Abstract
The mitochondrial adenine nucleotide carrier (Ancp) catalyzes the transport of ADP and ATP across the mitochondrial inner membrane, thus playing an essential role in cellular energy metabolism. During the transport mechanism the carrier switches between two different conformations that can be blocked by two toxins: carboxyatractyloside (CATR) and bongkrekic acid. Therefore, our understanding of the nucleotide transport mechanism can be improved by analyzing structural differences of the individual inhibited states. We have solved the three-dimensional structure of bovine carrier isoform 1 (bAnc1p) in a complex with CATR, but the structure of the carrier-bongkrekic acid complex, and thus, the detailed mechanism of transport remains unknown. Improvements in sample processing in the hydrogen/deuterium exchange technique coupled to mass spectrometry (HDX-MS) have allowed us to gain novel insights into the conformational changes undergone by bAnc1p. This paper describes the first study of bAnc1p using HDX-MS. Results obtained with the CATR-bAnc1p complex were fully in agreement with published results, thus, validating our approach. On the other hand, the HDX kinetics of the two complexes displays marked differences. The bongkrekic acid-bAnc1p complex exhibits greater accessibility to the solvent on the matrix side, whereas the CATR-bAnc1p complex is more accessible on the intermembrane side. These results are discussed with respect to the structural and biochemical data available on Ancp.
Keywords: Mass Spectrometry (MS), Membrane Enzymes, Membrane Proteins, Metabolism, Mitochondria, Mitochondrial Transport, ADP/ATP Carrier, Conformational Dynamics, H/D Exchange, Mitochondrial Carrier
Introduction
Import and export of metabolites across mitochondrial membranes are vital processes that are highly controlled and regulated at the level of the inner mitochondrial membrane. Proteins of the mitochondrial carrier family (MCF)5 are embedded in this membrane, and each member of the family achieves the selective transport of specific metabolites (1). Among these, the ADP/ATP carrier (Ancp) transports ADP into the mitochondrial matrix and exports newly synthesized ATP toward the cytosol. Mainly due to its natural abundance, the ADP/ATP carrier is the best characterized within the MCF (2). The mitochondrial ADP/ATP exchange process can be blocked by two specific inhibitors, namely carboxyatractyloside (CATR) and bongkrekic acid (BA). CATR and BA bind with high affinity to two distinct pre-existing conformations of the carrier referred to as CATR and BA conformations, respectively, resulting in the formation of stable CATR- and BA-carrier complexes. It was suggested that the transition between the CATR and BA conformations is similar to that involved in the ADP/ATP transport (2).
Members of MCF share common features, i.e. similar molecular masses of about 30 kDa, a so-called tripartite organization consisting of three sequence repeats of about 100 amino acid residues each, and the presence of the conserved motif PX(D/E)XX(K/R) in each repeat (1). Additionally, the Ancp have a common signature sequence, RRRMMM, that is absent in other MCF members (3). The intrinsic mechanisms of ADP/ATP transport, nucleotide recognition, and Ancp conformational changes have been widely investigated mainly with the bovine isoform 1 (bAnc1p) and the Saccharomyces cerevisiae isoform 2 (ScAnc2p). The large body of biochemical and biophysical data available demonstrated that CATR- and BA-carrier complexes display distinct structural features (4).
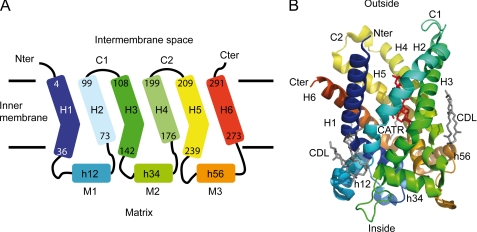
Understanding the mechanism of transport proteins in biological membranes at the molecular level requires high resolution structural information that is usually obtained from x-ray crystallography and/or NMR spectroscopy studies. The three-dimensional structure of bAnc1p locked with CATR has been solved at 2.2 Å resolution by x-ray crystallography (5). In this structure the six transmembrane helices of bAnc1p called H1 to H6 form a cavity with a deep, cone-shaped depression accessible only from the cytosolic side (see Fig. 1). Binding of CATR in the cavity blocks Ancp, and numerous studies have suggested that CATR- and ADP-binding sites overlap at least partially (4). In each odd-numbered helix, the proline of the MCF motif introduces a sharp kink, which is suggested to act as a hinge in the straightening out the helices when bAnc1p is open to the matrix side (5). The connections between the even- and odd-numbered helices are made by the intermembrane space (IMS) loops C1 and C2 and by matrix loops M1, M2, and M3 including short α-helical stretches h12, h34, and h56, respectively (see Fig. 1). The latter are parallel to the membrane surface and strengthen the closed conformation of the CATR-carrier complex on the matrix side (Fig. 1). In contrast to the CATR-bound bAnc1p, the structure of the bovine BA inhibited form still remains unknown. Its characterization, however, would significantly extend our understanding of the ADP/ATP transport mechanism.
FIGURE 1.
Architecture of bAnc1p in the membrane adapted from Pebay-Peyroula et al. (5). A, shown is a schematic diagram of the carrier secondary structure. Transmembrane helices, matrix helices, IMS loops, and matrix loops are labeled H, h, c and M, respectively. The helices comprise the following residues: 4–36 (H1), 53–63 (h12), 73–99 (H2), 108–142 (H3), 156–166 (h34), 176–199 (H4), 209–238 (H5), 253–263 (h56), 273–291 (H6). Odd-numbered helices are kinked by the presence of prolines (Pro27, Pro132, Pro229). B, shown is a ribbon diagram of the carrier viewed from the side. The structure is colored according to the sequence, blue (N terminus) to red (C terminus). Membrane boundaries are drawn in agreement with the hydrophobic segments of the helices. The three-dimensional structure of bAnc1p covers residues 2–293 (Protein Data Bank code 1OCK). Three cardiolipin molecules (CDL) and one molecule of CATR are represented as sticks in gray and red, respectively. The ribbon diagram was drawn with the program PyMOL Version 0.99, DeLano Scientific LLC.
