Abstract
Polysialic acid is a developmentally regulated, anti-adhesive polymer that is added to N-glycans on the fifth immunoglobulin domain (Ig5) of the neural cell adhesion molecule (NCAM). We found that the first fibronectin type III repeat (FN1) of NCAM is required for the polysialylation of N-glycans on the adjacent Ig5 domain, and we proposed that the polysialyltransferases recognize specific sequences in FN1 to position themselves for Ig5 N-glycan polysialylation. Other studies identified a novel FN1 acidic surface patch and α-helix that play roles in NCAM polysialylation. Here, we characterize the contribution of two additional FN1 sequences, Pro510-Tyr511-Ser512 (PYS) and Gln516-Val517-Gln518 (QVQ). Replacing PYS or the acidic patch dramatically decreases the O-glycan polysialylation of a truncated NCAM protein, and replacing the α-helix or QVQ shifts polysialic acid to FN1 O-glycans in full-length NCAM. We also found that the FN1 domain of the olfactory cell adhesion molecule, a homologous but unpolysialylated protein, could partially replace NCAM FN1. Inserting Pro510-Tyr511 eliminated N-glycan polysialylation and enhanced O-glycosylation of an NCAM- olfactory cell adhesion molecule chimera, and inserting other FN1 sequences unique to NCAM, predominantly the acidic patch, created a new polysialyltransferase recognition site. Taken together, our results highlight the role of the FN1 α-helix and QVQ sequences in N-glycan polysialylation and demonstrate that the acidic patch primarily functions in O-glycan polysialylation.
Keywords: Carbohydrate Biosynthesis, Glycoprotein Biosynthesis, Glycosylation, Neurobiology, Polysaccharide, Adhesion Molecule, Cell Adhesion, Neural Cell Adhesion Molecule, Polysialylation, Polysialyltransferase
Introduction
The neural cell adhesion molecule (NCAM)2 is a member of the immunoglobulin superfamily. It is expressed on the cell surface where it engages in homophilic and heterophilic interactions, resulting in cell adhesion and modulation of signal transduction cascades (reviewed in Refs. 1, 2). In the developing embryo and neonate, NCAM is modified by the addition of long chains of α2,8-polysialic acid (3, 4). The presence of this highly hydrated and negatively charged carbohydrate polymer disrupts overall cell adhesion and allows cell migration (5–7). Polysialylation is catalyzed by the polysialyltransferases (polySTs), ST8SiaII and ST8SiaIV (8–11). NCAM polysialylation is critical during embryogenesis and early post-natal development. Mice that are null for both polySTs are born at Mendelian ratios but have severe neuronal defects, fail to thrive, and the majority die within 4 weeks of birth (12). Simultaneous deletion of NCAM and the polySTs rescues the lethal phenotype, indicating that polysialic acid is required to down-regulate the adhesive properties of NCAM during development (12). In addition, using varying allelic combinations of ST8SiaIV, ST8SiaII, and NCAM, Hildebrandt et al. (13) demonstrated that an aberrant increase in the level of unpolysialylated NCAM caused defects in brain connectivity in postnatal day 1 mice.
NCAM is mainly unpolysialylated in the adult brain (3, 4). However, polysialylated NCAM does persist in specific regions of the central nervous system, including the olfactory bulb and hippocampus, where it functions in cell migration, synaptic plasticity, and repair (14–17). Highly polysialylated NCAM is also re-expressed in several cancers, including neuroblastoma, glioma, small cell and non-small cell lung tumors, and Wilms' tumor, where it has been suggested to promote cancer invasiveness (18–23). Recently, polysialylated NCAM has been shown to enhance metastasis of a murine model of lung cancer (24) and has been implicated in colorectal carcinoma progression (25). In addition, polysialic acid has been demonstrated to be involved in hematopoietic development (26, 27). For example, ST8SiaIV−/− mice have reduced thymocyte numbers due to fewer progenitor cells mobilizing to the thymus (27).
Although NCAM is by far the most abundant and well documented polysialylated protein, it is striking that only six other polysialylated glycoproteins have been identified. These are the α subunit of the voltage-dependent sodium channel (28), the scavenger receptor CD36 found in milk (29), neuropilin-2 expressed on dendritic cells (30), and the polySTs themselves (autopolysialylation) (31, 32). Recently, polysialic acid was reported to be present on another member of the immunoglobulin superfamily of proteins, the synaptic cell adhesion molecule SynCAM 1 (33). The very small number of polysialylated proteins, and the fact that the polySTs add polysialic acid to N-glycans associated with NCAM much more efficiently than to free N-glycans or other typically unpolysialylated proteins in vitro (34, 35), led to the hypothesis that polysialylation may be a protein-specific modification, requiring an initial interaction between the polyST and protein substrate.
NCAM consists of five immunoglobulin domains (Ig1–5), two fibronectin type III repeats (FN1 and FN2), and either a transmembrane region (TM) and cytosolic tail (NCAM140 and NCAM180) or a glycosylphosphatidylinositol anchor (NCAM120) (36). The majority of polysialic acid is added to N-glycans on Asn449 (Asn5) and Asn478 (Asn6), the fifth and sixth N-glycosylation sites located on Ig5 (37). We demonstrated that a truncated NCAM140 protein consisting of Ig5-FN1-TM-tail is fully polysialylated when co-expressed with a polyST, whereas an Ig5-TM-tail protein remains unpolysialylated (38). In addition, deletion of the FN1 domain from full-length NCAM eliminates Ig5 N-glycan polysialylation (39). From these and other results, we hypothesized that the polySTs recognize and bind the FN1 domain, and this positions them to polysialylate N-glycans on the adjacent Ig5 domain (38–40).
Molecular modeling and structural analysis of the Ig5-FN1 unit has given us insights into the FN1 sequences that may play a role in NCAM recognition (39, 40). Molecular modeling led to the identification of a unique acidic patch on the surface of the FN1 domain (39). Replacing these sequences with alanine residues slightly reduced NCAM polysialylation, and replacing these sequences with arginine residues eliminated polysialylation (39). These results suggested that some or all of the acidic patch residues may be part of a larger polyST recognition region. We solved the crystal structure of NCAM FN1 (40). The structure confirmed the presence of the acidic patch (Asp506, Asp520, Glu521, and Glu523) and also revealed a unique α-helix linking strands 4 and 5 of the FN1 β-sandwich (40). Replacing the α-helix with two threonine residues, as found in the Robo2 FN1 structure, or two alanine residues did not prevent polysialylation. However, polysialic acid was found on FN1 O-glycans rather than on Ig5 N-glycans (40). These data suggested that the α-helix has a role in positioning the polySTs and/or mediating an Ig5-FN1 interaction that allows the polySTs to access Ig5 N-glycans.
To understand the relationship between the NCAM Ig5 and FN1 domains, we solved the crystal structure of the Ig5-FN1 tandem (41). No interaction between the Ig5 and FN1 domains was observed, but surprisingly, Asn5 and Asn6 that carry the N-glycans modified with polysialic acid were not aligned with the FN1 α-helix and acidic patch. To evaluate how flexible the polysialylation process is, we engineered glycosylation sites at different surface positions in the Ig5 domain and evaluated their polysialylation in the absence of Asn5 and Asn6. Remarkably, several of these sites that circled the Ig5 domain and one that was positioned further away from the FN1 domain than Asn5 or Asn6 were polysialylated. These results suggested that, either because the relationship of the two domains is flexible or because the polySTs can engage different sites on FN1, N-glycans at a number of positions can be polysialylated (41).
In this study, to further evaluate the role of the FN1 acidic patch and α-helix in polyST recognition, and to identify other polyST recognition sequences, we decided to compare NCAM FN1 to a similar domain in the unpolysialylated olfactory cell adhesion molecule, OCAM, and then use the OCAM FN1 domain to create a chimeric protein for gain-of-polysialylation studies. We report the identification of two sequences unique to NCAM FN1, Pro510-Tyr511-Ser512 and Gln516-Val517-Gln518, that are particularly important for O-glycan and N-glycan polysialylation, respectively. In the creation of an NCAM-OCAM chimera, we made two surprising observations. First, we found that placing three amino acids between the Ig5 and FN1 domains eliminates Ig5 N-glycan polysialylation. Second, we found that OCAM FN1 can partially replace NCAM FN1 in the chimera to allow the polysialylation of Ig5 N-glycans. Inserting NCAM FN1 sequences (acidic patch, α-helix, PYS, and QVQ) does not substantially enhance the polysialylation of the chimeras, but instead it shifts the addition of polysialic acid to O-glycans. Further dissection of the contribution of inserted NCAM FN1 residues demonstrated that the FN1 α-helix and QVQ sequences are critical for polyST positioning and Ig5 N-glycan polysialylation, whereas the acidic patch and PYS sequences play a primary role in FN1 O-glycan polysialylation.
