Abstract
How cell morphology and the cell cycle are coordinately regulated is a fundamental subject in cell biology. In fission yeast, 2 germinal center kinases (GCKs), Sid1 and Nak1, play an essential role in septation/cytokinesis and cell separation/cell polarity control, respectively, as components of the septation initiation network (SIN) and the morphogenesis Orb6 network (MOR). Here we show that a third GCK, Ppk11, is also required for efficient cell separation particularly, at a high temperature. Although Ppk11 is not essential for cell division, this kinase plays an auxiliary role in concert with MOR in cell morphogenesis. Ppk11 physically interacts with the MOR component Pmo25 and is localized to the septum, by which Ppk11 is crucial for Pmo25 targeting/accumulation to the septum. The conserved C-terminal WDF motif of Ppk11 is essential for both septum accumulation of Pmo25 and efficient cell separation. In contrast its kinase activity is required only for cell separation. Thus, both interaction of Ppk11 with Pmo25 and Ppk11 kinase activity are critical for efficient cell separation.
Keywords: Cell Cycle, Cell Division, Protein Kinases, Signal Transduction, Yeast, Cell Morphogenesis
Introduction
Cell morphogenesis and the cell cycle are coordinately regulated. The fission yeast Schizosaccharomyces pombe is an excellent model system in which to study this coordinated regulation (for reviews, see Refs. 1, 2), because growth polarity dynamically changes at 3 stages during the cell cycle (3, 4), i.e. the initiation of growth upon cell division, NETO (new end take off) (5), and septum formation. Upon cell division, cortical F-actin moves from the new end, which is newly produced by division, to the old end, which existed in the previous cell cycle; and the cell growth is initiated from only the old end. Upon NETO at about 0.34 of the cell cycle, F-actin localization shifts from the old end to both ends, thereby changing growth polarity from monopolar to bipolar. This growth polarity is maintained during the following interphase. At the onset of mitosis, the cell growth ceases; and the cortical F-actin patches at both ends disappear, having been translocated to the medial ring, which corresponds to the following division site. The onset of cytokinesis is triggered by the septation initiation network (SIN),4 analogous to the budding yeast mitotic exit network (MEN), that comprises Spg1 GTPase and a downstream kinase cascade (Cdc7, Sid1, and Sid2; Ref. 6). SIN promotes actomyosin ring constriction and septum formation in the middle of the cell (6).
The Ste20 group of kinases has been implicated in various cellular events including the regulation of cell morphogenesis, cytoskeletal rearrangements, and apoptosis (7, 8). This group includes germinal center kinases (GCKs) and p21-activated kinases (PAKs). GCKs have an N-terminal kinase domain followed by less conserved C-terminal putative regulatory regions but lack the conserved G-protein binding sites possessed by PAKs. Genome sequencing shows that fission yeast cells have 3 GCKs, i.e. Sid1, Nak1, and Ppk11. Sid1 and Nak1 are essential for cell growth, and play a critical role in septation/cytokinesis and cell morphogenesis (cell separation and cell polarity control), respectively, as components of SIN and the morphogenesis Orb6 network (MOR) (6, 9). However, a function of Ppk11, that is non-essential for cell growth, remains unclear.
Members of the MO25 family are evolutionally conserved proteins that are structurally related armadillo-repeat scaffold proteins (10, 11). Accumulating evidence indicates that MO25 proteins are important for regulation of cell polarity and are also related functionally to GCK (11–13). In fission yeast, MO25/Pmo25 interacts with Nak1 and functions as an upstream component of the Drosophila Furry-like Mor2 and the NDR kinase Orb6 in the MOR pathway (9, 14). The Pmo25 localization to the spindle pole bodies (SPBs) and the Nak1-Orb6 kinase activities in early interphase following cytokinesis are under the control of Cdc7-Sid1, indicating that Pmo25 plays a connecting role between SIN and MOR by interacting functionally with 2 GCKs, Sid1 and Nak1 (9, 15). The budding yeast MO25/Hym1 is involved in the RAM (regulation of Ace2p activity and cellular morphogenesis) pathway that consists of the budding yeast homologs (Kic1, Tao3, and Cbk1) of fission yeast Nak1, Mor2, and Orb6 (16). RAM controls not only cell separation via regulation of the Ace2 transcription factor but also polarized cell growth (16). In Drosophila, Mo25 and the GCK Fray act in concert to regulate asymmetric division in embryonic neuroblasts (17). In vertebrates, MO25 functionally interacts with the GCK family pseudokinase STRAD (Ste-20-related adaptor) to activate serine/threonine kinase LKB1, which is implicated as a regulator of multiple biological processes, including the control of cell-cycle arrest and cell polarity control, and whose mutation causes the Peutz-Jeghers cancer syndrome (18–20). ATP and MO25 regulate the conformational state of STRAD and activation of LKB1 (21). LKB1 activation by STRAD can establish complete polarity in single epithelial cells through the induction of brush borders at the apical domain (19). Recently, it has been shown that human MO25 interacts with the GCK Mst4 and that the active MO25/STRAD/LKB1 complex induces the brush-border formation through Mst4 and the actin remodeler Ezrin (22).
In this present study, we characterized the third GCK, Ppk11, in detail and obtained data indicating that Ppk11 interacted with Pmo25 and was important for efficient cell separation under high temperature conditions.
EXPERIMENTAL PROCEDURES
Strains, Media, and General Methods
The S. pombe strains used in this study are listed in Table 1. Standard methods for S. pombe, media, and genetic manipulations were followed as described previously (23). Cell number was measured using Sysmex F-820. Epitope tagging and preparation of the truncated ppk11 mutant were performed by the PCR-based method (24).
