Abstract
The Slit-Robo GTPase-activating proteins (srGAPs) are critical for neuronal migration through inactivation of Rho GTPases Cdc42, Rac1, and RhoA. Here we report that srGAP2 physically interacts with protein arginine methyltransferase 5 (PRMT5). srGAP2 localizes to the cytoplasm and plasma membrane protrusion. srGAP2 knockdown reduces cell adhesion spreading and increases cell migration, but has no effect on cell proliferation. PRMT5 binds to the N terminus of srGAP2 (225–538 aa) and methylates its C-terminal arginine residue Arg-927. The methylation mutant srGAP2-R927A fails to rescue the cell spreading rate, is unable to localize to the plasma membrane leading edge, and perturbs srGAP2 homodimer formation mediated by the F-BAR domain. These results suggest that srGAP2 arginine methylation plays important roles in cell spreading and cell migration through influencing membrane protrusion.
Keywords: Cell Migration, Plasma Membrane, Protein Domains, Protein Methylation, Protein-Protein Interactions, Rho, F-BAR Domain, PRMT5, Cell Spreading, srGAP2
Introduction
Cell migration is essential for a wide variety of physiological and pathological processes such as embryonic development, wound tissue repair, and tumor metastasis (1–2). Classically, the major driving force for migration in mammalian cells is thought to be provided by actin polymerization at the cell protrusion of the front edge and F-actin depolymerization to retract cell body at the rear (3). These coordinated processes are regulated by small Rho GTPases, especially Cdc42, Rac1, and RhoA (4–5). Similar to other GTPases, Rho GTPases act as intracellular molecular switches, cycling between a GDP-bound inactive form and a GTP-bound active form. The two interconvertible forms are controlled by two classes of proteins, guanine nucleotide exchange factors (GEFs),2 which promote the exchange of GDP for GTP, and GTPase-activating proteins (GAPs), which increase the GTPase activity of Rho GTPases through converting GTP to GDP, thereby closing the switch (6–7).
srGAP is a family of RhoGAP proteins, identified in the Slit-Robo signal pathway. The srGAP family consists of three members, srGAP1, -2, and -3. All of them possess three conserved domains, F-BAR, RhoGAP, and SH3 (8). The function of RhoGAP domain of srGAP family protein is extensively studied because of its role in the negative regulation of Rho GTPase activities important for cytoskeleton rearrangement (8–9). In addition to the RhoGAP domain, studies have found that the F-BAR domain-containing proteins function as key regulators in membrane remodeling in eukaryotes. Each of these proteins shares a similar quaternary “banana-like” structure, which forms elongated dimers mediated by the anti-parallel association of α helices in each monomer (10–11). Moreover, positive charges within the “banana-like” structure are aligned to interact with negative charges of the membrane via electrostatic interactions (12). Therefore, the F-BAR domains act as membrane-associated scaffolds and deform cell membrane independently of its F-actin bundling activity (13–14).
Based on structural characteristics and phylogenetic analysis, the BAR domain super family includes “classical” BAR domain, F-BAR domain (FCH domain, followed by a coiled-coil region) (15–17) and I-BAR (Inverse-BAR) domain (11). Classical BAR and F-BAR domains promote the formation of plasma membrane invaginations in endocytosis (14–16). In contrast, I-BAR domains induce plasma membrane protrusions to generate filopodia in vitro and in living cells (16, 18–22). Interestingly, unlike canonical F-BAR, the F-BAR domain of srGAP2 deforms membrane like an I-BAR domain to induce filopodia formation. F-BAR is necessary and sufficient for srGAP2 to directly regulate membrane deformation observed in COS7 cells and in neuronal migration and morphogenesis (23). However, the detail mechanism by which the auto-inhibited srGAP2 monomers are transformed into the functional homodimers via the F-BAR domain remains somewhat unclear.
S-Adenosylhomocysteine hydrolase (SAHH) has been shown to enrich at the leading edge of motile Dictyostelium amoebae and human neutrophils (24). SAHH promotes SAM-dependent methylation in eukaryotes (25). Although in general, SAM-dependent methylation functions at the front of migrating cells, the key methyltransferase(s) and critical protein substrate(s) remain unknown. Our previous study has shown that srGAP2 co-exists in a complex with the protein arginine methyltransferase PRMT5 (26). PRMT5 is critical for cell proliferation (27–28) and differentiation (29–31) through modification of histones (14, 32–33) and RNA splicing factors (34). We find recently that PRMT5 dominantly localizes to cytoplasm in somatic cells, and methylates golgin GM130 and ribosomal protein Rps10 (26, 35). In this report, we show that srGAP2 homodimerization depends on its arginine methylation catalyzed by PRMT5. The formation of srGAP2 homodimers promotes its binding and deformation of cell membrane, leading to accelerated cell membrane protrusion and cell spreading.
EXPERIMENTAL PROCEDURES
DNA Constructs and Antibodies
Human srGAP2 full-length cDNA clone (KIAA0456) was generously provided by Riken. PRMT5 (BC025979) was purchased from Open Biosystems and its subcloning as described (26, 35). Reps1N(1–373 aa), Reps1C (367–743 aa), srGAP2, srGAP2N (2–538 aa), srGAP2N1 (2–538 aa), srGAP2N2 (2–118 aa), srGAP2N3 (225–538 aa), srGAP2C (485–1068 aa), srGAP2C2 (673–1068 aa), srGAP2C3 (783–1068 aa), and srGAP2C4 (783–854 aa) were subcloned into pET-28b (Novagen), pGEX4T-1/3 (GE Healthcare), pCMV-Tag 2B (Stratagene), pEGFPN3/C1 (Clontech), and pcDNA-3×HA-B (a gift of Dr. Zhiheng Xu) in-frame by PCR (as specified in each experiment). The U6 promoter-driven shRNA expression vector pDsU6 and control shRNA (siLuc) were previously described (36). Two short hairpin-based shRNA silencing vectors were generated, which were specific for target srGAP2 mRNA codon sequence. The target sites in the srGAP2 coding region were 466–485 and 721–741 (+1 being the A residue in the initiating codon) and were verified in the human genome sequence data base (NCBI) as unique sequences. Two high-efficiency vectors pDs466 and pDs721 were named as U1 and U2, respectively. The shRNA in U1 was subcloned into a GFP-containing shRNA expression vector, and the construct was named GFP-U1. srGAP2 shRNA-resistant mutant (srGAP2-R-GFP) construct was prepared by introducing two silent mutations within the U1-targeting sequence using site-directed mutagenesis with the sequence 5′-GTCCTGAACGAaCTgTACTCG-3′. srGAP2 methylation mutant (R927A, abbreviated with m) was generated with site-directed mutagenesis, and srGAP2-dm-GFP was constructed from the plasmids of srGAP2-R-GFP and srGAP2-m-GFP. HA-Rac1V12 construct was a generous gift from Dr. Kozo Kaibuchi (Nagoya University), and GST-PBD was a kind gift from Dr. Xueliang Zhu (SIBS). All constructs were verified by double-stranded DNA sequencing. The polyclonal antibody against PRMT5 was purchased from Upstate. The monoclonal antibodies against Actin, GST, Flag, and HA were products of Sigma. Monoclonal antibody against GFP was purchased from Clontech. Anti-Rac1 antibody was from Millipore. The polyclonal and monoclonal antibodies raised against the C terminus of srGAP2 (783–1068) were made by Animal Center of IGDB.