Hydrogen/deuterium exchange (HDX) experiments provide information on the local accessibility of the solvent to proteins. Their monitoring with NMR spectroscopy (HDX-NMR) or mass spectrometry (HDX-MS) has proven particularly valuable in characterizing the structure of partially folded states of soluble proteins during a folding reaction (6–8). HDX experiments have also been recently performed on soluble amphitropic proteins interacting with lipid vesicles, allowing characterization of different conformations of the proteins bound to lipid vesicles and localization of the interacting regions of the proteins at a resolution of a single amino acid residue for HDX-NMR (9–11) or a peptide scale for HDX-MS (9, 12–15). Compared with NMR, MS provides information at lower resolution but has no constraints with respect to the molecular weight of the protein. The resolution can be improved significantly if the protein is digested by different acidic proteases (12, 16–18). Up to now, HDX-MS studies of intrinsic membrane proteins in conditions close to the native state have remained anecdotal because of the difficulty to handle detergents. To overcome this problem, the detergent needs to be separated from the peptide mixture obtained after the proteolysis step. In non-native experiments, strong ionic detergent, such as sodium dodecyl sulfate, could be precipitated by adding potassium salt in the medium (19). However, in native experiments, the detergent used to maintain the membrane protein in solution cannot be easily precipitated. The detergent could be separated from the peptides during the reversed phase chromatography (20, 21) or replaced by liposomes or more recently by lipid nanodiscs that mimic the lipid bilayer (22). Nevertheless, the residual detergent peaks may overlap with the peptides peaks and alter the collected data. To fully remove the detergent, we recently developed a method based on solid-phase extraction with chlorinated solvents (23). Its main advantages are its efficiency, its compatibility with HDX-MS, and the ease of automation.
In this paper we have employed HDX-MS and this new method to study the functional dynamics of an integral membrane protein, bAnc1p, extracted in Triton X-100 by analyzing CATR- and BA-complexes. Not only are our results consistent with previous structural and biochemical work on bAnc1p, but they also provide new insights into the conformational state of the BA-bAnc1p complex in detergent solution and, thus, into the ADP/ATP translocation mechanism across the inner membrane.
EXPERIMENTAL PROCEDURES
Chemicals
BA was prepared as previously described (24). Hydroxylapatite was from Bio-Rad. The polyclonal antibodies used in this work were generated in rabbits against bovine SDS-treated bAnc1p (25). CATR, commercial proteases (pepsin, type XVIII protease, and type XVIII protease), and bovine serum albumin (BSA) were obtained from Sigma. Recombinant type XVIII protease was obtained as described in Rey et al. (16). Trifluoroacetic acid, acetonitrile, and dichloromethane were purchased from Sigma, Carlo Erba Reagenti, and Riedel de Haën, respectively.
Purification of bAnc1p with Triton X-100
The ADP/ATP carrier was isolated from beef heart mitochondria as a CATR-carrier complex or a BA-carrier complex in the presence of Triton X-100 detergent (1% (w/v) final concentration) according to the procedure described by Brandolin et al. (26).
bAnc1p Digestion
All protein digestions in solution were performed in an ice bath at 0 °C. Protease solutions were prepared in 20 mm glycine, pH 2.5, and cooled to 0 °C. The ADP/ATP carrier was digested in 20 mm glycine, pH 2.5, for 2 min using a protease/substrate ratio of 1:1 (w/w) for pepsin and recombinant type XVIII protease and 10:1 (w/w) and 17:1 (w/w) for commercial proteases type XIII and XVIII, respectively. Online digestion of bAnc1p in the presence of 1 m guanidinium chloride (final concentration) was performed in an ice bath at 0 °C using a column packed with pepsin immobilized on a POROS-20AL resin (16).
SDS-PAGE and Western Blotting
The protein concentrations were determined using a BCA protein assay kit (Sigma) and BSA as a standard. For SDS-PAGE, samples were prepared as described in Brandolin et al. (25). Antibodies directed against SDS-treated bAnc1p were used at a 1/1000 final dilution. Immunodetection was performed using HRP-coupled protein A and the enhanced chemiluminescence (ECL) system (Amersham Biosciences).
H/D Exchange of bAnc1p Complexes
After purification and concentration of the ADP/ATP carrier-inhibitor complexes above 5 mg protein/ml, the HDX reaction was initiated by a 10× dilution into deuterated buffer containing 10 mm MOPS-NaOH, 10 mm NaCl, 1 mm EDTA, pH 6.8, and performed at 4 °C. Time-course of the H/D exchange was followed over a 10,000-s period by sequential withdrawing of 80 μl of deuterated samples that were immediately added to 20 μl of quenching buffer (1 m glycine-HCl, pH 2.5), rapidly mixed, and flash-frozen in liquid nitrogen. Samples were stored in liquid nitrogen until they were analyzed.
High Pressure Liquid Chromatography Peptide Separation
Peptides obtained by protein digestion in solution or online procedures were loaded onto a peptide MicroTrap column (Michrom Bioresources, Auburn, CA). Samples were desalted by washing with solution A, and the detergent and lipids were removed (23). After re-equilibration of the trap column, gradient elution was started. The peptides were separated on a C18 reversed phase column (1 × 100 mm, Jupiter; Phenomenex) using a linear gradient from 15 to 40% (v/v) solution B in 20 (deuterated samples) to 40 min (peptide mapping) followed by 40% (v/v) solution B for 5 min at a flow rate of 50 μl·min−1. Before the separation the column was equilibrated with 15% (v/v) B. Solution A was 0.03% (v/v) trifluoroacetic acid in water; solution B was 95% (v/v) CH3CN, 0.03% (v/v) trifluoroacetic acid in water. To minimize back exchange, the valves, trap column, and analytical column were cooled to 0 °C by immersion in an ice-water bath.
MS Analyses
The liquid chromatography tandem MS (LC-ESI-MS/MS) analyses were performed on an ion trap mass spectrometer (Esquire 3000+, Bruker Daltonics). The HPLC system was connected directly through a splitting T-piece to the ESI source of the spectrometer. The settings were described in detail in Rey et al. (23). Data were processed using DataAnalysis 3.0, and MS/MS spectra were searched with MASCOT against single protein data base containing bAnc1p sequence. All assignments were verified manually and by accurate mass measurements. The analyses of deuterated samples and accurate mass measurements were done on a liquid chromatography electrospray time-of-flight MS (LC ESI-TOF-MS) 6210 system (Agilent Technologies). Isotope envelopes were extracted with Mass Hunter Qualitative Analyses software (Agilent Technologies), and the centroid of the isotopic distribution was measured with MagTran software (Zhongqi Zhang, Amgen, Thousand Oaks, CA) (27). Representation of carrier mapping was done with the DrawMap program available online as a part of the software suite MSTools.