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 972 | h− | Lab stock |
| DH1520-1B | h− Δppk11::ura4+ura4 | This study |
| TG174-20 | h− Δppk11::ura4+ura4 leu1-32::ppk11-K35R | This study |
| TG3-3 | h−ppk11+:13Myc:kanr | This study |
| DH1247-4A | h−ppk11+:GFP:kanr sad1+:dsRED:LEU2 leu1 | This study |
| DH1231-3C | h−ppk11+:GFP:kanr | This study |
| DH1255-6B | h−ppk11+:GFP:kanr cdc7-24 | This study |
| DH1256-3B | h−ppk11+:GFP:kanr sid1-239 ura4 | This study |
| DH1258-1C | h−ppk11+:GFP:kanr sid2-250 ura4 | This study |
| TG68-1D | h−ppk11+:GFP:kanrsad1+:dsRED:LEU2 cdc15-140 leu1 | This study |
| DH842-1D | h−pmo25-35:ura4+ura4 | Kanai et al. (9) |
| KP1-6D | h−mor4-125/nak1-125/orb3-125 leu1 | Kanai et al. (9) |
| DH107-4C | h−mor2-786 leu1 | Kanai et al. (9) |
| DH433-12C | h−orb6-25 leu1 | Kanai et al. (9) |
| DH1517-4C | h− Δppk11::ura4+pmo25-35:ura4+ura4 | This study |
| DH1357-1A | h− Δppk11::ura4+mor4-125/nak1-125/orb3-125 ura4 leu1 | This study |
| DH1398-4D | h− Δppk11::ura4+mor2-786 ura4 leu1 | This study |
| DH1391-1A | h− Δppk11::ura4+orb6-25 ura4 leu1 | This study |
| KSP1-1 | h−pmo25+:GFP:kanr | Kanai et al. (9) |
| TG2-2 | h−ppk11+:3HA:kanr | This study |
| TG65-11D | h−ppk11+:3HA:kanr pmo25+:GFP:kanr | This study |
| TG62-2C | h−ppk11+:GFP:kanr pmo25+:tdTomato:kanr | This study |
| DH1249-1A | h+ppk11+:GFP:kanr pmo25-35:ura4+ura4 his2 | This study |
| DH1250-3A | h+ppk11+:GFP:kanr mor4-125/nak1-125/orb3-125 leu1 his2 | This study |
| DH1251-1B | h−ppk11+:GFP:kanr mor2-786 leu1 ura4 | This study |
| DH1252-7B | h−ppk11+:GFP:kanr orb6-25 | This study |
| KL284 | h−nak1+:G5:GFP:kanr leu1-32 ura4-D18 ade6-M210 | Kanai et al. (9) |
| DH288-3A | h−kanr:nmt1:GFP:mor2+ura4 | Lab Stock |
| KK6-1 | h−orb6+:GFP:kanr | Kanai et al. (9) |
| DH1335-3C | h− Δppk11::ura4+ pmo25+:GFP:kanrura4 leu1 | This study |
| DH1333-3B | h− Δppk11::ura4+nak1+:G5:GFP:kanr ura4 leu1 | This study |
| DH1356-4B | h− Δppk11::ura4+kanr:nmt1:GFP:mor2+ura4 | This study |
| DH1334-4A | h− Δppk11::ura4+orb6+:GFP:kanr ura4 | This study |
| DH1780-5C | h−pmo25+:GFP:kanr crn1+:tdTomato:kanr | This study |
| TG143-2 | h−ppk11Δ8:GFP:kanr | This study |
| TG242-3 | h−ppk11Δ8:pmo25+:GFP:kanr | This study |
| TG218-1 | h− Δppk11::ura4+ura4 leu1-32::ppk11-WDF/AAA | This study |
| TG64-5A | h−ppk11+:GFP:kanr crn1+:tdTomato:kanr ura4 | This study |
| TG241-4 | h− Δppk11::ura4+ura4 leu1-32::ppk11-WDF/AAA:GFP:kanr | This study |
| TG184-1 | h− Δppk11::ura4+ura4 leu1-32::ppk11-K35R:GFP:kanr | This study |
| TG170-4B | h−pmo25+:GFP:kanr ppk11Δ8:13Myc:kanr | This study |
| TG215-1 | h−pmo25+:GFP:kanr Δppk11::ura4+ura4 leu1-32::ppk11-WDF/AAA | This study |
| TG177-36 | h−pmo25+:GFP:kanr Δppk11::ura4+ura4 leu1-32::ppk11-K35R | This study |
Construction of a Ppk11Δ8-Pmo25-GFP Strain
A Ppk11Δ8-Pmo25-GFP strain was created by the PCR-based method (24). DNA fragments containing the pmo25+:GFP:kanr gene were amplified by PCR from a pmo25+:GFP:kanr strain with the primers containing 5′-extensions with ppk11+-specific DNA sequences and integrated into the ppk11+ locus in WT cells.
Plasmids for Expression of the ppk11+ Gene
All plasmids were constructed by using standard molecular biology techniques. In general, gene fragments were cloned as PCR products from genomic DNA with primers containing 5′-extensions with specific restriction sites. The yeast 2-hybrid system was used to detect possible interactions between Ppk11 and MOR components. DNA fragments containing the ppk11+ gene were cloned in frame with the GAL4-DNA binding domain (pGBKT7) and the GAL4-transcription activating domain (pACT2). Interaction was tested in the Saccharomyces cerevisiae strain Y190 co-transformed with the 2-hybrid plasmids.
Construction of ppk11 Kinase-dead and WDF Motif Mutants
Site-directed mutagenesis was performed by using pJK148-ppk11+ (pTG24) as a template and a QuikChange kit (Stratagene) to introduce the point mutation, according to the manufacturer's instructions. Successful mutagenesis for each mutation was confirmed by DNA sequencing. The mutated ppk11 gene was integrated into the leu1–32 locus of the ppk11-deleted cells.
Immunological Assay
Preparation of whole-cell extracts, immunoprecipitation, and immunoblotting were performed as previously described (25–27). Protein samples were electrophoresed on SDS-PAGE gels and electrophoretically transferred onto nitrocellulose filters. The primary antibodies, mouse monoclonal anti-GFP (8362–1; Clontech), anti-HA (HA.11; BabCO), anti-Myc (9E10, BabCO), anti-PSTAIRE (sc-53; Santa Cruz Biotechnology), and anti phospho-Cdc2 (Tyr15) (9111S; Cell Signaling Technology), were used for detection of the epitope-tagged proteins, Cdc2, and Cdc2 phosphorylated on its Tyr-15. Immunoprecipitation was done by using anti-HA (HA.11, BabCO) or anti-GFP antibody (Roche), and magnetizable beads conjugated to protein A (Dynabeads, DYNAL). Horseradish peroxidase-conjugated sheep anti-rabbit IgG or anti-mouse IgG (Amersham Biosciences) was used as the second antibody. Enhanced chemiluminescence (ECL; Amersham Biosciences) was used to detect bound antibody.
Microscopy Techniques
Cytological techniques were performed as described previously (9, 26). Calcofluor white (Sigma) was used to monitor cell wall growth, and 4,6-diamidino-2-phenylindole (DAPI; Sigma) and Hoechst 333342 (Calbiochem) were used for observing chromatin regions. For the observation of Ppk11-GFP, the cells expressing Ppk11-GFP were fixed with methanol (−20 °C) for 20 min and washed three times with sterile water. Fixed-cell images were collected with an Axiophot 2 MOT (ZEISS) and AxioCam MRm CCD camera; and the images were further processed with AxioVision software. Live-cell images were collected with an IX70 (OLYMPUS) and DeltaVision sectioning system. GFP intensity was measured with ImageJ software (National Institutes of Health).