Cell Culture, Transfection, and Immunofluorescence Microscopy
HCT116, HEK293, CHO, HeLa, and U2OS cells were maintained at 37 °C, and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) plus 10% fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin (Invitrogen). Cells were transfected using Lipofectamine 2000 (Invitrogen) following manufacturer's recommendations. For immunofluorescence staining, cells grown on coverslips were either fixed to stain with indicated antibodies or fluorescent dyes, or transfected with GFP, srGAP2-R-GFP, or srGAP2-dm-GFP, and then fixed to stain with TRITC 545-conjugated phalloidin (Sigma). The stained cells were visualized under the Spectral Imaging Confocal Microscope DIGITAL ECLIPSE C1Si (Nikon, Japan). Images were acquired using a 100× Plan Apochromat VC NA 1.40 oil objective and analyzed using the NIS-Elements AR software provided by Nikon.
Colony Formation Assay and Stable Cell Lines Setup
HCT116 cells were plated on 60-mm dishes (6 × 105 cells/dish), cultured for 24 h at 37 °C, and then transfected with 3 μg of siLuc, U1 and U2, respectively. Cells were reseeded on 100-mm dishes 24 h after transfection (3 × 104 cells/dish), and cultured in 10% FBS of DMEM with 800 μg/ml G418 (Calbiochem). After a 2-week selection, the cell colonies were photographed using an inverted microscope for colony formation assay. For stable cell lines setup, the colonies above were picked into 24-well plate, cultured in 10% FBS of DMEM with 200 μg/ml G418, and then screened with Western blot. The positive cell lines were freezed or used for other specific experiments.
Cell Proliferation Assay
5 × 103 cells in 100 μl of culture medium in each well of 96-well assay plate were maintained for 5 days. At every time point, 20 μl of a modified MTT reagent, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) mixed with an electron coupling reagent (phenazine ethosulfate) from Promega, was added to each well with gentle pipetting. After incubation in the incubator for 90 min at 37 °C, in a humidified, 5% CO2 atmosphere, 25 μl of 10% SDS was added the well with gentle pipetting to stop the reaction, and then absorbance at 490 nm was recorded.
Wound Healing Assay
2 × 106 cells were seeded in 6-well dishes marked a line with a needle along the diameter of the well on the bottom of the dish, allowed to form a confluent monolayer, and then starved in DMEM with 0.5% FBS overnight. The monolayer was scratched perpendicular to the marked line with a 200 μl pipette tip, and the medium was replaced with the complete medium containing 10% FBS. When the destroyed cells in the cell-free area had been repaired after culturing for 12 h, the same wounded areas were photographed using an inverted microscope (Nikon Eclipse TE2000-S) equipped with a digital camera (Nikon DXM1200C) with 10× objective at the intersection of the marked line and wound edge at 0 h and at desired time intervals, and the areas of wound closure were analyzed using the NIH Image J software. Each experiment was carried out three times in duplicate.
Cell Spreading Assay
Cells of ∼80% confluence were detached using EDTA-trypsin (Invitrogen), and the DMEM with 10% FBS was added to terminate trypsinization immediately. The cells were then spun down, resuspended in DMEM containing 10% FBS, and seeded on coverslips in 24-well dishes to culture in the incubator. At various time points, cells were fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton X-100/phosphate-buffered saline (PBS), blocked in 5% BSA/PBS, and then stained with TRITC 545-conjugated phalloidin (Sigma) and DAPI (Vector Laboratories). Fluorescent images from multiple fields in each coverslip were captured with a CCD camera. To obtain quantitative data of the extent of cell spreading, the surface areas of cells were measured with the NIH Image J software, and cells were scored blindly as “spreading” (cell area>250 μm2). At least 100 cells were analyzed per sample. Results were from three independent experiments. Data shown are mean ± S.D./Data are mean ± S.E.
Live Cell Imaging
U2OS cells grown to 20–30% confluency on chambered coverglass (Thermo Scientific Nunc) were transfected with LipofectamineTM 2000 and imaged at 37 °C, with 5% CO2 24 h later. Time-lapse fluorescence images were acquired with the Spectral Imaging Confocal Microscope DIGITAL ECLIPSE C1Si equipped with a T-PFS Perfect Focus Unit and a 20× Plan Apochromat VC NA 0.75 DIC Dry objective. Images were captured with the EZ-C1 Spectral software (Nikon). Single-channel images were acquired at 8-min intervals. Images were exported as TIFF files using NIS-element AR (Nikon).
Protein Purification and Methylation Analysis
GST, GST-fused proteins, and His-fused proteins expression, purification, and methylation assays were carried out as previously described (37).