RESULTS
Purification and Pepsin Digestion of the bAnc1p
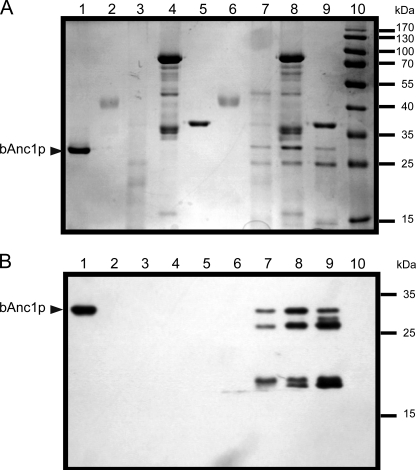
bAnc1p was isolated as a CATR- or BA-carrier complex in Triton X-100 (Fig. 2, lane 1) for which we found a suitable way of removal after the pepsin digestion step. Next, the purified complex was concentrated to a 5 mg/ml protein concentration leading to a detergent concentration close to 10% (w/v). Digestion of bAnc1p as a CATR-carrier complex was first assayed in solution in parallel experiments using three different acidic proteases, and the corresponding lysates were subjected to SDS-PAGE and Western blotting (Fig. 2). Pepsin, protease type XIII, and protease type XVIII from Sigma were used at enzyme/bAnc1p ratios (w/w) of 1, 10, and 17, respectively (Fig. 2, lanes 6–8). The results showed unequivocally that pepsin was the most efficient protease to digest bAnc1p as no residual fragment of a size greater than 15 kDa was detected by SDS-PAGE after Coomassie Blue staining (Fig. 2A, lane 6) and Western blotting using antibodies raised against SDS-treated bAnc1p (Fig. 2B, lane 6). In contrast to pepsin, type XIII and type XVIII proteases were under our experimental conditions unable to digest the carrier completely, and large-size peptides were observed between 15 and 30 kDa (Fig. 2, lanes 7 and 8). Similar results were obtained with the BA-carrier complex (not shown). Increasing the protease/bAnc1p ratio did not significantly improve the digestion of the bovine carrier (not shown). The recombinant type XVIII protease, which we have characterized recently in our laboratory (16), was also evaluated but with no significant improvement (Fig. 2, lanes 9). Combinations of proteases were also tested without better success (not shown). Thus, only pepsin was used in the following experiments. As was shown previously, the use of immobilized proteases offers several advantages in HDX-MS experiments (16, 28, 29). Alongside higher protease stability and the absence of autodigestion, more important advantages are the much higher local concentration of protease leading to better digestion efficiency and the possible use of chaotropic reagent such as guanidinium chloride. Thus, to reach a fast, efficient, and highly reproducible digestion of bAnc1p, we performed online the proteolysis steps using a column packed with immobilized pepsin.
FIGURE 2.
Proteolysis of bAnc1p. Pepsin, type XIII, and type XVIII proteases (commercial and recombinant) were tested, and the corresponding lysates were analyzed by SDS-PAGE (12.5% acrylamide) and revealed by Coomassie Blue staining (A) or Western blot with anti-SDS-bAnc1p (B). Lanes 1–5, 2 μg of bAnc1p, 2 μg of pepsin, 26 μg of type XIII protease, 32 μg of commercial type XVIII protease, and 2 μg of activated recombinant type XVIII protease were incubated in acidic buffer for 2 min at 4 °C. Lanes 6–9, digestion assays were performed with 2 μg of bAnc1p and protease/substrate ratio of 1:1 (w/w) for pepsin, 10:1 (w/w) for type XIII protease, 17:1 (w/w) for commercial type XVIII protease, and 1:1 (w/w) for recombinant type XVIII protease in acidic buffer during 2 min at 4 °C, respectively. Lane 10, prestained molecular weight markers. The position of bAnc1p is indicated by an arrowhead.
Peptide Mapping of bAnc1p Covers Two-thirds of the Carrier
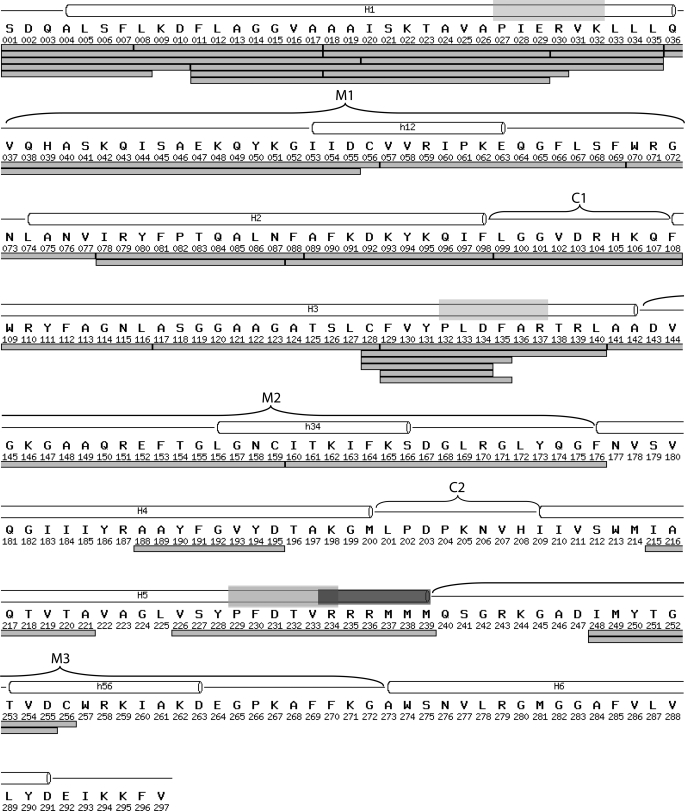
We digested bAnc1p as a CATR- or BA-carrier complex as described above, and LC ESI- MS/MS analyses were performed. On the basis of the MS/MS spectra, we identified 56 peptides covering 94% of the protein sequence (23). An acetylserine at the N-terminal extremity and a trimethyllysine at position 51 were also identified. We did not detect phosphorylation of bAnc1p tyrosines 190 and 194, in contrast to what has been described for isoforms 1 and 3 of the rodent ADP/ATP carrier (30–32). Thus, the bAnc1p primary amino acid sequence we determined was identical to the one obtained by Aquila et al. (33) and, therefore, to the one used to solve the three-dimensional structure of the CATR-bAnc1p complex (5). Protein coverage fell to 72% (37 peptides) in HDX-MS due to a low signal-to-noise or overlapping signals corresponding mainly to the C-terminal part of the carrier (Fig. 3). As a consequence, the N-terminal half of bAnc1p, covering residue 1–176, reached 100% sequence coverage with 32 peptides, some of them overlapping, thus, increasing the spatial resolution of the peptide regions mainly between residues 1 to 35 and residues 128 to 140 (Fig. 3). The other part of the carrier (from residues 177 to 297) presented lower sequence coverage (32%) with only 5 peptides. The digestion pattern gave six pairs of overlapping peptides that differed by only one residue, allowing us to get precise HDX information for the following individual amino acids: Cys56, Phe129, and Cys256 (Fig. 3).
