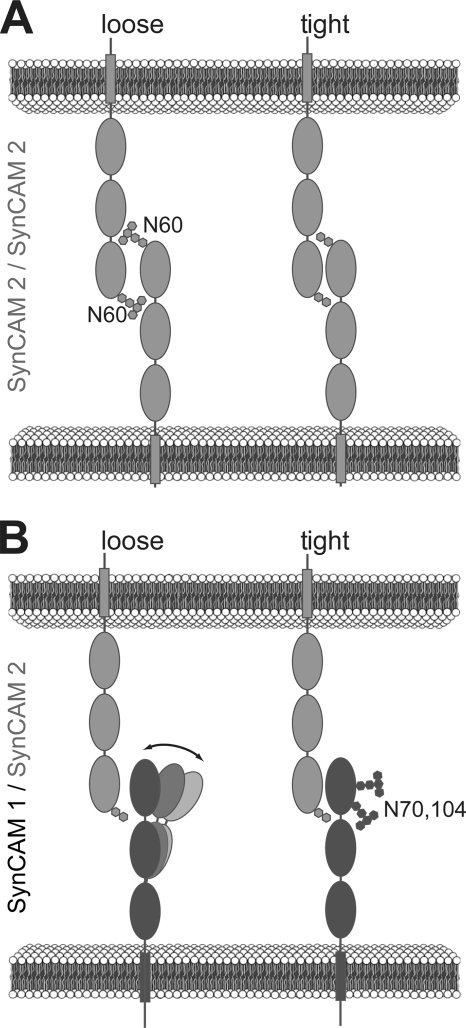
The post-translational modification of synaptic adhesion molecules could be an attractive mechanism to regulate them. Indeed, an inhibitory effect of N-glycosylation on neuroligin 1 binding to neurexin 1β has been previously reported (27). How can N-glycosylation differentially reduce SynCAM 2 Ig1 binding and promote SynCAM 1 adhesion? Our crystal structure of SynCAM 2 Ig 1 and homology model of SynCAM 1/SynCAM 2 Ig1 show that these adhesive trans-dimer interfaces consist mainly of hydrophobic interactions. The location of Asn60 at this SynCAM 2 dimer interface leaves little room for a bulky glycan in the crystal structure at this site. Glycans at Asn60 of SynCAM 2 may therefore weaken the dominant hydrophobic interactions at the dimer interface and reduce SynCAM 2 adhesion (Fig. 8A).
FIGURE 8.
Model of differential SynCAM modulation by N-glycosylation of the first Ig domain. A, N-glycans may reduce SynCAM 2 adhesion through steric hindrance within the Ig1 binding interface. B, in contrast, N-glycans facing away from the SynCAM 1 Ig1 domain may restrict its conformational freedom and position it toward binding. Note that we do not exclude additional interactions between SynCAM Ig domains. Dark gray, SynCAM 1; light gray, SynCAM 2.
In contrast, the glycans at Asn70 and Asn104 in SynCAM 1 do not participate in Ig1 dimer contacts and face away from the adhesive dimer interface. Why then is the N70Q,N104Q mutant deficient in adhesive dimer formation? We consider it possible that N-glycans of SynCAM 1 favor adhesive binding through limiting the conformational space available to the protein or by inhibiting nonspecific protein clustering. Both mechanisms have been previously proposed for other Ig superfamily adhesion proteins (46). Specifically, the glycans on Asn70 and Asn104 may bias or restrict the relative orientations of the SynCAM 1 Ig1 domain to favor adhesive dimer formation, for example by limiting the conformational space available to the Ig1 domain (Fig. 8B).
Our results complement a body of studies characterizing the role of glycosylation for Ig superfamily members. These studies have established that carbohydrates can modulate homophilic adhesion and function, such as shown for L1 and NCAM, and that specific carbohydrate structures on Ig proteins can regulate extracellular interactions as demonstrated for polysialylated NCAM (47–49). Interestingly, a fraction of SynCAM 1 also carries polysialic acids, making it only the second protein next to NCAM that exhibits this modification in the brain (45). This polysialylation of SynCAM 1 occurs at the third N-glycosylation site, which was not analyzed in our study, and may serve as an additional mechanism regulating adhesive strength. Sialic acids can also specify protein interactions as shown for the Siglec family of Ig-like lectins (50, 51). However, SynCAMs do not conform to conserved sequence motif in Siglecs (52) and appear unlikely to belong to this protein family. The potential roles of carbohydrates in binding specificity and carbohydrate-carbohydrate interactions (53) can now be addressed in future studies of adhesive SynCAM recognition.
The significant developmental changes in the post-translational modification of SynCAM 1 indicate that specific, presently unknown glycosyltransferases modify it in the brain. In contrast, only a minor fraction of SynCAM 2 may undergo regulated carbohydrate modification. Functionally, this differential glycosylation could modulate SynCAM interactions between neuronal populations, refining the potential for adhesive coding provided by the distinct SynCAM gene expression patterns (20, 21). The modification of SynCAMs with glycans may not only adjust their synaptic adhesive strength during brain development. Glycosylation could also change the structural organization of SynCAM complexes in the synaptic cleft, analogous to the role of N-glycans in patterning the trans-adhesion arrays formed by L1 (54). Future studies will determine whether glycans on residues other than those analyzed here further modulate SynCAM structure and function, including the O-glycans at the stalk of the SynCAM 1 extracellular domain (19).