Immunoprecipitation, in Vitro Binding Assay, and Western Immunoblotting
For reciprocal immunoprecipitation, 800 μg of cell (transfected with foreign DNA or not) extract protein was incubated overnight with 2 μg of antibody and protein A/G-agarose beads (OncogeneScience) at 4 °C, or incubated with monoclonal anti-HA-agarose (Sigma) for 2 h. For in vitro binding assay, a series of GST-tagged peptides were incubated with Flag-PRMT5 protein purified from CHO cell lysates with Flag beads at 4 °C for 1 h. After extensive washing, all associated proteins were eluted and subjected to Western immunoblotting (as specific experiment). Cell lysis buffer I (50 mm Tris-HCl, pH 7.4, 50 mm NaC1, 1% Triton X-100, 1 mm EDTA, 10 μg/ml leupeptin, and 10 μg/ml aprotinin, Roche) was used for srGAP2-PRMT5 reciprocal immunoprecipitation, cell lysis buffer II (20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 0.1% Triton X-100, 10 μg/ml leupeptin, and 10 μg/ml aprotinin,) was for srGAP2-Rac1 reciprocal immunoprecipitation, cell lysis buffer III (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 15 mm EGTA, 0.1% Triton X-100, 1 mm DTT, 1 mm PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) was for srGAP2 dimer formation assay, and cell lysis buffer IV (50 mm Tris-HCl, 150 mm NaCl, 0.5% Triton X-100, pH 7.4, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) was for in vitro binding assay.
Detection of GTP-bound Rac1 by Use of GST-PBD
The cells were lysed in binding buffer (50 mm Tris, pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 500 mm NaC1, 10 mm MgC12, 10 μg/ml leupeptin, and aprotinin, and 1 mm PMSF). The cleared lysates were then transferred quickly into tubes with GST-PBD (20 μg) bound to glutathione-Sepharose 4B beads and rotated at 4 °C for 45 min. The beads were washed four times with washing buffer containing 1% Triton X-100, 150 mm NaC1, 10 mm MgC12, 10 μg/ml leupeptin and aprotinin, and 0.1 mm PMSF and analyzed for bound protein by Western immunoblotting with anti-Rac1 antibody.
RESULTS
Loss of srGAP2 Increases Cell Migration
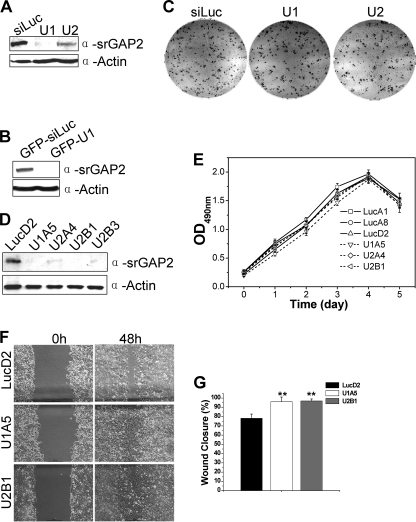
To examine the physiological function of srGAP2 in cells, we generated two short hairpin-based interfering RNA (shRNA) constructs named U1 and U2, which knock down srGAP2 expression efficiently (Fig. 1A) and the shRNA in U1 was subcloned into a GFP-containing shRNA expression vector (GFP-U1) (Fig. 1B). To assess whether srGAP2 is essential for cell proliferation, colony formation assay was performed, and the results (Fig. 1C) showed that srGAP2 knockdown did not cause any measurable reduction when compared with control knockdown cells (siLuc), suggesting that srGAP2 is not essential for cell survival. We then generated four stable cell lines in HCT116 background (U1A5, U2A4, U2B1, and U2B3), which srGAP2 proteins were depleted by U1 or U2, respectively. A control cell line (LucD2) was also generated in parallel (Fig. 1D). Consistent with the clonogenic assay result, depletion of srGAP2 from HCT116 cells did not affect cell viability as determined by the MTS assay (Fig. 1E). However, the srGAP2-depleted cells healed the wound track much faster than control cells (Fig. 1, F and G). Because lack of srGAP2 has no detectable impact on cell growth, most likely the increased wound healing is a result of increased cell migrating rate, suggesting that srGAP2 may be linked to cell migration.
FIGURE 1.
srGAP2 influences cell migration. A, Western blot with anti-srGAP2 and anti-actin antibodies, 100 μg of whole cell lysates from HCT116 cells transfected with shRNA. U1 and U2 are two shRNAs of srGAP2 and siLuc as control. B, 100 μg of whole cell lysates from HCT116 cells transfected with either GFP-siLuc (as control) or GFP-U1 probed with anti-srGAP2 and anti-actin antibodies. C, colony formation assay: HCT116 cells transfected with shRNAs as indicated. D, Western blot showing the srGAP2 expression levels of HCT116 stable cell lines. LucD2 was stably expressing siLuc, U1A5 or U2A4, U2B1 and U2B3 represented the clone number stably expressing srGAP2-shRNA U1 or U2. Actin shows equal loading. E, growth curves of control cells (LucA1, LucA8, and LucD2) and srGAP2-depleted cells in D by MTS analysis. F, wound healing assay. The confluent monolayers of LucD2 (control), U1A5, and U2B1 cells were scratched to remove cells, and images of the cells were collected as the 0′ hour time point (left panels). The cells were then cultured for 48 h, and captured at the same point of 0′ hour time points as the 48′ hour time point (panels). Scale bar, 50 μm. G, quantitative analysis of cell migration showing the areas of wound closure (%) compared with initial wound areas (0 h) using NIH image J analysis. Data are mean ± S.E. (n = 3; **, p < 0.001; *, p < 0.05; Student's t test).
srGAP2 Is Required for Protrusion Formation and Cell Spreading
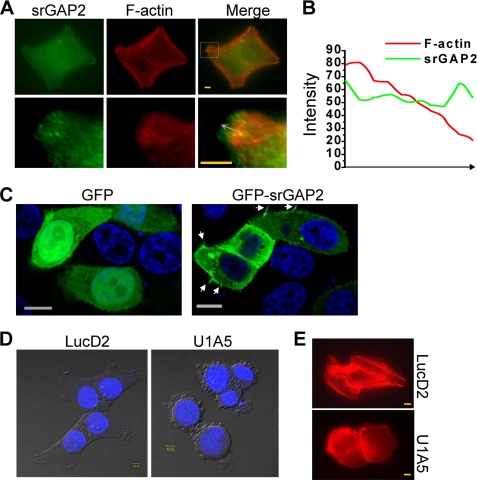
To find out how srGAP2 affects the cell migration, we examined its subcellular localization in HCT116 cells by immunostaining and observed that srGAP2 was enriched at the leading edge of membrane protrusions (Fig. 2A), and the distribution of srGAP2 located to the front of F-actin bundles in the membrane protrusions (Fig. 2B). In addition, GFP-srGAP2 fusion protein was also localized into the protrusional membrane (Fig. 2C). When compared with control cells spreading efficiently on the matrix and showing lamellar protrusions and stress fiber-like actin bundles, the srGAP2-depleted cells displayed much reduced lamellar protrusions and appeared round in shape with a reduction in stress fibers (Fig. 2, D and E).