FIGURE 3.
Peptide mapping of bAnc1p after immobilized-pepsin digestion used in HDX-MS experiments. All peptides were identified by ESI-Trap-MS/MS analyses and are represented under the amino acid primary sequence by light gray bars. Transmembrane helices (H1-H6), matrix loops (M1, M2, and M3), encompassing matrix helices (h12, h34, and h56), and IMS loops (C1 and C2) as found in the bAnc1p three-dimensional structure (see the legend of Fig. 1) are shown above the sequence. The PX(D/E)XX(K/R) sequence characteristic of mitochondrial carriers and the RRRMMM signature of the ADP/ATP carriers is indicated respectively as a transparent light gray and dark gray rectangles above the sequence.
HDX Kinetics Performed on the CATR-Carrier Complex Correlate with the Three-dimensional Structure
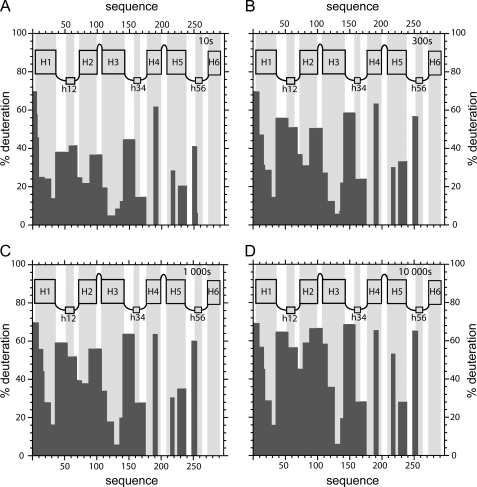
To monitor local HDX kinetics of individual parts of bAnc1p as a CATR-carrier complex, we digested the protein after 3–10,000 s of HDX. Incubation times of 10, 300, 1,000, and 10,000 s were the most representative to show that the deuterium exchange agrees with the organization of the CATR-carrier complex in the detergent micelles (Fig. 4). As bAnc1p has a fold made up of about 70% α-helix and 30% coil, H/D exchange kinetics vary according to the secondary structure. This information was taken into account in our analysis. The results obtained were unambiguous for the first half of the carrier. After only 10 s of HDX, region 1–35 fully covering helix H1 presents deuterium incorporation levels ranging from 14 to 70 % (Fig. 4). These results probably reflected a negative gradient of deuteration from the top to the bottom and can account for the funnel defined by the cavity evidenced in the three-dimensional structure of bAnc1p. Helices H2 (region 71–88) and H3 (118–140) presented behavior similar to that of helix H1 throughout the HDX experiment (Figs. 3 and 4). Moreover, we have shown that regions 20–35 (helix H1) and 110–140 (helix H3) remained strongly shielded from the deuteration (<40% of exchange) even after 10,000 s of HDX (Fig. 4). They include regions 31–35 and 130–135, which contain part of the MCF motif (Fig. 3). Based on the three-dimensional structure, the low HDX rates recorded for these two regions could be explained by their limited accessibility at the bottom of the cavity. Indeed, it should be remembered that H1 and H3 are longer and tightened to close the cavity via several salt bridges between conserved amino acid residues of the MCF motif (5). Interestingly, helix H2 was much less protected from deuteration (Fig. 4), which is consistent with its lower hydrophobicity as compared with helices H1 and H3 (34). Regions 1–8, 37–69, 89–108, and 142–159 (Fig. 3), corresponding to the N terminus, the matrix loop M1, a region including the C1 loop, and the N-terminal half of the matrix loop M2, respectively, are quickly and highly deuterated throughout the time course of the experiment (Fig. 4). Despite the presence of several secondary structures, these results would be consistent with exposure of these regions toward the solvent. In summary, the data collected from HDX account fully for the three-dimensional structure of the bovine carrier complexed to CATR in detergent.
FIGURE 4.
Topography of bAnc1p-CATR complex revealed by HDX experiments. Deuteration levels of peptides obtained from pepsin digestion of the bAnc1p-CATR complex after 10 (A), 300 (B), 1000 (C). and 10000 (D) seconds of HDX are plotted as black bars. Architecture of bAnc1p deduced from the three-dimensional structure (see legend of the Fig. 1) is shown in the panels (gray bars) in agreement with the primary sequence of bAnc1p.
Conformational-dependent Accessibility of Cysteine Residues Analyzed by Local HDX Is in Agreement with Chemical Modification Data
Three cysteine residues, Cys56, Cys159, and Cys256, are located at similar positions in the three matrix short α-helical stretches h12, h34, and h56, respectively (Fig. 3). The fourth (Cys128) is located in the helix H3. Chemical modifications of SH groups with N-ethylmaleimide or eosin-5-maleimide were used to probe conformational changes of bAnc1p (35–37). As confirmed by the three-dimensional structure, Cys128 remained inaccessible to these reagents because its SH group is shielded by the peptide chain of helix H3. N-Ethylmaleimide was shown to target in mitochondria only Cys56 in the presence of BA, whereas CATR prevented its labeling. This might be explained by the low solvent accessibility of Cys56 in the CATR-carrier complex.
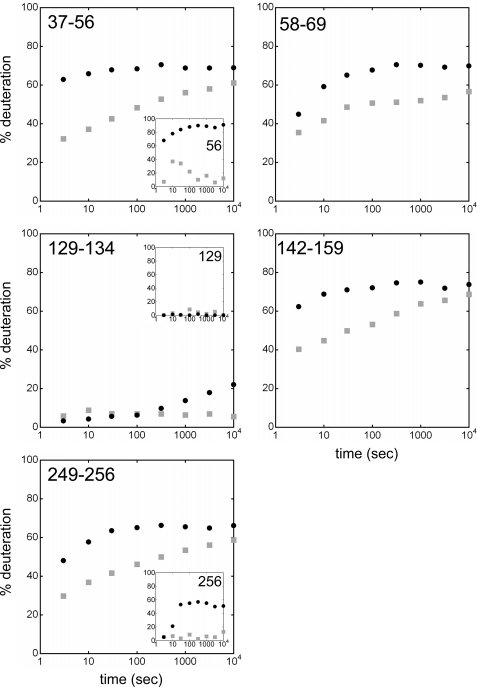
Pepsin digestion led to four pairs of overlapping peptides, i.e. 36–55/36–56, 128–134/129–134 or 128–135/129–135, and 248–255/248–256, which differed from each other by only one residue, i.e. Cys56, Phe129 (close to Cys128), and Cys256, respectively (Fig. 3). We can, therefore, indirectly determine the deuteration rate of the amidic proton of these three amino acids according to the inhibitor used. Deuterium exchange kinetics for regions 37–56 and 249–256 depend on the complex analyzed (Fig. 5). An increase in exchange was observed for both regions in the BA-carrier complex. Analyses of the single residues Cys56 and Cys256 amides showed a higher deuterium uptake in the presence of BA (Fig. 5, insets). In contrast, regions 129–134 and 129–135 were exchanged very slowly in both complex analyzed (Figs. 5 and 6). Deuterium incorporation in Phe129 was, therefore, close to 0 (Fig. 5, inset). In summary, HDX-MS data obtained for the amidic proton of Cys56 and for Phe129 in detergent-solution corroborate the biochemical results published earlier about the labeling of the SH groups of Cys56 and Cys128 in the mitochondrial inner membrane (35, 37). Moreover, the better deuterium labeling of Cys56 and Cys256 assessed by HDX kinetics in the BA-carrier complex suggests the unmasking of theses residues to the solvent, which could be due to a repositioning of helices h12 and h56.