RESULTS
Ppk11 Is Required for Efficient Cell Separation
The genome of fission yeast contains 3 ORFs that encode GCKs Nak1, Sid1, and Ppk11 (Fig. 1A), in which there is a distinct peptide sequence, (v/i)GTPyWMAPEv (a small letter indicates less conservation), termed the Ste20 signature sequence (8), in the kinase subdomain VIII (Fig. 1B). Nak1 and Sid1 are essential for cell growth, and their roles have been established as a signaling component of MOR and SIN, respectively. In contrast, the role of Ppk11 remains elusive, though this GCK is known to be non-essential for cell division at least under normal growth conditions (28). To investigate the potential role of Ppk11 in cell division, we examined defective phenotypes of ppk11-deleted (Δppk11) cells at various temperatures (18–36 °C). The growth rates of Δppk11 cells at the lower temperatures (18–30 °C) were indistinguishable from those of wild-type (WT) cells, but its growth rate at the high temperature (36 °C) was reduced by 50% compared with that for the WT cells (data not shown). After incubation at 36 °C for 4 h, the population of monopolar cells in Δppk11 was significantly increased to 50% (data not shown), as previously described (29). In addition, after a prolonged incubation at 36 °C for 12 h, the population of septated cells in Δppk11 was increased by 2-fold (∼40%) compared with that in WT cells (Fig. 2A), indicating that Ppk11 was important for cell separation at the high temperature.
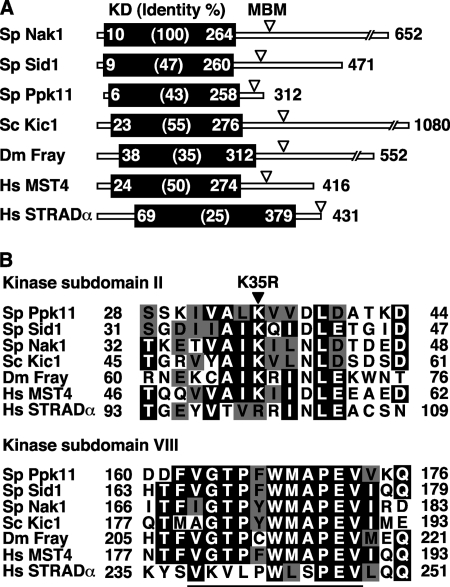
FIGURE 1.
Structure of GCKs. A, identity of KD and MBM of GCKs. S. pombe Nak1, Sid1, and Ppk11, S. cerevisiae Kic1, Drosophila melanogaster Fray, and Homo sapiens MST4 and STRADα are shown. The identities (%) between Nak1 and other GCKs and the amino acid number of the beginning and the end of the KD are indicated. Arrowheads indicate the position of the putative MBM. B, alignment of the kinase subdomain of GCKs. Identical or similar residues are represented in black or gray, respectively. The kinase domain is subdivided into 11 conserved subdomains, and subdomain II is essential for the kinase activity by acting as the ATP binding site. The solid arrowhead indicates the conserved lysine that was mutated to arginine to construct the Ppk11 kinase dead mutant (ppk11-K35R). In subdomain VIII of Ste20 group kinases, there is a distinct peptide sequence, (v/i)GTPyWMAPEv (a small letter indicates less conservation), termed the Ste20 signature sequence. This sequence is underlined. The sequences were aligned by using the software ClustalW provided by PBIL (Pole Bio-Informatique Lyonnais).
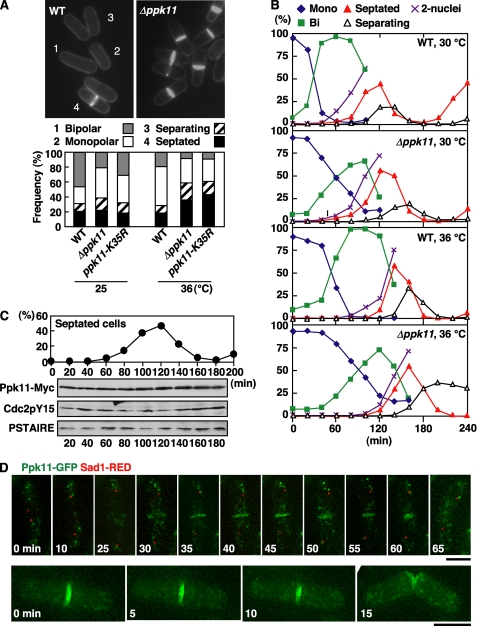
FIGURE 2.
Phenotype of ppk11-deletion mutant and localization of Ppk11. A, wild type (WT, 972), ppk11-deleted (Δppk11, DH1520-1B), and ppk11-kinase dead mutant (ppk11-K35R, TG174-20) cells grown in YPD medium at 25 °C were shifted to 36 °C and incubated for 12 h. Cell morphology was observed by staining with Calcofluor (n > 200 cells). In fission yeast, Calcofluor stains septa most brightly and also stains newly synthesized cell wall in the growing cell end, which can be easily distinguished from the non-growing dark cell end. The data on the frequency of septated cells were analyzed by statistical processing (chi-squared tests). The Δppk11 and ppk11-K35R cells showed significant increases (p < 0.01) in frequency of septated cells. B, cell morphology of ppk11-deletion mutant during the cell cycle. The elutriated early G2 cells were cultured at 30 or 36 °C and taken at the indicated times for observation of morphology and nuclear number. C, Ppk11 protein level during the cell cycle. The elutriated early G2 cells having ppk11+:13Myc:kanr (Ppk11-Myc, TG3-3) were cultured at 30 °C and taken at the indicated times for observation of cell morphology and for immunoblotting. Total cell extracts prepared from the samples of the same cultures were immunoblotted with anti-Myc (for Ppk11-Myc), anti-Cdc2 phosphorylated on its tyrosine-15 (for Cdc2pY15), and PSTAIR (for Cdc2). D, live image of Ppk11-GFP and Sad1-RED during the cell cycle. WT cells having ppk11+:GFP:kanr and sad1+:dsRED:LEU2 (DH1247-4A) grown in YES5 medium at 25 °C were used. Bars indicate 5 μm.
To further investigate cell morphogenesis defects in Δppk11 cells during the cell cycle, we examined the cell shape of Δppk11 by using synchronous cultures with elutriation (Fig. 2B). At 36 °C, the onset of mitosis in Δppk11 cells was delayed by 20 min compared with that for WT cells, suggesting that the ppk11-deletion induced a modest G2-delay. Further, separating cells in Δppk11 cells accumulated (27%) after mitosis (open triangles in Fig. 2B), indicating that Ppk11 is required for efficient cell separation at the high temperature. In addition to the defect in cell separation, the transition from monopolar to bipolar in Δppk11 cells was delayed by 20–40 min compared with that in WT cells (green squares in Fig. 2B), indicating that Ppk11 played an important role in NETO progression, as previously suggested (29).
To investigate whether the kinase activity of Ppk11 was important for cell separation, we constructed the kinase dead ppk11-K35R mutant, in which the conserved lysine of the ATP binding site in the kinase subdomain II (Fig. 1B) was substituted with arginine, and this ppk11-K35R mutated gene was integrated into the leu1 locus in Δppk11 cells. The ppk11-K35R mutant cells showed septation defects indistinguishable from those of the Δppk11 cells (Fig. 2A), indicating that the kinase activity of Ppk11 was required for efficient cell separation.