FIGURE 2.
srGAP2 localizes to sites of membrane protrusion and is required for cell morphology. A, HCT116 cells at 24 h after seeding on the coverslip were double labeled with mouse anti-srGAP2 antibody, followed by anti-mouse FITC-conjugated secondary antibody and TRITC 545-phalloidin. Bars, 10 μm. The lower figures are the high-magnification from the upper selected inset, showing that srGAP2 localized to leading edge of F-actin at the cell membrane protrusion. B, graphs correspond to intensities in arbitrary units of the green (srGAP2) and red (F-actin) labeling for each pixel of the arrow drawn through the axis in A. C, HeLa cells transfected with GFP or GFP-srGAP2. The arrow indicated filopodia-like protrusion. Bars, 10 μm. D and E, srGAP2 regulates the cell morphology. Graphs taken at 33 h after seeding, LucD2 (control) showed well-pronounced protrusions and spread well, U1A5 (srGAP2 depleted) became rounded. DNA was stained with DAPI (blue) and visualized by DIC (D). Cells were stained with TRITC 545-phalloidin to visualize F-actin (red) (E). Bars, 10 μm.
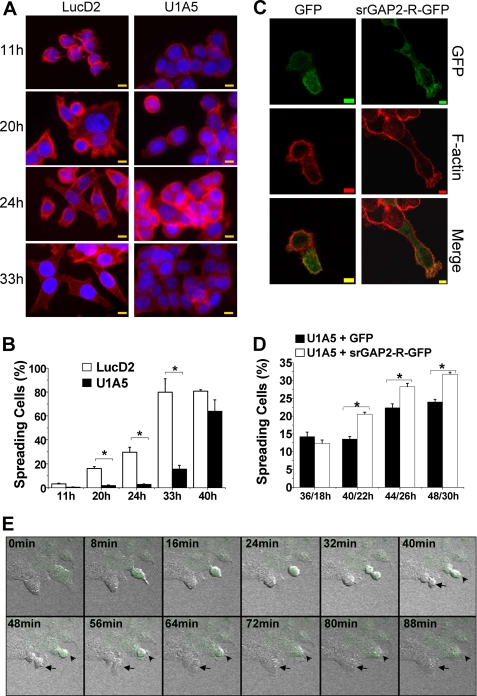
To gain further insights into srGAP2 in cell spreading, we tested F-actin distribution as an indicator of cell spreading at different time intervals after plating the control and srGAP2-depleted cells. We found that only about 1.9, 2.8, and 16% of srGAP2-depleted cells (U1A5) spread at 20, 24, 33 h after plating, respectively. In contrast, about 16, 30, and 80% of control cells spread completely at the respective time points (Fig. 3, A and B). At longer incubation time such as 40 h after plating, the difference of cell spreading rate between srGAP2 lesion and control cells was much less, suggesting that loss of srGAP2 slows down cell spreading process significantly but does not abolish it. To exclude the possibility of the result of nonspecific activation of the RNAi machinery or off-target effects, we performed rescue experiments. The srGAP2-depleted cells (U1A5) transfected with a shRNA-resistant form of srGAP2 (srGAP2-R-GFP) complemented the defects of cell spreading (Fig. 3, C and D).
FIGURE 3.
srGAP2 is required for cell spreading. A, cells were stained at indicated times with TRITC 545-phalloidin to visualize F-actin (red) and DAPI to indicate nuclei (blue). Bars, 10 μm. B, cell spreading of either LucD2 or U1A5 at each time point was counted using the cell spreading assay. Data are mean ± S.E. (n = 3; *, p < 0.05, Student's t test). C, rescuing assay: U1A5 transfected with GFP or srGAP2-R-GFP and stained with TRITC 545-phalloidin to visualize F-actin distribution 22 h after transfection. Bars, 5 μm. D, cell spreading situation of either U1A5 or U1A5 transfected with GFP or srGAP2-R-GFP at each indicated time point (n1/n2: n1 means the length of time after plating the cells, and n2 means the length of time after transfection) was counted using the cell spreading assay; see “Experimental Procedures” for details. Data are mean ± S.E. (n = 3; *, p < 0.05; Student's t test.). E, time-lapse confocal microscopy analysis showing the slowness of extension and spreading after mitosis. Living U2OS cells transfected with the GFP-U1 plasmid was indicated with an arrowhead, and the untransfected control cell shown with an arrow. Living cells were imaged at 8-min intervals.
Adherent eukaryotic cells round up during prophase and metaphase and reacquire their extended and flattened shape during cytokinesis (38–39). Consistently, time-lapse confocal microscopy analysis confirmed that cells transfected with GFP-U1 spread much slower than the untransfected cells (Fig. 3E). This observation suggests that srGAP2 influences cell migration through regulating lamelipodia-like membrane protrusions.
Interaction of srGAP2 and PRMT5
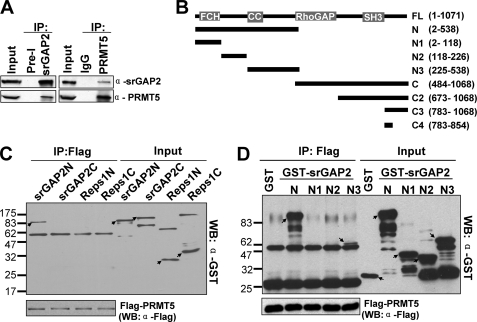
Our previous study has shown that srGAP2 is a component of the PRMT5 protein complex (26). To further verify potential interactions between the endogenous srGAP2 and PRMT5 proteins, we performed co-immunoprecipitation with anti-srGAP2 and -PRMT5 antibodies. As shown in Fig. 4A, reciprocal interactions were confirmed between srGAP2 and PRMT5.