FIGURE 5.
HDX kinetics for bAnc1p matrix loops. The time units for the HDX kinetics are in seconds. Deuterium exchange is expressed as percentages relative to the maximum theoretical deuteration level. HDX kinetics are in gray-filled squares (CATR-carrier complex) and black-filled circles (BA-carrier complex). Inset, HDX kinetics for amide proton 56 (Cys56), 129 (Phe129), and 256 (Cys256) deduced from the couples of overlapping peptides 36–55/36–56, 128–134/129–134 and 248–255/248–256, respectively.
FIGURE 6.
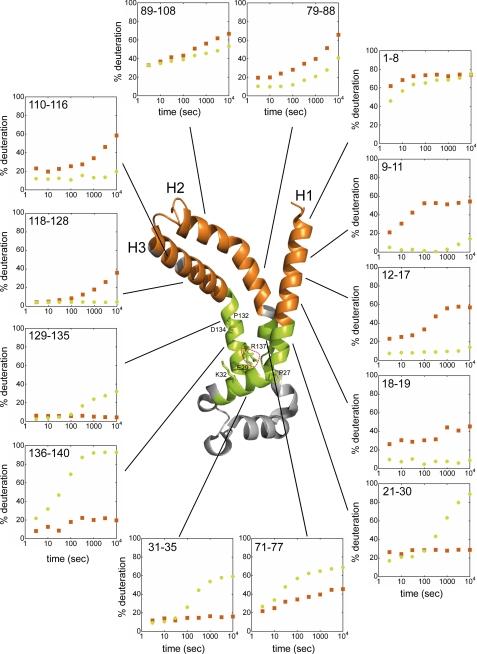
HDX kinetics for bAnc1p helices H1 to H3 mapped on the crystal structure of bAnc1p complexed to CATR. The orange ribbons represent the regions covered by eight peptides showing that the CATR-carrier complex is more exposed to the solvent than the BA-carrier complex at 300 s of HDX, and vice versa for the green ribbon diagrams corresponding to five other regions. Transmembrane α-helices are labeled, and the linker region connecting helices H1 to H2 (loop M1) is colored in gray. Residues involved in the MCF signature motives (Pro27, Glu29, Lys32, Pro132, Asp134, and Arg137) are labeled. The salt bridge Glu29–Arg137 involved in stabilizing the bottom of the cavity is surrounded by red dots. Time units for the HDX kinetics are in seconds. HDX is expressed as percentages relative to the maximum theoretical deuteration level. HDX kinetics for regions coming from the CATR- and the BA-carrier complexes are in orange-filled squares and green-filled circles, respectively.
Topography of the Matrix Loops in bAnc1p Complexed with Either CATR or BA as Assessed by HDX-MS Experiments
Accessibility of the bAnc1p matrix loops to Lys- or Arg-specific proteases was previously investigated in our laboratory (25, 38). Only the BA-carrier complex was cleaved in inside-out submitochondrial particles at Arg30 and/or Arg59, Lys42, Lys146, and Lys244, the CATR-carrier complex being insensitive. These residues, except Arg30, were located in the matrix loops identified in the three-dimensional structure of the CATR-bAnc1p complex (5).
Helix h12 contains Arg59. Residues corresponding to Lys42, Lys146, and Lys244 are located in the N-terminal part of matrix loops M1, M2, and M3, respectively. As we can see in Fig. 3, these parts of bAnc1p were covered partially or totally by the regions 37–56, 58–69, 142–159, and 249–256. Their level of deuteration was found to be quite high after 10,000s of HDX (≥50% exchange) whatever the complex analyzed (Fig. 5), in agreement with their exposure to the solvent. However, consistent with the results described above, all these regions were more deuterium-exchanged in the BA-carrier complex than in the CATR-carrier complex (Fig. 5). Even though structural loss of the matrix helices could partly contribute to this increase of deuterium incorporation, our data provide evidence for the unmasking of the matrix loops (comprising unstructured regions) to the solvent. This could be interpreted as the opening of the carrier toward the matrix in the BA-carrier complex.
Transmembrane Helices Are Involved in the Conformational Dynamics of bAnc1p
HDX experimental analyses gave insights into the conformational dynamics of helices H1, H2, and H3. Analysis of the N terminus of bAnc1p via the region 1–8 showed a strong and fast deuteration (>60% exchange after only 30s) whatever the nature of the complex (Fig. 6). Nevertheless, there was a significant and reproducible time-lag delay in the deuteration of this region in the BA-carrier complex (Fig. 6). The differences in deuteration between the complexes increased in the first transmembrane segment. Indeed, regions 9–11, 12–17, and 18–19 localized in helix H1 presented higher and faster deuteration in the CATR-carrier complex, unlike the BA-carrier complex, which remained weakly deuterated throughout the time course of the experiment (Fig. 6). These results showed that the N-terminal half of helix H1 is much less exposed to the solvent in the BA-carrier complex. Conversely, the C-terminal half of helix H1, which contains the first MCF signature motif (regions 21–30 and 31–35), is more deuterated in the presence of BA (Fig. 6). Helix H3 exhibited behavior similar to that of helix H1 with respect to its level of deuteration in both complexes. The N-terminal half of helix H3 covered by regions 110–116 and 118–128 was more accessible to the solvent in the CATR-carrier complex than in the BA-carrier complex (Fig. 6). Opposite results were found for the C-terminal half, which contains the second MCF signature motif, i.e. the regions 129–135 and 136–140 (Fig. 6).
