Abstract
Galectins are defined by a conserved β-galactoside binding site that has been linked to many of their important functions in e.g. cell adhesion, signaling, and intracellular trafficking. Weak adjacent sites may enhance or decrease affinity for natural β-galactoside-containing glycoconjugates, but little is known about the biological role of this modulation of affinity (fine specificity). We have now produced 10 mutants of human galectin-3, with changes in these adjacent sites that have altered carbohydrate-binding fine specificity but that retain the basic β-galactoside binding activity as shown by glycan-array binding and a solution-based fluorescence anisotropy assay. Each mutant was also tested in two biological assays to provide a correlation between fine specificity and function. Galectin-3 R186S, which has selectively lost affinity for LacNAc, a disaccharide moiety commonly found on glycoprotein glycans, has lost the ability to activate neutrophil leukocytes and intracellular targeting into vesicles. K176L has increased affinity for β-galactosides substituted with GlcNAcβ1–3, as found in poly-N-acetyllactosaminoglycans, and increased potency to activate neutrophil leukocytes even though it has lost other aspects of galectin-3 fine specificity. G182A has altered carbohydrate-binding fine specificity and altered intracellular targeting into vesicles, a possible link to the intracellular galectin-3-mediated anti-apoptotic effect known to be lost by this mutant. Finally, the mutants have helped to define the differences in fine specificity shown by Xenopus, mouse, and human galectin-3 and, as such, the evidence for adaptive change during evolution.
Keywords: Carbohydrate Function, Galactose, Mutant, Neutrophil, Trafficking, Galectin, Specificity
Introduction
Galectins are a family of small soluble proteins that are defined by a conserved β-galactoside binding site, but they also have adjacent binding sites within their ∼130-amino acid β-sandwich carbohydrate recognition domains (CRD)3 that define the carbohydrate-binding fine specificity of each galectin for larger natural saccharides (1, 2). Galectins are attracting rapidly increasing interest because of apparent important regulatory functions in immunity, inflammation (3, 4), and cancer (5, 6), but how they function at the cellular level has been difficult to understand because of some unusual properties. Galectins are synthesized as cytosolic proteins and may function in the cytosol and nucleus, but they are also released extracellularly by a non-classical mechanism (7, 8). There they encounter galactoside-containing glycoconjugates that through galectin-mediated cross-linking may result in the formation of ordered lattices (9), the regulation of trafficking and receptor turnover (10), and/or the induction of signaling (3–6). These effects must depend on a match between the structural context of the galactoside in the particular glycoprotein and the fine specificity of the galectin, but this relationship has been studied to a very limited extent so far (11–15).
The galectin CRD is folded into two slightly bent anti-parallel β-sheets (F1–5 and S1–6), with the concave surface of the S-sheet forming a groove that is long enough to accommodate a tetrasaccharide (16) and, therefore, may be divided into sites A, B, C, and D (2, 17) (Fig. 1a). Site C is the galectin-defining β-galactose-binding site made up of approximately six conserved amino acids (white in Fig. 1) and is the only site providing binding affinity for a monosaccharide strong enough to be measured by itself, albeit weak, with a Kd in the 5–20 mm range. The other sites can be described as modifiers and enhance or decrease affinity depending on what saccharides are linked to the galactose moiety bound in site C. The hydroxyl groups at positions 4 or 6 of the Gal provide key interactions in the core binding site, and substitutions at these positions preclude binding to all galectins (Fig. 1, b and c); the inability of galectin-1 to bind α2–6-linked sialic acids, for example, makes T-cells resistant to the apoptosis inducing effect of galectin-1, which in turn regulates the Th1/Th2 balance of the immune response (13). A saccharide moiety in site D (i.e. one linked to position 1 of the Gal) (Fig. 1, b and c) is also required to boost the affinity to a biologically significant range (Kd μm or better). However, the nature of the monosaccharide in site D as well as the substitutions at positions 2 and 3 of the Gal are tolerated or preferred in different ways by different galectins, giving each its unique fine specificity for larger β-galactose-containing saccharides (17–19).
FIGURE 1.
Galectin-3 CRD in complex with saccharide ligands. a, the galectin-CRD is shown as a solvent-accessible surface with bound LacNAc as a stick model based on the x-ray crystal structure (PDB code 1KJL) (38). The positions of sites A-D are indicated above the binding groove and the loosely defined site E to the right. The conserved six amino acids making up site C are shown in white (with black text for Trp-181), and the surrounding mutated residues (Arg-144, Ala-146, Lys-176, Asn-180, Gly-182, Arg-186) are in black with white text; Asp-148 is labeled for reference but was not mutated. The Gal residue of LacNAc is shown in black with position 3 labeled, and the GlcNAc is shown in white with position 1 labeled. b and c, shown is a close-up of the core carbohydrate recognition site (C-D) with bound LacNAc (PDB code 1KJL) (b) or a model of Galβ1–3GlcNAc (c). The white numbers indicate the positions where interactions with the conserved binding site are made and where substitutions would sterically hinder binding. Other labeling is as for panel a. Note the difference in orientation of the reducing hydroxyl in b–c (labeled 1), which would direct structures linked here in a different way. The images were made by PyMOL (DeLano Scientific LLC, Palo Alto, CA).
Galectin-3 is one of the best studied galectins with many proposed functions in the immune system and cancer, including for example regulation of cell adhesion, apoptosis, differentiation, and migration (3–5, 20–22) and neutrophil activation as studied here (23). Extensive analysis of the binding of small saccharides to human (17, 24), rat (18), mouse (25), and hamster (26, 27) galectin-3 have revealed an unusually multifaceted fine specificity. Three types of monosaccharide residues linked at the 3 position of the Gal in site C are tolerated, thus, providing additions to the saccharide chain that project into site B (Fig. 1). The addition of Siaα2-3 changes binding affinity only slightly. The addition of GalNAcα1–3 or Galα1–3, as in blood group A or B determinants, enhance binding affinity by 10–25-fold. The addition of GlcNAcβ1–3, as in poly-N-acetyl-lactosaminoglycans, is tolerated and with an additional Gal residue, presumably in site A, may give enhanced affinity. The binding to poly-N-acetyl-lactosaminoglycans or similar structures has been proposed to be involved in the galectin-3-mediated modulation of cell adhesion (26), intracellular trafficking (28), receptor turnover (10), and the interaction with microbes (29, 30). However, the biological effects associated with Siaα2-3 or GalNAcα1–3/Galα1–3 in site B have not been defined for galectin-3. For galectin-8, the strong preference for NeuAcα2–3 in site B of its N-CRD (19) determines its intracellular trafficking after endocytosis (14), and recently the affinity of the galectin-4 and -8 C-CRDs for blood group A and B saccharides was shown to confer specific anti-bacterial activity (31).
Regarding the saccharide linked at position 1 of the Gal in site C, galectin-3, like galectin-1, prefers 4-linked GlcNAc as in Galβ1–4GlcNAc (LacNAc), which is commonly found in the N-glycans of glycoproteins, molecules known to be important galectin ligands (11). In contrast, many other galectins seem to prefer disaccharides that put a 4-linked Glc (as in lactose), a 3-linked GlcNAc, or a 3-linked GalNAc in site D (18, 32). Galβ1–3GalNAc (the T-antigen) has been proposed to act as a functional ligand of galectin-3 (33), although additional interactions would likely be required because the affinity of galectin-3 for this disaccharide is weak, i.e. 10–50-fold lower than for LacNAc (17, 18, 24, 26).
A fifth loosely defined site E (2) was added next to site D as a landmark to indicate possible interaction with additional parts of the glycoprotein (or glycolipid) carrying the saccharide in sites A–D. When added to an established interaction, even a weak interaction immeasurable by itself can boost affinity in a biologically significant way as discussed in Carlsson et al. (19). Therefore, such interactions in site E may also contribute to the different fine specificity for natural ligands of galectins.
From the above discussion it follows that a more detailed analysis of the relationship between galectin-3 carbohydrate-binding fine specificity and biological function is needed. To address this we have mutated amino acid residues in or near sites B–D (black in Fig. 1a), with the goal of creating galectin-3 proteins that have lost one or more facets of its binding specificity but that have retained others. The mutant proteins were then tested for biological effects to establish structure-function relationships. Finally we have analyzed whether orthologues of galectin-3 from other species also have altered fine specificity as predicted from differences in their sequence.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were from Saveen Werner AB (Malmö, Sweden) unless stated otherwise. Primers for mutagenesis were ordered from MWG-Biotech (Ebensberg, Germany) or from Invitrogen (Lidingö, Sweden). All the fluorescein-tagged saccharides have previously been described (19). Plasmids encoding human galectin-3 in pET-3c were as described (34), Xenopus galectin-3 (galectin VIIa (35)) in pET-11a was kindly provided by Dr. Hiroki Shoji, Kagawa University, Japan, and mouse galectin-3 in pIN III ompA2 (36) was kindly provided by Professor John L. Wang, Michigan State University.