FIGURE 4.
srGAP2 interacts with PRMT5. A, endogenous PRMT5 and srGAP2 interact with each other. HEK293 cell lysates were subjected to immunoprecipitation with anti-srGAP2 or preimmunoserum (Pre-I), and anti-PRMT5 or IgG antibodies as indicated. The immunocomplexes were separately and probed for PRMT5 and srGAP2. B, diagrams of different fragment of srGAP2. C, pull-down assay to test interaction of Flag-tagged PRMT5 with GST-tagged N (1–538 aa)/C (484–1068 aa)-terminal srGAP2 and N (1–373aa)/C (367–743) -terminal Reps1. Western blot analysis was performed with GST and Flag antibodies. D, pull-down assay to test the interaction of Flag-tagged PRMT5 with different fragments of GST-tagged srGAP2-N-terminal. Each protein is indicated with an arrow. Western blot analysis was performed with GST and Flag antibodies.
To localize which region of srGAP2 is required for its interaction with PRMT5, in vitro protein binding analysis was carried out. To avoid the influence of anti-Flag antibody cross reaction with endogenous PRMT5 in human cells, Flag-tagged PRMT5 was purified from Chinese hamster ovarian (CHO) cells with stable expression of Flag-PRMT5 and then incubated with recombinant GST-tagged srGAP2N (1–538 aa), srGAP2C (484–1068 aa), and two GST-tagged fragments (Reps1N (1–373 aa) and Reps1C (367–743 aa)) of Reps1, another component of the PRMT5 complex. As shown (Fig. 4, B–D), Flag-PRMT5 could pull down GST-srGAP2N, but not GST-srGAP2C or GST-Reps1. The PRMT5-srGAP2 interaction domain was further defined to the 225–538 aa N-terminal region of srGAP2, whereas other regions of N-terminal srGAP2 did not interact with Flag-PRMT5.
PRMT5 Methylates srGAP2
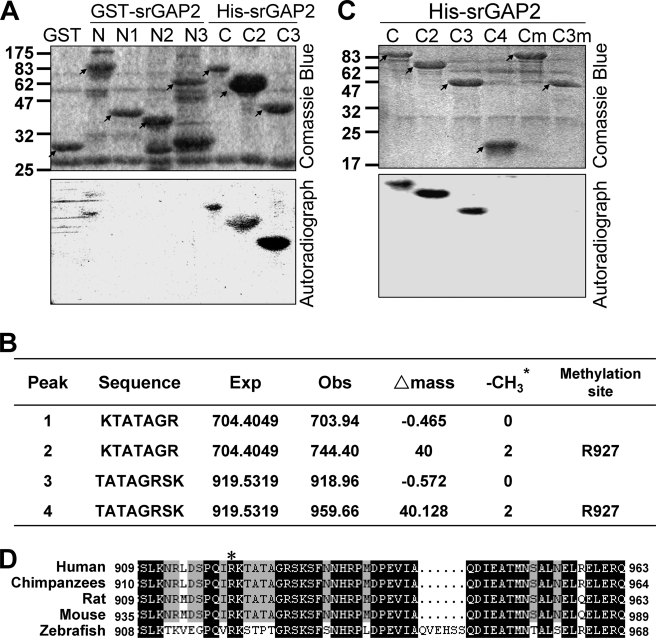
Because srGAP2 interacts with PRMT5, it is reasonable to consider whether srGAP2 is a substrate of PRMT5. We subsequently examined whether PRMT5 can methylate arginine residues of srGAP2. A series of recombinant fragments in srGAP2 purified from Escherichia coli were incubated with Flag-PRMT5 purified from CHO cells in the presence of tritium-labeled SAM. In vitro methylation results indicated that the C terminus of srGAP2 was methylated by PRMT5, and the signal from the srGAP2C3 fragment (783- 1068 aa) was the most prominent, whereas no methylation was observed with GST or the srGAP2N-terminal GST fusion (GST-srGAP2N, N1, N2, and N3). Thus, it appeared that PRMT5 most likely methylates arginine residue(s) in the srGAP2C3 region (Fig. 5A). Consistently, mass spectrometry analysis of the 30.4 kDa methylated srGAP2C3 polypeptide identified Arg-927 in srGAP2 as the residue methylated by PRMT5 (Fig. 5B).
FIGURE 5.
PRMT5 methylates srGAP2 in vitro. A, methylation assay in vitro. 2–5 μg of GST, GST-tagged srGAP2-N, N1, N2, and N3, and His-tagged C, C2, and C3 were purified from E. coli, and incubated with 1 μg of Flag-PRMT5, purified from CHO cells. Incorporation of 3H-labeled methyl groups (from the SAM donor) was visualized by autoradiography. Each protein is indicated with an arrow. B, 30.4 kDa methylated polypeptide band (srGAP2-C3) in Fig. 5A was fragmented and analyzed by mass spectrometry, and the result indicated the molecular weight of srGAP2 fragments and assignment of the methylated arginine. Note that the mass shifts reflect the tritium molecular weight. * represents 3H-labeled -CH3. C, arginine residues in srGAP2 C and C3 fragments were mutated to alanine (R927A). 2–5 μg of His-tagged C, C2, C3, C4, and the R927A mutants in C (Cm) and C3 (C3m) were purified from E. coli, and the methylation assay as in A. Each protein is indicated with an arrow. D, amino acid sequence alignment of human Arg-927 site of srGAP2, marked with *, in some eukaryotic species was performed using DNAMAN version 4.0 software. Conserved residues are highlighted with the following color type: black (=100%) and gray (≥75%).