The Ppk11 Is Constitutively Expressed and Localizes to the Site of Cytokinesis
To examine the protein level of Ppk11 during the cell cycle, we constructed WT cells expressing Ppk11-Myc or Ppk11-GFP under the chromosomal native promoter. These epitope-tagged strains were morphologically normal rods, and the frequency of septated cells was the same as for WT cells, indicating that the tagged proteins were fully functional. The level of Ppk11-Myc protein did not change during the cell cycle (Fig. 2C). Next we observed Ppk11 localization by living cell imaging of Ppk11-GFP with a DeltaVision RT system (note that an SPB marker, Sad1-RED, is also included; Fig. 2D). Interestingly, Ppk11-GFP was localized to the medial region of the cell in post-anaphase (Fig. 2D, upper panels, at 30 min). The localization of Ppk11-GFP to split rings was also observed (Fig. 2D, lower panels). This observation suggests that Ppk11 co-localized to the septa rather than to the contractile actomyosin ring. Upon cell separation, Ppk11-GFP disappeared within 5 min from the new ends of the cells, and no specific localization was evident afterward until the next mitosis.
Ppk11 Localization Requires SIN and a Functional Actomyosin Ring
To investigate whether the localization of Ppk11 required SIN function, we examined Ppk11 localization in various SIN kinase mutants (cdc7-24, sid1-239, and sid2-250). In these SIN mutant cells cultured at 36 °C for 4 h, no clear localization of Ppk11 to the medial region in post-anaphase cells was observed (Fig. 3A). This observation is consistent with our foregoing results that suggested Ppk11 to be co-localized with septa rather than with the actomyosin ring, as SIN is required for septum formation. SIN is required for 2 events upon mitotic exit. One is formation of the septum, while the other is constriction of the actomyosin ring, both of which are essential for successful cell division (6). To distinguish between the two possibilities that activation of SIN is required for Ppk11 localization to the medial region or formation of a functional actomyosin ring is essential, we examined Ppk11 localization in cdc15-140, a ts mutant defective in actomyosin ring assembly but nonetheless in which SIN is activated. As shown in Fig. 3B, no clear localization of Ppk11 to the medial region of the cell was observed (Fig. 3B). This result indicates that the localization of Ppk11 to the medial region requires a proper actomyosin ring assembly and function.
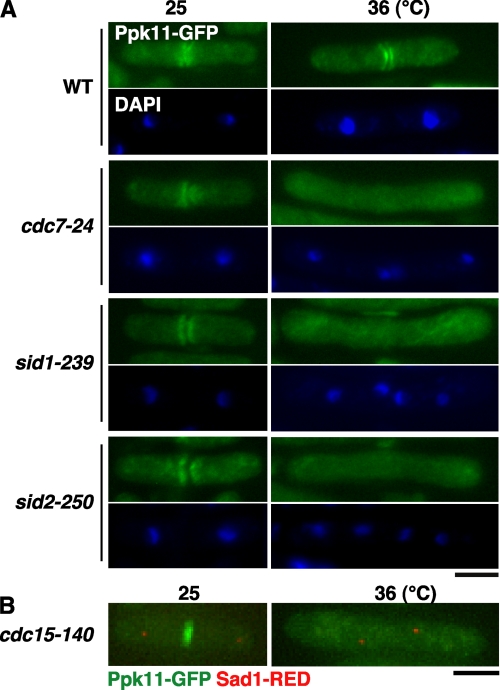
FIGURE 3.
Functional interaction between Ppk11 and SIN. A and B, localization of Ppk11 in the SIN mutant. The indicated SIN mutants having ppk11+:GFP:kanr grown in YES5 medium at 25 °C were shifted to 36 °C, incubated for 4 h, and fixed (A). Cells were fixed and stained with DAPI for visualizing the nucleus. The cdc15 mutant having ppk11+:GFP:kanr and sad1+:dsRED:LEU2 (TG68-1D) grown at 25 °C was shifted to 36 °C and incubated for 4 h (B). Bars indicate 5 μm.
Ppk11 Plays an Auxiliary Function in Concert with MOR in Cell Morphogenesis
To investigate a possible functional relationship between Ppk11 and MOR, we examined the cell morphology of the Δppk11MOR double mutants at the semi-permissive temperature of 33 °C. The percentage of septated cells in the double mutants between Δppk11 and 3 MOR mutants, nak1-125, mor2-786, and orb6-25, was higher than that in each single mutant (Fig. 4A), indicating that Ppk11 plays an auxiliary role in concert with MOR in cell separation. In addition we measured the cell size of septated cells in these double mutants. The roundness (diameter/length, D/L in Fig. 4B) of the Δppk11MOR double mutant was greater than that of the each single MOR mutant (Fig. 4B), indicating that Ppk11 also has an auxiliary role in concert with MOR in cell polarity control. Thus Ppk11 and MOR are functionally related in not only cell separation but also cell polarity control.
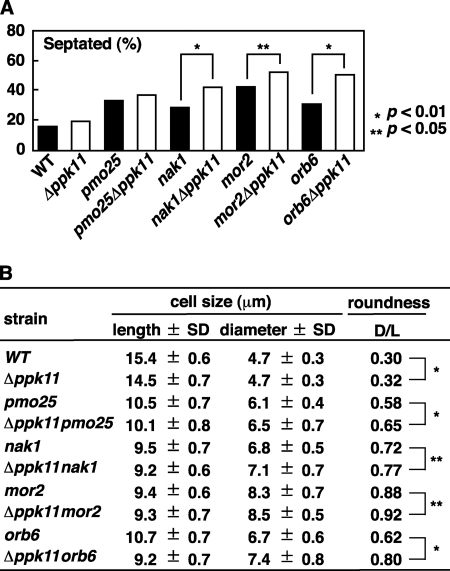
FIGURE 4.
Functional interaction between Ppk11 and MOR. A and B, phenotypes of the double mutants between ppk11 and MOR. The indicated strains grown in YPD medium at 25 °C were shifted to the semi-permissive temperature of 33 °C. After incubation at 33 °C for 12 h, the cells were taken for observation of cell morphology. Frequency of the septated cells was examined (n > 200, A). The cell size (Length and Diameter) and roundness (D/L) of the septated cells (n = 50) were measured (B). The data were analyzed by statistical processing (chi-squared tests for A, Student's t test for B). The p values are indicated as *, p < 0.01 and **, p < 0.05.