EXPERIMENTAL PROCEDURES
Tissue culture materials, including Dulbecco's modified Eagle's medium (DMEM), Opti-MEM I, Lipofectin, and fetal bovine serum (FBS), were purchased from Invitrogen. Oligonucleotides, restriction enzymes, PCR supermix, and anti-V5 epitope tag antibody were obtained from Invitrogen. The cDNA for human NCAM140 was a gift from Dr. Nancy Kedersha (Brigham and Women's Hospital, Boston). The cDNA for rat GPI-linked OCAM was provided by Drs. John A. Hamlin and James E. Schwob (Tufts University, Boston). The cDNA for human ST8SiaIV was obtained from Dr. Minoru Fukuda (Burnham Institute, La Jolla, CA). The QuikChangeTM site-directed mutagenesis kit and Pfu DNA polymerase were purchased from Stratagene. DNA purification kits were purchased from Qiagen. Protein A-Sepharose was purchased from GE Healthcare. Protein N-glycosidase F (PNGase F) and T4 DNA ligase were obtained from New England Biolabs. Neuraminidase purified from Clostridium perfringens and neuraminidase purified from Vibrio cholerae were purchased from Roche Applied Science. Phage PK1E endo-N-acylneuraminidase (Endo N) cloned in the pEndo-N expression plasmid was a kind gift from Dr. Eric Vimr (University of Illinois, Champagne-Urbana). Precision Plus ProteinTM standard was purchased from Bio-Rad. Nitrocellulose membranes were purchased from Schleicher & Schuell. Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from The Jackson Laboratories. Supersignal West Pico chemiluminescence reagent was obtained from Pierce. Other chemicals and reagents were purchased from Sigma and Fisher.
Construction of NCAM-OCAM Chimeras
The OCAM FN1 domain was PCR-amplified from rat GPI-linked OCAM using PCR supermix and the following primers: 5′-TCTAGACGATGTCCCCTCTAGTCCCC-3′ and 5′-TCTAGACCCTCACGAACTGGCAGTGTC-3′. These primers introduced an XbaI site at both ends of the amplified OCAM FN1 sequence. An NCAM construct cloned upstream of the epitope tag in the pcDNA3.1 V5/HisB vector, which lacks the FN1 domain (NCAM ΔFN1 (39)) and contains an XbaI site between the Ig5 and FN2 domains, was digested with XbaI and gel-purified. The OCAM FN1 PCR product was cut with XbaI, gel-purified, and ligated into linearized NCAM ΔFN1. In this way, OCAM FN1 was ligated between Ig5 and FN2 of NCAM to form the NCAM-OCAM chimera. The correct orientation of the OCAM FN1 insert was confirmed by DNA sequencing. A consensus glycosylation site within OCAM FN1 was mutated (N562N/T563T/T564A) using the following primers: 5′-GAACCAAATACTGCATATGAAGTCAGGG-3′ and 5′-CCCTGACTTCATATGCAGTATTTGGTTC-3′. The 5′ XbaI site between Ig5 and FN1 and the 3′ XbaI site between FN1 and FN2 of the chimera were subsequently removed using the following primers: 5′-CCTTGTTCAAGCAGATGTCCCCTCTAG-3′/5′-CTAGAGGGGACATCTGCTTGAACAAGG-3′ and 5′-CTGCCAGTTCGTGAGCCCAGTGCACCTAAG-3′/5′-CTTAGGTGCACTGGGCTCACGAACTGGCAG-3′, generating NCAM-OCAM + GLE and NCAM-OCAM + ALD, respectively.
Mutagenesis of NCAM and NCAM-OCAM Chimeras
Insertion and mutagenesis reactions were performed using the Stratagene QuikChangeTM site-directed mutagenesis kit according to the manufacturer's protocol. Insertion of the acidic patch involved replacement of OCAM Lys503 with Asp, Asn517 with Asp, and Lys518 with Glu. Insertion of the α-helix involved replacement of OCAM Lys543 to Ser558 with the corresponding His546 to Gly565 of the β4-αH-β5 strands of NCAM FN1. The PYS and QVQ insertions replaced OCAM Leu507-Ser508-Gln509 and Lys513-Ile514-Ser515, respectively. Primers used for insertions are listed in supplemental Table 1. All replacements and mutations were confirmed by DNA sequencing performed by the DNA Sequencing Facility of the Research Resources Center at the University of Illinois, Chicago.
Transfection of COS-1 Cells with NCAM or NCAM-OCAM Chimera and ST8SiaIV cDNAs
COS-1 cells maintained in DMEM, 10% FBS were plated on 100-mm tissue culture plates and grown at 37 °C, 5% CO2 until 50–70% confluent. Cells were transfected using 30 μl of Lipofectin in 3 ml of Opti-MEM 1 and 10 μg of both V5-tagged NCAM and ST8SiaIV-Myc cDNA, according to the manufacturer's protocol. The NCAM or NCAM-OCAM chimera cDNAs were cloned into the pcDNA3.1 V5/HisB vector. ST8SiaIV cDNA was cloned upstream of the Myc tag in the pcDNA3.1 Myc/HisB vector, in which a stop codon was placed before the His6 coding sequence. Cells were incubated with transfection mixture for 6 h and then 7 ml DMEM, 10% FBS was added to bring the mixture to a final volume of 10 ml.
Immunoprecipitation of NCAM and NCAM-OCAM Chimeric Proteins
Eighteen hours post-transfection, cells were washed with 10 ml of phosphate-buffered saline (PBS) and lysed in 1 ml of immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1% SDS). Lysates were pre-cleared with 50 μl of protein A-Sepharose beads (50% suspension in PBS) for 1 h at 4 °C. NCAM or NCAM-OCAM chimera proteins were immunoprecipitated with 3 μl of anti-V5 epitope tag antibody for 2 h at 4 °C, followed by incubation for 1 h with 50 μl of protein A-Sepharose beads. Beads were washed four times with immunoprecipitation buffer and once with immunoprecipitation buffer containing 1% SDS. For PNGase F treatment, transfections were performed in duplicate, and after immunoprecipitation and washing, identical samples were resuspended in 77 μl of distilled H2O, 10 μl of Nonidet P-40, 10 μl of G7 buffer (0.5 m sodium phosphate, pH 7.5), with or without 3 μl of PNGase F and incubated with shaking at 37 °C overnight. Samples were then resuspended in 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol, heated at 65 °C for 10 min, and separated on a 3% stacking, 5% resolving SDS-polyacrylamide gel. To evaluate relative NCAM and NCAM-OCAM protein expression levels, an aliquot of cell lysate was removed prior to immunoprecipitation, and an equal volume of Laemmli sample buffer, 5% β-mercaptoethanol was added. Samples were boiled at 110 °C for 10 min and separated on a 5% stacking, 7.5% resolving SDS-polyacrylamide gel.
Immunoblot Analysis of Expression and Polysialylation of NCAM and NCAM-OCAM Chimeric Proteins
Following gel electrophoresis, proteins were transferred to a nitrocellulose membrane at 500 mA overnight. Membranes were blocked for 1 h at room temperature in blocking buffer (5% nonfat dry milk in Tris-buffered saline, pH 8.0, 0.1% Tween 20). To detect polysialic acid, membranes were incubated overnight with a 1:50–1:250 dilution of OL.28 anti-polysialic acid antibody in 2% nonfat dry milk in Tris-buffered saline, pH 8.0, and for 1 h with HRP-conjugated goat anti-mouse IgM, diluted 1:4000 in blocking buffer. To test relative NCAM and NCAM-OCAM expression levels, membranes were incubated for 2 h or overnight with a 1:5000 dilution of anti-V5 epitope tag antibody diluted in blocking buffer and for 1 h with HRP-conjugated goat anti-mouse IgG, diluted 1:4000 in blocking buffer. Membranes were washed with Tris-buffered saline, pH 8.0, 0.1% Tween 20 for 15 min two or four times before and after secondary antibody incubation, respectively. Immunoblots were developed using the SuperSignal West Pico chemiluminescence kit and BioExpress Blue Ultra Autorad film.
Pulse-Chase Analysis of NCAM-OCAM Chimera Polysialylation
COS-1 cells were transfected with V5-tagged NCAM-OCAM or mutants along with ST8SiaIV-Myc, as described above. Eighteen hours post-transfection, cell media were removed, and 5 ml of Met/Cys-free DMEM was added to the plates for 1 h. Cells were then labeled with 100 μCi/ml 35S-Express protein labeling mix (PerkinElmer Life Sciences) diluted in 4 ml of fresh Met/Cys-free DMEM. The labeling mixture was removed after a 1-h incubation, and 4 ml of DMEM, 10% FBS, was added to each plate. Following a 3-h chase, media were removed, and cell lysates were collected. NCAM-OCAM proteins were immunoprecipitated with anti-V5 epitope tag antibody as described above. Following immunoprecipitation and washing, proteins bound to protein A-Sepharose beads were subjected to enzyme digestion. To cleave N-linked glycans, PNGase F treatment was performed as described above. To cleave N-linked glycans and α2,8-linked sialic acid chains of 8 units and higher, beads were incubated with 67 μl of distilled H2O, 10 μl of Nonidet P-40, 10 μl of G7 buffer, 10 μl of Endo N, and 3 μl of PNGase F. To digest N-linked glycans and all α2,3-, α2,6-, and α2,8-linked sialic acid, beads were incubated with 67 μl of distilled H2O, 10 μl of Nonidet P-40, 10 μl of G7 buffer, 5 μl of V. cholerae neuraminidase, 5 μl of C. perfringens neuraminidase, and 3 μl of PNGase F. For untreated samples, 80 μl of distilled H2O, 10 μl of Nonidet P-40, and 10 μl of G7 buffer was added to the beads. All samples were incubated overnight at 37 °C, with rotation, and then 50 μl of Laemmli sample buffer containing 5% β-mercaptoethanol was added. Samples were heated at 65 °C for 10 min and resolved on a 3% stacking, 5% separating SDS-polyacrylamide gel. The gel was treated with 10% 2,5-diphenyloxazole in dimethyl sulfoxide to visualize radiolabeled proteins by fluorography and exposure to BioExpress Blue Ultra Autorad film.