To confirm the mass spectrometry results, we generated Arg-927 → Ala substitution (R927A) in srGAP2. In vitro methylation assay showed that the methylation signals drastically reduced in the R927A srGAP2 C or C3 mutants (Fig. 5C). This result suggests that Arg-927 in srGAP2 is the primary target site of PRMT5. Amino acid sequence alignment of vertebrate srGAP2 suggests that Arg-927 is highly conserved in evolution (Fig. 5D). It raises the possibility that the function of Arg-927 methylation in srGAP2 is significant.
srGAP2 Arginine Methylation Affects Cell Spreading
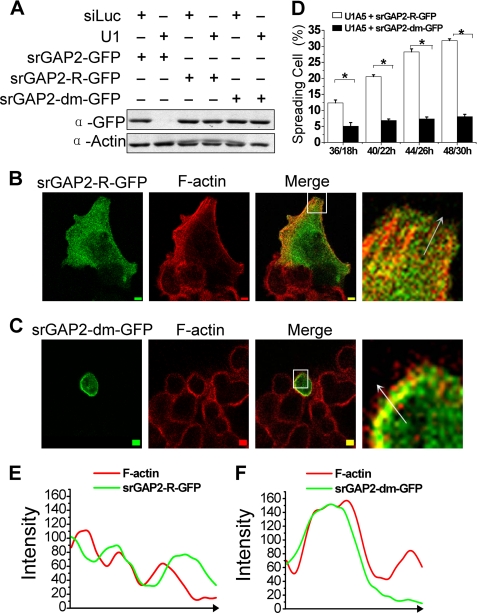
To test whether the function of srGAP2 methylation by PRMT5 is involved in cell spreading, we first generated a GFP-tagged srGAP2 R927A mutant (srGAP2-dm-GFP), which is also resistant to srGAP2 shRNA (Fig. 6A). Cell spreading assay indicated that srGAP2-R-GFP rescued the cell spreading defect in srGAP2-depleted cells (Fig. 6B), while the srGAP2 methylation mutant failed to do so (Fig. 6, C and D). Furthermore, we found the R927A methylation mutant could not be localized at membrane protrusions, compared with wild-type srGAP2, which is found far away from F-actin in the membrane protrusions (Fig. 6, B, C, E, F). Therefore, srGAP2 methylation at arginine 927 is required for cell spreading and srGAP2 localization in the cell membrane protrusion.
FIGURE 6.
The functional analysis of srGAP2 arginine methylation. A, HCT116 cells cotransfected with shRNA and GFP-tagged srGAP2 as indicated, and whole cell extracts were probed with antibodies against GFP or actin (loading control). As shown, srGAP2-R-GFP and srGAP2-dm-GFP were resistant to U1 (srGAP2 shRNA). B and C, U1A5 transfected with srGAP2-R-GFP (B) or srGAP2-dm-GFP (C) were stained with TRITC 545-phalloidin to visualize F-actin distribution 22 h after transfection. The right figures are the high-magnification from the left selected inset, showing the distribution srGAP2 and F-actin at the cell membrane protrusion. Bars, 5 μm. D, cell spreading situation of either srGAP2-depleted HCT116 cells (U1A5) transfected with srGAP2-R-GFP or srGAP2-dm-GFP at each indicated time point (n1/n2: n1 means the length of time after plating the cells, and n2 means the length of time after transfection) was counted using the cell spreading assay; see “Experimental Procedures” for details. Data are mean ± S.E. (n = 3; *, p < 0.05, Student's t test). E and F, graphs correspond to intensities in arbitrary units of the green and red labels for each pixel of the arrow drawn through the axis. Graph E is from the arrow in B, and graph F is from the arrow in C.
srGAP2 Arginine Methylation Is Not Associated with Its RhoGAP Activity
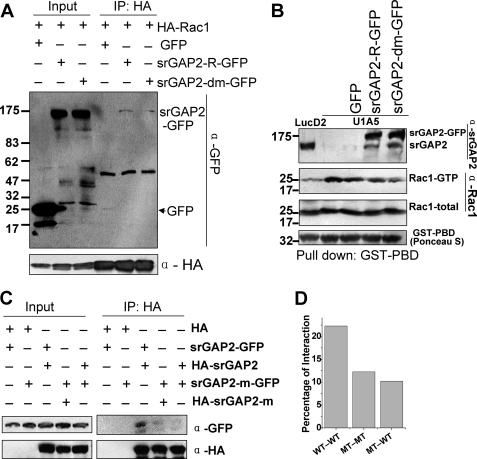
srGAP2 is a Rac1-specific activating protein. Rac1 activates the Arp2/3 complex via WAVE to initiate F-actin polymerization and promote lamellipodia formation (4, 40). Thus, it is possible that arginine methylation of srGAP2 regulates cell spreading by affecting its RhoGAP activity. To verify this notion, we first tested whether srGAP2 methylation mutant could interact with constitutively active form of Rac1. Immunoprecipitation assay was performed using HEK293 cells co-transfected with HA-tagged constitutively active form of Rac1 (HA-Rac1) and GFP-tagged srGAP2 wt (srGAP2-R-GFP) or srGAP2 methylation mutant (srGAP2-dm-GFP). The IP results showed that both srGAP2 wt and methylation mutant maintained interaction with the constitutively active form of Rac1 (Fig, 7A), suggesting that the RhoGAP activity was unaltered by the R927A mutation.
To further confirm that srGAP2 methylation mutant does not affect the RhoGAP activity, we applied GST pull-down assay to test the level of GTP- bound form Rac1 between srGAP2-depleted cells transfected with srGAP2-R-GFP and srGAP2dm-GFP, using a GST fusion protein of the p21 binding domain of in PAK1 (GST-PBD) (41–42). The binding activity of srGAP2-dm-GFP was the same with srGAP2-R-GFP-transfected cells. Both decreased the level of active form Rac1, compared with srGAP2-depleted cells (Fig. 7B). Collectively, these data ruled out that srGAP2 methylation mediated by PRMT5 is associated with its RhoGAP activity.