Ppk11 Physically Interacts with Pmo25
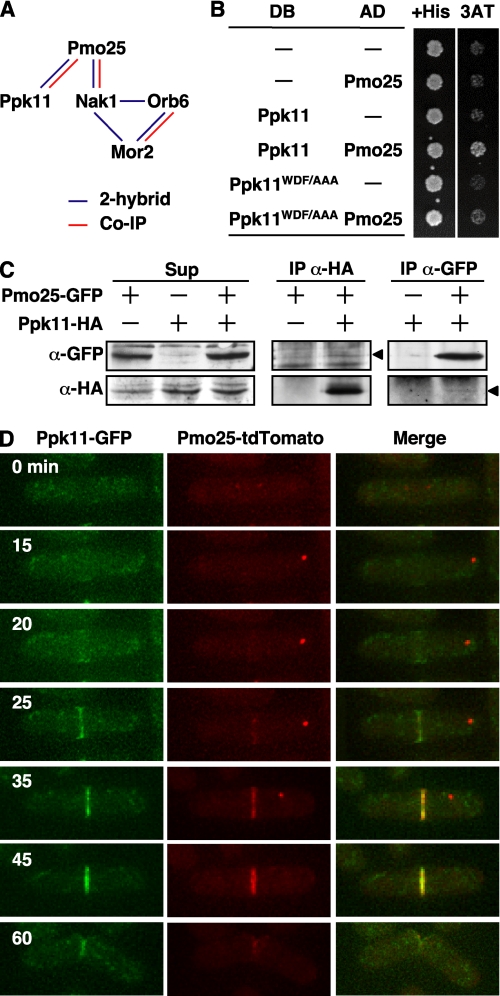
We have previously shown the interactions among the MOR components by using the yeast 2-hybrid system and co-immunoprecipitation (Fig. 5A) (9). To investigate whether Ppk11 interacts with the MOR components, we first performed the yeast 2-hybrid assay between Ppk11 and individual MOR components, Pmo25, Nak1, Orb6, and Mor2. Among these components examined, we detected an interaction, albeit modest, with Pmo25 (Fig. 5B). To substantiate this result, we performed co-immunoprecipitation using a strain expressing Ppk11-HA and Pmo25-GFP from the chromosomal native promoters. As shown in Fig. 5C, Ppk11-HA and Pmo25-GFP did exhibit co-precipitation in a reciprocal fashion.
FIGURE 5.
Physical interaction between Ppk11 and MOR. A, summary of the physical interactions between Ppk11 and MORs. The interactions detected by yeast 2-hybrid or by co-immunoprecipitation (Co-IP) are indicated as blue or red lines, respectively. B, interaction of Ppk11/Ppk11-mt with Pmo25 in yeast-2-hybrid assay. S. cerevisiae Y190 cells (104 cells) transformed with Gal4-DNA binding domain-fused (DB) and Gal4 activation domain-fused (AD) genes were spotted on SD containing histidine (His) or 3-aminotriazole (3AT). C, co-IP of Ppk11 with Pmo25. Extracts of S. pombe cells expressing Pmo25-GFP and/or Ppk11-HA from the chromosomal native promoter were immunoprecipitated with anti-HA or anti-GFP antibodies. Precipitated materials were then immunoblotted using antibodies against HA and GFP. The arrowheads indicate the position of the co-immunoprecipitated proteins. D, co-localization of Ppk11 and Pmo25 in cytokinesis. WT cells having ppk11+:GFP:kanr and pmo25+:tdTomato:kanr (TG62-2C) grown in YES5 medium at 25 °C were used. The localization in the living cells was observed by time-lapse cinemicrography.
Next we explored co-localization between Ppk11 and Pmo25 in live imaging using a strain containing Ppk11-GFP and Pmo25-tdTomato expressed from each endogenous native promoter (Fig. 5D). As previously shown (9), Pmo25 was found in 2 SPBs during early mitosis (Fig. 5D, cell at 0 min), 1 SPB during anaphase B (15–35 min), and the medial region (25–45 min) and in both new cell ends upon cell division (60 min). Although Ppk11 was not localized to the mitotic SPB(s), it was localized to the medial region during anaphase B (20–45 min) and both new ends of the cell, indicating partial overlapping localization between Pmo25 and Ppk11. It is noteworthy that Ppk11 appeared to be localized to the medial region slightly earlier than Pmo25 (20–25 min, see below).
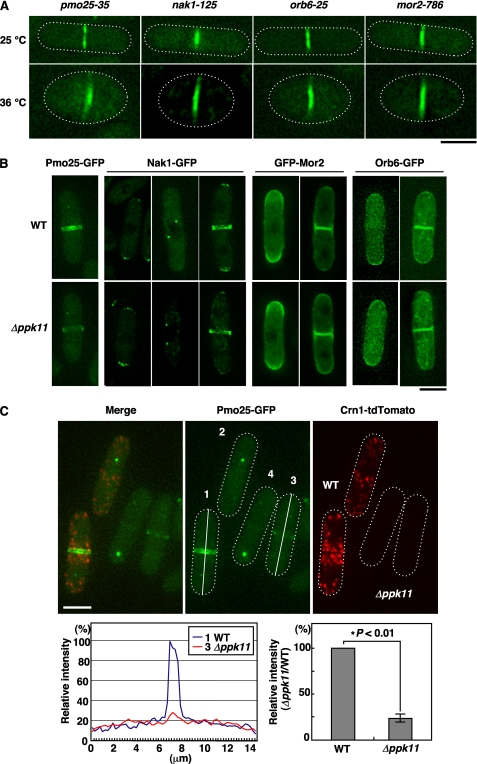
Ppk11 Is Important for Accumulation of Pmo25 at the Site of Cytokinesis
To investigate whether the localization of Ppk11 is dependent on MOR function, we examined Ppk11 localization in various ts MOR mutants (pmo25-35, nak1-125, mor2-786, and orb6-25). In these MOR mutants cultured at 36 °C for 4 h, clear localization of Ppk11 to the medial region was observed (Fig. 6A), indicating that the localization of Ppk11 to the medial region did not require the MOR components. We next examined conversely whether localization of the MOR components is dependent on Ppk11 function. In Δppk11 cells, Pmo25 was normally localized to the mitotic SPB(s), but the localization of Pmo25 to the medial region was substantially compromised (Fig. 6B, Pmo25-GFP). On the other hand, other MOR components, Nak1, Mor2, and Orb6, in Δppk11 cells were all localized normally (Fig. 6B). To quantify Pmo25 accumulation at the septum, we compared the intensity of the Pmo25-GFP localized to the septum between WT and Δppk11 cells in the same visual field. To distinguish Pmo25-GFP signals from WT and Δppk11 cells, we mixed 2 strains, one being WT cells expressing both Pmo25-GFP and Crn1-tdTomato, and the other, Δppk11 cells expressing only Pmo25-GFP. Quantification of the septum-localized Pmo25-GFP signals in the same visual field indicated that, as expected, the Pmo25-GFP intensity in the medial region of Δppk11 cells was decreased to 25% compared with that for the WT (Fig. 6C). The total amount of Pmo25-GFP protein in the Δppk11 cells was the same as that in the WT cells (data not shown). These results indicate that Ppk11 is required for accumulation of Pmo25 at the septum.