Site-directed Mutagenesis
Mutants of human galectin-3 in the pET-3c plasmid were made using the QuikChange® II site-directed mutagenesis kit from Stratagene (Amsterdam, The Netherlands), with template-DNA isolated from Escherichia coli XL1Blue (Stratagene). Mutagenic primers for PCR were: Gal-3A146Q sense (5′-GCC CAA TGC AAA CAG AAT TCA GTT AGA TTT CCA AAG AGG G-3′) and antisense (5′-CCC TCT TTG GAA ATC TAA CTG AAT TCT GTT TGC ATT GGG C-3′), Gal-3K176L sense (5′- GTC ATT GTT TGC AAT ACA CTG CTG GAT AAT AAC TGG GGA AGG-3′) and antisense (5′-CAG TAA CAA ACG TTA TGT GAC GAC CTA TTA TTG ACC CCT TCC-3′), Gal-3K176L/N180T sense (5′-GTC ATT GTT TGC AAT ACA CTG CTG GAT AAT ACC TGG GGA AGG-3′) and antisense (5′-CAG TAA CAA ACG TTA TGT GAC GAC CTA TTA TGG ACC CCT TCC-3′), Gal-3R186S sense (5′-CTG GGG AAG GGA AGA AAG CCA GTC GGT TTT CCC- 3′) and antisense (5′-GGG AAA ACC GAC TGG CTT TCT TCC CTT CCC CAG-3′), Gal-3R86I sense (5′-CTG GGG AAG GGA AGA AAT ACA GTC GGT TTT CCC-3′) and antisense (5′-GGG AAA ACC GAC TGT ATT TCT TCC CTT CCC CAG-3′), Gal-3R144S sense (5′-GAA GCC CAA TGC AAA CAG CAT TGC TTT AGA TTT CCA AAG AG-3′) and antisense (5′-CTC TTT GGA AAT CTA AAG CAA TGC TGT TTG CAT TGG GCT TC- 3′), and Gal-3G182A sense (5′-GCT GGA TAA TAA CTG GGC AAG GGA AGA AAG ACA GTC G-3′) and antisense (5′-CGA CTG TCT TTC TTC CCT TGC CCA GTT ATT ATC CAG C-3′). Successful mutagenesis was confirmed by sequencing by GATC Biotech (Konstanz, Germany) in the forward direction from the T7 promotor primer and in the reverse direction from the pET-RP primer.
Production of Recombinant Galectins
Recombinant human galectin-3 and mutants and Xenopus galectin-3 were produced in E. coli BL21Star (DE3) cells (Invitrogen) and purified by affinity chromatography on lactosyl-Sepharose essentially as described for wild type human galectin-3 (34) but with some variation to optimize yield for each protein. The initial yields ranged from 3 mg/liter culture for G182A to 80 mg/liter for R144S, but, for example, lowering the temperature of the isopropyl 1-thio-β-d-galactopyranoside induction from 37 to 30 °C increased the yield of G182A galectin-3 about 10-fold. Mouse galectin-3 was produced from vector pIN III ompA2 in E. coli JA221 cells as previously described (36), except that Tryptone soy broth was used. The bacteria were processed and galectin-purified in the same way as for human galectin-3 described above. The galectins, in phosphate-buffered saline (118 mm NaCl, 67 mm Na+/K+-phosphate, pH 7.2) containing 4 mm β-mercaptoethanol, 2 mm EDTA and 150 mm lactose, were dialyzed against 2 liters of water that was changed once every 2 h 7 times and lyophilized and stored at −20 °C until use. This procedure does not completely deplete the galectin-bound lactose, which helps to preserve stability. Before use, the galectins were dissolved in the appropriate buffer, and any remaining lactose was removed by repeated ultrafiltration and concentration in a Centricon Plus-70, Ultracel PL10, or Centriprep Y10 ultrafiltration cell (Millipore AB, Sundbyberg, Sweden). Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad, Sundbyberg, Sweden). Protein size and integrity was determined by SDS-PAGE using 4–20% PreciseTM Protein Gels from Pierce (Nordic Biolabs, Täby, Sweden) in Tris/HEPES running buffer.
Differential scanning calorimetry measurements were performed on a MicroCal differential scanning calorimeter (MicroCal Inc., Northampton, MA) with a cell volume of 0.5072 ml. All samples were degassed for 15 min at room temperature before scanning. Protein samples (0.5 mg/ml) in PBS buffer with or without ligand were scanned in the temperature range 25 to 80 °C at a rate of 1 °C/min.The reversibility of the calorimetric traces was assessed by the reproducibility of scans upon rapid cooling to 25 °C followed by rescanning. Base-line scans were collected with buffer in both the reference and sample cells.
FITC Labeling of Galectins
About 2 mg of freeze-dried galectin was suspended in 1.3 ml of 0.1 m NaHCO3 and 0.9% NaCl, pH 9.3. A 20 mm stock solution of fluorescein isothiocyanate (FITC) (Sigma, St. Louis, MO) prepared in dimethyl sulfoxide (DMSO) was added in a 10-fold excess to the protein. The solution was rocked at 8 °C in the dark overnight. Labeled protein was separated from unbound fluorescein by buffer exchange in PBS using PD10 desalting columns (Amersham Biosciences).
Glycan Array
The glycan microarray consisting of 406 glycans in replicates of six (Version 3.2) were printed on N-hydroxysuccinimide-activated glass microscope slides (SCHOTT Nextrerion, Louisville, KY). FITC-labeled galectins diluted to 200 μg/ml in Tris-buffered saline (20 mm Tris, 150 mm NaCl, pH 7.4) containing 2 mm CaCl2, 2 mm MgCl2, 1% bovine serum albumin, and 0.05% Tween 20 were used for analysis on the glycan microarray. 70 μl of each galectin was added to separate microarray slides and incubated under a coverslip for 60 min in a dark, humidified chamber at room temperature. After incubation, the coverslips were gently removed in a solution of Tris-buffered saline containing 0.05% Tween 20 and washed by gently dipping the slides 4 times in each of three successive washes in Tris-buffered saline containing 0.05% Tween 20, Tris-buffered saline alone, and deionized water. After the last water wash, the slides were spun in a slide centrifuge for ∼15 s to dry them; they were immediately scanned in a ProScanArray MicroArray Scanner (PerkinElmer Life Sciences) using an excitation wavelength of 488 nm and analyzed using ImaGene software (BioDiscovery, Inc., El Segundo, CA) to quantify fluorescence, reported as RFU. The maximum measurable value was 40,000 RFU. Each saccharide on the array is represented by six spots. The highest and lowest values were omitted, and the data are reported as the average of the remaining four.
Fluorescence Anisotropy (FA)
An FA assay was done as described previously (19, 37), with a fixed concentration (0.1 μm) of fluorescein-labeled saccharide (probe) and 0.05–80 μm galectin in a final volume of 180–200 μl; anisotropy (A) was measured at room temperature and after incubation on ice (about 4 °C) using the instrument POLARstar (excitation 485 nm/emission 520 nm) with software FLUOstar Galaxy Version 4.11–0 (BMG Lab-Technologies, Offenburg, Germany). Anisotropy data (y axis) was plotted against galectin concentration (x axis). From the resulting binding curve, Kd was calculated using measured anisotropy values of the probe alone (A0) and the probe saturated with galectin (Amax, representing the probe-galectin complex), as previously described. For low affinity ligands, when only partial curves were obtained, Amax was estimated by extrapolation, and from that an estimated Kd could be calculated. If the anisotropy had not exceeded 50 mA at a galectin concentration of 50 μm, it was considered to be not detected.