RESULTS
In this study we have used the FN1 domain from the unpolysialylated olfactory cell adhesion molecule, OCAM, for both sequence comparisons and gain-of-polysialylation experiments. OCAM (42), also known as Rb-8 neural cell adhesion molecule (RNCAM) (44), NCAM2 (44), or mammalian fasciclin II (mamFasII) (45), was cloned by three groups in 1997 (42–44). It is an adhesion protein and in adults is expressed on the surface of specific neurons within the olfactory system and in other neural tissues, such as the retina (42, 43). There are two isoforms of OCAM, a transmembrane form and a GPI-linked form, and both have the same extracellular domain structure as NCAM (42, 43). In addition, there are consensus N-linked glycosylation sites on OCAM Ig5 in positions equivalent to those that are polysialylated on NCAM Ig5. Notably, OCAM FN1 shares 37% identity with NCAM FN1 and is expressed at some of the same times and in some of the same places as NCAM (42). Despite these features in common with NCAM, OCAM is not polysialylated (42).
Modeling of the OCAM FN1 domain based on the structure of NCAM FN1 predicts that OCAM FN1 lacks the acidic patch, and α-helix that we have shown play roles in the polysialylation of NCAM Ig5 N-glycans. The deposited crystal structures of OCAM Ig4-Ig5-FN1 and Ig4-Ig5-FN1-FN2 confirmed these predictions (PDB codes 1JLK and 1JLL). Comparison of the NCAM and OCAM FN1 domains revealed two regions that are significantly different between the domains. Pro510-Tyr511-Ser512 (PYS) and Gln516-Val517-Gln518 (QVQ) of NCAM FN1 align with Leu507-Ser508-Gln509 and Lys513-Ile514-Ser515 of OCAM FN1 (numbered according to OCAM amino acid sequence, beginning at Met1 of signal sequence) (Fig. 1). We investigated the roles of these sequences in the N-glycan polysialylation of NCAM and the O-glycan polysialylation of a truncated NCAM protein, NCAM7, by mutation and immunoblot analysis.
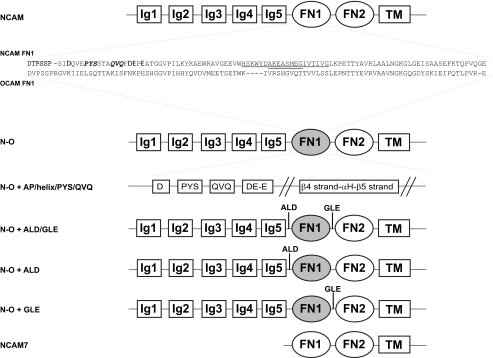
FIGURE 1.
Schematic of NCAM and NCAM-OCAM chimeras used in the study. The FN1 domain of NCAM140 was replaced with OCAM FN1 to generate an NCAM-OCAM chimera. Specific NCAM sequences inserted into the OCAM FN1 domain of the chimera are indicated. Residues of the acidic patch are in boldface, PYS and QVQ are in italicized boldface, and the β4-αH-β5 region is underlined with residues of the α-helix double-underlined. XbaI restriction sites flanking the FN1 domain of the chimera introduced during cloning are Ala-Leu-Asp (ALD) between Ig5 and FN1 and Gly-Leu-Glu (GLE) between FN1 and FN2. These sites were individually removed from the N-O + ALD/GLE chimera to generate N-O + ALD and N-O + GLE. The N-O chimera used in the majority of the work has both restriction sites removed (N-O). A truncated NCAM construct, NCAM7, consisting of FN1-FN2-TM-tail is also shown.
FN1 QVQ Sequence Plays a Role in PolyST Positioning for Ig5 N-Glycan Polysialylation, and FN1 PYS Sequence Is Critical for the O-Glycan Polysialylation of a Truncated NCAM Protein
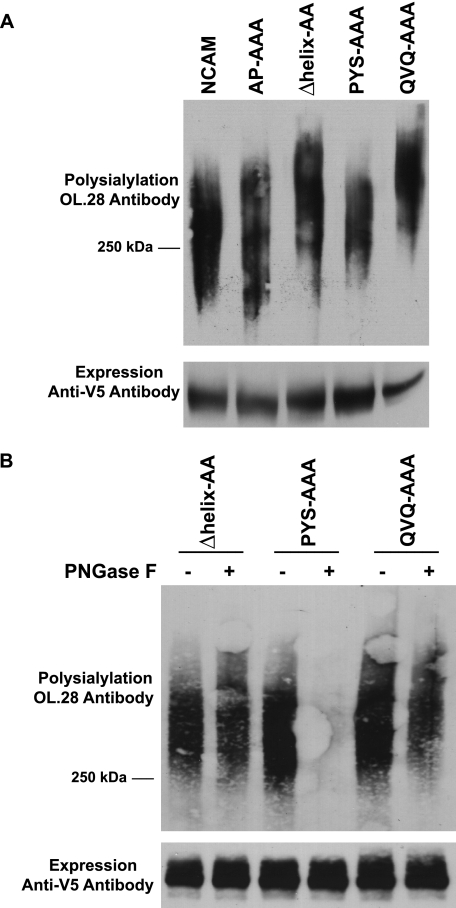
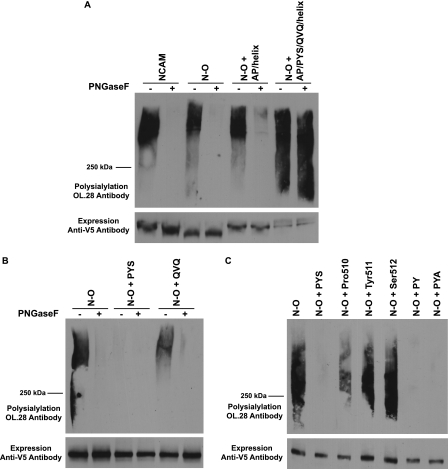
PYS and QVQ were individually replaced with three alanine residues in NCAM. Mutated or wild type NCAM was co-expressed with the polyST, ST8SiaIV, in COS-1 cells. Cell lysates from expressing cells were collected and NCAM proteins immunoprecipitated, and their polysialylation was evaluated by immunoblotting with the anti-polysialic acid antibody, OL.28. Mutation of the core acidic patch residues to alanine had no effect on NCAM polysialylation as observed previously (Fig. 2A) (39). Replacement of PYS led to only a slight decrease in the polysialylation of NCAM (Fig. 2A). However, we noticed that like the Δhelix-AA protein, the QVQ-AAA mutant migrated with a higher molecular mass than NCAM (Fig. 2A). We therefore used PNGase F treatment to determine whether polysialylation was occurring on N- or O-glycans. PNGase F is a glycosidase that cleaves N-glycans from the protein backbone but leaves O-glycans intact (46). We found that replacing QVQ resulted in polysialic acid being added to PNGase F-insensitive O-glycans rather than N-glycans (Fig. 2B), suggesting that QVQ, like the α-helix, may have a role in positioning the polySTs to polysialylate Ig5 N-glycans.
FIGURE 2.
Replacing PYS in NCAM has no effect on polysialylation, and replacing QVQ causes a switch from N- to O-glycan polysialylation. A, COS-1 cells were co-transfected with wild type NCAM or NCAM proteins with AP-AAA, PYS-AAA, QVQ-AAA, or Δhelix-AA mutations and ST8SiaIV-Myc. NCAM proteins were immunoprecipitated from cell lysates with anti-V5 epitope tag antibody. B, COS-1 cells were co-transfected in duplicate with mutated NCAM proteins and ST8SiaIV-Myc. After immunoprecipitation, one sample was treated with PNGase F, to remove N-linked glycans, and the second sample was left untreated. Upper panels, the polysialylation of immunoprecipitated proteins was measured by immunoblotting with the anti-polysialic acid antibody OL.28. Lower panels, relative NCAM protein expression levels were determined by immunoblotting with anti-V5 epitope tag antibody after boiling samples to remove polysialic acid.