Helix H2, which is shorter than helices H1 and H3, was covered in our experiment by regions 71–77, 79–88, and 89–108 (Fig. 3). The first region, which corresponds to the N-terminal part of helix H2, was more deuterated in the BA-carrier complex than in the CATR-carrier complex (Fig. 6). Moreover, the deuteration of region 79–88 was higher in the CATR-carrier complex than in the BA-carrier complex (Fig. 6). Detailed analyses of region 89–108 were hampered by its length (20 amino acids) and by the presence of two secondary structures, i.e. a α-helix (the C-terminal half of the helix 2) and the C1 loop. However, for incubation times longer than 100 s, this region was overall more deuterated in the CATR-carrier complex than in the BA-carrier complex (Fig. 6). Consequently, helices H1, H2, and H3 display similar behaviors in the conformational dynamics of bAnc1p in Triton-X100 micelles; first, the regions of these three helices toward the IMS were much more deuterated in the CATR-carrier complex than in the BA-carrier complex (Fig. 6), and second, the deuterium exchange reversal evidenced in regions 21–30, 71–77, and 129–135, was localized at the level of the salt bridges that close the cavity in the CATR-carrier complex (Fig. 6).
The parts of the carrier corresponding to helices H5 and H4 were covered by two peptides and one peptide, respectively (Fig. 3). As reported for helices H1 and H3, the N-terminal half of helix H5, covered partially by region 216–221, presented higher deuteration incorporation throughout the time course of the experiment in the CATR-carrier complex than in the BA-carrier complex (supplement Fig. 1). Surprisingly, the region 227–239 corresponding to the third MCF motif and to the Ancp signature was characterized by a low accessibility to the solvent (∼20–30% exchange) in both complexes (supplement Fig. 1). Thus, this signature motif behaves differently from that of the first two. Furthermore, no difference in solvent accessibility was found after 100 s of HDX for the region 189–195, which corresponds to the C-terminal half of helix H4 (supplement Fig. 1). These two regions do not appear to undergo conformational or accessibility changes. The same suggestion holds for region 161–176, which covers the C-terminal half of the matrix loop M2, just before helix H4 (supplement Fig. 1).
DISCUSSION
The physiological role of Ancp is to export ATP from the mitochondria in exchange for external ADP. Transport takes place once ADP binds to a monomer from the outside and ATP to another monomer from the inside (39). During the transport mechanism, the carrier switches between two different conformations that can be blocked by two toxins: CATR and BA. Therefore, our understanding of the nucleotide transport mechanism can be improved by studying the structural differences between the individual inhibited states. The three-dimensional structure of bAnc1p complexed to CATR shows a cavity that is only accessible from the outside (5). It was suggested that CATR binding occurs at a site identical to or overlapping the ADP-binding site, thus preventing ADP/ATP transport (5). In this work we investigated the bAnc1p conformer complexed to BA using the HDX-MS approach. It was possible to gain insights into the BA-bAnc1p complex by comparing the deuteration levels and kinetics of both complexes.
The large body of evidence available in the literature is consistent with the idea that the CATR- and the BA-carrier complexes correspond to different conformations of the protein (2). However, it is now accepted that the two conformers inserted in the membrane or in detergent solution have basically a similar three-dimensional structure formed by six transmembrane helices and three short helices toward the solvent. The CATR- and the BA-bAnc1p complexes solubilized in Triton X-100 are predominantly monomeric as assessed recently by analytical ultracentrifugation sedimentation velocity experiments (40). Therefore, our results are discussed assuming monomeric structures.
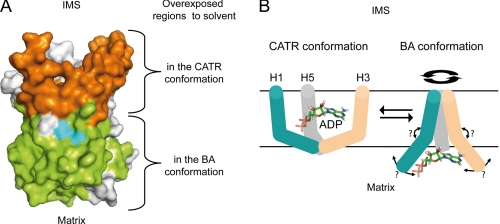
Analysis of our results shows that in the BA-carrier complex, the IMS-exposed side was less accessible than the matrix-exposed side. A reverse situation is observed for the CATR-carrier complex (Fig. 7A). These results might reflect a slight translocation motion of bAnc1p in the membrane, as was suggested 30 years ago from freeze-fracture electron microscopy experiments (26) and more recently from immunological and biochemical data obtained with bAnc1p (25) and ScAnc2p (41, 42). Therefore, the transition from the CATR conformation to the BA conformation, trapping the inhibitors, might be related to a pulling motion of Ancp toward the matrix in presence of BA, the regions of bAnc1p facing the IMS becoming partially buried in the phospholipid bilayers in the BA conformation. The reverse motion would occur when the carrier adopts the CATR conformation. However, this mechanistic model hardly explains why the peptide regions of ScAnc2p equivalent to the loops C1 and C2 facing the IMS in bAnc1p are easily accessible to the solvent in mitochondria in an inhibitor-independent manner (41, 42). Thus, the transition from one conformation to another is probably more complex and not confined to this simple phenomenon.
FIGURE 7.
Topography of bAnc1p as assessed by HDX-MS after 300 s of deuteration and ADP-dependent structural transition between conformers. A, the regions exposed to the solvent in the CATR- or in the BA-carrier complexes are colored in orange or in green, respectively. The regions that display no differences in the deuteration level whatever complex analyzed and the uncovered regions are shown in gray. Conserved signature motives PX(D/E)XX(K/R) are in cyan. B, shown is a hypothetical model of ADP transport mechanism across the mitochondrial inner membrane from the IMS to the matrix mimicked by the BA and CATR conformations. Only the odd-numbered helices are drawn for the sake of clarity, and ADP is represented in stick. ADP is trapped in the cavity of the CATR conformation closed from the matrix side. As a diaphragm, the N-terminal half of the helices H1 (blue), H3 (gray), and H5 (pink) might associate in the BA conformation to close the cavity toward the IMS as a peptide plug. Opening the cavity toward the matrix would then be performed via an undetermined movement of C-terminal half of the odd-numbered helices, thus, breaking the salt bridges, repositioning the matrix loops, and finally, releasing the ADP.