Molecular Modeling
Molecular modeling was performed with MacroModel (MacroModel; 9.7 edition; Schrödinger, LLC: New York, 2009) using the MMFFs force field and the GB/SA solvation model for water. A model of the A-tetrasaccharide bound to galectin-3 was constructed based on the x-ray crystal structure of galectin-3 in complex with LacNAc (Protein Data Bank code 1KJL (38)). The lactose part of the A-tetrasaccharide was initially superimposed on the LacNAc of the galectin complex. A Monte Carlo-Stochastic Dynamics conformational search of the A-trisaccharide complex showed that the φ and ψ dihedral angles of GalNAcα1–3Gal are sterically constrained to a single minimum, similar to those of Protein Data Bank structure 1ULF (39). A Monte Carlo-Stochastic Dynamics conformational search of the Arg-144 side chain and the full A-tetrasaccharide resulted in a single conformation. Models of sialyllactose (NeuAcα2–3Galβ1–4Glc) bound to galectin-3 were also constructed based on the x-ray crystal structure 1KJL. The lactose part of the sialyl lactose was initially superimposed on the LacNAc of the galectin complex. The flexibility of the Arg-144 side chain, and of the NeuAcα2 residue (15 rotatable bonds) prohibited the exploration of conformational space in a single run. Therefore, two low energy conformers of the ligand were manually constructed and used as a starting point for a Monte Carlo-Stochastic Dynamics search. Trajectory snapshots were energy minimized, resulting in three distinct binding modes. The least stable binding mode with an energy of −13,074 kJ/mol was obtained with the ligand in a conformation very similar to that of Protein Data Bank structure 1WW4 of a fungal galectin (40). The two more stable binding modes (−13,093 and −13,098 kJ/mol) had a ligand conformation similar to a model in complex with galectin-8N (19).
Neutrophil Purification, Priming, and Measurement of NADPH Oxidase Activity
Peripheral blood neutrophils were isolated as described by Böyum (41) from buffy coats from healthy volunteers using dextran sedimentation and Ficoll-Paque (Fisher Scientific, Göteborg, Sweden) gradient centrifugation. The cells were resuspended in Krebs-Ringer phosphate buffer containing glucose (10 mm), Ca2+ (1 mm), and Mg2+ (1.5 mm) (pH 7.3) and stored on melting ice until use. The neutrophils (1 × 107cells/ml) were primed by incubating the cells at 37 °C for 20 min in the presence of TNFα (10 ng/ml). The NADPH oxidase activity was measured using a luminol/isoluminol-amplified chemiluminescence system (42). The chemiluminescence was measured in a Biolumat LB 9505 (Berthold Co., Wildbad, Germany) using polypropylene tubes with a 900-μl reaction mixture containing 106 neutrophils. The tubes were equilibrated for 5 min in the Biolumat at 37 °C before the addition of 100 μl of stimulus. The light emission was recorded continuously. To quantify the intracellularly and extracellularly generated reactive oxygen species, respectively, two different reaction mixtures were used. The extracellular release of superoxide anion was measured in tubes containing neutrophils, HRP (a cell-impermeable peroxidase; 4 units) and isoluminol (a cell-impermeable chemiluminescence substrate; 6 × 10−5 m). The intracellular production of reactive oxygen species was measured in tubes containing neutrophils, superoxide dismutase (a cell impermeable scavenger for O2⨪; 50 units), catalase (a cell impermeable scavenger for H2O2; 2000 units), and luminol (a cell-permeable chemiluminescence substrate; 2 × 10−5 m).
Galectin Binding to Neutrophils
To study cell surface binding of galectin, neutrophils were purified as above. TNFα-primed or non-primed cells were then incubated with FITC-labeled galectin-3 and mutants (40 μg/ml) for 1 h at 4 or 37 °C and washed twice with FACSwash (PBS, 0.02% NaN3, EDTA 10−4 m). The analysis of the fluorescent markers was performed by FACScan (BD Biosciences).
Transfection of Enhanced Green Fluorescent Protein (EGFP)-conjugated Galectin-3 and Mutants
Mutants (R186S and G182A) were made on the pEGFP-C1 vector (Clontech) carrying the galectin-3 gene using the QuikChange® II site-directed mutagenesis kit from Stratagene as described above. Plasmids were isolated from E. coli XL1Blue and transfected into the human breast cancer cell line SKBR3 using LipofectamineTM 2000 reagent (Invitrogen). Cells were cultured in RPMI 1640 supplemented with 10% FBS, 1× nonessential amino acids, 1× sodium pyruvate, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Cell were cultured with 1 mg/ml G418 at 37 °C in 5% CO2 for at least 2 weeks to get stable transfectants. For experiments, cells then were cultured on coverslips for 1–2 days, fixed in 4% paraformaldehyde (VWR), and mounted with ProLong Antifade Reagent with DAPI (Invitrogen) onto glass slides. Samples were left to cure overnight and examined with a NIKON Eclipse TE2000-U epifluorescence microscope.
RESULTS AND DISCUSSION
To probe the relationship between carbohydrate-binding fine specificity and biological function, seven amino acids in the galectin-3 CRD were mutated (Fig. 1a) alone (6 mutants) or in different combinations with each other (4 mutants). All of the mutants could be purified in good yield by affinity chromatography on lactosyl-Sepharose, and they all bound lactose with affinity within the same order of magnitude as wild type galectin-3. Based on this we conclude that they are all properly folded, a conclusion further supported by thermal stability measurements made using differential scanning calorimetry as described in supplemental Results (Fig. S1 and Table S1).
Evaluation of Glycan Binding
To evaluate differences in carbohydrate-binding fine specificity between wild type galectin-3 and the mutants, we used two different complementary methods; a 406-element immobilized glycan array to assess binding trends (Fig. 2 and supplemental Table S2, a and b) and for 15 selected saccharides a solution-based FA assay to give more accurate affinities (Fig. 3, supplemental Fig. S2 and Table S3).
FIGURE 2.
Glycan array analysis of wild type galectin-3 and mutants. Each diagram shows binding (as thin bars) of fluorescein-tagged galectin (wild type or mutant, final concentration 200 μg/ml) to each of the 179 potential galectin binding glycans, expressed as RFU ×10−4 on y axis. The binding data are shown twice, with the glycans grouped by the disaccharide that is likely to be in site C-D of the galectin (panel a) or the monosaccharide that is likely to be in site B, i.e. that is linked to the 3-position of the galactose in site C (panel b). In both panels the glycans are ordered by their intensity of binding to wild type galectin-3 (from left to right). The features by which glycans are grouped are shown under the x axis using the symbols recommended by the Consortium for Functional Glycomics. For site C-D they are, from left to right, Galβ1–4GlcNAc, Galβ1–3GlcNAc, Galβ1–4Glc, Galβ1–3GalNAc, and other. For site B they are sulfate-3, NeuAcα2–3, GalNAcα1–3, Galα1–3, GlcNAcβ1–3, and nothing (that is the Gal in site C is terminal). The bottom right diagram of panel a shows the very low background binding of wild type galectin-3 and all of the mutants to the remaining 227 glycans on the array, which are not expected to bind galectins either because they do not contain β-galactose or all of the β-galactoside-containing disaccharides that they do contain are blocked by substitutions sterically preventing binding in site C and D (2) as explained in Fig. 1, b and c. The R186I mutant gave higher background (five peaks >1000 RFU, maximum 2674) than the others (four peaks >500 RFU, maximum 810) but not high enough to confound interpretation. The numerical values of binding, glycan structures and original glycan numbering are given in supplemental Tables S2, a and b.
FIGURE 3.
Fluorescence anisotropy analysis of wild type galectin-3 and mutants. Each panel shows a bar diagram of the relative affinity ((Ka mutant)/(Ka galectin-3 wt) = (Kd galectin-3 wt/Kd mutant) of 1 mutant for 15 different fluorescein-labeled saccharide probes (1–15). Average values of relative affinities are based on analysis using a fluorescence anisotropy assay at chilled conditions (∼ 4 °C), n ≥ 4, as described in more detail in supplemental Fig. S2 and Table S3. The saccharide probes are shown in a table below the figure with the disaccharide most likely to bind in site C-D shown in bold.
On the array, 227 glycans served as non-binding controls either because they lack β-galactose or all the β-galactose containing disaccharides they do possess are blocked by substitutions preventing binding in site C and D as explained in Fig. 1, b and c. All such glycans showed very low binding by wild type galectin-3 and all of the mutants (Fig. 2a, bottom, right panel).
The remaining 179 potential galectin-binding glycans were those having at least one β-galactoside residue not blocked from binding in site C by the criteria described above. These are shown twice in Fig. 2. In panel a they are grouped into one of the four disaccharide types found within the glycans of the array that would be capable of binding to site C-D of wild type galectin-3, and in panel b they are grouped based on the nature of the moiety likely to be found in site B (i.e. linked to the 3-position of the Gal residue in site C). The glycans within each group shown in Fig. 2 were furthermore ordered by their binding intensity for wild type galectin-3 (from high left to low right).