FIGURE 6.
Hierarchy of localization between Ppk11 and MOR. A, localization of Ppk11 in MOR mutants. The MOR mutants having ppk11+:GFP:kanr grown in YES5 medium at 25 °C were shifted to 36 °C and incubated for 4 h. Bars indicate 5 μm. B, localization of MORs in ppk11-deletion mutants. The indicated cells having pmo25+:GFP:kanr (DH1335-3C), kanr:nmt1:GFP:mor2+ (DH1356-4A), nak1+:G5:GFP:kanr (DH1333-3B) or orb6+:GFP:kanr (DH1334-4A) were grown in YES5 medium at 25 °C. The bar indicates 5 μm. C, relative intensity of Pmo25-GFP signal in WT and ppk11-deletion cells. WT having pmo25+:GFP:kanr and crn1+:tdTomato:kanr (DH1780-5C) and Δppk11 having pmo25+:GFP:kanr (DH1335-3C) were grown in YES5 medium at 25 °C. The fluorescence intensity of GFP in the cells was measured along the long axis (indicated on the Pmo25-GFP-labeled cells #1 and #3) by using the ImageJ software, and the relative intensity was calculated by comparison with the intensity in WT (n = 11). The data were analyzed by statistical processing (Student's t test).
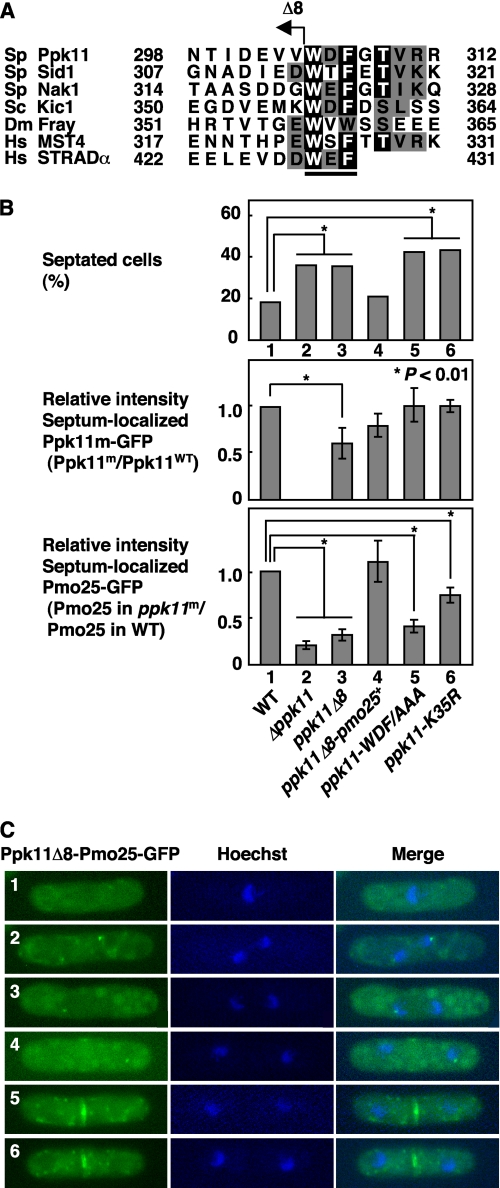
The WDF Motif of Ppk11 Is Essential for Both Its Interaction with Pmo25 and Efficient Cell Separation
The C-terminal Trp-Glu-Phe residues (WEF motif) of STRAD bind to MO25, and mutations of these residues abolish this interaction (18, 30). We searched for a similar sequence in GCKs and found that the GCKs functionally related to MO25 possess the MO25 binding motif (MBM) at their C-terminal regulatory regions (Figs. 1A and 7A). To investigate whether the C-terminal WDF motif of Ppk11 is important for Ppk11 function, we constructed the ppk11Δ8 mutant, in which 8 residues from the C terminus, containing the MBM (WDF), was truncated, and examined the mutant phenotype (the frequency of septated cells; Fig. 7B, top), localization of the Ppk11Δ8-GFP protein to the septum (Fig. 7B, middle), and localization of Pmo25-GFP to the septum in the ppk11Δ8 mutant (Fig. 7B, bottom). As before, to investigate septum localization of Ppk11Δ8-GFP, we mixed 2 strains, WT cells expressing both Ppk11-GFP and Crn1-tdTomato and the mutant cells expressing only the Ppk11Δ8-GFP, and quantified GFP signals in the same field (Fig. 7B, middle). In a similar way, to investigate septum localization of Pmo25-GFP in the ppk11Δ8 mutant, we used 2 strains, WT cells expressing both Pmo25-GFP and Crn1-tdTomato and the mutant cells only expressing Pmo25-GFP (Fig. 7B, bottom). The ppk11Δ8 mutant showed the defective cell separation (the increase in septated cells; Fig. 7B, top); and on this mutant background, the accumulation of Pmo25 to the septum decreased to 31% compared with that in WT cells (Fig. 7B, bottom), indicating that the WDF motif of Ppk11 was important for not only efficient cell separation but also accumulation of Pmo25 to the septum. The septum localization of the Ppk11Δ8 mutant protein was also slightly reduced compared with that of the Ppk11-wt protein (Fig. 7B, middle), though the amount of Ppk11Δ8 mutant protein was indistinguishable from that of the Ppk11 wild-type protein (data not shown), suggesting that the C-terminal 8 residues of Ppk11 was partially important for its own localization to the septum.
FIGURE 7.
Motif analysis of Ppk11 for interaction with Pmo25. A, alignment of MBM in GCKs. Identical and similar residues are represented as black and gray letters, respectively. The MBM of the human STRAD is underlined. The C-terminal truncated mutation of Ppk11Δ8 is indicated. B, phenotype of the ppk11 mutants. The indicated strains grown at 25 °C were shifted to 36 °C and incubated for 12 h. The cells were taken for observation of cell morphology (n > 200, upper panel). WT having ppk11+:GFP:kanr and crn1+:tdTomato:kanr (TG64–5A) and the ppk11 mutants having ppk11-m:GFP:kanr (TG143-2, TG242-3, TG241-4, TG184-1) were grown in YES5 medium at 25 °C and examined for the relative intensity of the septum localization of the GFP-fused Ppk11 mutant proteins (middle panel). WT having pmo25+:GFP:kanr and crn1+:tdTomato:kanr (DH1780-5C) and the ppk11 mutants having pmo25+:GFP:kanr (DH1335-3C, TG170-4B, TG242-3, TG215-1, TG177-36) were grown in YES5 medium at 25 °C and examined for the relative intensity of the septum localization of Pmo25-GFP (lower panel). The fluorescence intensity of GFP in the cells was measured along the long axis with the ImageJ software, and the relative intensity was calculated by comparison with the intensity in WT (n > 5). The data were analyzed by statistical processing (chi-squared tests for upper panel, Student's t test for middle and lower panels). C, localization of the Ppk11Δ8-Pmo25 fusion protein. The cells having ppk11Δ8:pmo25:GFP:kanr (TG242–3) were grown in YES5 medium at 25 °C.