The study described in this paper affords new insights into the conformational transition of bAnc1p. First, it is consistent with the fact that in the CATR conformation the cavity defines a deep cone-shaped depression open only to the IMS. This explains why an increasing gradient of deuteration is observed in this complex for helices H1, H2, and H3 from bottom to top of the cavity. Thus, as we can expect, regions 21–35 (helix H1), 129–140 (helix H3), and 227–239 (helix H5), encompassing the three MCF signature motives, were weakly deuterated in the CATR-carrier complex because of their localization at the bottom of the cavity, which is also closed from the matrix side by a peptide plug of 10 Å (5). Such a gradient of deuteration was not observed in the case of the BA-carrier complex. We showed that the regions of helices H1, H2, H3, and H5 toward the IMS were overall much less open in the BA-carrier complex than in the CATR-carrier complex. These results are in agreement with the lower accessibility to SH reagents of the C-terminal and N-terminal ends of helices H2 and H5 in the BA-ScAnc2p complex assessed by cysteine scanning (41, 42). Taken together, these results suggest a coordinated movement of at least four of the six transmembrane segments hypothesized to close the cavity from the IMS. The limited deuteration (≤20% exchange) measured throughout regions 9–19 (helix H1), 110–128 (helix H3), and 216–221 (helix H5) suggests in the structure of the BA-carrier complex the presence of a peptide plug of ∼15 Å, isolating the cavity from the IMS. However, we have no evidence on the formation of a second network of salt bridges to stabilize the closure of the cavity in the BA conformation as recently proposed (43). Moreover, our data do not seem consistent with the presence in the BA-carrier complex of a cavity similar to that of the CATR-carrier complex but open toward the matrix (44).
Regions 21–35 (helix H1) and 129–140 (helix H3) encompassing the first two MCF motives were much more deuterated in the BA-carrier complex than in the CATR-carrier complex, unlike regions 1–19 (Helix H1) and 110–128 (Fig. 6). These results strongly suggest intramolecular rearrangements of helices H1 and H3, underlining a pivotal role of the first two signature motives in the opening and closing mechanism of the cavity mimicked by the inhibitor-carrier complexes (Fig. 7A). Interestingly, helix H5 distinguishes from the other odd-numbered helices. Indeed, unlike its N-terminal half, which presented a similar behavior to that of helices H1 and H3, the C-terminal half showed a similar accessibility to the solvent whatever the complex analyzed. Although its analysis is limited to two peptide regions (Fig. 3), we can hypothesize that helix H5, which displays the conserved signature RRRMMM of the Ancp, is less flexible than the first three helices H1 to H3 during the conformational transition of the carrier. This hypothesis is reinforced by its greater rigidity compared with the five others helices in bAnc1p, as assessed by the recent molecular dynamics simulation studies performed on the opening of bAnc1p toward the matrix during the ADP transport (45).
In summary, our results are in agreement with conformational movements of the odd-numbered helices, first locking the cavity from the IMS side by a peptide plug and then opening it on the matrix side in the BA conformer and vice versa in the CATR conformer, leading to ADP translocation as depicted in Fig. 7B. This mechanism may be coupled to the disruption of the salt bridge network at the bottom of the cavity in the CATR conformer. As previously hypothesized by Brandolin and co-workers (5), a change would occur in the bend angles that defines the degree of straightening of the odd-numbered helices supported by the presence of conserved prolines in the MCF motives, which could act as hinges. More recently, the structural dynamics of the ADP/ATP carrier revealed by molecular dynamics simulation studies showed that prolines also provide a hinge wobbling motion of the odd numbered helices (46). This suggests a more complex mechanism to close and open the cavity that we could liken to the movement of a diaphragm (Fig. 7B).
The mobility of the three loops connecting the transmembrane helices on the matrix side in bAnc1p seems to be especially high. This was already suggested from probing their basic and cysteine residues (25, 35–38). Our results underline a concomitant rearrangement of loops M1 to M3 in presence of BA for a better solvent accessibility of the cavity to the matrix. Interestingly, amide protons of Cys56 and Cys256 localized in helices h12 and h56, respectively, present a higher deuteration level in the BA-carrier complex. These two short helices probably have a similar position in the matrix but are different enough to allow the only labeling of Cys56, and not that of Cys256, by N-ethylmaleimide or eosin-5-maleimide (35, 37). This observation probably deviates from a symmetrical positioning of helices h12 and h56 with respect to the solvent in the BA-carrier complex, unlike what is described for the CATR-carrier complex (5).
In conclusion, we describe for the first time the study of bAnc1p, a member of the MCF, by HDX-MS, providing new insights into the conformational dynamics of the ADP/ATP carrier through the analysis of the two conformers trapped by CATR and BA. Our results are consistent for the bovine carrier with an ADP translocation mechanism from the IMS to the matrix in which the intrinsic flexibility of the odd-numbered helices would be coupled to a conformational transition of the matrix loops leading to an opening of the cavity toward the matrix. A closing of the cavity on the IMS side is proposed to explain the limited deuterium incorporation of bAnc1p toward this mitochondrial compartment in the BA conformation (Fig. 7B). This step is an important prerequisite to allowing the substrate to cross the membrane without leakage of protons. Although our model sheds light on some features of the ADP/ATP mechanism via the comparative study of the BA and the CATR conformations, the transport mechanism itself and the organization of bAnc1p in the membrane need additional investigation. In this context the study of the conformational dynamics of the carrier in mitochondria in the presence of substrates translocated or not across the mitochondrial inner membrane would be a major step forward.
Supplementary Material
This work was supported by grants from the University Joseph Fourier, the CNRS, and the Commissariat à l'Energie Atomique (programme Signalisation et Transport Membranaires).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- MCF
- mitochondrial carrier family
- CATR
- carboxyatractyloside
- BA
- bongkrekic acid
- IMS
- intermembrane space
- Ancp
- ADP/ATP carrier
- bAnc1p
- bovine isoform 1 Ancp
- ScAnc2p
- yeast isoform 2 Ancp
- HDX
- hydrogen/deuterium exchange
- ESI
- electrospray ionization.