Wild type galectin-3 gave clear binding to representatives of three structural groups in Fig. 2a (site C-D), but it also did not bind to some of the glycans from each group. The latter tend to be those with short saccharide chains, e.g. di-, tri-, or tetrasaccharides, which apparently are not well recognized on the array even if they are known to bind galectin-3. The glycans with a LacNAc available for binding in site C-D bound the best and most frequently (45 of 94 glycans on the array), and those with available Galβ1–3GlcNAc also bound well (15 of 36), whereas those with only lactose available bound weaker (6 of 24), and those with only Galβ1–3GalNAc available did not bind at all. Wild type galectin-3 also gave clear binding to representatives of each group in Fig. 2b (site B), demonstrating its broad multi-faceted fine specificity.
The affinities (Kd) of 15 different fluorescein-tagged saccharide probes were determined by an FA assay (19, 37) for each mutant and compared with wild type galectin-3 as summarized in Fig. 3 and presented in detail in supplemental Fig. S2 and Table S3. The relative affinities of the different probes was again consistent with the galectin-3 specificity described in the Introduction, e.g. with a preference for LacNAc over lactose (probe 2 versus 1), tolerance for 2–3 sialylation (probes 6–8) of β-Gal, tolerance or preference for extension with GlcNAcβ1–3 (probes 9–15), and a strong preference for A-tetrasaccharide (probe 5).
This FA assay also measures the anisotropy of the galectin-probe complex (as Amax) that reports indirectly on the position of the bound saccharide in the galectin binding groove, as explained in the supplemental Results and Sörme et al. (37). The Amax values of wild type galectin-3 were relatively high (110–140 mA) for all probes except 3 and 8, indicating that with the exception of probes 3 and 8, the probe fluorophores experience similar segmental motion when bound to the galectin and thereby provided evidence that the internal Gal (bold in the Table of Fig. 3) in probes 10–15 is in site C; had the terminal Gal of these probes been in site C, the fluorophore would have been further removed from the protein surface, and Amax would be much lower as found for galectin-1.4 Further support for the suggestion that the internal Gal of probes 10–15 is found in site C stems from the fact that they have similar Amax values to those of probes 11, 12, and 15, saccharides where the terminal GalβGlcNAc residue is prevented from binding in site C-D by fucosylation on the GlcNAc (probe 11 and 15) or 6-sialylation on the Gal (probe 12) (Fig. 1, b and c). Probes 3 and 8, which place Galβ1–3GlcNAc in site C-D, possess Amax values that are considerably lower (by about 40 mA) than those of the other probes where Galβ1–4GlcNAc is in site C-D. Presumably this is explained by a difference in the way that the fluorophore projects into site E (Fig. 1c) such that the segmental motion of the fluorophore is less restricted when Galβ1–3GlcNAc is in the site C-D.
Point Mutations in Site D
The preference of galectin-3 for Galβ1–4GlcNAc (LacNAc) over Galβ1–4Glc (Lac), Galβ1–3GlcNAc, and Galβ1–3GalNAc may be explained by the fact that its N-acetyl group stacks against a π-electron surface formed by the conserved Arg-186 in ion-pairs with carboxylates of Glu-165 and Glu-184 (32) (Fig. 1b); lactose instead presents an OH, and the two 3-linked disaccharides present a CHOH at this position (Fig. 1c), apparently providing weaker interactions. We, therefore, made two mutants replacing Arg-186, one with Ser (R186S) and the other with Ile (R186I) as found in galectin-8N, which has the reverse preference for the disaccharides mentioned above compared with galectin-3. In the FA assay, both mutants had lost binding to the glycans where LacNAc or Galβ1–3GlcNAc is likely to be in site C-D (e.g. 2, 3, 7, and 8 in Fig. 3), whereas they retained reduced affinity for glycans with Lac in site C-D; the affinity of R186S for soluble LacNAc decreased by a factor of 70 (32), whereas its affinity for lactose was reduced only about four times. On the array, galectin-3 R186S bound only one glycan (Fig. 2), an N-glycan with Galβ1–3GlcNAc on the antennae and capped by a blood group B-determinant. The R186I mutant bound a few more glycans on the array (Fig. 2), most also capped by blood group A or B determinants, substituents that perhaps compensate for the lower affinity to the disaccharide in site C-D.
Point Mutation Near Site C
One mutation, G182A, near site C was included because it has lost two interesting tumor growth promoting effects shown by galectin-3; that is, the intracellular anti-apoptotic effect (43) and the induction of a shift from N-Ras to K-Ras activation (44). This mutant was originally made because the sequence 180NWGR183 in galectin-3 resembles a conserved sequence in the Bcl-2 family of proteins (43), but its carbohydrate binding specificity has not been reported. The most striking effect of the G182A mutation was that it preferentially, although not exclusively, lost binding to Galβ1–3GlcNAc containing glycans on the array (Fig. 2, panel a). Modeling did not suggest a mechanism, as neither Gly-182 nor Ala-182 would be in contact with the disaccharide in site C-D (Fig. 1). However, disruptive interactions with structures at the reducing end (i.e. the remainder of the glycan) of the Galβ1–3GlcNAc disaccharide in site C-D may occur in the mutant as these structures would be directed more toward Gly-182 (Fig. 1c). In the FA assay, the Amax values with the G182A mutant increased (to ∼110 mA) for the Galβ1–3GlcNAc-containing probes 3 and 8 relative to those obtained for wild type galectin-3 and all of the other mutants (mA ∼60–90) (supplemental Table S3), indicating an effect (e.g. reduced mobility) on the structure at the reducing end (i.e. the linker and fluorescein moiety).
Asn-180 is also part of the sequence 180NWGR183, and its side chain points out into solution, away from site C, in the direction opposite to the side chain of Trp-181 (Fig. 1a). The double mutant K176L/N180T was obtained by a mistake in primer design, and as expected it did not show much difference in carbohydrate binding specificity compared with K176L (described below), with the exception that it had a slightly increased affinity for some of the longer GlcNAcβ-extended galactosides (Fig. 3).
Point Mutations in Site B
A number of different substitutions at OH3 of the galactose in site C can be tolerated by galectin-3, as described in the Introduction. X-ray crystallography and modeling suggest that these additions to the saccharide chain may interact with the side chains of Arg-144, Ala-146, Asp-148, and Lys-176, together making up site B as shown in Fig. 4, a–c.
FIGURE 4.
Modeling of interactions in site B. The monosaccharide attached to position 3 of Gal in site C and selected amino acid side chains are shown as stick models, with some positions and functional groups labeled and hydrogen bonds denoted by dotted lines. A transparent surface of the galectin-3 CRD is shown as background. a, shown is one possible position of NeuAc in NeuAca2–3Galβ1–4Glc based on the complex of NeuAcα2–3Lac with the N-CRD of galectin-8 (19); an alternative similar position was found in an x-ray crystal structure of a complex of NeuAcα2–3Lac with a fungal galectin (40), although the structure of site B is different in this case. b, shown is the position of GlcNAcβ in lacto-N-neotetraose (Galβ1–4GlcNAcβ1–3Galβ1–4Glc) and the amino acid side chains from the x-ray crystal structure of a galectin-3 CRD complex with lacto-N-neotetraose.5 c, shown is the position of GalNAcα in a blood group A tetrasaccharide (GalNAcα1–3(Fucα1–2)Galβ1–4Glc) based on a complex with a fungal galectin (39). The models were generated by MacroModel (9.7 edition), and the images were generated by PyMOL.
Modeling suggested a few alternative positions of NeuAcα in site B with similar energy minima, with the best shown in Fig. 4a. In all of them the C1 carboxyl group is oriented toward the side chain of Arg-144, which is flexible and can adapt to permit a hydrogen bond. Arg-144 is clearly important, as the R144S mutant lost affinity for the sialylated (and sulfated) galactosides on the array (Fig. 2b) and probes 6–9 used in the FA-assay (Fig. 3), an effect also found for the corresponding mutant of hamster galectin-3 (27).