To confirm the role of the WDF motif in Ppk11 function, we constructed the ppk11-WDF/AAA mutant, in which the residues WDF were substituted with alanines. The ppk11-WDF/AAA mutant cells showed defects in both cell separation and the accumulation of Pmo25 to the septum (Fig. 7B, top and bottom). Indeed, the interaction of Ppk11 with Pmo25 was abolished by the WDF/AAA mutation in the yeast 2-hybrid assay (Fig. 5B). If the WDF motif is essential for both its interaction with Pmo25 and accumulation of Pmo25 to the septum, the requirement of the WDF motif would be bypassed by the fusion of Pmo25 to Ppk11Δ8. To address this point, we constructed the ppk11Δ8-pmo25+ strain, in which the ppk11Δ8 gene (WDF-less, see above) was fused in-frame with pmo25+. As expected, defects in both cell separation and accumulation of Pmo25 to the septum in the ppk11Δ8 mutant cells were rescued by the Pmo25 fusion (Fig. 7B, compare ppk11Δ8 and ppk11Δ8-pmo25+). Like Pmo25, the Ppk11Δ8-Pmo25 fusion protein was localized normally to the mitotic SPB(s) and the septum (Fig. 7C). These results indicate that the major, if not sole, role of the WDF motif of Ppk11 lies in the interaction with Pmo25, thereby accumulating/targeting this protein to the septum site.
Finally we examined whether the kinase activity of Ppk11 is essential for accumulation of Pmo25 at the septum. In the ppk11-K35R mutant, the septum localization of Pmo25-GFP was slightly reduced but localized at a relatively normal level (78% of that in wt cells, Fig. 7B), suggesting that the kinase activity of Ppk11 was not essential for accumulation of Pmo25 to the septum.
DISCUSSION
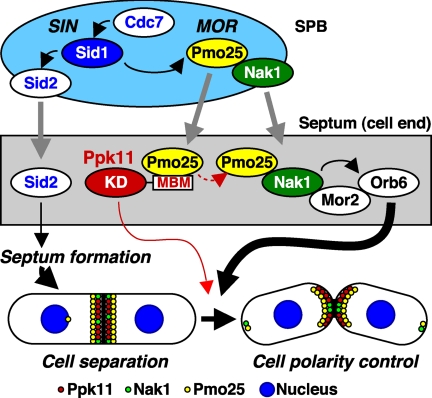
GCKs play pivotal roles in various aspects of cell signaling in eukaryotes, and normally constitute kinase cascades like MAPK pathways (7, 8). Fission yeast contains 3 GCKs and previous work including our laboratory shows that Sid1 and Nak1 are components of SIN and MOR, and play essential roles in septum formation/cytokinesis and cell morphogenesis, respectively (6, 9). In this study, we analyzed the role of the third GCK, Ppk11, in the cell morphogenesis. Ppk11 is not essential for cell growth but is important for efficient cell separation at the high temperature. Ppk11 is localized to the medial region after mitosis, more specifically probably to the septum, as it shows double-ring patterns upon cytokinesis. Localization of Ppk11 to the medial region requires a functional actomyosin ring, rather than SIN activation per se. Like other GCKs, Ppk11 does not exert its function on its own, but instead it acts at least in part with MOR, as this kinase physically interacts with Pmo25, one of the MOR components. Importantly Pmo25 accumulation/targeting to the medial region substantially depends on the Ppk11 protein. The absence of Ppk11 in MOR mutants exaggerates cell morphogenesis defects. These results lead us to conclude that Ppk11 plays an auxiliary but independent function in cell separation and cell polarity control coordinately with MOR. A model for functional interaction between Ppk11 and SIN/MOR in cell morphogenesis is depicted in Fig. 8.
FIGURE 8.
A model for functional interaction between Ppk11 and SIN/MOR in cell morphogenesis. Ppk11 has an N-terminal KD and the conserved C-terminal MBM. The kinase activity of Ppk11 is essential for efficient cell separation at high temperature (red line). The MBM of Ppk11 is crucial for Pmo25 targeting/accumulation to the septum. Septum localization of the proteins, Ppk11, Nak1, or Pmo25, is indicated by red, green, or yellow circle, respectively. Translocation of proteins from the mitotic spindle pole body (SPB) to the septum is also indicated by gray line.
Role of Ppk11 in Cell Separation
Ppk11 was important for efficient cell separation particularly at the high temperature. The Ppk11 kinase dead mutant cells, in which Pmo25 localized to the septum relatively normally, showed a defect in cell separation. These results suggest that Ppk11 protein plays a dual role in cell separation, one as a protein kinase, while the other acts as a structural platform for the Pmo25 accumulation/recruitment, which would contribute to the activation of other MOR component kinases such as Nak1 and Orb6, especially under the high temperature condition (Fig. 8, red dotted line). Consistent with this notion, the localization of Ppk11 to the septum precedes that of Pmo25. We envision that, by interacting with and accumulating Pmo25 to the medial region, Ppk11 is further activated at the site of cytokinesis, thereby in turn activating a downstream substrate(s)/kinase(s) for efficient cell separation. In Δppk11 cells, a certain amount of Pmo25 was still localized at the septum, indicating that Ppk11 per se is not essential for the localization of Pmo25 to the septum. Under a stringent condition such as the high temperature, a larger amount of Pmo25 protein at the septum, mediated by Ppk11, would be important for completion of cell separation through full activation of GCKs Nak1 and Ppk11. The Ppk11 protein level was not changed by the high temperature-shift (data not shown). It is possible that a target molecule(s) of Ppk11 might be induced under the high temperature condition.
Functional Interactions among MO25 and 3 GCKs
The partnership between MO25 and GCK appears to be evolutionally conserved among eukaryotes (11–13). We showed that in fission yeast Pmo25 physically/functionally interacts with 3 GCKs, i.e. Sid1, Nak1, and Ppk11 (9, 15). In other organisms, it has been shown that MO25 interacts with just 1 GCK. Our result is thus the first evidence showing the interaction of MO25 with multiple GCKs. The biological significance of this Pmo25 network is still not fully understood. To clarify this point, it would be important for construction of the pmo25 mutant that lacks the ability to interact with GCKs or the double/triple GCK mutants that lack the ability to interact with Pmo25.