FIGURE 7.
srGAP2 arginine methylation promotes the dimerization of srGAP2. A, co-immunoprecipitation was performed in HEK293 cells cotransfected with two plasmids, as indicated. IP: antibodies used in immunoprecipitation; Western blot with the anti-GFP antibody first, then blotted with anti-HA antibody after stripping away GFP antibody. B, GST-PBD pull-down assay. GST-PBD was performed to pull down the associated proteins from cell lysates, and followed by Western blot using anti-srGAP2 and Rac1 antibodies. GST-PBD was indicated with Ponceau S staining. Lanes from left to right: LucD2, U1A5, U1A5 transfected with GFP or srGAP2-R-GFP and/or srGAP2-m-GFP. C, co-immunoprecipitation was performed in HEK293 cells cotransfected with the indicated plasmids. IP: antibodies used in immunoprecipitation. Western blot with the anti-GFP and -HA antibodies. D, GFP-tagged srGAP2 binding relative to HA-tagged srGAP2, normalized from C, lanes 3–5, analyzed using the NIH Image J software. WT-WT: srGAP2-GFP pulled by HA-srGAP2; MT-MT: srGAP2-m-GFP pulled by HA-srGAP2-m; MT-WT: srGAP2-m-GFP pulled by HA-srGAP2.
srGAP2 Arginine Methylation Promotes the Dimerization of srGAP2
Light scattering analysis indicated that srGAP2 forms homodimer through its F-BAR domains in solution (23). The homodimer mediated by F-BAR domain possesses a quaternary “banana-like” structure, which can deform the cell membrane (10, 12, 14, 43). F-BAR domain of srGAP2 induces filopodia in COS7 cells (23). We therefore tested whether srGAP2 R927 methylation was required for the formation of a homodimer. HA or GFP-tagged srGAP2 wild type or its arginine methylation mutant were co-expressed in cells. Immunoprecipitation showed that HA-srGAP2 binds srGAP2-GFP with high affinity as expected. However, the association between HA-srGAP2 and srGAP2-m-GFP or HA-srGAP2-m and srGAP2-m-GFP was reduced drastically (Fig. 7, C and D), suggesting that srGAP2 Arg-927 methylation is required for its homodimer formation.
DISCUSSION
Results from this report suggest that srGAP2 is a novel substrate of PRMT5, and arginine methylation of srGAP2 regulates membrane protrusion, cell spreading, and migration. Our findings that srGAP2 depletion in cultured cells led to promotion of cell migration are consistent with a recent report that srGAP2 negatively regulates neuronal migration (23).
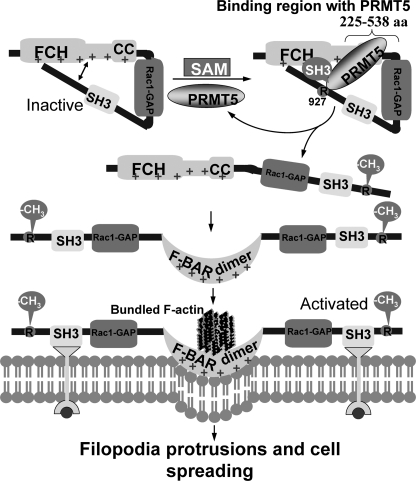
It has been proposed that the F-BAR domain of srGAP2, resembling I-BAR domains, induces filopodia-like membrane protrusion, and this process is dependent on srGAP2 homodimerization. srGAP2 might normally be in an auto-inhibited conformation through interaction between the N-terminal F-BAR domain and the C-terminal regions (including the SH3 domain). Given that the SH3 domain mutant completely abrogated the srGAP2 ability to induce filopodia formation and that deletion of the C terminus induces filopodia, it is expected that the C terminus of srGAP2 harbors a regulatory domain(s) critical for its function (23). In this work, we find that the srGAP2-N3 region (225 to 538 aa) interacts with PRMT5 and srGAP2 is methylated by PRMT5 at R927 (Fig. 4D). The methylation site and the interacting region of srGAP2 are distant in the primary structure. srGAP2-N3 contains the CC motif of F-BAR domain, and Arg-927 site is in the C terminus outside the SH3 domain (Fig. 4B), this is congruent with the notion that there is an auto-inhibitory conformation through structural interaction between the N-terminal F-BAR domain and the C-terminal region (23). However, it remains unclear as to how the auto-inhibitory conformation transforms into the active form of the homodimer. Our findings show that the srGAP2 methylation mutant fails to bind to the cell membrane or spread the cell (Fig. 6) or form the dimer (Fig. 7, C and D). Therefore, we propose a model (Fig. 8) for the role of srGAP2 methylation. In this model, the F-BAR domain of srGAP2 is restricted by the C-terminal region when Arg-927 is unmodified, constituting steric hindrance for the F-BAR region. Upon PRMT5-mediated Arg-927 methylation, the C-terminal region is disengaged from the F-BAR domain, allowing dimerizaton to take place, with the result that the active form of srGAP2 translocates into the plasma membrane and enacts membrane deformation and protrusion necessary for cell spreading.
FIGURE 8.
Methylation of srGAP2 is required for filopodia protrusions and cell spreading. Hypothetical model of the molecular mechanisms illustrating that methylation of srGAP2 promotes the dimerization of srGAP2, plasma membrane protrusions, and cell spreading. See text for details.
Acknowledgments
We thank Dr. Kozo Kaibuchi (Nagoya University) for HA-Rac1V12 and Dr. Xueliang Zhu (SIBS) for GST-PBD. We thank Drs. Lei Li (The University of Texas, M. D. Anderson Cancer Center) and Yingchun Wang (IGDB) for comments and suggestions during the preparation of this manuscript. We thank Zhongwei Zhou and Pei Wei, former members in our group, for technical assistance and suggestions.
This work was supported by the National Basic Research Program of China (2005CB22400), National High Technology Development Program (2006AA02Z140), Chinese Academy of Sciences (KSCX1-YW-R-59), and National Natural Sciences Foundation of China (30670479).
- GEF
- guanine nucleotide exchange factor
- aa
- amino acids
- srGAP
- Slit-Robo GTPase-activating protein
- SAHH
- S-adenosylhomocysteine hydrolase
- SAM
- S-adenosylmethionine.