The A146Q mutant bound the 3-sialylated galactosides with slightly reduced affinity. This mutant was made to explore the basis for the strongly enhanced affinity shown by galectin-8N for 3-sialylated galactosides (19). The larger side chain of the corresponding residue, Gln-47, in galectin-8N forces the side chain of Arg-45 (corresponding to Arg-144 of galectin-3) into a different conformation (PDB code 2YXS) relative to that shown in Fig. 4a and similar to that shown in Fig. 4c, such that hydrogen bonds are possible between the side chains of both Gln-47 and Arg-45 and the sialic acid carboxyl group (Ref. 19 not shown). Similar interactions may explain why the galectin-3 A146Q mutant retains binding to 3-sialylated galactosides (Fig. 3), but the decrease in affinity clearly shows that they alone do not account for the strong preference for 3-sialylated galactosides shown by galectin-8N.
Interactions with the NeuAcα moiety in site B may also be made by Asp-148 and/or Lys-176 as shown in Fig. 4a and in an alternative conformation by Lys-176 with OH8 (not shown). Lys-176 is clearly important for binding NeuAcα in site B, as the K176L mutant lost affinity for the sialylated galactosides (Figs. 2b and 3). The model also explains why galectin-3 binds galactosides with 3-linked N-glycoloyl neuraminic acid (NeuGc) equally well (supplemental Table S2); as shown in Fig. 4a, the oxidized methyl group (of NAc in NeuAc) points into solution.
In the model of GlcNAcβ in site B (Fig. 4b), the OH6 may interact with both Arg-144 and Asp-148 via hydrogen bonds. The former appears to be important as the R144S mutant lost affinity for all of the GlcNAcβ1–3-substituted galactosides (9–15 in Fig. 3). The A146Q mutant retained affinity, albeit lower, for the GlcNAcβ1–3-substituted galactosides (Fig. 3) probably because of the alternative position of Arg-144 forced by the Gln, as mentioned above. In agreement with this, the reverse mutation of galectin-8N, Q47A, has increased affinity for the GlcNAcβ1–3-substituted galactosides (14). The K176L mutant binds the GlcNAcβ1–3-substituted galactosides significantly better than wild type galectin-3, but no direct interaction with the side chain is apparent in the model shown in Fig. 4b. Instead, indirect interactions/effects are possible (e.g. water mediated) as there is a particularly strong change in mobility (45) and position5 of the Lys-176 side chain upon ligand binding. The orientation of OH3 and OH4 of GlcNAcβ away from the protein (Fig. 4b) explains why the addition of Fucα here (as in compound 11 and 15 in Fig. 3) does not block interaction with the galectins, as it would if the GlcNAc had been in site D (Fig. 1, b and c).
In the model of GalNAcα in site B (Fig. 4c), its OH6 interacts with Trp-181 via a hydrogen bond. The same would be expected for Galα and may explain why both GalNAcα1–3 and Galα1–3 galactosides bind galectin-3 with enhanced activity. Arg-144 is forced into the alternative conformation by the NAc of GalNAc but may interact with it by a hydrogen bond. The conformational change and the additional hydrogen bond apparently does not give any net increase in affinity, as the R144S mutant in fact has increased affinity for the GalNAcα extended galactoside (5 in Fig. 3). The A146Q mutant showed a selective loss of affinity for glycans with GalNAcα in site B but not those with Galα (blood group B type) (Fig. 2b). A likely explanation is that the Gln side chain would come into steric conflict with the NAc of GalNAcα but not with Galα in site B. Similarly, galectin-8N has very low affinity for GalNAcα-extended galactosides but retains significant affinity for some of the Galα extended galactosides found in the glycan array (supplemental Table S1 in Carlsson et al. (19).
Combined Mutations
To further increase the resemblance between galectin-3 and the galectin-8N CRD in site B, we combined the A146Q mutation with the K176L, K176L/N180T, and R186I mutations. This resulted in a combination of the lost binding properties of each component mutant, with no evidence for other alterations in binding (Fig. 3).
Evolutionary Change of Galectin-3 Fine Specificity
The sequence and unique domain organization of galectin-3 permits certain identification of a single galectin-3 orthologue in each of the vertebrate species studied, including fish, amphibians, birds, and mammals (46). The amino acids making up site C-D have been completely conserved, but among the amino acids in site B studied here, there are differences in different species, and additional differences are seen in site A. To explore how this changes carbohydrate binding specificity, we analyzed Xenopus galectin-3 and mouse galectin-3 by the FA assay in the same way as that described for the mutants above.
In Xenopus galectin-3 two amino acids in site B differ relative to human galectin-3; Lys-176 to Met and Asn-180 to Arg. The specificity of Xenopus galectin-3 indeed resembled the K176L mutants in that its relative affinity for the sialylated galactosides (no. 6, 7, and 8 in Fig. 5, left) was much lower compared with that shown by human galectin-3, whereas its relative affinity for some of those extended with GlcNAcβ (no. 9–15) was higher. Xenopus galectin-3 also had a clear preference for Galβ1–3GlcNAc in site C-D over lactose and LacNAc that may be due to other differences in the amino acid sequence, e.g. the presence of Arg in place of Asn-180.
FIGURE 5.
Fluorescence anisotropy analysis of the affinity of Xenopus (left) and mouse (right) galectin-3 relative to human galectin-3. Analysis was done, and results are presented as for Fig. 3, showing bar diagrams of the relative affinity of Xenopus and mouse galectin-3 ((Ka Xenopus or mouse galectin-3)/(Ka human galectin-3 wt)) for 15 different fluorescein labeled saccharide probes (1–15). Below each diagram is a model of human galectin-3 made as in Fig. 1a but colored black for amino acids that differ in the respective animal galectin and white for those that are identical. The residues in site B that differ are labeled.
In mouse galectin-3 the only surface-exposed changes in the whole carbohydrate binding groove are Ala-146 to Val in site B (Fig. 5, right) and Gln-150 to Arg in site A and two more at the far end of site A. Mouse galectin-3 has lower affinity for all the saccharide probes tested and most significantly for the ones extended by GlcNAcβ (i.e. “polylactosamine” type) into site B. These differences may in part be due to the larger Val side chain (compared with Ala-146 in human galectin-3) hindering extensions into site B, but the differences in site A may also contribute.
In rat galectin-3 there are three changes compared with human galectin-3: Arg-144 to Ser, Ala-146 to Thr, and Asp-148 to Asn. Rat galectin-3 was not available for study here, but previous detailed studies (18, 24) showed that it has a specificity similar to that of human galectin-3, except for an approximate 5-fold lower affinity for NeuAcα2–3Lac. The latter is consistent with the specificity of the R144S mutant, which also shows a loss in affinity for α2–3-sialylated galactosides as described above. With regard to the binding of other saccharides, the loss of interaction with Arg-144 (Fig. 4, b and c) may have been compensated for by the presence of Thr instead of Ala-146. The results described above clearly show that there are species specific sequence differences in site B of galectin-3 that alter carbohydrate binding specificity and may represent an adaptation to the carbohydrate structures that each species produces or encounters.
Biological Effects of Extracellularly Added Galectin-3 Mutants; Activation of and Binding to Neutrophils
As the final part of the study, we wanted to use the galectin-3 mutants to gain an understanding of the correlation between carbohydrate binding specificity and biological function. We first studied the activation of NADPH-oxidase in primed neutrophils, a well characterized effect of galectin-3 (23, 47–49). Neutrophils were purified from peripheral blood and preincubated with TNFα (primed) and then activated by the addition of galectin-3 or mutants; fresh non-primed neutrophils are not activated by galectin-3 (23). For each galectin concentration, the intracellular and extracellular production of superoxide anions was measured over time (supplemental Fig. S3) using a chemiluminescence assay. The peak value from the time curve was used to compare the mutants with wild type galectin-3, which was run in parallel as control in each experiment (Fig. 6).
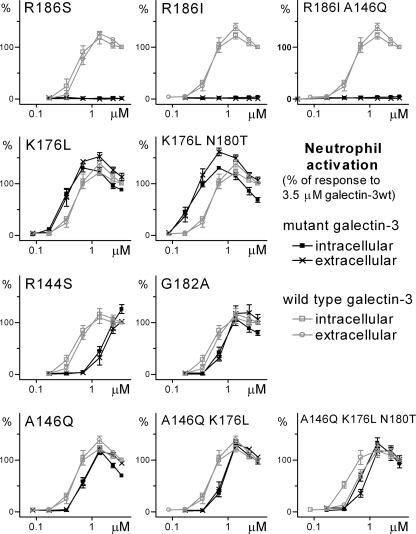
FIGURE 6.