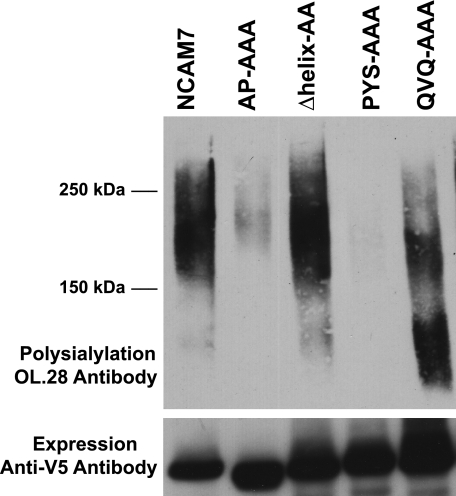
Previous work demonstrated that NCAM7, a truncated NCAM protein lacking all the Ig domains and consisting of the FN1-FN2-TM-tail (Fig. 1), is polysialylated exclusively on O-glycans (38). We also showed that replacing the acidic patch with either alanine or arginine residues substantially decreases or eliminates NCAM7 polysialylation, although replacing the α-helix has no effect (40). We evaluated whether replacing PYS and QVQ in the simpler NCAM7 protein might reveal roles for these sequences not observed in full-length NCAM. Just like replacing the acidic patch, replacing PYS also substantially reduced O-glycan polysialylation (Fig. 3, AP-AAA and PYS-AAA). Replacing QVQ did not eliminate polysialylation but caused polysialylated NCAM7 to migrate as a somewhat smaller molecular mass protein suggesting that this sequence might have some impact on the efficiency of polyST recognition (Fig. 3, QVQ-AAA).
FIGURE 3.
Acidic patch and PYS sequences are required for optimal O-linked polysialylation of the truncated NCAM protein NCAM7. Wild type or mutated NCAM7 proteins were co-expressed with ST8SiaIV-Myc in COS-1 cells. NCAM7 proteins were immunoprecipitated from cell lysates, and polysialylation was evaluated by immunoblotting with the anti-polysialic acid antibody OL.28. Lower panel, the relative expression levels of NCAM7 proteins were determined by immunoblotting with anti-V5 epitope tag antibody.
In sum, these results showed that the acidic patch plays a more major role in the O-glycan polysialylation of NCAM7 than it does in the N-glycan polysialylation of full-length NCAM, where other FN1 domain sequences, and even the Ig5 domain, may be involved in polyST recognition. The striking effect of replacing either the acidic patch or PYS on NCAM7 O-glycan polysialylation was a bit puzzling. It was clear that the presence of the acidic patch could not compensate for the absence of PYS and vice versa and suggested that although both these sequences are required for the polysialylation of O-glycans, they may be functioning in different ways. The absence of a significant effect of PYS replacement on the N-glycan polysialylation of full-length NCAM made us wonder whether the presence of PYS was particularly important for the polysialylation of O-glycans. Finally, these results supported the idea that the QVQ sequence, in addition to having a positioning role in full-length NCAM to ensure Ig5 N-glycan polysialylation, may also have a second role in polyST recognition.
Replacing Either the FN1 α-Helix or QVQ Sequence Leads to Polysialylation of O-Glycans, Including Thr562
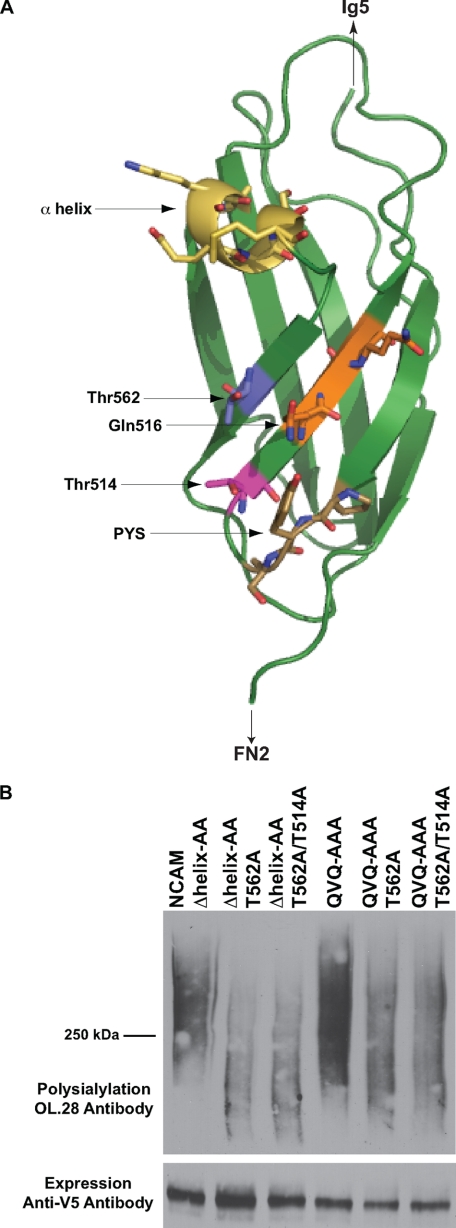
Our results suggest that the FN1 α-helix and QVQ sequence are important for positioning the polyST to modify Ig5 N-glycans. We reasoned that if replacing QVQ or the α-helix causes the same type of positioning defect, then the same O-linked glycans would be polysialylated in both mutant proteins. We previously demonstrated that the sites of polysialylation in the NCAM Δhelix-AA mutant are located in the FN1 domain on O-glycans attached to Thr514, Thr525, and Thr562, with the O-glycan on Thr562 being the major site of modification (40). The structure of the NCAM FN1 domain reveals that both Thr514 and Thr562 are positioned very close to Gln516 of QVQ, and interestingly, Thr562 is located between both the α-helix and Gln516 (Fig. 4A). We mutated Thr562 to alanine in both the Δhelix-AA and QVQ-AAA backgrounds, alone or in combination with a T514A mutation. The T562A mutation caused a dramatic drop in polysialylation of NCAM Δhelix-AA and a noticeable drop in polysialylation of NCAM QVQ-AAA (Fig. 4B). The additional T514A mutation did not cause a further decrease in polysialylation in either protein. The fact that the same O-linked glycan on the FN1 domain is most prominently polysialylated when either the α-helix or QVQ is replaced suggests that both sequences may play a similar role in simultaneously blocking FN1 O-glycan polysialylation and promoting Ig5 N-glycan polysialylation.
FIGURE 4.
O-Glycan on Thr562 is the major site of O-glycan polysialylation for the Δhelix-AA and QVQ-AAA mutants. A, schematic diagram of NCAM FN1 showing the relative positions of Thr514 (pink), Thr562 (blue), PYS (brown), QVQ (orange), and the α-helix (yellow) (41) (PDB code 3MTR). B, COS-1 cells were co-transfected with NCAM Δhelix-AA, NCAM QVQ-AAA, or mutants with indicated threonine replacements and ST8SiaIV-Myc. NCAM proteins were immunoprecipitated from cell lysates with anti-V5 epitope tag antibody, and polysialylation was determined by immunoblotting with OL.28 antibody. Lower panel, relative expressions of NCAM proteins were determined by immunoblotting with anti-V5 epitope tag antibody.
OCAM FN1 Supports Recognition and Polysialylation of an NCAM-OCAM Chimera
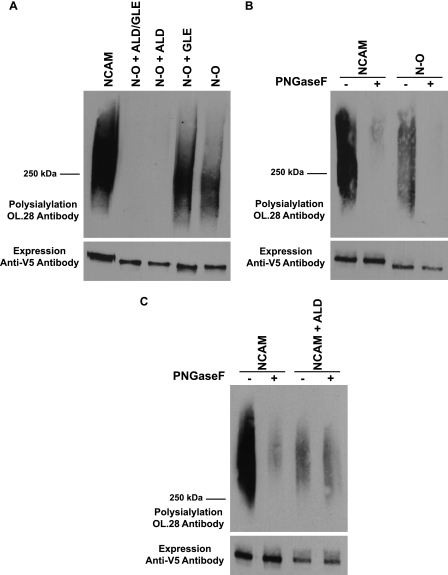
To determine whether the FN1 sequences we identified were sufficient for polyST recognition and NCAM polysialylation, we wanted to create an unpolysialylated NCAM-OCAM chimera in which NCAM FN1 was replaced with OCAM FN1. To remove NCAM FN1 and insert the OCAM FN1 into the NCAM sequence, additional XbaI sites were created that generated the sequence Ala-Leu-Asp between the Ig5 and FN1 domains and the sequence Gly-Leu-Glu between the FN1 and FN2 domains (N-O + ALD/GLE) (Fig. 1). A consensus N-glycosylation site within OCAM FN1 that is not present in NCAM FN1 was eliminated to avoid any potential adverse effect on polyST recognition. Subsequently, either the Ala-Leu-Asp or Gly-Leu-Glu sequences were removed, generating N-O + GLE and N-O + ALD, respectively (Fig. 1). Finally, both sequences were removed to generate a chimera with no linkers flanking the FN1 domain (N-O).