With respect to the roles of modulated adhesion, it is interesting to note that the glycosylation sites Asn60 of SynCAM 2 and Asn70 of SynCAM 1 are evolutionarily conserved between human and murine orthologs and that the Asn104 site of mammalian SynCAM 1 is even present in the avian and fish orthologs (19). This indicates that the ability to modify these sites in SynCAM Ig1 domains is functionally relevant. Together, N-glycosylation alters the adhesive interactions and synapse-inducing functions of SynCAMs, demonstrating that this modification modulates trans-synaptic SynCAM interactions. Our findings support the notion that glycosylation plays important roles in synaptic surface interactions (55, 56).
Supplementary Material
Acknowledgments
We thank the members of the Biederer and Modis laboratories for helpful discussions. We also thank Edward Voss (W. M. Keck Foundation Biotechnology Resource Laboratory) and Michael Easterling (Bruker Daltonics, Inc.) for running some of the samples on the FT-ICR MS. We thank Howard Robinson, Annie Héroux, and other staff at the X25 and X29A beamlines of the National Synchrotron Light Source at Brookhaven National Laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DA018928 (to T. B.). This work was also supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease grant (to Y. M.), National Institutes of Health Predoctoral Program in Cellular and Molecular Biology Grant T32 GM007223, and by National Institutes of Health Grant P30 DA018343 from the National Institute on Drug Abuse. Use of the National Synchrotron Light Source is supported by the Offices of Biological and of Basic Energy Sciences of the U.S. Department of Energy.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S3.
- SynCAM
- synaptic cell adhesion molecule
- GPI
- glycosylphosphatidylinositol
- PNGase F
- peptide:N-glycosidase F
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- Pn
- postnatal day n
- FT-ICR
- Fourier transform ion cyclotron resonance.
REFERENCES
- 1.Palay S. L. (1956) J. Biophys. Biochem. Cytol. 2, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray E. G. (1959) J. Anat. 93, 420–433 [PMC free article] [PubMed] [Google Scholar]
- 3.Lucić V., Yang T., Schweikert G., Förster F., Baumeister W. (2005) Structure 13, 423–434 [DOI] [PubMed] [Google Scholar]
- 4.Jin Y., Garner C. C. (2008) Annu. Rev. Cell Dev. Biol. 24, 237–262 [DOI] [PubMed] [Google Scholar]
- 5.Biederer T., Stagi M. (2008) Curr. Opin. Neurobiol. 18, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giagtzoglou N., Ly C. V., Bellen H. J. (2009) Cold Spring Harbor Perspect. Biol. 1, a003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Südhof T. C. (2008) Nature 455, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z. J., Scheiffele P. (2008) Curr Opin Neurobiol 18, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig A. M., Kang Y. (2007) Curr. Opin. Neurobiol. 17, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biederer T., Sara Y., Mozhayeva M., Atasoy D., Liu X., Kavalali E. T., Südhof T. C. (2002) Science 297, 1525–1531 [DOI] [PubMed] [Google Scholar]
- 11.Woo J., Kwon S. K., Choi S., Kim S., Lee J. R., Dunah A. W., Sheng M., Kim E. (2009) Nat. Neurosci. 12, 428–437 [DOI] [PubMed] [Google Scholar]
- 12.Linhoff M. W., Laurén J., Cassidy R. M., Dobie F. A., Takahashi H., Nygaard H. B., Airaksinen M. S., Strittmatter S. M., Craig A. M. (2009) Neuron 61, 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Wit J., Sylwestrak E., O'Sullivan M. L., Otto S., Tiglio K., Savas J. N., Yates J. R., 3rd, Comoletti D., Taylor P., Ghosh A. (2009) Neuron 64, 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko J., Fuccillo M. V., Malenka R. C., Südhof T. C. (2009) Neuron 64, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayser M. S., McClelland A. C., Hughes E. G., Dalva M. B. (2006) J. Neurosci. 26, 12152–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim B. K., Matsuda N., Poo M. M. (2008) Nat. Neurosci. 11, 160–169 [DOI] [PubMed] [Google Scholar]
- 17.Takeichi M. (2007) Nat. Rev. Neurosci. 8, 11–20 [DOI] [PubMed] [Google Scholar]
- 18.Kwiatkowski A. V., Weis W. I., Nelson W. J. (2007) Curr. Opin. Cell Biol. 19, 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biederer T. (2006) Genomics 87, 139–150 [DOI] [PubMed] [Google Scholar]