Dose response of activation of TNFα-primed neutrophils by wild type galectin-3 and mutants. The dose (x axis) response (y axis) of wild type and mutants are shown in gray and black, respectively, in all panels. Extra and intracellular release of reactive oxygen species was measured using a luminescence assay. Each data point is the peak (reactive oxygen species) response from a time curve (as given in supplemental Fig. S3), normalized by defining 100% as the peak response to the highest concentration (3.5 μm) of wild type galectin-3 run in parallel. n = 3.
The most striking effect was seen for the LacNAc binding-deficient mutants (R186S, R186I, and A146Q/R186I), which did not induce any release of oxygen radicals from the primed neutrophils, an observation demonstrating that binding to LacNAc, as found in glycoproteins, is required for neutrophil activation and that binding to lactose, as found in glycolipids, is not enough. Consistent with this is the fact that the R186S mutant also completely lost the ability to bind human serum glycoproteins (50) and, as another consequence, the ability to promote the phagocytosis of apoptotic cells by neutrophils (49).
Among the specificities of the other mutants, the ability to bind saccharides containing GlcNAcβ1–3Gal, as found in repeating polylactosamine structures, correlates best with the ability to activate primed neutrophils. Thus, the mutants that had gained affinity for saccharides, placing GlcNAcβ1–3 in site B galactosides (K176L and K176L/N180T), had a clearly increased potency, shown by a left shift of the dose-response curve (Fig. 6); even K176L/N180T concentrations as low as 0.2 μm induced significant neutrophil activation. On the other hand, the R144S mutant, which had lost affinity for GlcNAcβ1–3-substituted galactosides, had a much reduced ability to activate neutrophils, as shown by a right shift of the dose-response curve, even though it had increased affinity for GalNAcα2–3 substituted galactosides. Binding to α2–3-linked sialylated galactosides did not correlate with the ability to activate neutrophils, as binding to these glycans was lost for the best activating mutants (K176L and K176L/N180T) as well as the poorly activating mutant R144S. The other mutants had a slightly decreased ability to activate the primed neutrophils and, for the remaining combined (A146Q/R186I, A146Q/K176L, A146Q/K176L/N180T) mutants, the potencies were similar to that of the least active single mutant of the combination (Fig. 6). The ability to bind a single non-extended LacNAc, which is maintained in most mutants, also did not correlate with the ability to activate the neutrophils.
These results suggest that the best activating receptors of galectin-3 on neutrophils contain a repeating LacNAcβ1–3LacNAc moiety, where the second LacNAc must be available for binding in site C-D (e.g. non-fucosylated on the GlcNAc), although the first LacNAc can be modified by e.g. Fucα1–3 on the GlcNAc. Such structures are prominent and typical on neutrophil N- and O-glycans (51–53), and they have also been found in some purified neutrophil glycoproteins (54–57). Moreover, a neutrophil N-glycopeptide fraction was found to bind tightly to galectin-3 (granulocyte LAG glycopeptide in Table 2 of Sparrow et al. (24)). In what may be a related phenomenon, Stowell et al. (17) concluded, based on the treatment of cells with neuraminidases and endo-β-galactosidases, that the galectin-3 induced exposure of phosphatidylserine in differentiated HL60-cells (a widely used neutrophil model) was also mediated by polylactosamine structures.
The priming event, required to make neutrophils sensitive to activation by galectin-3, entails, among other effects, the mobilization of intracellular granule components to the plasma membrane, which may act as galectin-3 receptors (23, 47, 58). Therefore, we compared the binding of wild type galectin-3 and five mutants to primed and non-primed neutrophils. The neutrophils bound wild type galectin-3 and the K176L mutant about equally well and in both cases 30–50% more after TNFα priming (Fig. 7). The R186S mutant did not bind any of the neutrophil preparations, whereas the other mutants showed intermediate binding without significant increase after priming. Taken together these observations suggest that neutrophil priming leads to an increase in polylactosamine-like glycans at the cell surface. Consistent with this suggestion is the fact that galectin-3 receptors have been found in gelatinase and specific granules (23). The glycans of the intracellular granules have not been analyzed in detail, but one report on α-1-acid glycoprotein from neutrophils (59) and another on granule glycolipids (60) do suggest an increased level of structures with repeating LacNAc residues.
FIGURE 7.
Binding of wild type galectin-3 and mutants to unprimed and TNFα-primed neutrophils. FITC-labeled galectin was incubated with freshly prepared neutrophils that had been pretreated by incubation for 30 min at 37 °C without (white bars) or with TNFα (gray bars) and at 4 °C without TNFα (black bars) as a control to prevent granule mobilization. The amount of bound galectin was measured by flow cytometry and is given as the mean and S.E. of the geometric mean of fluorescence intensity (y axis) from three experiments on different batches of neutrophils. The p values were calculated from paired one-tailed t tests; paired, to discount the variability between the neutrophils, and one-tailed, to specifically test the hypothesis of an increased binding after incubation with TNFα.
Biological Effect of Intracellularly Expressed Mutants; Intracellular Trafficking
Galectin-3 has been shown to be involved in directing intracellular trafficking of glycoproteins by entering selected vesicles from the cytosol and then binding to and cross-linking the glycoproteins (28, 61), and it associates with different intracellular vesicles in a dynamic way dependent on cell density and maturation (62, 63). To begin exploring the relationship of galectin-3 carbohydrate binding specificity to this, we examined wild type galectin-3 and two of the mutants (R186S and G182A), expressed as fusion proteins with EGFP, in the human breast cancer cell line SKBR3, which does not express its own galectin-3 (64). Galectin-3 was present in the cytosol and possibly nucleus of all cells, seen as a diffuse staining covering the cells, albeit of different intensity. Galectin-3 clearly also entered vesicles in some of the cells, seen as strongly fluorescent dots inside the cell (Fig. 8a). This was a dynamic process, with dots seen only in a few cells, estimated to be about 15–30% of cells, depending on how confluent the cells were. There was also a great difference in the number of dots per cell, ranging from 1 to around 30 (bar diagram in Fig. 8a). The predominant pattern with relatively few large dots is consistent with them being recycling endosomes and related compartments as shown for e.g. COS-1 cells and MDCK cells (63).
FIGURE 8.
Expression and localization of GFP-galectin-3 wt and two mutants in SKBR3 breast carcinoma cells. The human breast cancer cell line SKBR3 was stably transfected with the pEGFP-C1 vector carrying the gene for EGFP-galectin-3 wild type (panel a), EGFP-R186S (panel b), and EGFP-G182A (panel c), respectively. Cells were cultured on coverslips, fixed in 4% paraformaldehyde, mounted with Prolong Antifade Reagent with DAPI, and analyzed by fluorescence microscopy. The number of galectin containing vesicles (seen as fluorescent dots) per cell was counted in 300 cells for each case and are represented as bar diagrams under each sample microscope panel.
The R186S mutant was chosen as a potential negative control because it does not bind common glycoproteins, as described above. Expressed as an EGFP fusion protein in MDCK cells, it did not enter vesicles and did not direct apical targeting of glycoproteins (61), and it did not accumulate around invading Shigella bacteria (65) as did wild type galectin-3. Under the present conditions it also was not visible in vesicles in the breast carcinoma cell line, SKBR3 (Fig. 8b), whereas galectin-3 wild type was.
The G182A mutant was included in these experiments because it has lost the cytosol-based tumor growth-promoting effects of wild type galectin-3 (as discussed above), and as shown in this study, it has altered carbohydrate binding specificity. The G182A mutant did accumulate in SKBR3 vesicles, but they were much fewer in number per cell (Fig. 8c) compared with that seen with wild type galectin-3. The pattern with one dot in some cells resembles the transient association of galectin-3 with centrosomes in Madin-Darby canine kidney cells (62), although there may be other explanations. In either case, it is clear that the G182A mutation alters the intracellular trafficking of galectin-3. As such, there may be a link between galectin-3 carbohydrate binding specificity and its role in intravesicular sorting on the one hand and its previously proposed cytosolic anti-apoptotic and Ras-regulating effect on the other (43, 44). Further evidence for such a link is the reversal of galectin-3 anti-apoptotic effect by an inhibitor of its carbohydrate binding activity (66).