As a measure of the folding of the NCAM-OCAM chimeras, we evaluated their subcellular localization by immunofluorescence microscopy and found that like wild type NCAM, the chimeras were efficiently transported out of the endoplasmic reticulum, through the Golgi, and to the cell surface (data not shown). This suggested that they were not recognized as misfolded by the quality control system of the cell. Next, we determined whether the chimeras could be polysialylated by ST8SiaIV. As shown in Fig. 5A, N-O + ALD/GLE containing both linkers was not polysialylated. However, although removing the Gly-Leu-Glu linker between FN1 and FN2 had no effect (Fig. 5A, N-O + ALD), removing the Ala-Leu-Asp linker between Ig5 and FN1 restored polysialylation (Fig. 5A, N-O + GLE). Additional deletion of the Gly-Leu-Glu linker did not further enhance chimera polysialylation and may have even reduced polysialylation somewhat (Fig. 5A, N-O). PNGase F analysis demonstrated that the N-O chimera is polysialylated on N-linked glycans (Fig. 5B). Elimination of the Asn5 and Asn6 consensus glycosylation sites in N-O demonstrated that glycans on these sites were polysialylated in the chimera (data not shown). We also noticed that the N-O chimera was not polysialylated as efficiently as wild type NCAM, exhibiting only 50% of the polysialylation of the wild type molecule (Fig. 5, A and B). This suggested that NCAM FN1 is more effective in promoting polyST recognition than OCAM FN1. In sum, these results surprisingly demonstrated that the FN1 domain of OCAM allows polysialylation of the NCAM Ig5 N-glycans, albeit more weakly than NCAM FN1, and demonstrate that a precise spacing of the Ig5 and FN1 domains in the chimera is critical for Ig5 N-glycan polysialylation to occur.
FIGURE 5.
FN1 domain of OCAM allows N-linked polysialylation of an N-O chimera, and correct spacing between the Ig5 and FN1 domains is critical for the polysialylation of both NCAM and N-O N-glycans. A, polysialylation of NCAM, N-O, and N-O chimeras with an ALD sequence between Ig5 and FN1, a GLE sequence between FN1 and FN2, or both ALD and GLE insertions were compared. NCAM and N-O proteins were immunoprecipitated from the lysates of COS-1 cells also expressing ST8SiaIV-Myc. B, COS-1 cells were co-transfected in duplicate with NCAM or N-O, along with ST8SiaIV-Myc. After immunoprecipitation, one sample was treated with PNGase F, and the second sample was left untreated. C, lysates from COS-1 cells co-expressing ST8SiaIV-Myc and either NCAM or NCAM with the three amino acids Ala-Leu-Asp inserted between the Ig5 and FN1 domains were either treated with PNGase F or left untreated after immunoprecipitation. Upper panels, the polysialylation of immunoprecipitated proteins was analyzed by immunoblotting with the OL.28 anti-polysialic acid antibody. Lower panels, relative NCAM protein expression levels were determined by immunoblotting with anti-V5 epitope tag antibody.
Spacing between Ig5 and FN1 Is Critical for NCAM Ig5 N-Glycan Polysialylation
To determine whether maintaining a precise spacing of Ig5 and FN1 domains in full-length NCAM is also critical for Ig5 N-glycan polysialylation, and not a specific requirement for N-O chimera polysialylation, we inserted the Ala-Leu-Asp linker between the Ig5 and FN1 domains of NCAM. We found that this eliminated NCAM N-glycan polysialylation while retaining a lower level polysialylation on O-glycans (Fig. 5C, NCAM + ALD). To rule out the possibility that it was the specific amino acids in the linker, rather than changes in spacing between domains, that caused this effect, we inserted an Ala-Ala-Ala linker between Ig5 and FN1 of NCAM and observed even further reduction with retention of O-glycan polysialylation (data not shown). These results demonstrated that a precise relationship between the Ig5 and FN1 domains in NCAM must be maintained to allow the polysialylation of N-glycans on Ig5. They suggest that when Ig5 N-glycans are not accessible to the polySTs due to linker insertion, O-linked glycans on NCAM but not the chimera, are polysialylated.
Inserting NCAM FN1 Sequences into the Polysialylated NCAM-OCAM Chimera Shifts Polysialylation from N-Glycans to O-Glycans
The data in Fig. 5, A and B, demonstrate that OCAM FN1 does not support the polysialylation of the NCAM Ig5 N-glycans to the same extent as NCAM FN1. We considered the possibility that we might be able to enhance the polysialylation of the N-O chimera by inserting NCAM FN1 sequences in OCAM FN1. We found that replacing corresponding OCAM FN1 sequences with the NCAM FN1 acidic patch and α-helix did not enhance N-glycan polysialylation and only weakly enhanced PNGase F-insensitive O-glycan polysialylation (Fig. 6A, N-O + AP/helix). Strikingly, further replacing LSQ and KIS of OCAM with the corresponding PYS and QVQ sequences of NCAM FN1 shifted polysialic acid addition to O-glycans (Fig. 6A, N-O + AP/PYS/QVQ/helix). The polysialylation of the N-O + AP/PYS/QVQ/helix protein was particularly robust compared with other proteins we had seen with O-glycan polysialylation (NCAM7 and NCAM + ALD), and it migrated with a much broader molecular mass that included smaller species in the <250-kDa range not found, or found at lower levels, in these other polysialylated proteins (Fig. 6A).
FIGURE 6.
Insertion of specific NCAM FN1 sequences into the N-O chimera causes a switch from N-linked to O-linked polysialylation. A, COS-1 cells were transfected in duplicate with NCAM, N-O, N-O with NCAM acidic patch and α-helix sequences inserted, or N-O with acidic patch, α-helix, PYS, and QVQ insertions, along with ST8SiaIV-Myc. One sample was treated with PNGase F, and the other sample was left untreated. B, COS-1 cells were transfected in duplicate with N-O, N-O + PYS, and N-O + QVQ proteins along with ST8SiaIV-Myc. One sample was treated with PNGase F, and the other sample was left untreated. C, COS-1 cells were co-transfected with N-O or PYS mutants and ST8SiaIV-Myc. Upper panels, after immunoprecipitation of N-O proteins with anti-V5 epitope tag antibody, polysialylation was measured by immunoblotting with the anti-polysialic acid antibody, OL.28. Lower panels, relative N-O protein expression levels.
Additional analysis of the N-O chimera showed that replacing OCAM FN1 KIS with NCAM FN1 QVQ somewhat decreased the extent of polysialylation but did not significantly change the location of polysialic acid (Figs. 6B and 8A, N-O + QVQ). In contrast, replacing OCAM FN1 LSQ with NCAM FN1 PYS caused a dramatic decrease in N-glycan polysialylation (Fig. 6B, N-O + PYS). Further analysis revealed that replacing OCAM Leu507-Ser508 with NCAM Pro510-Tyr511 is sufficient to cause this dramatic drop in polysialylation (Fig. 6C). Individually replacing OCAM Leu507 with NCAM Pro510 (N-O + Pro510) causes about 50% decrease in N-O polysialylation, whereas replacing OCAM Ser508 with NCAM Tyr511 (N-O + Tyr511) or Gln509 with Ser512 (N-O + Ser512) had little effect. In contrast, replacing OCAM LSQ with NCAM PYS (N-O + PYS) or PYA (N-O + PYA) or replacing OCAM LS with NCAM PY (N-O + PY) all cause a similar dramatic decrease in N-O polysialylation (Fig. 6C).
FIGURE 8.
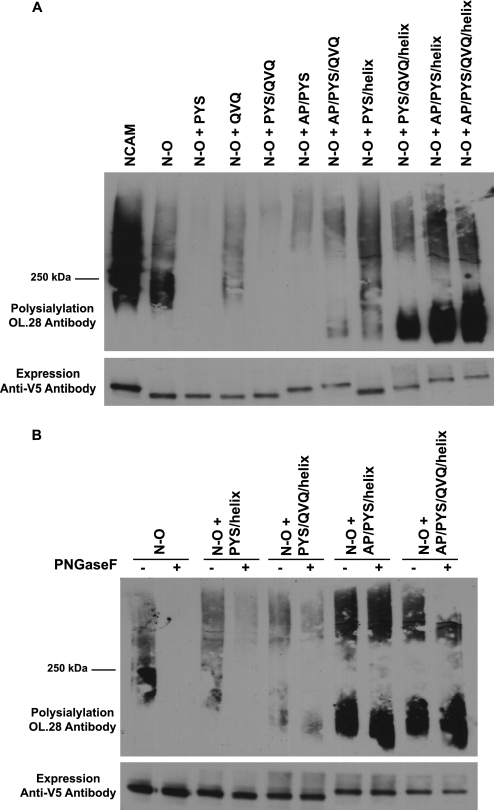
The Acidic patch is largely responsible for the O-linked polysialylation of N-O + AP/PYS/QVQ/helix. A, NCAM or the indicated N-O chimeras were immunoprecipitated from the lysates of COS-1 cells also expressing ST8SiaIV-Myc. B, COS-1 cells were co-transfected in duplicate with N-O or N-O with various combinations of NCAM FN1 sequence insertions, along with ST8SiaIV-Myc. N-O proteins were immunoprecipitated with anti-V5 epitope tag antibody and either treated with PNGase F or left untreated. Upper panels, polysialylation of immunoprecipitated proteins was analyzed by immunoblotting with the anti-polysialic acid antibody OL.28. Lower panels, relative N-O protein expression levels were determined by immunoblotting with anti-V5 epitope tag antibody.
The robust O-glycan polysialylation observed for the N-O + AP/PYS/QVQ/helix chimera was unexpected, as the N-O chimera with or without the ALD linker is not significantly polysialylated on O-glycans (Fig. 5A). Likewise, although full-length NCAM exhibits some weak O-glycan polysialylation that is slightly enhanced in the presence of the ALD linker, it is not observed at the same level as the N-glycan polysialylation of the wild type protein (see Fig. 5C, + PNGase F). Taking these observations into consideration, we predicted that inserting Pro510-Tyr511 into the OCAM FN1 domain in the N-O chimera not only disrupted OCAM FN1 polyST recognition, but also may have allowed additional O-glycans to be added or existing O-glycans to become more accessible to the polyST.