- 20.Fogel A. I., Akins M. R., Krupp A. J., Stagi M., Stein V., Biederer T. (2007) J. Neurosci. 27, 12516–12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas L. A., Akins M. R., Biederer T. (2008) J. Comp. Neurol. 510, 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dresbach T., Neeb A., Meyer G., Gundelfinger E. D., Brose N. (2004) Mol. Cell. Neurosci. 27, 227–235 [DOI] [PubMed] [Google Scholar]
- 23.Graf E. R., Kang Y., Hauner A. M., Craig A. M. (2006) J. Neurosci. 26, 4256–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boucard A. A., Chubykin A. A., Comoletti D., Taylor P., Südhof T. C. (2005) Neuron 48, 229–236 [DOI] [PubMed] [Google Scholar]
- 25.Chih B., Gollan L., Scheiffele P. (2006) Neuron 51, 171–178 [DOI] [PubMed] [Google Scholar]
- 26.Koehnke J., Jin X., Trbovic N., Katsamba P. S., Brasch J., Ahlsen G., Scheiffele P., Honig B., Palmer A. G., 3rd, Shapiro L. (2008) Structure 16, 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comoletti D., Flynn R., Jennings L. L., Chubykin A., Matsumura T., Hasegawa H., Südhof T. C., Taylor P. (2003) J. Biol. Chem. 278, 50497–50505 [DOI] [PubMed] [Google Scholar]
- 28.Biederer T., Scheiffele P. (2007) Nat. Protoc. 2, 670–676 [DOI] [PubMed] [Google Scholar]
- 29.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W., Carter C. W., Jr. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Dong X., Xu F., Gong Y., Gao J., Lin P., Chen T., Peng Y., Qiang B., Yuan J., Peng X., Rao Z. (2006) J. Biol. Chem. 281, 10610–10617 [DOI] [PubMed] [Google Scholar]
- 32.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 34.Araç D., Boucard A. A., Ozkan E., Strop P., Newell E., Südhof T. C., Brunger A. T. (2007) Neuron 56, 992–1003 [DOI] [PubMed] [Google Scholar]
- 35.Bai Y., Auperin T. C., Tong L. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandel C. R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J. L., Tong L. (2006) Nature 444, 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bork P., Holm L., Sander C. (1994) J. Mol. Biol. 242, 309–320 [DOI] [PubMed] [Google Scholar]
- 38.Kakunaga S., Ikeda W., Itoh S., Deguchi-Tawarada M., Ohtsuka T., Mizoguchi A., Takai Y. (2005) J. Cell Sci. 118, 1267–1277 [DOI] [PubMed] [Google Scholar]
- 39.Maurel P., Einheber S., Galinska J., Thaker P., Lam I., Rubin M. B., Scherer S. S., Murakami Y., Gutmann D. H., Salzer J. L. (2007) J. Cell Biol. 178, 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiegel I., Adamsky K., Eshed Y., Milo R., Sabanay H., Sarig-Nadir O., Horresh I., Scherer S. S., Rasband M. N., Peles E. (2007) Nat. Neurosci. 10, 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper C. A., Joshi H. J., Harrison M. J., Wilkins M. R., Packer N. H. (2003) Nucleic Acids Res. 31, 511–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morelle W., Canis K., Chirat F., Faid V., Michalski J. C. (2006) Proteomics 6, 3993–4015 [DOI] [PubMed] [Google Scholar]
- 43.Fiala J. C., Feinberg M., Popov V., Harris K. M. (1998) J. Neurosci. 18, 8900–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drescher B., Witte T., Schmidt R. E. (2003) Immunology 110, 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galuska S. P., Rollenhagen M., Kaup M., Eggers K., Oltmann-Norden I., Schiff M., Hartmann M., Weinhold B., Hildebrandt H., Geyer R., Mühlenhoff M., Geyer H. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10250–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudd P. M., Dwek R. A. (1997) Crit. Rev. Biochem. Mol. Biol. 32, 1–100 [DOI] [PubMed] [Google Scholar]
- 47.Rutishauser U., Landmesser L. (1996) Trends Neurosci. 19, 422–427 [DOI] [PubMed] [Google Scholar]
- 48.Acheson A., Sunshine J. L., Rutishauser U. (1991) J. Cell Biol. 114, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadmon G., Kowitz A., Altevogt P., Schachner M. (1990) J. Cell Biol. 110, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varki A. (2007) Nature 446, 1023–1029 [DOI] [PubMed] [Google Scholar]
- 51.Crocker P. R. (2002) Curr. Opin. Struct. Biol. 12, 609–615 [DOI] [PubMed] [Google Scholar]
- 52.Zaccai N. R., May A. P., Robinson R. C., Burtnick L. D., Crocker P. R., Brossmer R., Kelm S., Jones E. Y. (2007) J. Mol. Biol. 365, 1469–1479 [DOI] [PubMed] [Google Scholar]
- 53.Bucior I., Burger M. M. (2004) Curr. Opin. Struct. Biol. 14, 631–637 [DOI] [PubMed] [Google Scholar]
- 54.He Y., Jensen G. J., Bjorkman P. J. (2009) Structure 17, 460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleene R., Schachner M. (2004) Nat. Rev. Neurosci. 5, 195–208 [DOI] [PubMed] [Google Scholar]
- 56.Martin P. T. (2002) Glycobiology 12, 1R–7R [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.