REFERENCES
- 1.Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 2.Lauffenburger D. A., Horwitz A. F. (1996) Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 3.Webb D. J., Parsons J. T., Horwitz A. F. (2002) Nat. Cell Biol. 4, E97–100 [DOI] [PubMed] [Google Scholar]
- 4.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 5.Hall A. (2005) Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 6.Lamarche N., Hall A. (1994) Trends Genet. 10, 436–440 [DOI] [PubMed] [Google Scholar]
- 7.Narumiya S. (1996) J. Biochem. 120, 215–228 [DOI] [PubMed] [Google Scholar]
- 8.Wong K., Ren X. R., Huang Y. Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L., Wu J. Y., Xiong W. C., Rao Y. (2001) Cell 107, 209–221 [DOI] [PubMed] [Google Scholar]
- 9.Yang Y., Marcello M., Endris V., Saffrich R., Fischer R., Trendelenburg M. F., Sprengel R., Rappold G. (2006) Exp. Cell Res. 312, 2379–2393 [DOI] [PubMed] [Google Scholar]
- 10.Henne W. M., Kent H. M., Ford M. G., Hegde B. G., Daumke O., Butler P. J., Mittal R., Langen R., Evans P. R., McMahon H. T. (2007) Structure 15, 839–852 [DOI] [PubMed] [Google Scholar]
- 11.Frost A., De Camilli P., Unger V. M. (2007) Structure 15, 751–753 [DOI] [PubMed] [Google Scholar]
- 12.Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. (2004) Science 303, 495–499 [DOI] [PubMed] [Google Scholar]
- 13.Lim K. B., Bu W., Goh W. I., Koh E., Ong S. H., Pawson T., Sudhaharan T., Ahmed S. (2008) J. Biol. Chem. 283, 20454–20472 [DOI] [PubMed] [Google Scholar]
- 14.Shimada A., Niwa H., Tsujita K., Suetsugu S., Nitta K., Hanawa-Suetsugu K., Akasaka R., Nishino Y., Toyama M., Chen L., Liu Z. J., Wang B. C., Yamamoto M., Terada T., Miyazawa A., Tanaka A., Sugano S., Shirouzu M., Nagayama K., Takenawa T., Yokoyama S. (2007) Cell 129, 761–772 [DOI] [PubMed] [Google Scholar]
- 15.Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005) Dev. Cell 9, 791–804 [DOI] [PubMed] [Google Scholar]
- 16.Tsujita K., Suetsugu S., Sasaki N., Furutani M., Oikawa T., Takenawa T. (2006) J. Cell Biol. 172, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aspenström P. (1997) Curr. Biol. 7, 479–487 [DOI] [PubMed] [Google Scholar]
- 18.Mattila P. K., Pykäläinen A., Saarikangas J., Paavilainen V. O., Vihinen H., Jokitalo E., Lappalainen P. (2007) J. Cell Biol. 176, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suetsugu S., Murayama K., Sakamoto A., Hanawa-Suetsugu K., Seto A., Oikawa T., Mishima C., Shirouzu M., Takenawa T., Yokoyama S. (2006) J. Biol. Chem. 281, 35347–35358 [DOI] [PubMed] [Google Scholar]
- 20.Mattila P. K., Salminen M., Yamashiro T., Lappalainen P. (2003) J. Biol. Chem. 278, 8452–8459 [DOI] [PubMed] [Google Scholar]
- 21.Yamagishi A., Masuda M., Ohki T., Onishi H., Mochizuki N. (2004) J. Biol. Chem. 279, 14929–14936 [DOI] [PubMed] [Google Scholar]
- 22.Woodings J. A., Sharp S. J., Machesky L. M. (2003) Biochem. J. 371, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W. L., Frost A., Polleux F. (2009) Cell 138, 990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu S., Mahadeo D. C., Liu X., Liu W., Parent C. A., Korn E. D. (2006) Proc. Natl. Acad. Sci.U.S.A. 103, 19788–19793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De La Haba G., Cantoni G. L. (1959) J. Biol. Chem. 234, 603–608 [PubMed] [Google Scholar]
- 26.Zhou Z., Sun X., Zou Z., Sun L., Zhang T., Guo S., Wen Y., Liu L., Wang Y., Qin J., Li L., Gong W., Bao S. (2010) Cell Res. 20, 1023–1033 [DOI] [PubMed] [Google Scholar]
- 27.Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. (2004) Mol. Cell. Biol. 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. (2008) Nat. Cell Biol. 10, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 29.Ancelin K., Lange U. C., Hajkova P., Schneider R., Bannister A. J., Kouzarides T., Surani M. A. (2006) Nat. Cell Biol. 8, 623–630 [DOI] [PubMed] [Google Scholar]
- 30.Gonsalvez G. B., Rajendra T. K., Tian L., Matera A. G. (2006) Curr. Biol. 16, 1077–1089 [DOI] [PubMed] [Google Scholar]
- 31.Dacwag C. S., Bedford M. T., Sif S., Imbalzano A. N. (2009) Mol. Cell. Biol. 29, 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak Y. T., Guo J., Prajapati S., Park K. J., Surabhi R. M., Miller B., Gehrig P., Gaynor R. B. (2003) Mol. Cell 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 33.Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., Jane S. M. (2009) Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008) Cell 135, 497–509 [DOI] [PubMed] [Google Scholar]
- 35.Ren J., Wang Y., Liang Y., Zhang Y., Bao S., Xu Z. (2010) J. Biol. Chem. 285, 12695–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao S., Lu T., Wang X., Zheng H., Wang L. E., Wei Q., Hittelman W. N., Li L. (2004) Oncogene 23, 5586–5593 [DOI] [PubMed] [Google Scholar]
- 37.Rho J., Choi S., Seong Y. R., Cho W. K., Kim S. H., Im D. S. (2001) J. Biol. Chem. 276, 11393–11401 [DOI] [PubMed] [Google Scholar]
- 38.Boucrot E., Kirchhausen T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7939–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boucrot E., Kirchhausen T. (2008) PLoS One 3, e1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takenawa T., Miki H. (2001) J. Cell Sci. 114, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 41.Benard V., Bohl B. P., Bokoch G. M. (1999) J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 42.Sander E. E., van Delft S., ten Klooster J. P., Reid T., van der Kammen R. A., Michiels F., Collard J. G. (1998) J. Cell Biol. 143, 1385–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E. H., De Camilli P., Unger V. M. (2008) Cell 132, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]