The N-O Chimera Containing NCAM FN1 PYS Sequences Is Modified by Additional Sialylated O-Glycans
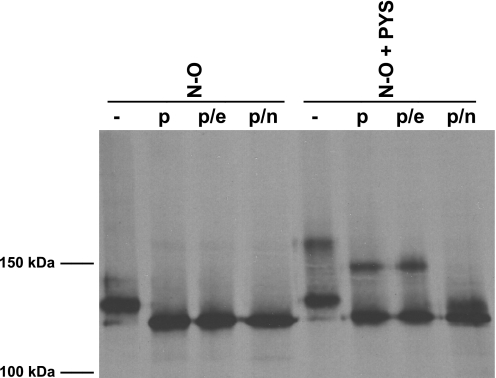
To test the idea that inserting PYS into the N-O chimera may have changed its O-glycosylation, we evaluated the glycosylation status of the N-O and N-O + PYS chimeras following co-expression with ST8SiaIV. Expressing cells were labeled with 35S-Express protein labeling mix for 1 h and chased with media containing unlabeled amino acids for 3 h. N-O proteins were immunoprecipitated using the anti-V5 epitope tag antibody. The types of glycans found on the immunoprecipitated proteins were evaluated using PNGase F, Endo N (removes α2,8-polysialic acid) (47), and a combination of neuraminidases that remove α2,3-, α2,6-, and α2,8-linked sialic acid. Treated and untreated proteins were subjected to SDS-PAGE and fluorography, as described under “Experimental Procedures.”
As observed previously, transient co-expression of ST8SiaIV and NCAM proteins leads to a partial polysialylation of the NCAM proteins due to the absence of the polyST from some cells and the incomplete processing of the N-glycans in the overexpressed proteins (41). This was even more pronounced for the N-O chimera that exhibits lower polysialylation than wild type NCAM (Fig. 7, N-O (−)). Nevertheless, what was most striking was the difference in molecular mass of the N-O and N-O + PYS proteins (Fig. 7). The N-O chimera migrated as a 130-kDa band that collapsed upon PNGase F treatment to ∼110–115 kDa and remained at that molecular mass following PNGase F + Endo N treatment, and PNGase F + combined neuraminidase treatment (Fig. 7, N-O). In contrast, N-O + PYS migrated as two bands as follows: one band of ∼130-kDa molecular mass that is identical to the major unpolysialylated band observed for the N-O chimera, and a second higher molecular mass band of 160 kDa. PNGase F treatment of the N-O + PYS protein led to a collapse of the 160-kDa band to 140–145 kDa and a collapse of the 130-kDa band to ∼110–115 kDa. Although the 130-kDa bands of both the N-O and N-O + PYS proteins appeared similarly modified by N-glycans, the 160-kDa band contained these N-glycans and was additionally modified. Treatment with both PNGase F and Endo N, expected to remove not only N-glycans but also any polysialic acid attached to O-glycans, did not decrease the molecular mass of N-O + PYS suggesting that O-glycan α2,8-polysialylation was not responsible for the molecular mass increase (Fig. 7, N-O + PYS, p/e). In contrast, PNGase F and combined neuraminidase treatment did decrease the molecular mass of the larger band to 115–120 kDa suggesting that O-glycans modified with α2,3- or α2,6-linked sialic acids contribute substantially to the increase in molecular mass for a population of the N-O + PYS protein (Fig. 7, N-O + PYS, p/n). We cannot completely rule out that ST8SiaIV may add mono-, di-, or short α2,8-oligosialic acid chains to O-glycan termini that would be removed by the combined neuraminidases but not Endo N. However, we observed a similar high molecular mass band of N-O + PYS in cells not expressing ST8SiaIV, suggesting that this is not likely (data not shown). Because we did not observe polysialylated O-glycans in the N-O + PYS chimera, we conclude that PYS itself is not promoting polysialylation per se, but instead is causing an increase in sialylated O-glycans on the N-O + PYS chimera. Some or all of these glycans are then polysialylated when other NCAM FN1 sequences are present and allow engagement by the polyST.
FIGURE 7.
Pulse-chase analysis reveals that inserting PYS into N-O leads to increased levels of sialylated O-glycans. COS-1 cells expressing N-O or N-O + PYS along with ST8SiaIV-Myc were labeled for 1 h with 35S-Express protein labeling mix followed by a 3-h chase in unlabeled media. N-O proteins were immunoprecipitated from cell lysates with anti-V5 epitope tag antibody and either left untreated or incubated with PNGase F to remove N-glycans (p), PNGase F and Endo N to remove N-glycans and all α2,8-polysialic acid (p/e), or PNGase F and neuraminidases to remove N-glycans and all sialic acid (p/n). Samples were resolved by SDS-PAGE and visualized by fluorography.
The α-Helix and Acidic Patch Are Primarily Responsible for Promoting Polysialylation in the N-O + AP/PYS/QVQ/Helix Chimera
To further dissect the contributions of the acidic patch, α-helix and QVQ sequences to N-O + PYS chimera O-glycan polysialylation, we tested different combinations of these sequences in the N-O + PYS background. We found that the presence of QVQ, the acidic patch, or both did little to increase the polysialylation of the N-O + PYS chimera (Fig. 8A, N-O + PYS/QVQ, N-O + AP/PYS, N-O + AP/PYS/QVQ). However, an N-O + PYS/helix chimera exhibited a substantial increase in polysialylation that included the lower molecular mass species (<250 kDa) observed in the N-O + AP/PYS/QVQ/helix chimera in Fig. 6A. Addition of QVQ to the N-O + PYS/helix protein decreased the higher molecular mass polysialylated species while enhancing the <250-kDa lower molecular mass species. In contrast, when the acidic patch was added to the N-O + PYS/helix chimera, both high and low molecular mass species were enhanced, and the lower molecular mass polysialylated species was further enhanced when the QVQ sequences were then added to generate the complete N-O + AP/PYS/QVQ/helix chimera (Fig. 8A).
PNGase F treatment demonstrated that in the presence of the α-helix, the PYS-containing chimera was primarily polysialylated on N-glycans, although there was also an increase in O-glycan polysialylation compared with N-O (Fig. 8B, N-O + PYS/helix). QVQ allowed additional low molecular weight O-glycan polysialylation (Fig. 8B, N-O + PYS/QVQ/helix). Interestingly, the further addition of the acidic patch resulted in a striking enhancement of O-glycan polysialylation of both chimeras (N-O + AP/PYS/helix and N-O + AP/PYS/QVQ/helix). Taken together, the α-helix supports both N- and O-glycan polysialylation of the chimera in the presence of PYS, whereas the acidic patch is the major mediator of robust O-glycan polysialylation. These results highlight the importance of the PYS and acidic patch sequences in O-glycan polysialylation and correspond with the observed decrease or elimination of O-glycan polysialylation when these sequences are replaced in NCAM7 (Fig. 3).
Analysis of Potential Sites of O-Glycan Polysialylation in the N-O + AP/PYS/QVQ/Helix Chimera
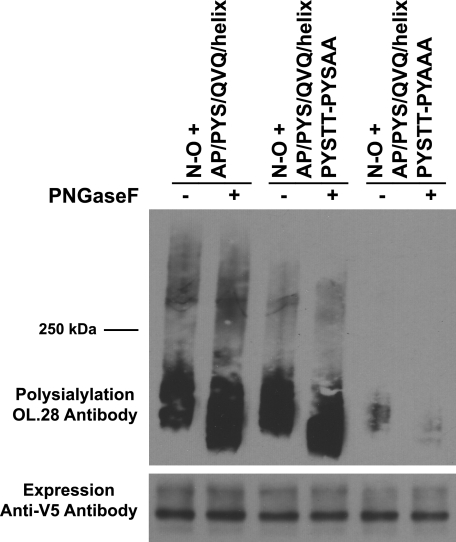
We reasoned that the changes in O-glycosylation and polysialylation may have occurred close by the PYS insertion. Indeed, O-glycosylation sites are frequently found near proline residues (48). We first focused on potential O-glycosylation sites in the loop adjacent to the PYS sequences. Within this loop, PYS is followed by Ser513-Thr514 in NCAM FN1 and two threonine residues in OCAM FN1. We found that replacing the two threonines with alanine residues in the N-O + AP/PYS/QVQ/helix chimera only slightly decreased the higher molecular mass O-linked polysialylated species (Fig. 9, N-O + AP/PYS/QVQ/helix PYSTT-PYSAA). However, additionally replacing the serine residue contributed by PYS caused a dramatic drop in total O-glycan polysialylation, suggesting that the bulk of polysialic acid is found on an O-glycan on Ser512.
FIGURE 9.