REFERENCES
- 1.Walker J. E., Runswick M. J. (1993) J. Bioenerg. Biomembr. 25, 435–446 [DOI] [PubMed] [Google Scholar]
- 2.Klingenberg M. (2008) Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 3.Klingenberg M. (1989) Arch. Biochem. Biophys. 270, 1–14 [DOI] [PubMed] [Google Scholar]
- 4.Trézéguet V., Pélosi L., Lauquin G. J., Brandolin G. (2008) J. Bioenerg. Biomembr. 40, 435–443 [DOI] [PubMed] [Google Scholar]
- 5.Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G. J., Brandolin G. (2003) Nature 426, 39–44 [DOI] [PubMed] [Google Scholar]
- 6.Wales T. E., Engen J. R. (2006) Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]
- 7.Redfield C. (2004) Methods Mol. Biol. 278, 233–254 [DOI] [PubMed] [Google Scholar]
- 8.Krishna M. M., Hoang L., Lin Y., Englander S. W. (2004) Methods 34, 51–64 [DOI] [PubMed] [Google Scholar]
- 9.Pinheiro T. J., Cheng H., Seeholzer S. H., Roder H. (2000) J. Mol. Biol. 303, 617–626 [DOI] [PubMed] [Google Scholar]
- 10.Halskau Ø., Frøystein N. A., Muga A., Martínez A. (2002) J. Mol. Biol. 321, 99–110 [DOI] [PubMed] [Google Scholar]
- 11.Chenal A., Vernier G., Savarin P., Bushmarina N. A., Gèze A., Guillain F., Gillet D., Forge V. (2005) J. Mol. Biol. 349, 890–905 [DOI] [PubMed] [Google Scholar]
- 12.Man P., Montagner C., Vernier G., Dublet B., Chenal A., Forest E., Forge V. (2007) J. Mol. Biol. 368, 464–472 [DOI] [PubMed] [Google Scholar]
- 13.Hsu Y. H., Burke J. E., Li S., Woods V. L., Jr., Dennis E. A. (2009) J. Biol. Chem. 284, 23652–23661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke J. E., Karbarz M. J., Deems R. A., Li S., Woods V. L., Jr., Dennis E. A. (2008) Biochemistry 47, 6451–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke J. E., Hsu Y. H., Deems R. A., Li S., Woods V. L., Jr., Dennis E. A. (2008) J. Biol. Chem. 283, 31227–31236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rey M., Man P., Brandolin G., Forest E., Pelosi L. (2009) Rapid Commun. Mass Spectrom. 23, 3431–3438 [DOI] [PubMed] [Google Scholar]
- 17.Mazon H., Marcillat O., Forest E., Vial C. (2005) Biochimie 87, 1101–1110 [DOI] [PubMed] [Google Scholar]
- 18.Cravello L., Lascoux D., Forest E. (2003) Rapid Commun. Mass Spectrom. 17, 2387–2393 [DOI] [PubMed] [Google Scholar]
- 19.Joh N. H., Min A., Faham S., Whitelegge J. P., Yang D., Woods V. L., Bowie J. U. (2008) Nature 453, 1266–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busenlehner L. S., Codreanu S. G., Holm P. J., Bhakat P., Hebert H., Morgenstern R., Armstrong R. N. (2004) Biochemistry 43, 11145–11152 [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Chien E. Y., Chalmers M. J., Pascal B. D., Gatchalian J., Stevens R. C., Griffin P. R. (2010) Anal. Chem. 82, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebling C. M., Morgan C. R., Stafford D. W., Jorgenson J. W., Rand K. D., Engen J. R. (2010) Anal. Chem. 82, 5415–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rey M., Mrázek H., Pompach P., Novák P., Pelosi L., Brandolin G., Forest E., Havlícek V., Man P. (2010) Anal. Chem. 82, 5107–5116 [DOI] [PubMed] [Google Scholar]
- 24.Lauquin G. J., Vignais P. V. (1976) Biochemistry 15, 2316–2322 [DOI] [PubMed] [Google Scholar]
- 25.Brandolin G., Boulay F., Dalbon P., Vignais P. V. (1989) Biochemistry 28, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 26.Brandolin G., Doussiere J., Gulik A., Gulik-Krzywicki T., Lauquin G. J., Vignais P. V. (1980) Biochim. Biophys. Acta 592, 592–614 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z., Marshall A. G. (1998) J. Am. Soc. Mass Spectrom. 9, 225–233 [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Kaveti S., Engen J. R. (2006) Anal. Chem. 78, 1719–1723 [DOI] [PubMed] [Google Scholar]
- 29.Hamuro Y., Coales S. J., Molnar K. S., Tuske S. J., Morrow J. A. (2008) Rapid Commun. Mass Spectrom. 22, 1041–1046 [DOI] [PubMed] [Google Scholar]
- 30.Lewandrowski U., Sickmann A., Cesaro L., Brunati A. M., Toninello A., Salvi M. (2008) FEBS Lett. 582, 1104–1110 [DOI] [PubMed] [Google Scholar]
- 31.Feng J., Zhu M., Schaub M. C., Gehrig P., Roschitzki B., Lucchinetti E., Zaugg M. (2008) Cardiovasc. Res. 80, 20–29 [DOI] [PubMed] [Google Scholar]
- 32.Feng J., Lucchinetti E., Enkavi G., Wang Y., Gehrig P., Roschitzki B., Schaub M. C., Tajkhorshid E., Zaugg K., Zaugg M. (2010) Am. J. Physiol. Cell Physiol. 298, C740–C748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aquila H., Misra D., Eulitz M., Klingenberg M. (1982) Hoppe Seylers Z Physiol. Chem. 363, 345–349 [PubMed] [Google Scholar]
- 34.Saraste M., Walker J. E. (1982) FEBS Lett. 144, 250–254 [DOI] [PubMed] [Google Scholar]
- 35.Majima E., Shinohara Y., Yamaguchi N., Hong Y. M., Terada H. (1994) Biochemistry 33, 9530–9536 [DOI] [PubMed] [Google Scholar]
- 36.Majima E., Koike H., Hong Y. M., Shinohara Y., Terada H. (1993) J. Biol. Chem. 268, 22181–22187 [PubMed] [Google Scholar]
- 37.Boulay F., Vignais P. V. (1984) Biochemistry 23, 4807–4812 [DOI] [PubMed] [Google Scholar]
- 38.Marty I., Brandolin G., Gagnon J., Brasseur R., Vignais P. V. (1992) Biochemistry 31, 4058–4065 [DOI] [PubMed] [Google Scholar]
- 39.Block M. R., Vignais P. V. (1984) Biochim. Biophys. Acta 767, 369–376 [DOI] [PubMed] [Google Scholar]
- 40.Nury H., Manon F., Arnou B., le Maire M., Pebay-Peyroula E., Ebel C. (2008) Biochemistry 47, 12319–12331 [DOI] [PubMed] [Google Scholar]
- 41.Kihira Y., Majima E., Shinohara Y., Terada H. (2005) Biochemistry 44, 184–192 [DOI] [PubMed] [Google Scholar]
- 42.Iwahashi A., Kihira Y., Majima E., Terada H., Yamazaki N., Kataoka M., Shinohara Y. (2006) Mitochondrion 6, 245–251 [DOI] [PubMed] [Google Scholar]
- 43.Robinson A. J., Overy C., Kunji E. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17766–17771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmieri F., Pierri C. L. (2010) FEBS Lett. 584, 1931–1939 [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Tajkhorshid E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9598–9603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falconi M., Chillemi G., Di Marino D., D'Annessa I., Morozzo della Rocca B., Palmieri L., Desideri A. (2006) Proteins 65, 681–691 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.