Downstream Kinase(s) of Ppk11
MO25 physically interacts with GCK and activates downstream kinases. Sid1 and Pmo25 are important for the activities of the downstream kinases Nak1 and Orb6. In the MOR pathway, Pmo25 and Nak1 activate the downstream NDRK Orb6. The identification of a putative downstream kinase of Ppk11, other than Orb6, would be important for understanding a role of Ppk11 in cell morphogenesis. As in general GCK activates NDRK, we searched for a putative downstream NDRK(s) whose deletion exhibits the defect in cell separation similar to that seen in Δppk11 cells. Our homology search indicates that fission yeast has 5 non-essential NDRKs (Orb6-homologous NDRK-like kinases) on the genome that might act together with Ppk11. However, so far we have not found any NDRKs whose deletion results in defective cell separation, at least a single deletion.5 It is possible that some of these NDRKs share functions in cell separation in a redundant fashion. Multiple deletions would be necessary to address this point further. GCK Nak1 physically interacts with NDRK Orb6. To identify the downstream kinase of Ppk11, it would be intriguing to search for a kinase(s) that interacts with Ppk11.
Domain Analysis of GCK
In the ppk11-WDF/AAA mutant cells the septum localization of Pmo25 decreased significantly to 41% compared with that in WT cells; however, the Ppk11-WDF/AAA mutant protein was localized normally to the septum. This result indicates that the WDF motif of Ppk11 is important for the interaction with Pmo25 but not for the septum localization of Ppk11. A septum-targeting sequence might exist on Ppk11. It would be interesting to identify such a sequence on Ppk11 for the septum localization. Two GCKs, Sid1 and Nak1, also possess a MO25 binding motif (MBM) in their C-terminal regulatory regions. The importance of MBM in these GCKs for their function should be clarified. Further domain analysis should provide us with valuable information on the biological significance of the MO25-GCK network.
Acknowledgments
We thank M. Balasubramanian for the generous gift of the kinase deletion set used and I. Hagan and V. Simanis for strains. We thank D. McCollum, F. Verde, M. Balasubramanian, E. Tsuchiya, and I. Yamashita for helpful discussions. We are grateful to all other members of Hirata's laboratory for help.
This work was supported by grants from the Ministry of Education, Science, and Culture of Japan (to K. K. and D. H.) and the Cancer Research UK (to T. T.).
T. Goshima and D. Hirata, unpublished results.
- SIN
- septation initiation network
- GCK
- germinal center kinase
- MBM
- MO25 binding motif
- KD
- kinase domain
- MOR
- morphogenesis Orb6 network.
REFERENCES
- 1.Hayles J., Nurse P. (2001) Nat. Rev. Mol. Cell. Biol. 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 2.Chang F., Martin S. G. (2009) Cold Spring Harb. Perspect. Biol. 1, a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchison J. M. (1970) in Methods in Cell Physiology (Prescott D. M. ed) pp. 131–165, Vol. 4, Academic Press, New York [Google Scholar]
- 4.Marks J., Hagan I., Hyams J. S. (1986) J. Cell. Sci. 5, 229–241 [DOI] [PubMed] [Google Scholar]
- 5.Mitchison J. M., Nurse P. (1985) J. Cell. Sci. 75, 357–376 [DOI] [PubMed] [Google Scholar]
- 6.Simanis V. (2003) J. Cell. Sci. 116, 4263–4275 [DOI] [PubMed] [Google Scholar]
- 7.Daniels R. H., Bokoch G. M. (1999) Trends Biochem. Sci. 24, 350–355 [DOI] [PubMed] [Google Scholar]
- 8.Dan I., Watanabe N. M., Kusumi A. (2001) Trends Cell. Biol. 11, 220–230 [DOI] [PubMed] [Google Scholar]
- 9.Kanai M., Kume K., Miyahara K., Sakai K., Nakamura K., Leonhard K., Wiley D. J., Verde F., Toda T., Hirata D. (2005) EMBO J. 24, 3012–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyamoto H., Matsushiro A., Nozaki M. (1993) Mol. Reprod Dev. 34, 1–7 [DOI] [PubMed] [Google Scholar]
- 11.Nozaki M., Onishi Y., Togashi S., Miyamoto H. (1996) DNA Cell. Biol. 15, 505–509 [DOI] [PubMed] [Google Scholar]
- 12.Karos M., Fischer R. (1999) Mol. Gen. Genet. 260, 510–521 [DOI] [PubMed] [Google Scholar]
- 13.Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. (2006) Nat. Rev. Mol. Cell. Biol. 7, 253–264 [DOI] [PubMed] [Google Scholar]
- 14.Mendoza M., Redemann S., Brunner D. (2005) Eur. J. Cell. Biol. 84, 915–926 [DOI] [PubMed] [Google Scholar]
- 15.Kume K., Goshima T., Miyahara K., Toda T., Hirata D. (2007) Biosci. Biotechnol. Biochem. 71, 615–617 [DOI] [PubMed] [Google Scholar]
- 16.Nelson B., Kurischoko C., Horecka J., Mody M., Nair P., Pratt L., Zougman A., McBroom L. D., Hughes T. R., Boone C., Luca F. C. (2003) Mol. Biol. Cell. 14, 3782–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto Y., Izumi Y., Matsuzaki F. (2008) Biochem. Biophys. Res. Commun. 366, 212–218 [DOI] [PubMed] [Google Scholar]
- 18.Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. (2003) EMBO J. 22, 5102–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baas A. F., Smit L., Clevers H. (2004) Trends Cell. Biol. 14, 312–319 [DOI] [PubMed] [Google Scholar]
- 20.Alessi D. R., Sakamoto K., Bayascas J. R. (2006) Annu. Rev. Biochem. 75, 137–163 [DOI] [PubMed] [Google Scholar]
- 21.Zeqiraj E., Filippi B. M., Goldie S., Navratilova I., Boudeau J., Deak M., Alessi D. R., van Aalten D. M. (2009) PLoS Biol. 7, e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Klooster J. P., Jansen M., Yuan J., Oorschot V., Begthel H., Di Giacomo V., Colland F., de Koning J., Maurice M. M., Hornbeck P., Clevers H. (2009) Dev. Cell. 16, 551–562 [DOI] [PubMed] [Google Scholar]
- 23.Moreno S., Klar A., Nurse P. (1991) Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 24.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 25.Bähler J., Nurse P. (2001) EMBO J. 20, 1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsusaka T., Hirata D., Yanagida M., Toda T. (1995) EMBO J. 14, 3325–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang T. Y., Markley N. A., Young D. (2003) J. Biol. Chem. 278, 991–997 [DOI] [PubMed] [Google Scholar]
- 28.Bimbó A., Jia Y., Poh S. L., Karuturi R. K., den Elzen N., Peng X., Zheng L., O'Connell M., Liu E. T., Balasubramanian M. K., Liu J. (2005) Eukaryot. Cell 4, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koyano T., Kume K., Konishi M., Toda T., Hirata D. (2010) Biosci. Biotechnol. Biochem. 74, 1129–1133 [DOI] [PubMed] [Google Scholar]
- 30.Boudeau J., Scott J. W., Resta N., Deak M., Kieloch A., Komander D., Hardie D. G., Prescott A. R., van Aalten D. M., Alessi D. R. (2004) J. Cell. Sci. 117, 6365–6375 [DOI] [PubMed] [Google Scholar]