Ser512 of PYS is a major site of polysialylation in the N-O + AP/PYS/QVQ/helix chimera. Cell lysates from COS-1 cells expressing N-O + AP/PYS/QVQ/helix or its serine/threonine mutants along with ST8SiaIV-Myc were collected, and N-O proteins were immunoprecipitated with anti-V5 epitope tag antibody. Samples were either subject to PNGase F digestion or left untreated, and polysialylation was determined by immunoblotting with the OL.28 antibody. Lower panel, immunoblot analysis of relative N-O protein expression levels.
DISCUSSION
In this study, we further investigated the role of the FN1 domain in the protein-specific polysialylation of NCAM and provided evidence that its relationship to the Ig5 domain, as well as specific sequences within the domain, plays roles in both N- and O-glycan polysialylation. The most surprising finding of these studies was that OCAM FN1 could replace NCAM FN1 to promote polysialylation of the N-O chimera. Because OCAM itself is not polysialylated, this implies that other aspects of the OCAM protein are preventing polysialylation. It also suggests that sequences common to NCAM and OCAM FN1 domains may be required for polysialylation and are recognized by the polySTs. We are currently evaluating these possibilities.
Inserting a three-amino acid linker between the Ig5 and FN1 domains of both NCAM and N-O dramatically reduced polysialylation, although some O-linked polysialylation of NCAM did occur. The most straightforward explanation is that increasing the distance between the domains disrupts polyST access to N-glycans on Ig5, which would suggest a relatively rigid relationship between Ig5 and FN1. We recently solved the crystal structure of NCAM Ig5-FN1, and although we observed some flexibility in the location of N-glycans that can be polysialylated (41), the relatively short linker region and the fact that the insertion of three amino acids has such a profound effect on polysialylation would seem to imply minimal flexibility. In contrast, the structures of OCAM Ig4-Ig5-FN1 (PDB code 2JLK) and Ig4-Ig5-FN1-FN2 (PDB code 2JLL) differ in the lengths of the unstructured region between the Ig5 and FN1 domains, and they exhibit a sharp difference in the bend of the linker region between the domains depending on the presence of FN2 in the structure. This suggests that the relationship between the Ig5 and FN1 domains in OCAM is somewhat flexible. Although this may in part explain the lack of OCAM polysialylation, we cannot rule out the possibility that the NCAM Ig5-FN1 relationship might be different in the presence of additional domains.
Comparing the sequence of NCAM FN1 to OCAM FN1, we identified two nonconserved regions, PYS and QVQ, that play roles in O-glycan polysialylation and polyST positioning, respectively. Replacing QVQ, like replacing the α-helix, shifts polysialic acid addition to O-glycans on FN1 with the major site of polysialylation in both cases being an O-glycan on Thr562. Based on the location of these sequences and the location of Thr562, we suggest that both the α-helix and QVQ function to block the access of the polyST to this glycan, which may be the better or closer acceptor if available.
The role of PYS was initially more difficult to explain but was later facilitated by our studies of the N-O chimera. We observed little change in NCAM polysialylation when PYS was replaced. In contrast, replacing PYS essentially eliminated O-glycan polysialylation of NCAM7. Further analysis of an N-O chimera plus and minus NCAM FN1 sequences suggested that PYS and, more specifically, the Pro510-Tyr511 sequence functions to maintain a particular conformation of the FN1 domain that eliminates N-O N-glycan polysialylation and promotes the generation of sialylated O-glycans in N-O. How did these changes occur? PYS was inserted at the carboxyl terminus of the first strand of OCAM FN1 (Fig. 4A), which is immediately adjacent to an unstructured region, including the Ig5-FN1 junction. One possibility is that the presence of Pro510-Tyr511 caused a conformational change in the first strand of OCAM FN1 that altered the Ig5-FN1 interface and possibly made the Ig5 N-glycans inaccessible to the polyST engaged with the FN1 domain. Alternatively, this possible conformational change could have altered the site in OCAM FN1 that was being recognized by the polyST. In addition, the structure of the loop between the first and second strands of FN1 could have been altered promoting O-glycosylation of the serine and threonine residues in the loop and/or enhanced the terminal sialylation of existing O-glycans.
O-Glycan polysialylation is not unique to the N-O + AP/PYS/QVQ/helix chimera. It was first observed on salmonid egg polysialoglycoprotein, PSGP (49), and later found on neuropilin-2, CD36, unidentified proteins in the RBL-1 rat basophilic leukemia cell line, and several NCAM mutants, such as those lacking the FN1 α-helix or QVQ sequences, and the truncated NCAM protein NCAM7 (29, 30, 38, 40, 50). In addition, we frequently observe a small amount of O-glycan polysialylation in full-length NCAM (see Fig. 5). Our earlier work and this study support a function for the acidic patch in polyST recognition and O-glycan polysialylation. In NCAM7, Asp520, Glu521, and Glu523 of the acidic patch appear to be a primary recognition site for O-glycan polysialylation, and in NCAM they may be part of a recognition site or one of multiple recognition sites for N-glycan polysialylation. We have previously demonstrated that replacing the acidic patch with alanines severely disrupts N-linked polysialylation of a truncated Ig5-FN1-TM-tail construct (39). It appears that the polySTs interact with the acidic patch within FN1 in a different manner depending on what other domains (Ig5 or FN2) are present. However, in the context of full-length NCAM, the acidic patch has a lesser role. We show in this work that the acidic patch residues (including Asp506) function in enhancing the O-polysialylation of both the N-O + AP/PYS/helix and N-O + AP/PYS/QVQ/helix chimeras, consistent with the acidic patch playing a larger role in O-glycan polysialylation.
The deposited OCAM domain structures (PDB codes 2JLK and 2JLL) demonstrate that the NCAM and OCAM FN1 domains are very similar except for the absence of the α-helix in OCAM FN1. Despite this similarity in structure, the two domains are only 37% identical. OCAM engages in homophilic anti-parallel interactions in cell aggregation assays (42). NCAM is believed to engage in not only homophilic anti-parallel interactions to mediate cell adhesion but also parallel homophilic and heterophilic interactions (reviewed in Ref. 1). For example, NCAM interacts with the fibroblast growth factor receptor (FGFR), promoting signaling events that lead to neurite outgrowth (51, 52). In non-neuronal cells, the binding of soluble NCAM to FGFR1 has been shown to induce receptor internalization and recycling, thereby prolonging signaling and promoting cell migration (53). Two peptides derived from the NCAM FN1 and FN2 domains have been identified that can stimulate FGFR signaling and neurite outgrowth (52, 54). The putative FGFR activation motif (FRM) within NCAM FN1 includes residues Asp506 of the acidic patch, PYS, and Gln516 of QVQ (55), residues not conserved in OCAM FN1. Therefore, specific residues in NCAM FN1 appear to have a dual function, in both binding FGFR and mediating or modulating NCAM modification. These roles are not evolutionarily conserved in OCAM, and this may have led to distinct structural differences between these domains.
In conclusion, our results have highlighted the importance of proper Ig5-FN1 spacing in NCAM Ig5 N-glycan polysialylation and revealed the contributions of four NCAM sequences to N-glycan and O-glycan polysialylation. QVQ, like the α-helix, is required for Ig5 N-glycan polysialylation. For both sequences, this is likely achieved by positioning the polyST to polysialylate N-glycans and at the same time preventing the polysialylation of a nearby O-glycan. PYS maintains a conformation of the FN1 domain that promotes the generation of sialylated O-glycans, and its presence is particularly important for O-glycan polysialylation. Unexpectedly, our results strongly indicate that the FN1 acidic patch residues are primarily responsible for the polysialylation of FN1 O-glycans, leaving us to consider which sequences mediate polyST recognition leading to Ig5 N-glycan polysialylation. The discovery that OCAM FN1 can replace NCAM FN1 to allow Ig5 N-glycan polysialylation now provides us with a new approach to understand the mechanism of polysialylation by allowing us to address whether FN1 sequences common to both proteins are mediating NCAM N-glycan polysialylation and the factors that could prevent OCAM polysialylation.
Supplementary Material
Acknowledgment
We thank Dr. Arnon Lavie for many helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM063843 (to K. J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- NCAM
- neural cell adhesion molecule
- polyST
- polysialyltransferase
- FN1
- first fibronectin type III repeat of NCAM
- FN2
- second fibronectin type III repeat of NCAM
- OCAM
- olfactory cell adhesion molecule
- N-O
- NCAM-OCAM chimera
- AP
- acidic patch
- PNGase F
- peptide N-glycosidase F
- Endo N
- endo-N-acylneuraminidase
- FGFR
- fibroblast growth factor receptor
- TM
- transmembrane
- PDB
- Protein Data Bank.