Studies of Mutants of Other Galectins
A few studies have analyzed the biological effects of galectin mutants, but most cases related it only to simple carbohydrate binding activity (or lack thereof), and the role of fine specificity may have been overlooked. Mutation of a residue corresponding to Arg-186 in galectin-1 (R73H) resulted in lost binding to asialofetuin, which could be due to selective loss of LacNAc affinity as found here, although affinity for lactose was not tested (67). In a recent structural and mutational study of a mushroom galectin (68), the corresponding mutant (R85A) had lost extracellular apoptosis-inducing activity but retained lactose binding activity, a result that could also be explained by a selective loss of affinity for LacNAc and as a consequence lost binding to cell surface glycoproteins. This and other studies have also analyzed mutants outside the carbohydrate recognition site, potentially disrupting other functional interactions of the galectin, such as dimer formation. The purpose of the present study, however, was to specifically link the effects on carbohydrate-binding fine specificity with biological function to begin to answer the following question.
What Is the Role of Galectin-3 Multifaceted Specificity?
Here we show that the preference for 4-linked GlcNAc in site D, dependent on Arg-186, is required for the binding of galectin-3 to glycoproteins, cell surfaces, activation of neutrophil leukocytes, and entry into vesicles. The optimal activation of neutrophils also required the accommodation of GlcNAcβ in site B and, hence, an internal LacNAc in site C-D, suggesting binding to polylactosaminoglycans. Even if the binding of terminal LacNAc residues, the tolerance for NeuAcα2–3 substituents, and a preference for GalNAcα (and Galα) in site B are not required for activation of neutrophils by galectin-3, these properties may be important in other contexts. Serum glycoproteins, for example, bind galectin-3 with μm affinity or better even if they do not contain polylactosamine structures (50). Hence, galectin-3 must bind the relatively rare terminal or α2–3-sialylated LacNAc residues found on these molecules as it does not bind to the more common NeuAcα2–6-capped structures. The binding to GalNAcα or Galα containing blood group determinants may be important in intestinal epithelial cells where N-glycans are capped by these structures instead of sialic acid (69) and where galectin-3 plays an important role in directing the apical targeting of certain glycoproteins (70). Roles in binding microbes are also possible (4, 31).
Although not yet fully characterized, it seems clear that the multifaceted specificity shown by galectin-3 serves to confer on it different properties for different cell types and/or cell compartments, depending on the match with the glycan structures present, a property already shown to be important in the intracellular sorting of galectin-8 (14). A similar analysis of the mutants reported here is ongoing in our group, and it is now clear that, relative to wild type galectin-3, they too show differences in intracellular sorting after endocytosis.
Supplementary Material
Acknowledgments
The glycan-array analysis was performed by the Consortium for Functional Glycomics (CoreH, Emory School of Medicine, Atlanta, GA) funded by NIGMS, National Institutes of Health Grant GM62116.
This work was funded by the Lund University Research School in Pharmaceutical Sciences, the Swedish Research Council, the programs “Glycoconjugates in Biological Systems” and “Chemistry for the Life Sciences” sponsored by the Swedish Strategic Research Foundation, and the Royal Physiographic Society in Lund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Results, Figs. S1–S3, and Tables S1–S3.
E. Salomonsson and H. Leffler, unpublished information.
H. Leffler and J. M. Rini, unpublished information.
- CRD
- carbohydrate recognition domain
- FA
- fluorescence anisotropy
- LacNAc
- N-acetyllactosamine (Galβ1–4GlcNAc)
- Lac
- lactose (Galβ1–4Glc)
- RFU
- relative fluorescence unit
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Barondes S. H., Cooper D. N., Gitt M. A., Leffler H. (1994) J. Biol. Chem. 269, 20807–20810 [PubMed] [Google Scholar]
- 2.Leffler H., Carlsson S., Hedlund M., Qian Y., Poirier F. (2004) Glycoconj. J. 19, 433–440 [DOI] [PubMed] [Google Scholar]
- 3.Rabinovich G. A., Toscano M. A. (2009) Nat. Rev. Immunol. 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 4.Vasta G. R. (2009) Nat. Rev. Microbiol. 7, 424–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F. T., Rabinovich G. A. (2005) Nat. Rev. Cancer 5, 29–41 [DOI] [PubMed] [Google Scholar]
- 6.Yang R. Y., Rabinovich G. A., Liu F. T. (2008) Expert Rev. Mol. Med. 10, e17. [DOI] [PubMed] [Google Scholar]
- 7.Seelenmeyer C., Wegehingel S., Tews I., Künzler M., Aebi M., Nickel W. (2005) J. Cell Biol. 171, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickel W. (2003) Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 9.Garner O. B., Baum L. G. (2008) Biochem. Soc. Trans. 36, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau K. S., Dennis J. W. (2008) Glycobiology 18, 750–760 [DOI] [PubMed] [Google Scholar]
- 11.Patnaik S. K., Potvin B., Carlsson S., Sturm D., Leffler H., Stanley P. (2006) Glycobiology 16, 305–317 [DOI] [PubMed] [Google Scholar]
- 12.Cabrera P. V., Amano M., Mitoma J., Chan J., Said J., Fukuda M., Baum L. G. (2006) Blood 108, 2399–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toscano M. A., Bianco G. A., Ilarregui J. M., Croci D. O., Correale J., Hernandez J. D., Zwirner N. W., Poirier F., Riley E. M., Baum L. G., Rabinovich G. A. (2007) Nat. Immunol. 8, 825–834 [DOI] [PubMed] [Google Scholar]
- 14.Carlsson S., Carlsson M. C., Leffler H. (2007) Glycobiology 17, 906–912 [DOI] [PubMed] [Google Scholar]
- 15.Amano M., Galvan M., He J., Baum L. G. (2003) J. Biol. Chem. 278, 7469–7475 [DOI] [PubMed] [Google Scholar]
- 16.Seetharaman J., Kanigsberg A., Slaaby R., Leffler H., Barondes S. H., Rini J. M. (1998) J. Biol. Chem. 273, 13047–13052 [DOI] [PubMed] [Google Scholar]
- 17.Stowell S. R., Arthur C. M., Mehta P., Slanina K. A., Blixt O., Leffler H., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leffler H., Barondes S. H. (1986) J. Biol. Chem. 261, 10119–10126 [PubMed] [Google Scholar]
- 19.Carlsson S., Oberg C. T., Carlsson M. C., Sundin A., Nilsson U. J., Smith D., Cummings R. D., Almkvist J., Karlsson A., Leffler H. (2007) Glycobiology 17, 663–676 [DOI] [PubMed] [Google Scholar]
- 20.Hsu D. K., Chen H. Y., Liu F. T. (2009) Immunol. Rev. 230, 114–127 [DOI] [PubMed] [Google Scholar]
- 21.Dumic J., Dabelic S., Flögel M. (2006) Biochim. Biophys. Acta 1760, 616–635 [DOI] [PubMed] [Google Scholar]
- 22.Henderson N. C., Sethi T. (2009) Immunol. Rev. 230, 160–171 [DOI] [PubMed] [Google Scholar]
- 23.Karlsson A., Follin P., Leffler H., Dahlgren C. (1998) Blood 91, 3430–3438 [PubMed] [Google Scholar]