REFERENCES
- 1.Walmod P. S., Kolkova K., Berezin V., Bock E. (2004) Neurochem. Res. 29, 2015–2035 [DOI] [PubMed] [Google Scholar]
- 2.Ditlevsen D. K., Povlsen G. K., Berezin V., Bock E. J. (2008) J. Neurosci. Res. 86, 727–743 [DOI] [PubMed] [Google Scholar]
- 3.Rothbard J. B., Brackenbury R., Cunningham B. A., Edelman G. M. (1982) J. Biol. Chem. 257, 11064–11069 [PubMed] [Google Scholar]
- 4.Finne J., Finne U., Deagostini-Bazin H., Goridis C. (1983) Biochem. Biophys. Res. Commun. 112, 482–487 [DOI] [PubMed] [Google Scholar]
- 5.Rutishauser U., Acheson A., Hall A. K., Mann D. M., Sunshine J. (1988) Science 240, 53–57 [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto I., Bruses J. L., Rutishauser U. (2001) J. Biol. Chem. 276, 31745–31751 [DOI] [PubMed] [Google Scholar]
- 7.Johnson C. P., Fujimoto I., Rutishauser U., Leckband D. E. (2005) J. Biol. Chem. 280, 137–145 [DOI] [PubMed] [Google Scholar]
- 8.Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 9.Kojima N., Yoshida Y., Tsuji S. (1995) FEBS Lett. 373, 119–122 [DOI] [PubMed] [Google Scholar]
- 10.Nakayama J., Fukuda M. N., Fredette B., Ranscht B., Fukuda M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7031–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheidegger E. P., Sternberg L. R., Roth J., Lowe J. B. (1995) J. Biol. Chem. 270, 22685–22688 [DOI] [PubMed] [Google Scholar]
- 12.Weinhold B., Seidenfaden R., Röckle I., Mühlenhoff M., Schertzinger F., Conzelmann S., Marth J. D., Gerardy-Schahn R., Hildebrandt H. (2005) J. Biol. Chem. 280, 42971–42977 [DOI] [PubMed] [Google Scholar]
- 13.Hildebrandt H., Mühlenhoff M., Oltmann-Norden I., Röckle I., Burkhardt H., Weinhold B., Gerardy-Schahn R. (2009) Brain 132, 2831–2838 [DOI] [PubMed] [Google Scholar]
- 14.Rutishauser U. (2008) Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 15.Kiss J. Z., Troncoso E., Djebbara Z., Vutskits L., Muller D. (2001) Brain Res. Rev. 36, 175–184 [DOI] [PubMed] [Google Scholar]
- 16.Durbec P., Cremer H. (2001) Mol. Neurobiol. 24, 53–64 [DOI] [PubMed] [Google Scholar]
- 17.Gascon E., Vutskits L., Kiss J. Z. (2007) Brain Res. Rev. 56, 101–118 [DOI] [PubMed] [Google Scholar]
- 18.Scheidegger E. P., Lackie P. M., Papay J., Roth J. (1994) Lab. Invest. 70, 95–106 [PubMed] [Google Scholar]
- 19.Hildebrandt H., Becker C., Glüer S., Rösner H., Gerardy-Schahn R., Rahmann H. (1998) Cancer Res. 58, 779–784 [PubMed] [Google Scholar]
- 20.Suzuki M., Suzuki M., Nakayama J., Suzuki A., Angata K., Chen S., Sakai K., Hagihara K., Yamaguchi Y., Fukuda M. (2005) Glycobiology 15, 887–894 [DOI] [PubMed] [Google Scholar]
- 21.Tanaka F., Otake Y., Nakagawa T., Kawano Y., Miyahara R., Li M., Yanagihara K., Nakayama J., Fujimoto I., Ikenaka K., Wada H. (2000) Cancer Res. 60, 3072–3080 [PubMed] [Google Scholar]
- 22.Roth J., Zuber C., Wagner P., Taatjes D. J., Weisgerber C., Heitz P. U., Goridis C., Bitter-Suermann D. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth J., Zuber C., Wagner P., Blaha I., Bitter-Suermann D., Heitz P. U. (1988) Am. J. Pathol. 133, 227–240 [PMC free article] [PubMed] [Google Scholar]
- 24.Campodónico P. B., de Kier Joffé E. D., Urtreger A. J., Lauria L. S., Lastiri J. M., Puricelli L. I., Todaro L. B. (2010) Mol. Carcinog. 49, 386–397 [DOI] [PubMed] [Google Scholar]
- 25.Fernández-Briera A., García-Parceiro I., Cuevas E., Gil-Martín E. (2010) Oncology 78, 196–204 [DOI] [PubMed] [Google Scholar]
- 26.Drake P. M., Nathan J. K., Stock C. M., Chang P. V., Muench M. O., Nakata D., Reader J. R., Gip P., Golden K. P., Weinhold B., Gerardy-Schahn R., Troy F. A., 2nd., Bertozzi C. R. (2008) J. Immunol. 181, 6850–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drake P. M., Stock C. M., Nathan J. K., Gip P., Golden K. P., Weinhold B., Gerardy-Schahn R., Bertozzi C. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11995–12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuber C., Lackie P. M., Catterall W. A., Roth J. (1992) J. Biol. Chem. 267, 9965–9971 [PubMed] [Google Scholar]
- 29.Yabe U., Sato C., Matsuda T., Kitajima K. (2003) J. Biol. Chem. 278, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 30.Curreli S., Arany Z., Gerardy-Schahn R., Mann D., Stamatos N. M. (2007) J. Biol. Chem. 282, 30346–30356 [DOI] [PubMed] [Google Scholar]
- 31.Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) EMBO J. 15, 6943–6950 [PMC free article] [PubMed] [Google Scholar]
- 32.Close B. E., Colley K. J. (1998) J. Biol. Chem. 273, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 33.Galuska S. P., Rollenhagen M., Kaup M., Eggers K., Oltmann-Norden I., Schiff M., Hartmann M., Weinhold B., Hildebrandt H., Geyer R., Mühlenhoff M., Geyer H. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10250–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima N., Tachida Y., Yoshida Y., Tsuji S. (1996) J. Biol. Chem. 271, 19457–19463 [DOI] [PubMed] [Google Scholar]
- 35.Angata K., Suzuki M., McAuliffe J., Ding Y., Hindsgaul O., Fukuda M. (2000) J. Biol. Chem. 275, 18594–18601 [DOI] [PubMed] [Google Scholar]
- 36.Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. (1987) Science 236, 799–806 [DOI] [PubMed] [Google Scholar]
- 37.Nelson R. W., Bates P. A., Rutishauser U. (1995) J. Biol. Chem. 270, 17171–17179 [DOI] [PubMed] [Google Scholar]
- 38.Close B. E., Mendiratta S. S., Geiger K. M., Broom L. J., Ho L. L., Colley K. J. (2003) J. Biol. Chem. 278, 30796–30805 [DOI] [PubMed] [Google Scholar]
- 39.Mendiratta S. S., Sekulic N., Lavie A., Colley K. J. (2005) J. Biol. Chem. 280, 32340–32348 [DOI] [PubMed] [Google Scholar]
- 40.Mendiratta S. S., Sekulic N., Hernandez-Guzman F. G., Close B. E., Lavie A., Colley K. J. (2006) J. Biol. Chem. 281, 36052–36059 [DOI] [PubMed] [Google Scholar]
- 41.Foley D. A., Swartzentruber K. G., Lavie A., Colley K. J. (2010) J. Biol. Chem. 285, 27360–27371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshihara Y., Kawasaki M., Tamada A., Fujita H., Hayashi H., Kagamiyama H., Mori K. (1997) J. Neurosci. 17, 5830–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alenius M., Bohm S. (1997) J. Biol. Chem. 272, 26083–26086 [DOI] [PubMed] [Google Scholar]
- 44.Paoloni-Giacobino A., Chen H., Antonarakis S. E. (1997) Genomics 43, 43–51 [DOI] [PubMed] [Google Scholar]
- 45.Hamlin J. A., Fang H., Schwob J. E. (2004) J. Comp. Neurol. 474, 438–452 [DOI] [PubMed] [Google Scholar]
- 46.Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr. (1989) Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 47.Pelkonen S., Pelkonen J., Finne J. (1989) J. Virol. 63, 4409–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida A., Suzuki M., Ikenaga H., Takeuchi M. (1997) J. Biol. Chem. 272, 16884–16888 [DOI] [PubMed] [Google Scholar]
- 49.Asahina S., Sato C., Matsuno M., Matsuda T., Colley K., Kitajima K. (2006) J. Biochem. 140, 687–701 [DOI] [PubMed] [Google Scholar]
- 50.Martersteck C. M., Kedersha N. L., Drapp D. A., Tsui T. G., Colley K. J. (1996) Glycobiology 6, 289–301 [DOI] [PubMed] [Google Scholar]
- 51.Williams E. J., Furness J., Walsh F. S., Doherty P. (1994) Neuron 13, 583–594 [DOI] [PubMed] [Google Scholar]
- 52.Kiselyov V. V., Skladchikova G., Hinsby A. M., Jensen P. H., Kulahin N., Soroka V., Pedersen N., Tsetlin V., Poulsen F. M., Berezin V., Bock E. (2003) Structure 11, 691–701 [DOI] [PubMed] [Google Scholar]
- 53.Francavilla C., Cattaneo P., Berezin V., Bock E., Ami D., de Marco A., Christofori G., Cavallaro U. (2009) J. Cell Biol. 187, 1101–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson A. A., Kendal C. E., Garcia-Maya M., Kenny A. V., Morris-Triggs S. A., Wu T., Reynolds R., Hohenester E., Saffell J. L. (2005) J. Neurochem. 95, 570–583 [DOI] [PubMed] [Google Scholar]
- 55.Doherty P., Walsh F. S. (1996) Mol. Cell. Neurosci. 8, 99–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.