- 24.Sparrow C. P., Leffler H., Barondes S. H. (1987) J. Biol. Chem. 262, 7383–7390 [PubMed] [Google Scholar]
- 25.Knibbs R. N., Agrwal N., Wang J. L., Goldstein I. J. (1993) J. Biol. Chem. 268, 14940–14947 [PubMed] [Google Scholar]
- 26.Sato S., Hughes R. C. (1992) J. Biol. Chem. 267, 6983–6990 [PubMed] [Google Scholar]
- 27.Henrick K., Bawumia S., Barboni E. A., Mehul B., Hughes R. C. (1998) Glycobiology 8, 45–57 [DOI] [PubMed] [Google Scholar]
- 28.Delacour D., Koch A., Jacob R. (2009) Traffic 10, 1405–1413 [DOI] [PubMed] [Google Scholar]
- 29.John C. M., Jarvis G. A., Swanson K. V., Leffler H., Cooper M. D., Huflejt M. E., Griffiss J. M. (2002) Cell Microbiol. 4, 649–662 [DOI] [PubMed] [Google Scholar]
- 30.van den Berg T. K., Honing H., Franke N., van Remoortere A., Schiphorst W. E., Liu F. T., Deelder A. M., Cummings R. D., Hokke C. H., van Die I. (2004) J. Immunol. 173, 1902–1907 [DOI] [PubMed] [Google Scholar]
- 31.Stowell S. R., Arthur C. M., Dias-Baruffi M., Rodrigues L. C., Gourdine J. P., Heimburg-Molinaro J., Ju T., Molinaro R. J., Rivera-Marrero C., Xia B., Smith D. F., Cummings R. D. (2010) Nat. Med. 16, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cumpstey I., Salomonsson E., Sundin A., Leffler H., Nilsson U. J. (2008) Chemistry 14, 4233–4245 [DOI] [PubMed] [Google Scholar]
- 33.Yu L. G., Andrews N., Zhao Q., McKean D., Williams J. F., Connor L. J., Gerasimenko O. V., Hilkens J., Hirabayashi J., Kasai K., Rhodes J. M. (2007) J. Biol. Chem. 282, 773–781 [DOI] [PubMed] [Google Scholar]
- 34.Massa S. M., Cooper D. N., Leffler H., Barondes S. H. (1993) Biochemistry 32, 260–267 [DOI] [PubMed] [Google Scholar]
- 35.Shoji H., Nishi N., Hirashima M., Nakamura T. (2003) J. Biol. Chem. 278, 12285–12293 [DOI] [PubMed] [Google Scholar]
- 36.Agrwal N., Sun Q., Wang S. Y., Wang J. L. (1993) J. Biol. Chem. 268, 14932–14939 [PubMed] [Google Scholar]
- 37.Sörme P., Kahl-Knutsson B., Huflejt M., Nilsson U. J., Leffler H. (2004) Anal. Biochem. 334, 36–47 [DOI] [PubMed] [Google Scholar]
- 38.Sörme P., Arnoux P., Kahl-Knutsson B., Leffler H., Rini J. M., Nilsson U. J. (2005) J. Am. Chem. Soc. 127, 1737–1743 [DOI] [PubMed] [Google Scholar]
- 39.Walser P. J., Haebel P. W., Künzler M., Sargent D., Kües U., Aebi M., Ban N. (2004) Structure 12, 689–702 [DOI] [PubMed] [Google Scholar]
- 40.Ban M., Yoon H. J., Demirkan E., Utsumi S., Mikami B., Yagi F. (2005) J. Mol. Biol. 351, 695–706 [DOI] [PubMed] [Google Scholar]
- 41.Böyum A. (1968) Scand J. Clin. Lab. Invest. Suppl 97, 77–89 [PubMed] [Google Scholar]
- 42.Dahlgren C., Karlsson A. (1999) J. Immunol. Methods 232, 3–14 [DOI] [PubMed] [Google Scholar]
- 43.Akahani S., Nangia-Makker P., Inohara H., Kim H. R., Raz A. (1997) Cancer Res. 57, 5272–5276 [PubMed] [Google Scholar]
- 44.Shalom-Feuerstein R., Cooks T., Raz A., Kloog Y. (2005) Cancer Res. 65, 7292–7300 [DOI] [PubMed] [Google Scholar]
- 45.Diehl C., Genheden S., Modig K., Ryde U., Akke M. (2009) J. Biomol. NMR 45, 157–169 [DOI] [PubMed] [Google Scholar]
- 46.Houzelstein D., Gonçalves I. R., Fadden A. J., Sidhu S. S., Cooper D. N., Drickamer K., Leffler H., Poirier F. (2004) Mol. Biol. Evol. 21, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 47.Feuk-Lagerstedt E., Jordan E. T., Leffler H., Dahlgren C., Karlsson A. (1999) J. Immunol. 163, 5592–5598 [PubMed] [Google Scholar]
- 48.Forsman H., Salomonsson E., Onnheim K., Karlsson J., Björstad A., Leffler H., Bylund J., Karlsson A., Dahlgren C. (2008) Glycobiology 18, 905–912 [DOI] [PubMed] [Google Scholar]
- 49.Karlsson A., Christenson K., Matlak M., Björstad A., Brown K. L., Telemo E., Salomonsson E., Leffler H., Bylund J. (2009) Glycobiology 19, 16–20 [DOI] [PubMed] [Google Scholar]
- 50.Cederfur C., Salomonsson E., Nilsson J., Halim A., Oberg C. T., Larson G., Nilsson U. J., Leffler H. (2008) Glycobiology 18, 384–394 [DOI] [PubMed] [Google Scholar]
- 51.Fukuda M., Carlsson S. R., Klock J. C., Dell A. (1986) J. Biol. Chem. 261, 12796–12806 [PubMed] [Google Scholar]
- 52.Fukuda M., Spooncer E., Oates J. E., Dell A., Klock J. C. (1984) J. Biol. Chem. 259, 10925–10935 [PubMed] [Google Scholar]
- 53.Fukuda M. N., Dell A., Oates J. E., Wu P., Klock J. C., Fukuda M. (1985) J. Biol. Chem. 260, 1067–1082 [PubMed] [Google Scholar]
- 54.Asada M., Furukawa K., Kantor C., Gahmberg C. G., Kobata A. (1991) Biochemistry 30, 1561–1571 [DOI] [PubMed] [Google Scholar]
- 55.Furukawa K., Funakoshi Y., Autero M., Horejsi V., Kobata A., Gahmberg C. G. (1998) Eur. J. Biochem. 251, 288–294 [DOI] [PubMed] [Google Scholar]
- 56.Rudd P. M., Mattu T. S., Masure S., Bratt T., Van den Steen P. E., Wormald M. R., Küster B., Harvey D. J., Borregaard N., Van Damme J., Dwek R. A., Opdenakker G. (1999) Biochemistry 38, 13937–13950 [DOI] [PubMed] [Google Scholar]
- 57.Sato T., Furukawa K., Autero M., Gahmberg C. G., Kobata A. (1993) Biochemistry 32, 12694–12704 [DOI] [PubMed] [Google Scholar]
- 58.Almkvist J., Fäldt J., Dahlgren C., Leffler H., Karlsson A. (2001) Infect. Immun. 69, 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Theilgaard-Mönch K., Jacobsen L. C., Rasmussen T., Niemann C. U., Udby L., Borup R., Gharib M., Arkwright P. D., Gombart A. F., Calafat J., Porse B. T., Borregaard N. (2005) J. Leukoc Biol. 78, 462–470 [DOI] [PubMed] [Google Scholar]
- 60.Karlsson A., Miller-Podraza H., Johansson P., Karlsson K. A., Dahlgren C., Teneberg S. (2001) Glycoconj J. 18, 231–243 [DOI] [PubMed] [Google Scholar]
- 61.Delacour D., Greb C., Koch A., Salomonsson E., Leffler H., Le Bivic A., Jacob R. (2007) Traffic 8, 379–388 [DOI] [PubMed] [Google Scholar]
- 62.Koch A., Poirier F., Jacob R., Delacour D. (2010) Mol. Biol. Cell 21, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider D., Greb C., Koch A., Straube T., Elli A., Delacour D., Jacob R. (2010) Eur. J. Cell Biol. 89, 788–798 [DOI] [PubMed] [Google Scholar]
- 64.Ruebel K. H., Jin L., Qian X., Scheithauer B. W., Kovacs K., Nakamura N., Zhang H., Raz A., Lloyd R. V. (2005) Cancer Res. 65, 1136–1140 [DOI] [PubMed] [Google Scholar]
- 65.Paz I., Sachse M., Dupont N., Mounier J., Cederfur C., Enninga J., Leffler H., Poirier F., Prevost M. C., Lafont F., Sansonetti P. (2010) Cell Microbiol. 12, 530–544 [DOI] [PubMed] [Google Scholar]
- 66.Lin C. I., Whang E. E., Donner D. B., Jiang X., Price B. D., Carothers A. M., Delaine T., Leffler H., Nilsson U. J., Nose V., Moore F. D., Jr., Ruan D. T. (2009) Mol. Cancer Res. 7, 1655–1662 [DOI] [PubMed] [Google Scholar]
- 67.Hirabayashi J., Kasai K. (1991) J. Biol. Chem. 266, 23648–23653 [PubMed] [Google Scholar]
- 68.Yang N., Li D. F., Feng L., Xiang Y., Liu W., Sun H., Wang D. C. (2009) J. Mol. Biol. 387, 694–705 [DOI] [PubMed] [Google Scholar]
- 69.Finne J., Breimer M. E., Hansson G. C., Karlsson K. A., Leffler H., Vliegenthart J. F., van Halbeek H. (1989) J. Biol. Chem. 264, 5720–5735 [PubMed] [Google Scholar]
- 70.Delacour D., Koch A., Ackermann W., Eude-Le Parco I., Elsässer H. P., Poirier F., Jacob R. (2008) J. Cell Sci. 121, 458–465 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.